Abstract
Opioid drugs play important roles in the clinical management of pain, as well as in the development and treatment of drug abuse. The mu opioid receptor is the primary site of action for the most commonly used opioids, including morphine, heroin, fentanyl, and methadone. By sequencing DNA from 113 former heroin addicts in methadone maintenance and 39 individuals with no history of drug or alcohol abuse or dependence, we have identified five different single-nucleotide polymorphisms (SNPs) in the coding region of the mu opioid receptor gene. The most prevalent SNP is a nucleotide substitution at position 118 (A118G), predicting an amino acid change at a putative N-glycosylation site. This SNP displays an allelic frequency of approximately 10% in our study population. Significant differences in allele distribution were observed among ethnic groups studied. The variant receptor resulting from the A118G SNP did not show altered binding affinities for most opioid peptides and alkaloids tested. However, the A118G variant receptor binds β-endorphin, an endogenous opioid that activates the mu opioid receptor, approximately three times more tightly than the most common allelic form of the receptor. Furthermore, β-endorphin is approximately three times more potent at the A118G variant receptor than at the most common allelic form in agonist-induced activation of G protein-coupled potassium channels. These results show that SNPs in the mu opioid receptor gene can alter binding and signal transduction in the resulting receptor and may have implications for normal physiology, therapeutics, and vulnerability to develop or protection from diverse diseases including the addictive diseases.
The mu opioid receptor is the primary site of action of several of the endogenous opioid peptides including β-endorphin, Met-enkephalin-Arg-Phe, and the recently identified endomorphins (1). This receptor is also the major target for clinically important opioid analgesic agents including morphine, methadone, fentanyl, and related drugs (2, 3). Activation of this receptor has diverse physiological effects (4, 5). Furthermore, it is the major molecular site of action for heroin (6, 7). Rapid activation of the mu opioid receptor, such as that which occurs in the setting of drug abuse, results in a euphoric effect, thus conferring the reinforcing or rewarding effects of the drug, contributing to the development of addiction. Clinical observations have suggested that individuals have varied sensitivity to opioids, suggesting potential variability in the receptor protein and gene.
Naturally occurring polymorphisms are well known to exist in human genes; some have been shown to produce profound effects on the function of the corresponding proteins. Molecular cloning of the mu opioid receptor (8–11) has made it possible to determine potential sequence polymorphism, as shown by two recent studies (12, 13). The mu opioid receptor is a member of the G protein-coupled receptor family (8, 14). There are a number of well documented cases where naturally occurring mutations in G protein-coupled receptors lead to malfunctioning proteins and disorders in humans. Examples include a single-nucleotide polymorphism (SNP) in the luteinizing hormone (LH) receptor gene that results in a constitutively activated LH receptor at the cellular level and the development of precocious puberty in young male children (15); SNPs in the thyrotropin receptor gene that cause constitutive activation of adenylyl cyclase and hyperfunctioning thyroid adenomas (16); and mutations in the vasopressin receptor gene that cause truncation of the receptor and alter its function (17).
To further identify SNPs in the mu opioid receptor and to characterize whether receptor activity may be altered by SNPs, we studied DNA samples from subjects including both former heroin addicts in methadone maintenance treatment and individuals with no history of opiate or nonopiate drug or alcohol dependence. Here we report the findings that, indeed, SNP polymorphism in the mu opioid receptor gene can affect the activity of the mu opioid receptor, changing its sensitivity to the endogenous agonist β-endorphin.
MATERIALS AND METHODS
Study Subjects and Procedures.
Addictive disease patients, who were long-term former heroin addicts currently in chronic methadone maintenance treatment, and also normal control subjects with no history of any drug or alcohol abuse, were included in this study. All study subjects were characterized extensively with respect to drug abuse, the addictive diseases, psychological and psychiatric profiles, and medical and ethnic family backgrounds. No study subjects were related.
Previously heroin-addicted patients admitted to the study conformed to the federally regulated criteria for admission to a methadone or l-alpha-acetyl-methadone (LAAM) maintenance program. These criteria are: (i) 1 or more years of daily multiple-dose self-administration of heroin or other opiates, (ii) the development of tolerance and dependence, and (iii) drug-seeking behavior (18). All opioid-dependent study subjects were former heroin addicts currently in treatment at methadone maintenance clinics in New York City, primarily, two clinics closely associated for more than 25 years with the Laboratory of the Biology of Addictive Diseases at The Rockefeller University, the Adolescent Development Program and Adult Clinic at the New York Hospital–Cornell Medical Center. Current or prior abuse of other drugs was not used as an exclusion criterion for this study group as long as long-term opiate addiction continued to be the primary diagnosis.
Healthy control subjects were recruited primarily through posting of notices and newspaper advertisements or by referral as possible research volunteers by physicians or staff at The Rockefeller University Hospital. Individuals with any ongoing drug or alcohol abuse, or prior periods of alcohol or drug abuse, were excluded from this category. The exclusion criteria were defined as follows: for current or continuing abuse of alcohol, at least one instance of drinking to intoxication during the previous 30 days; for opiates, cocaine, amphetamines, or other illicit drugs (excluding cannabis), any use during the previous 30 days. Users of nicotine or caffeine were not excluded, nor were individuals who had abused cannabis for up to 12 days during the previous 30 days. For prior abuse, subjects were excluded who had used illicit drugs (with the exception of cannabis) for at least three times a week for a period of at least 1 month at any point in their lifetime.
All study subjects were screened rigorously by specially trained research personnel, including psychiatrists and research nurses, to ensure appropriate characterization of addictive diseases, status of treatment, and presence or absence of any polydrug or alcohol abuse. Both addictive disease patients and normal volunteers admitted to the study were assessed by a psychiatrist or research nurse with several general and specific psychiatric and psychological instruments. The Addiction Severity Index was included, with previously established interobserver reliability for the trained clinical research staff involved in this study (19). Study subjects also were administered a detailed personal, medical, and specific addictive disease questionnaire. Urine specimens were obtained on all study subjects, both those in the addictive disease group (in whom regular, ongoing urine collections are made and analyzed both for clinical care and other research purposes), and in the healthy, normal volunteer group. Urine specimens were analyzed for multiple drugs of abuse. Although not utilized in the present study, a family medical history and addictive disease questionnaire, designed to provide information regarding any substance abuse and major mental illnesses of first- and second-degree relatives, was administered.
Study subjects provided detailed information regarding family origin and ethnic background, including country or geographic area of birth. This information was obtained for both the study subjects and their immediate ancestors (parents, grandparents, and great-grandparents) to the extent that the information was known by the study subjects. Study subjects were classified into five groups: African-American, Caucasian, Hispanic (Caribbean and Central or South American origin), Native North American, and Other.
Subjects entering the study were required to be competent to understand the study procedures and understand and sign the Institutional Review Board-approved informed consent. Patients with schizophrenia or other psychotic mental illnesses were excluded from the study by this criterion. The presence of any known serological markers for hepatitis B or C or HIV was not used as an exclusion criterion.
After the informed consent, psychiatric and behavioral assessment, and family history acquisition, venipuncture on each study subject was performed and a blood specimen was taken. Blood samples were processed for DNA extraction both at The Rockefeller University and at Indiana University. Epstein–Barr virus transformation was used to create stable cell lines that were stored at both institutions for future studies. All blood samples were coded for continued maintenance of full confidentiality. The psychiatrists and nurses who performed psychiatric and psychological assessments were blind to the ultimately determined genotypes of the study subjects. Similarly, the identity and categorization of the study subjects was and remained unknown to the laboratory research personnel.
Exon Amplification and Sequencing.
Sequences from the noncoding regions of the human mu opioid receptor gene were used to design PCR primers for use in the amplification of the coding regions. PCR primers were synthesized for three of the four exons of the gene; the fourth exon was not included in this study because this exon is small (4 or 12 aa residues) and alternative splicing in this exon has been shown to occur (20). Exon 1 forward primer sequences were based on the 5′ untranslated region of the receptor (11). Exon 1 reverse, exon 2 forward and reverse, and exon 3 forward primer sequences were based on partial intron sequence data obtained from inverse PCR of genomic DNA sequences for the receptor gene (data not shown). Exon 3 reverse primers were based on reported intron 3 sequence (20). Two sets of primers were designed for each exon to allow for nested PCRs to increase amplification specificity of the exonic regions. Only one reverse primer was used for exon 1. The PCRs were performed with 300–400 ng of genomic DNA, PCR products were separated on agarose gels, and the DNA fragments were purified for DNA sequencing. DNA polymorphisms were confirmed by both manual and automated sequencing.
Mutagenesis.
In vitro site-directed mutagenesis was performed to generate human mu opioid receptor (hMOR) cDNA containing one of two major variants found, the A118G SNP. Complementary oligonucleotides containing the desired mutation were synthesized and annealed to the pcDNA3 plasmid containing the most common allelic form of hMOR. Primer 1, TTGTCCCACTTAGATGGCGACCTGTCCGACCCA; and primer 2, ACCGCATGGGTCGGACAGGTCGCCATCTAAGTG. Primers were extended and the product was amplified by PCR using hMOR dsDNA as the template, and DpnI restriction enzyme was added afterward to digest the methylated, nonmutated, most common dsDNA. After transformation into Escherichia coli cells, DNA from individual colonies was examined by restriction enzyme digestion and DNA sequencing to confirm success of mutagenesis.
Cell Transfection and Binding Analysis.
Stable transfection of the A118G SNP plasmid into AV-12 cells was performed (21). Individual colonies were picked, expanded, and tested for expression levels by performing binding assays. Cells were harvested by washing with PBS at room temperature, scraped into homogenization solution (0.3 M sucrose/25 mM Tris⋅HCl, pH 7.4/0.05% BSA and protease inhibitor cocktail, including 0.5 mM phenylmethylsulfonyl fluoride/0.1 μg/ml leupeptin/0.01% aprotinin), transferred to Dounce homogenizer, and homogenized on ice. The suspension was centrifuged at 1,000 × g for 10 min, and the supernatant was saved in a clean tube. The cell pellet was resuspended in homogenization buffer, homogenized, and centrifuged as described above. The supernatants from both extractions were combined and centrifuged at 30,000 × g for 20 min. The pelleted membranes were resuspended in binding buffer (50 mM Tris⋅HCl, pH 7.4); binding assays were carried out using membrane protein preparations as described (11). It has been shown that N-linked glycosylation is carried out by AV-12 cells (22, 23).
Electrophysiology.
Preparation of Xenopus oocytes was as reported previously (11). Oocytes were injected with in vitro transcribed mRNAs for the most common or A118G variant mu opioid receptors together with the G protein-activated, inwardly rectifying K+ channels (GIRK1 and GIRK2). Two to 3 days after RNA injection, oocytes were voltage-clamped in ND96 solution (96 mM NaCl/2 mM KCl/1 mM MgCl2/1.8 mM CaCl2/5 mM Hepes, pH 7.6) by using a two-electrode voltage clamp (Axon Instruments). Cells then were superfused with a high potassium solution (98 mM KCl/1 mM MgCl2/1.8 mM CaCl2/5 mM Hepes, pH 7.6) and stimulated with opioid ligands to measure the resulting potassium current. Xenopus oocytes have been shown to carry out N-linked glycosylation of exogenously expressed proteins (24, 25).
RESULTS AND DISCUSSION
Polymorphisms in the Human mu Opioid Receptor Gene.
Molecular cloning of the mu opioid receptor (8–11) has made it possible to determine potential sequence polymorphism. To identify SNPs in the mu opioid receptor, we used a PCR-based strategy to amplify the coding regions of the mu opioid receptor gene and to determine the DNA sequence of the amplified exonic regions. Using this method, we sequenced DNA samples from 152 subjects, including both former heroin addicts in methadone maintenance treatment and individuals with no history of drug or alcohol dependence.
Inclusion criteria as defined in the Materials and Methods section were met by these 152 individuals. One hundred and thirteen of the study subjects (74.3%) were former heroin addicts in methadone maintenance treatment, with or without previous or current codependency for other substances; 39 study subjects (25.7%) had no history of drug or alcohol dependence or any ongoing illicit opiate or other drug use, or alcohol abuse. Study subjects included 69 females (45.4%) and 83 males (56.4%).
Within the group of former heroin addicts in methadone maintenance treatment, the mean years in treatment was 6.7 with a range from 2 months to 30 years (n = 112, only one patient’s history could not be verified by an additional interview or by medical chart records). Before treatment, the mean years of heroin addiction was 10.1 years, with a range from 1 to 30 years (n = 109; the exact duration of heroin use before entry into methadone treatment could not be verified for four subjects). The mean daily methadone dose of opioid dependent patients in stable treatment was 84 mg/day, with a range from 30 to 120 mg/day. Only patients (n = 106) with established stable methadone doses were included in this calculation; the other patients were undergoing induction, increasing, tapering, or elimination dose schedules.
The ethnic breakdown of the study subject populations was as follows: African-American, 31 (20.3%), Caucasian, 52 (34.2%), Hispanic, 67 (44.1%), Native North American, 1 (0.7%), and Other, 1 (0.7%). Several individuals reported that one parent’s ancestry was from one ethnic group and the other parent’s ancestry was from a second ethnic group, including four individuals (2.6%) who reported one African-American parent and one Caucasian parent, and five individuals (3.3%) who reported one Caucasian parent and one Hispanic. For the genotype calculations, the former study subjects were classified as African-American and the latter, as Hispanic.
By sequencing PCR-amplified DNA from the study subjects, we determined that the previously reported sequence for the human mu opioid receptor (10, 11) was the most common allele found in our study population. We also identified five different variants, all SNPs. For the purpose of this report, we use the term “most common” to denote the predominant mu opioid receptor allele and the corresponding receptor that was reported originally by cDNA cloning (10, 11), and the term “variant” to denote the allelic genes or the resulting receptors containing polymorphic variations. Table 1 shows these SNPs, with information on the position of amino acid substitutions, and overall frequency of the variant alleles in the study population. Two recent studies (12, 13) also have identified one or both of these five SNPs in the coding region (A118G and C17T), but not the other three SNPs (G24A, G779A, and G942A).
Table 1.
SNPs in the human mu opioid receptor gene
| Variant name | Nucleotide position | Exon location | Corresponding amino acid change | Protein domain | Allele frequency |
|---|---|---|---|---|---|
| A118G | 118 | 1 | Asn-40 → Asp (N40D) | N terminal | 10.5% (26 heterozygous individual and 3 homozygous individuals, n = 152) |
| C17T | 17 | 1 | Ala-6 → Val (A6V) | N terminal | 6.6% (14 heterozygous individuals and 3 homozygous individuals, n = 152) |
| G24A | 24 | 1 | Silent mutation | N terminal | 2% (six heterozygous individuals, n = 152) |
| G779A | 779 | 3 | Arg-260 → His (R260H) | CL3 | One heterozygous individual |
| G942A | 942 | 3 | Silent mutation | EL3 | One heterozygous individual |
Nucleotide position 1 is the first base of the start codon. Protein domains are based on the seven-transmembrane model for opioid receptors. EL, extracellular loop; CL, cytoplasmic loop.
Genotype and allele frequencies for the two most prevalent allelic variants in our population sample, the A118G and C17T polymorphisms, are shown in Table 2, with overall allelic frequencies of 10.5 and 6.6%, respectively. One of the two earlier reports, comparing healthy volunteers with cocaine- and/or opiate-dependent individuals, found an allelic frequency of 10 and 22% for the C17T receptor SNP in their two study groups, respectively (12). This study included Caucasian and African-American subjects. The A118G SNP was not identified (12). The second previously reported study included American Caucasian, Finnish Caucasian, and Native North American study subjects; an allelic frequency of 10.5–16.3% for the A118G variant among the three groups was found, but with only “rare” occurrence of the C17T polymorphism (13). Table 2 gives associations of each frequency in our study population broken down by ethnicity, gender, and opioid dependence. Because the number of individuals homozygous for the variant alleles was small, we used allele frequencies, rather than genotype frequencies, to test for significant differences.
Table 2.
Genotype and allele frequency associations
| A118G Genotypes
|
Total | A118G Alleles
|
Total | C17T Genotypes
|
Total | C17T Alleles
|
Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/A | A/G | G/G | A | G | C/C | C/T | T/T | C | T | |||||
| Ethnicity* | ||||||||||||||
| African-American | 30 (0.968) | 1 (0.032) | 0 — | 31 | 61 (0.984) | 1 (0.016) | 62 | 21 (0.677) | 7 (0.226) | 3 (0.097) | 31 | 49 (0.790) | 13 (0.210) | 62 |
| Caucasian | 41 (0.788) | 10 (0.192) | 1 (0.019) | 52 | 92 (0.885) | 12 (0.115) | 104 | 50 (0.962) | 2 (0.038) | 0 — | 52 | 102 (0.981) | 2 (0.019) | 104 |
| Hispanic | 50 (0.746) | 15 (0.225) | 2 (0.030) | 67 | 115 (0.858) | 19 (0.142) | 134 | 62 (0.925) | 5 (0.075) | 0 — | 67 | 129 (0.963) | 5 (0.037) | 134 |
| χ(2)2=7.15 (P=0.028) | χ(2)2=26.0 (P=0.000002) | |||||||||||||
| Gender | ||||||||||||||
| Female | 59 (0.855) | 8 (0.116) | 2 (0.029) | 69 | 126 (0.913) | 12 (0.087) | 138 | 58 (0.841) | 9 (0.130) | 2 (0.029) | 69 | 125 (0.906) | 13 (0.094) | 138 |
| Male | 64 (0.771) | 18 (0.217) | 1 (0.012) | 83 | 146 (0.880) | 20 (0.120) | 166 | 77 (0.928) | 5 (0.060) | 1 (0.012) | 83 | 159 (0.958) | 7 (0.042) | 166 |
| χ(1)2 = 0.90 (P = 0.343) | Yate’s corrected χ(1)2 = 2.53 (P = 0.112) | |||||||||||||
| Opioid Dependence | ||||||||||||||
| Dependent | 94 (0.832) | 18 (0.159) | 1 (0.009) | 113 | 206 (0.912) | 20 (0.088) | 226 | 97 (0.858) | 13 (0.115) | 3 (0.027) | 113 | 207 (0.916) | 19 (0.084) | 226 |
| Nondependent | 29 (0.744) | 8 (0.205) | 2 (0.051) | 39 | 66 (0.846) | 12 (0.154) | 78 | 38 (0.974) | 1 (0.026) | 0 — | 39 | 77 (0.987) | 1 (0.013) | 78 |
| Yale’s corrected χ(1)2 = 1.98 (P = 0.159) | Yate’s corrected χ(1)2 = 3.70 (P = 0.054) | |||||||||||||
The two individuals who were not classified into African-American, Caucasian, or Hispanic ethnic groups were not included in the analysis.
We tested for differences of allele frequencies among the three most common ethnic groups, in our study, African-American, Caucasian, and Hispanic, irrespective of opioid-dependency status. We found significant differences of allele frequencies among the ethnic groups studied for both the A118G [χ(2)2 = 7.15 (P = 0.028)] and the C17T [χ(2)2 = 26.0 (P = 0.000002)]. If the individuals who reported one parent from one ethnic group and one from another ethnic group were excluded from this analysis, similar significance levels were obtained for differences of both SNPs among ethnic groups. This result is not surprising because allele frequencies are known to vary among ethnic groups. However, it is important to consider these differences, which can confound association analyses. No significant association of gender with either polymorphism was observed.
For the A118G polymorphism, there was no significant difference in allele frequencies between opioid-dependent and nondependent study subjects with all ethnic groups combined. Similarly, no association between this allele and alcoholism or drug abuse of unspecified type was found in the study reported by Bergen et al. (13). However, within the Hispanic study subject group, we found that the A118G variant allele was present in a significantly higher proportion of non-opioid-dependent subjects compared with the opioid-dependent subjects [Yates-corrected Chi-square χ(1)2 = 8.22 (P = 0.0041)]. Although this finding could be explained by population admixture within this group, it suggests the possibility that the A118G SNP might confer a relative protection against opioid dependence. Further studies with a larger sample size would be necessary to test this hypothesis.
In contrast to the A118G allele, the C17T variant was present in a higher overall proportion of opioid-dependent persons in our sample at a marginal significance level [Yates-corrected Chi-square χ(1)2 = 3.70 (P = 0.054)]. This result is similar to that obtained by Berrettini et al. (12).
Table 3 shows data of this study stratified by ethnic group and opioid-dependency status for both the A118G and the C17T SNPs. The pooled Relative Risk (RR) and the Mantel–Haenszel Chi-square (26) were calculated. For the A118G polymorphism there was no significant difference in allele frequencies between former heroin addicts in treatment (cases) and those with no history of drug or alcohol abuse or dependence (controls) [RR = 0.48 χ(1)2 = 2.76 (P = 0.096)]. Although not significant, with respect to the Relative Risk there was evidence of heterogeneity among ethnic groups [χ(2)2 = 5.16 (P = 0.076)]. It should be noted that the direction of the Relative Risk, i.e., less than one, shows here that the A118G polymorphism was more frequent in control subjects than in opioid-dependent subjects and, again, suggests that the A118G polymorphism might confer some level of protection against opiate addiction, which is of particular interest given the differences in receptor activity (see below). There was a marginally significant difference in the allele frequencies for the C17T polymorphism between opioid-dependent and healthy control subjects [RR = 7.83 χ(1)2 = 3.73 (P = 0.05)]. The test for heterogeneity of the Relative Risk among ethnic groups for the C17T polymorphism was not significant [χ(2)2 = 3.95 (P = 0.14)].
Table 3.
Stratification of alleles of opioid-dependent and nondependent study subjects by ethnicity
| A118G
|
C17T
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Opioid-dependent (cases)
|
Nondependent (controls)
|
Opioid-dependent (cases)
|
Nondependent (controls)
|
|||||
| A | G | A | G | C | T | C | T | |
| African-American | 45 | 1 | 16 | 0 | 33 | 13 | 16 | 0 |
| Caucasian | 53 | 7 | 39 | 5 | 59 | 1 | 43 | 1 |
| Hispanic | 104 | 12 | 11 | 7 | 111 | 5 | 18 | 0 |
| Combined | 202 | 20 | 66 | 12 | 203 | 19 | 77 | 1 |
The two individuals who were not classified as African-American, Caucasian, or Hispanic ethnic groups were not included in the analysis.
The A118G SNP Affects β-Endorphin-Binding Affinity of the mu Opioid Receptor.
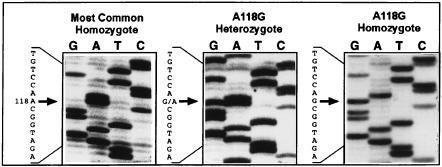
The most prevalent genetic polymorphism we identified is the A118G SNP with a substitution at the nucleotide position 118 with respect to the first base of the initiator codon for methionine (Fig. 1). This allele was observed in 29 of the 152 subjects, with 26 subjects being heterozygous and 3 being homozygous for the variant allele. Overall, this gives an allele frequency of 10.5% in the subject population that we have examined in this study. Nucleotide 118 is the first base in codon 40 of the human mu opioid receptor, and the A118G variant predicts an Asn-to-Asp change in amino acid residue 40 of the receptor (N40D). The Asn residue at amino acid position 40 in the most common sequence of the mu receptor is a putative site for N-glycosylation (11). Thus, the A118G variant would result in the loss of a putative N-glycosylation site. The position of amino acid 40 is in the N-terminal region of the mu opioid receptor (11). Based on sequence motif similarities with other G protein-coupled receptors (27), the N-terminal region of opioid receptors, including that of the mu opioid receptor, is predicted to be in the extracellular space (28). To explore any potential effects of the A118G polymorphism on the mu opioid receptor, we mutated the position 118 of the most common mu receptor cDNA by site-directed mutagenesis and generated a cDNA clone for the human mu opioid receptor containing the A118G variant. This way, both the most common and the A118G variant receptors could be expressed in cells to determine their cellular activity and their binding affinities.
Figure 1.
DNA sequences showing the A118G polymorphism. Examples of DNA sequence are shown from individuals homozygous for the most common allele (Left), heterozygous (Center), and homozygous for the A118G variant allele (Right). The arrows indicate the position of nucleotide 118, with the adjacent sequences shown.
We performed radioligand-binding assays with cell lines stably transfected with either the A118G variant or the most common mu receptor to determine whether the A118G polymorphism changes the receptor’s ability to bind opioid ligands, especially endogenous opioid peptides, because they are the physiological agonists for the mu opioid receptor. The A118G variant and the most common mu receptors yielded similar binding affinity values for most of the opioid ligands tested, including the small endogenous peptide agonists Met- and Leu-enkephalin, each with five aa residues; endomorphin-1 and -2, each with four residues; the mu-selective synthetic opioid peptide DAMGO, with five amino acid residues; the endogenous ligand for the kappa opioid receptor dynorphin A (1–17); as well as the mu-preferring opioid alkaloid agonists morphine, fentanyl, methadone, and the opioid antagonist naloxone (Fig. 2, and data not shown). These results suggest that the A118G polymorphism does not change the overall binding properties of the mu opioid receptor. This is not unexpected, because the predicted amino acid change as a result of the A118G SNP is a single residue substitution in the N-terminal region in the extracellular space and is unlikely to drastically affect the overall tertiary structure of the receptor.
Figure 2.
Binding of endogenous opioid peptides to the most common and A118G mu opioid receptors. Membrane preparations from cells expressing either the most common (■) or the A118G (▵) receptors were used in binding experiments to displace the [3H]DAMGO binding. Shown are examples of displacement binding for four endogenous peptides: Met-enkephalin, dynorphin A, β-endorphin, and endomorphin-1.
There was a substantial change for the A118G variant receptor binding of human β-endorphin, a much larger endogenous opioid peptide, which has 31 aa residues and which activates the mu opioid receptor. Whereas the other, smaller, endogenous opioid peptides and alkaloid agonists and antagonist displayed similar binding affinities for both receptors, the A118G variant receptor showed higher binding affinity for β-endorphin than the most common receptor (Fig. 2), with the ratio of the Ki of the most common allele to A118G variant SNP being 3.46 ± 0.31 (mean ± SEM, n = 3). These results indicate that although the A118G polymorphism did not alter the overall profile of ligand binding to the receptor, it specifically influenced the β-endorphin binding and resulted in much higher affinity for this large, endogenous opioid peptide.
The A118G SNP Changes the Agonist Potency of β-Endorphin for Activating the mu Opioid Receptor.
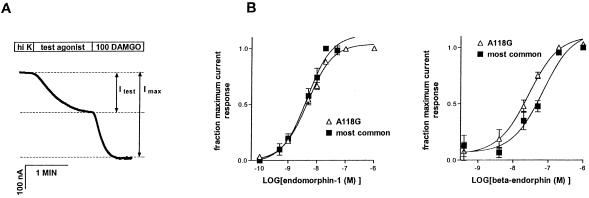
An important cellular activity of the mu opioid receptor is inhibition of neuronal excitability by receptor-mediated inhibition of presynaptic calcium channels and activation of postsynaptic potassium channels (29, 30). The major effector potassium channels for the mu receptor, as well as for many other G protein-coupled receptors, are the G protein-activated, inwardly rectifying K+ (GIRK) channels (31, 32). Coexpression studies have shown that the mu opioid receptor can readily activate GIRK channels via a G protein-mediated mechanism (11, 33, 34). To examine the effect of the A118G polymorphism, we used the Xenopus oocyte expression to compare the A118G variant receptor with the most common mu opioid receptor. Agonist stimulation of the A118G variant receptor activated a potassium current similar to that seen with the most common mu opioid receptor (11, 33). The EC50 values for endomorphin-1 are 4.6 nM for the most common receptor and 4.9 nM for the A118G variant receptor (Fig. 3), indicating that endomorphin-1 activated both receptors with similar potency. The EC50 values for β-endorphin, however, differed about 3-fold between the A118G variant and the most common mu opioid receptors (Fig. 3), consistent with the change in the binding affinity (Fig. 2). These data indicate that, as a result of the SNP in the receptor gene, the A118G variant receptor may be functionally different from the most common mu opioid receptor.
Figure 3.
Comparison of the most common and the A118G variant mu opioid receptors in coupling to G protein-activated, inwardly rectifying K+ (GIRK) channels. (A) Example of current trace showing the experimental protocol and calculation method for the agonist-induced response. Oocytes were clamped at a holding potential of −80 mV and superfused with different solutions as indicated. Imax, maximum K+ currents evoked by DAMGO at a saturating concentration (100 nM). ITest, K+ currents evoked by the test dose of agonists. The trace example shows the response of the A118G variant receptor to 20 nM β-endorphin. (B) Dose-response curves of receptor activation. The tested concentrations of agonists ranged from 0.1 nM to 1 μM. The response to a test dose is expressed as the fraction of the maximum activation by 100 nM DAMGO (ITest/Imax). Data are presented as mean ± SEM (n = 4–5). All oocytes were used only once to avoid desensitization.
An endogenous opioid with wide distribution in both the central nervous system (CNS) and the periphery, β-endorphin has been postulated to play a role in diverse biological functions (35, 36). As a neuropeptide, it can modulate neurotransmitter actions in the CNS to mediate antinociception. It is also a mediator in the stress response, of potential importance for the pathophysiology of the addictive diseases (37–42). β-endorphin can regulate the secretion of both stress and reproductive hormones, thereby influencing a variety of physiological functions. Given the diverse roles of β-endorphin, it is particularly interesting that the A118G polymorphism may change the receptor with respect to the binding affinity of β-endorphin and the potency of its cellular activity. The approximately 3-fold difference in binding affinity and potency of β-endorphin (Figs. 2 and 3) suggests that individuals carrying the variant receptor gene (A118G) may show differences in some of the functions mediated by β-endorphin action at the altered mu opioid receptors. Thus, for example, response to stress, reproductive function, and pain perception could be altered. Moreover, the data suggest that subtle changes, such as the SNP studied with respect to its binding and activity, could have significant effects on the susceptibility or vulnerability to develop multifactorial diseases such as specific addictions (7, 8, 43).
Acknowledgments
We thank W. Ai, L. S. Bye, J. Chen, A. Ho, Z. Lai, J. Ott, G. Perret, M. R. Vasko, L. Walsh, P. Young, and X. Yue for technical assistance, comments, and suggestions. We also thank Drs. E. Khuri and A. Wells, directors of the clinics that participated in the study. This research was supported in part by the National Institutes of Health Research Grants DA09444 (L.Y. and M.J.K.), DA05130 and DA00049 (M.J.K.), HG00008 (S.M.L.), and DA09116 and DA11891 (L.Y.), and by a General Clinical Research Center Grant (M01-RR00102) from the National Center for Research Resources at the National Institutes of Health.
ABBREVIATION
- SNP
single-nucleotide polymorphism
References
- 1.Zadina J E, Hackler L, Ge L J, Kastin A J. Nature (London) 1997;386:499–502. doi: 10.1038/386499a0. [DOI] [PubMed] [Google Scholar]
- 2.Basbaum A I, Fields H L. Annu Rev Neurosci. 1984;7:309–338. doi: 10.1146/annurev.ne.07.030184.001521. [DOI] [PubMed] [Google Scholar]
- 3.Pasternak G W. Clin Neuropharmacol. 1993;16:1–18. doi: 10.1097/00002826-199302000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Pasternak G W. The Opiate Receptors. Totowa, NJ: Humana; 1988. [Google Scholar]
- 5.Goldstein A. Trends Pharmacol Sci. 1987;8:456–459. [Google Scholar]
- 6.Kreek M J. Mol Psychiatry. 1996;1:232–254. [PubMed] [Google Scholar]
- 7.Kreek M J. Neurochem Res. 1996;21:1469–1488. doi: 10.1007/BF02532387. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Mestek A, Liu J, Hurley J A, Yu L. Mol Pharmacol. 1993;44:8–12. [PubMed] [Google Scholar]
- 9.Wang J B, Imai Y, Eppler C M, Gregor P, Spivak C E, Uhl G R. Proc Natl Acad Sci USA. 1993;90:10230–10234. doi: 10.1073/pnas.90.21.10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J B, Johnson P S, Persico A M, Hawkins A L, Griffin C A, Uhl G R. FEBS Lett. 1994;338:217–222. doi: 10.1016/0014-5793(94)80368-4. [DOI] [PubMed] [Google Scholar]
- 11.Mestek A, Hurley J H, Bye L S, Campbell A D, Chen Y, Tian M, Schulman H, Yu L. J Neurosci. 1995;15:2396–2406. doi: 10.1523/JNEUROSCI.15-03-02396.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berrettini W H, Hoehe M R, Ferrada T N, Gottheil E. Addiction Biol. 1997;2:303–308. doi: 10.1080/13556219772598. [DOI] [PubMed] [Google Scholar]
- 13.Bergen A W, Kokoszka J, Peterson R, Long J C, Virkkunen M, Linnoila M, Goldman D. Mol Psychiatry. 1997;2:490–494. doi: 10.1038/sj.mp.4000331. [DOI] [PubMed] [Google Scholar]
- 14.Kieffer B L. Cell Mol Neurobiol. 1995;15:615–635. doi: 10.1007/BF02071128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shenker A, Laue L, Kosugi S, Merendino J J, Jr, Minegishi T, Cutler G B., Jr Nature (London) 1993;365:652–654. doi: 10.1038/365652a0. [DOI] [PubMed] [Google Scholar]
- 16.Parma J, Duprez L, Van Sande J, Cochaux P, Gervy C, Mockel J, Dumont J, Vassart G. Nature (London) 1993;365:649–651. doi: 10.1038/365649a0. [DOI] [PubMed] [Google Scholar]
- 17.Rosenthal W, Antaramian A, Gilbert S, Birnbaumer M. J Biol Chem. 1993;268:13030–13033. [PubMed] [Google Scholar]
- 18.Rettig R A, Yarmolinsky A. Federal Regulation of Methadone Treatment. Washington, DC: Natl. Acad. Press; 1995. [PubMed] [Google Scholar]
- 19.McLellan A T, Luborsky L, Woody G E, O’Brien C P. J Nerv Ment Dis. 1980;168:26–33. doi: 10.1097/00005053-198001000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Bare L A, Mansson E, Yang D. FEBS Lett. 1994;354:213–216. doi: 10.1016/0014-5793(94)01129-x. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Liu J, Yu L. Addiction Biol. 1996;1:49–59. doi: 10.1080/1355621961000124686. [DOI] [PubMed] [Google Scholar]
- 22.Parkinson J F, Grinnell B W, Hoskins J, Vlahos C J, Bang N U. J Biol Chem. 1990;265:12602–12610. [PubMed] [Google Scholar]
- 23.Grinnell B W, Walls J D, Gerlitz B. J Biol Chem. 1991;266:9778–9785. [PubMed] [Google Scholar]
- 24.Hayes G, Busch A, Lotscher M, Waldegger S, Lang F, Verrey F, Biber J, Murer H. J Biol Chem. 1994;269:24143–24149. [PubMed] [Google Scholar]
- 25.Gurnett C A, De Waard M, Campbell K P. Neuron. 1996;16:431–440. doi: 10.1016/s0896-6273(00)80061-6. [DOI] [PubMed] [Google Scholar]
- 26.Mantel N, Haenszel W. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 27.O’Dowd B F, Lefkowitz R J, Caron M G. Annu Rev Neurosci. 1989;12:67–83. doi: 10.1146/annurev.ne.12.030189.000435. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Mestek A, Liu J, Yu L. Biochem J. 1993;295:625–628. doi: 10.1042/bj2950625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.North R A. In: Handbook of Experimental Pharmacology: Opioids I. Herz A, editor. Vol. 104. Berlin: Springer; 1993. pp. 773–797. [Google Scholar]
- 30.Chavkin C. In: The Opiate Receptors. Pasternak G W, editor. Totowa, NJ: Humana; 1988. pp. 273–303. [Google Scholar]
- 31.Kubo Y, Reuveny E, Slesinger P A, Jan Y N, Jan L Y. Nature (London) 1993;364:802–806. doi: 10.1038/364802a0. [DOI] [PubMed] [Google Scholar]
- 32.Dascal N, Schreibmayer W, Lim N F, Wang W, Chavkin C, DiMagno L, Labarca C, Kieffer B L, Gaveriaux-Ruff C, Trollinger D, et al. Proc Natl Acad Sci USA. 1993;90:10235–10239. doi: 10.1073/pnas.90.21.10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Yu L. J Biol Chem. 1994;269:7839–7842. [PubMed] [Google Scholar]
- 34.Kovoor A, Henry D J, Chavkin C. J Biol Chem. 1995;270:589–595. doi: 10.1074/jbc.270.2.589. [DOI] [PubMed] [Google Scholar]
- 35.Kreek M J, Hartman N. Ann N Y Acad Sci. 1982;398:151–172. doi: 10.1111/j.1749-6632.1982.tb39489.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Y, Spangler R, LaForge K S, Maggos C E, Ho A, Kreek M J. Peptides. 1996;17:435–441. doi: 10.1016/0196-9781(96)00014-9. [DOI] [PubMed] [Google Scholar]
- 37.Kreek M J, Wardlaw S L, Friedman J, Schneider B, Frantz A G. In: Advances in Endogenous and Exogenous Opioids. Simon E, Takagi H, editors. Tokyo: Kodansha; 1981. pp. 364–366. [Google Scholar]
- 38.Kreek M J, Ragunath J, Plevy S, Hamer D, Schneider B, Hartman N. Neuropeptides. 1984;5:277–278. doi: 10.1016/0143-4179(84)90081-7. [DOI] [PubMed] [Google Scholar]
- 39.Kosten T R, Kreek M J, Swift C, Carney M K, Ferdinands L. Life Sci. 1987;41:1071–1076. doi: 10.1016/0024-3205(87)90623-0. [DOI] [PubMed] [Google Scholar]
- 40.Kosten T R, Kreek M J, Ragunath J, Kleber H D. Life Sci. 1986;39:55–59. doi: 10.1016/0024-3205(86)90437-6. [DOI] [PubMed] [Google Scholar]
- 41.Kennedy J A, Hartman N, Sbriglio R, Khuri E, Kreek M J. Br J Addiction. 1990;85:1133–1140. doi: 10.1111/j.1360-0443.1990.tb03438.x. [DOI] [PubMed] [Google Scholar]
- 42.Culpepper-Morgan J A, Kreek M J. Metab Clin Exp. 1997;46:130–134. doi: 10.1016/s0026-0495(97)90289-4. [DOI] [PubMed] [Google Scholar]
- 43.Kreek M J. Pharmacol Biochem Behav. 1997;57:551–569. doi: 10.1016/s0091-3057(96)00440-6. [DOI] [PubMed] [Google Scholar]