Abstract
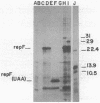
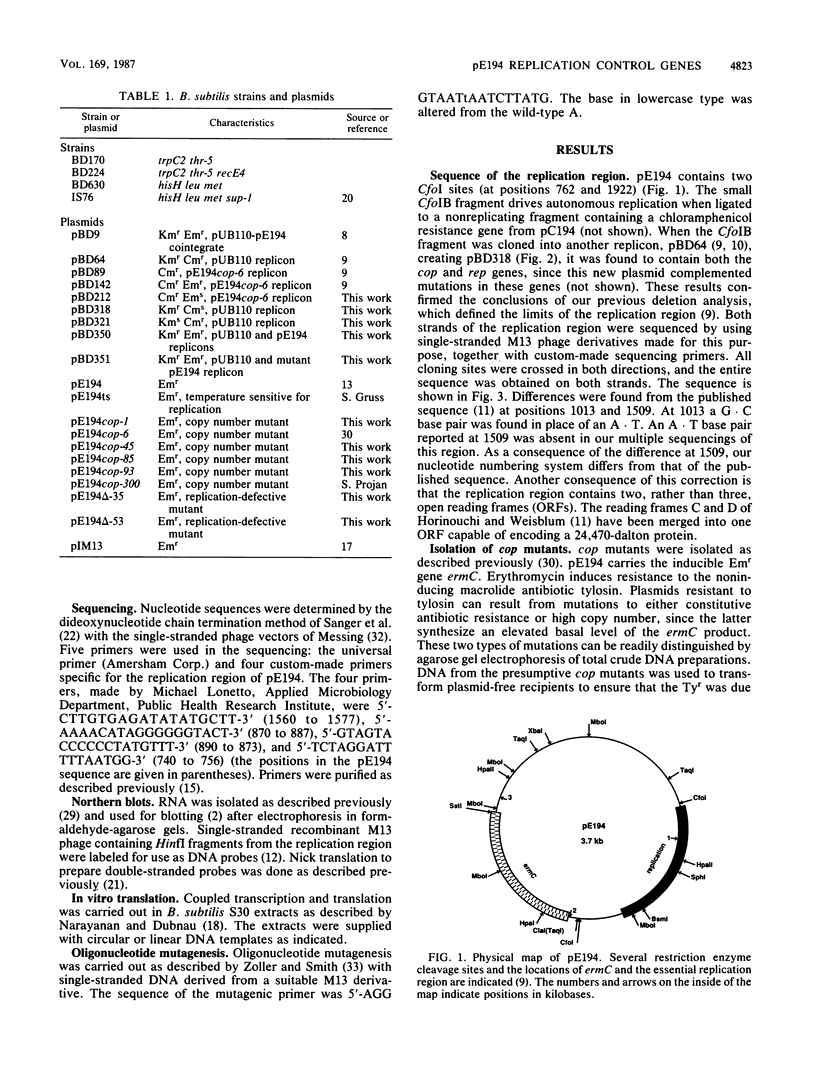
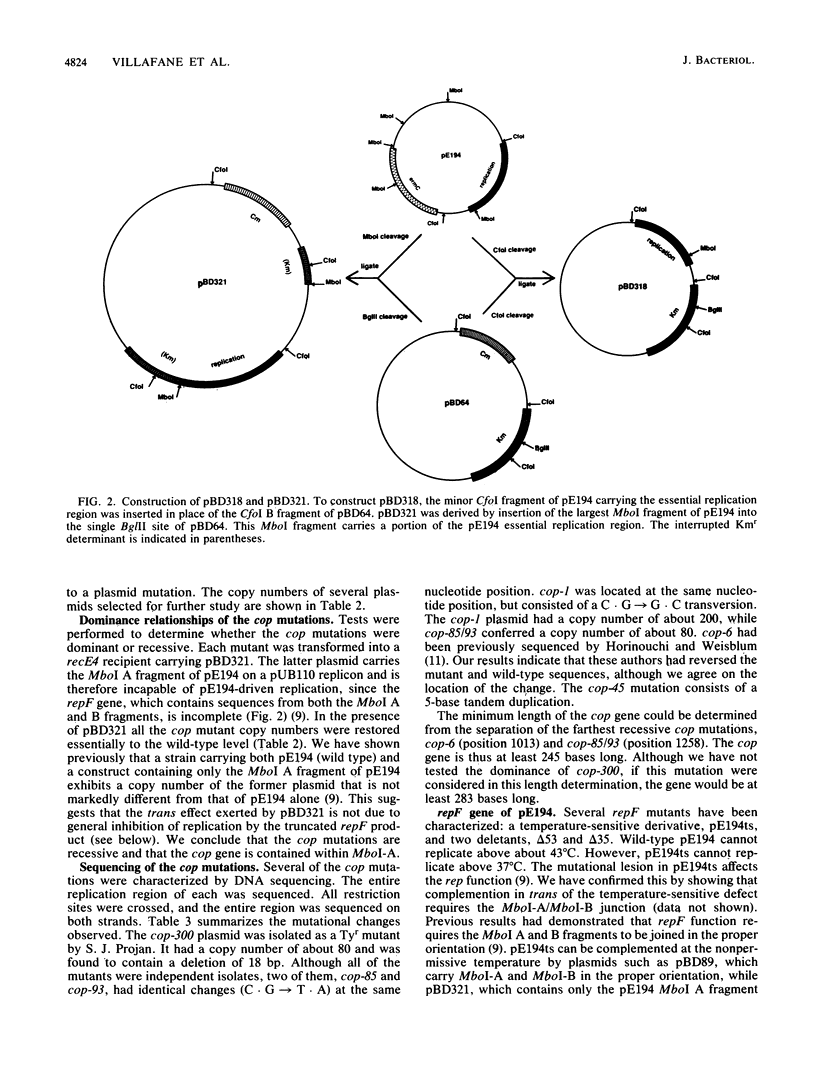
pE194, a 3.7-kilobase plasmid, confers resistance to macrolide, lincosamide, and streptogramin B antibiotics. The previously identified cop and repF genes of pE194 have been further localized by molecular cloning and mutational analysis together with DNA sequencing. The CfoIB fragment of pE194 is capable of autonomous replication and contains both genes. Most of this region has been resequenced, and two errors reported in a previous study have been corrected. The corrected sequence indicates that the replication region contains a single large open reading frame, which we propose encodes the repF product. Northern blot (RNA blot) analysis of this region detected six transcripts, all transcribed in the same direction as one another and opposite to repF. A 350-base transcript is synthesized from the region containing cop. No in vivo transcript for the repF gene was detected, but a protein was observed in an in vitro transcription-translation system which appears to be its product. An ochre mutation was inserted in the putative repF open reading frame, and a nonsense fragment was detected in the in vitro system. When carried passively on a pUB110 replicon, this mutant product appears capable of inhibiting pE194 replicons in trans. The pE194 origin of replication has been localized to within 200 bases.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Figurski D. H. Map location and nucleotide sequence of korA, a key regulatory gene of promiscuous plasmid RK2. Nucleic Acids Res. 1983 Nov 11;11(21):7453–7469. doi: 10.1093/nar/11.21.7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contente S., Dubnau D. Characterization of plasmid transformation in Bacillus subtilis: kinetic properties and the effect of DNA conformation. Mol Gen Genet. 1979 Jan 2;167(3):251–258. doi: 10.1007/BF00267416. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971 Mar 14;56(2):209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R., Scher B., Cirigliano C. Fate of transforming deoxyribonucleic acid after uptake by competent Bacillus subtilis: phenotypic characterization of radiation-sensitive recombination-deficient mutants. J Bacteriol. 1973 Apr;114(1):273–286. doi: 10.1128/jb.114.1.273-286.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Dubnau D. Construction and properties of chimeric plasmids in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1428–1432. doi: 10.1073/pnas.75.3.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Hahn J., Contente S., Dubnau D. Replication and incompatibility properties of plasmid pE194 in Bacillus subtilis. J Bacteriol. 1982 Nov;152(2):722–735. doi: 10.1128/jb.152.2.722-735.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T., Shivakumar A. G., Dubnau D. Characterization of chimeric plasmid cloning vehicles in Bacillus subtilis. J Bacteriol. 1980 Jan;141(1):246–253. doi: 10.1128/jb.141.1.246-253.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Iordănescu S. Three distinct plasmids originating in the same Staphylococcus aureus strain. Arch Roum Pathol Exp Microbiol. 1976 Jan-Jun;35(1-2):111–118. [PubMed] [Google Scholar]
- Light J., Molin S. Post-transcriptional control of expression of the repA gene of plasmid R1 mediated by a small RNA molecule. EMBO J. 1983;2(1):93–98. doi: 10.1002/j.1460-2075.1983.tb01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K. M., Jones S. S., Hackett N. R., Khorana H. G. Specific amino acid substitutions in bacterioopsin: Replacement of a restriction fragment in the structural gene by synthetic DNA fragments containing altered codons. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2285–2289. doi: 10.1073/pnas.81.8.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Monod M., Denoya C., Dubnau D. Sequence and properties of pIM13, a macrolide-lincosamide-streptogramin B resistance plasmid from Bacillus subtilis. J Bacteriol. 1986 Jul;167(1):138–147. doi: 10.1128/jb.167.1.138-147.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan C. S., Dubnau D. An in vitro study of the translational attenuation model of ermC regulation. J Biol Chem. 1987 Feb 5;262(4):1756–1765. [PubMed] [Google Scholar]
- Novick R. P., Projan S. J., Kumar C. C., Carleton S., Gruss A., Highlander S. K., Kornblum J. Replication control for pT181, an indirectly regulated plasmid. Basic Life Sci. 1985;30:299–320. doi: 10.1007/978-1-4613-2447-8_24. [DOI] [PubMed] [Google Scholar]
- Okubo S., Yanagida T. Isolation of a suppressor mutant in Bacillus subtilis. J Bacteriol. 1968 Mar;95(3):1187–1188. doi: 10.1128/jb.95.3.1187-1188.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer-Abramowitz J., Gryczan T. J., Dubnau D. Origin and mode of replication of plasmids pE194 and pUB110. Plasmid. 1981 Jul;6(1):67–77. doi: 10.1016/0147-619x(81)90054-8. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Regulation of plasmid replication. Microbiol Rev. 1984 Mar;48(1):1–23. doi: 10.1016/b978-0-12-048850-6.50006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar A. G., Gryczan T. J., Kozlov Y. I., Dubnau D. Organization of the pE194 genome. Mol Gen Genet. 1980;179(2):241–252. doi: 10.1007/BF00425450. [DOI] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Dubnau D. Studies on the synthesis of plasmid-coded proteins and their control in Bacillus subtilis minicells. Plasmid. 1979 Apr;2(2):279–289. doi: 10.1016/0147-619x(79)90046-5. [DOI] [PubMed] [Google Scholar]
- Shub D. A. Nature of the suppressor of Bacillus subtilis HA101B. J Bacteriol. 1975 May;122(2):788–790. doi: 10.1128/jb.122.2.788-790.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984 Oct;38(3):861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- Ulmanen I., Lundström K., Lehtovaara P., Sarvas M., Ruohonen M., Palva I. Transcription and translation of foreign genes in Bacillus subtilis by the aid of a secretion vector. J Bacteriol. 1985 Apr;162(1):176–182. doi: 10.1128/jb.162.1.176-182.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B., Graham M. Y., Gryczan T., Dubnau D. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J Bacteriol. 1979 Jan;137(1):635–643. doi: 10.1128/jb.137.1.635-643.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Dong X., Luckow V. A., Wu R. P., Rownd R. H. Analysis of the individual regulatory components of the IncFII plasmid replication control system. J Bacteriol. 1985 Feb;161(2):534–543. doi: 10.1128/jb.161.2.534-543.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]