Abstract
Mutations in superoxide dismutase 1 (SOD1), the only proven cause of amyotrophic lateral sclerosis (ALS), provoke disease through an unidentified toxic property. Neurofilament aggregates are pathologic hallmarks of both sporadic and SOD1-mediated familial ALS. By deleting NF-L, the major neurofilament subunit required for filament assembly, onset and progression of disease caused by familial ALS-linked SOD1 mutant G85R are significantly slowed, while selectivity of mutant-mediated toxicity for motor neurons is reduced. In NF-L-deleted animals, levels of the two remaining neurofilament subunits, NF-M and NF-H, are markedly reduced in axons but are elevated in motor neuron cell bodies. Thus, while neither perikaryal nor axonal neurofilaments are essential for SOD1-mediated disease, the absence of assembled neurofilaments both diminishes selective vulnerability and slows SOD1G85R mutant-mediated toxicity to motor neurons.
Amyotrophic lateral sclerosis (ALS), the most common adult motor neuron disease, is a progressive neurodegenerative disorder occurring in middle to late life, leading to paralysis and death within 3–5 years. Approximately 10% of ALS cases are inherited in an autosomal dominant fashion. Generally indistinguishable clinically, familial ALS (FALS) and sporadic ALS (SALS) are characterized by the selective degeneration and death of lower motor neurons in the brainstem and spinal cord, leading to generalized weakness and muscle atrophy. Disease in approximately 15–20% of familial ALS patients arises from missense mutations in the gene encoding Cu/Zn superoxide dismutase 1 (SOD1) (1, 2). Currently, more than 45 missense mutations are known (3, 4) in the 153-amino acid metalloenzyme that catalyzes the dismutation of superoxide anions to hydrogen peroxide (5).
Because SOD1 is expressed ubiquitously in all eukaryotic cells, providing protection against oxygen radical-induced cellular damage (6, 7), an obvious question is how mutations in SOD1 lead to late onset motor neuron degeneration. An initial hypothesis was that SOD1-linked FALS resulted from free radical damage due to decreased SOD1 activity (2, 8). However, sets of transgenic mice expressing three different FALS mutants show progressive motor neuron disease despite elevated (9, 10) or unchanged (11, 12) levels of SOD1 activity, demonstrating that the FALS mutations in SOD1 act through the gain of a novel, cytotoxic property, rather than increased or decreased SOD1 activity. The mechanism of this cytotoxicity is as yet unknown.
The expression of different familial ALS-linked SOD1 mutants in mice produces motor neuron degeneration and death, but with a surprising variation in the pathology arising during disease progression. For example, the SODG37R (glycine changed to arginine at position 37) mutation expressed at high levels in mice results in vacuole formation in axons and dendrites (10), apparently arising from degenerating mitochondria. In contrast, the SODG85R mice show neuronal and glial inclusions that stain intensely for SOD1 and ubiquitin (13). Mice expressing high levels of the SOD1G93A mutant display vacuolar pathology (14) similar to the SOD1G37R mice, except that it is more concentrated in cell bodies (15). These mice have also been reported to develop prominent neurofilamentous accumulations (16, 60). How any of these pathological hallmarks relate to the mechanism of disease remains unproved.
Neurofilament accumulations are a common pathological hallmark in ALS (17–21), leading to the hypothesis that abnormalities in neurofilament organization may be involved in the pathogenesis of ALS. Direct evidence for the ability of aberrant neurofilaments to cause motor neuron disease came initially from transgenic mice expressing 3–4 times the normal levels of wild-type mouse NF-L subunit (22) or wild-type human NF-H subunit (23). In both cases abnormal perikaryal and axonal accumulations of neurofilaments were accompanied by motor neuron dysfunction and neurogenic atrophy of skeletal muscle, although without extensive motor neuron death (22, 24). Expression of a point mutation in the highly conserved rod domain of NF-L at levels appropriate for a dominantly inherited disease does yield selective motor neuron death (and progressive disease) reminiscent of ALS (25). A particularly provocative finding was that only the largest neurofilament-rich motor axons are at risk in sporadic ALS (26), in SOD1-mediated disease in mice (12) and in mice expressing a neurofilament mutant (25).
Neurofilaments, assembled from the subunits NF-L (65 kDa), NF-M (95 kDa), and NF-H (115 kDa), are the major intermediate filaments in many types of mature neurons and are particularly abundant in large myelinated axons. All three neurofilament subunits can co-assemble into filaments, although assembly in vivo requires expression of NF-L and a substoichiometric amount of either NF-M or NF-H (27, 28, 61). Neurofilaments become abundant axonal components only after axons have reached their targets and are required for radial growth (29–31). Furthermore, mating of mice expressing an NF-H β-galactosidase fusion polypeptide (31) that blocks transport of all neurofilaments into axons to transgenic mice that express human FALS-linked SOD1G37R (10) has demonstrated that toxicity from this SOD1 mutant does not require axonal neurofilaments (32). However, the combination of neuronal cell bodies chronically distended with masses of neurofilaments, the absence of axonal neurofilaments, and the loss of ≈25% of motor axons during aging of the NF-H β-galactosidase mice (31) leaves untested how or whether axonal or perikaryal neurofilaments contribute to the timing of SOD1 mutant-mediated disease.
To investigate the mechanism of SOD1 mutant-mediated disease and the influence of neurofilaments on it, we now examine disease onset and progression in mice expressing the human FALS-linked SOD1 mutation G85R (12) in the presence or absence of neurofilaments. When neurofilaments are absent as a consequence of disruption of the NF-L gene (30), onset of disease is significantly delayed, even from this SOD1 mutant that does not produce prominent neurofilament misaccumulation (12). Moreover, because absence of neurofilaments significantly accelerates SOD1 mutant-mediated degeneration of other neurons, neurofilaments are one determinant of the selective vulnerability of motor neurons to SOD1 mutant toxicity.
METHODS
Screening of Mice Carrying Disrupted NF-L Gene and/or the SOD1G85R Transgene.
Mice containing the disrupted NF-L fragment were identified by screening blots of BamHI-digested genomic DNA by using a 3′ probe outside of the targeting construct. The normal NF-L gene results in a 10.7-kb fragment, whereas the disrupted gene yields a 4.6-kb fragment. Mice carrying the SOD1G85R transgene were identified by PCR screening of tail DNA with primers recognizing human SOD1 (10).
SDS/PAGE and Immunoblotting.
Mouse tissues were homogenized in a buffer containing 25 mM sodium phosphate at pH 7.2, 5 mM EDTA, 1% SDS, and 1 mM phenylmethylsulfonyl fluoride. A 20-μg sample of total protein for each tissue was separated on an SDS/PAGE gel (33) and stained with Coomassie blue or transferred to nitrocellulose. Proteins were detected by using appropriate primary antibodies followed by 125I-conjugated staphylococcal protein A (Amersham) for rabbit primary antibodies and rabbit anti-mouse IgG, followed by 125I-protein A for mouse monoclonal primary antibodies. NF-L and NF-H were detected with rabbit polyclonal antibodies against oligopeptides from the carboxyl terminus of each protein (22). SOD1 was detected by using a rabbit polyclonal antibody raised against a peptide common to human and mouse SOD1 (34), and peripherin was detected by using a mouse monoclonal antibody (Chemicon).
Histopathological Analysis.
Mice anesthetized with an intraperitoneal injection of 0.02 ml/g avertin (tribromoethanol and tert-amyl alcohol) were sacrificed by transcardiac perfusion with 0.1 M sodium phosphate, followed by 4% formaldehyde and 2.5% glutaraldehyde in 0.1 M sodium phosphate, pH 7.6. Tissues were incubated in 2% osmium tetroxide in 0.05 M sodium cacodylate for 3.5 hr, washed, dehydrated, and embedded in Epon/Araldite resin (Ernest F. Fullam, Latham, New York). Sections (0.75 μm) were stained with toluidine blue and examined by light microscopy.
Quantitation of Axon Number.
The total number of axons was counted in L5 ventral and dorsal roots from five end-stage animals of each genotype and age-matched controls. The total number of axons was also counted in L5 dorsal and ventral roots of two 12-month-old NF-L −/−, SOD1G85R mice. The number of degenerating axons was determined by counting the total number of axonal profiles undergoing Wallerian degeneration in the same L5 dorsal and ventral roots.
RESULTS
Production of SOD1 Mutant Mice That Carry a Disrupted NF-L Gene.
Mice carrying both the SOD1G85R transgene and a disrupted NF-L gene were produced in a two-step mating scheme (Fig. 1A). The G85R(148) line of SOD1 mutant mice was chosen because this line has been well characterized (12) and also because the mutant SOD1 protein levels in these mice are quite low [equal to endogenous (12)], making the condition of these mice similar to the human disease and precluding any potential changes due to increased SOD1 activity found in other FALS-linked SOD1 mouse models (9, 10). SOD1G85R mutant transgenic mice were first mated to homozygous NF-L knockout mice to produce NF-L heterozygous mice, with or without the SOD1 transgene. Neither the NF-L −/− nor the SOD1G85R transgenics were fully inbred, but both were predominantly C57B6, minimizing concern over possible confounding influences of genetic background. Heterozygous NF-L mice were then mated to heterozygous NF-L mice carrying the SOD1G85R transgene. Of the offspring of this mating 12.5% are predicted to have the NF-L −/−, SOD1G85R genotype, and all of the relevant control genotypes are predicted to be represented at the same or higher levels. The number of progeny actually obtained for each genotype was as expected for Mendelian transmission (Fig. 1A).
Figure 1.
(A) The two-step mating scheme used to produce SOD1G85R, NF-L −/− mice. (B) Immunoblot of 20-μg spinal cord extracts made by using an antibody that recognizes mouse and human SOD1G85R with equal affinity. (C) Parallel Coomassie blue-stained gel demonstrating comparable protein loading. (D and E) Coomassie blue-stained SDS/PAGE gel (D) and immunoblots (E) to measure NF-H and NF-L in 20 μg of sciatic nerve or spinal cord from wild-type mice and heterozygous NF-L and homozygous NF-L knockout mice.
Quantitative immunoblotting of spinal cord extracts was used to confirm the levels of accumulated neurofilament proteins in NF-L −/−, NF-L +/−, and NF-L +/+ mice. All three neurofilament subunits were decreased by about half in spinal cord and sciatic nerve tissue from animals heterozygous for the NF-L disruption, and none could be detected in nerves of homozygously disrupted mice (Fig. 1 D and E). Very low levels of NF-H (Fig. 1E) were detected in spinal cord. These results not only demonstrate that NF-L protein expression was eliminated in the NF-L −/− mice but also show that NF-M and NF-H are highly unstable in axons and/or cell bodies. Similar results have been found in cortex, where mRNA levels for NF-M and NF-H have been shown to be normal (9, 10).
Complete Absence of Neurofilaments Slows Onset of SOD1G85R-Mediated ALS in Mice.
To test how the absence of neurofilaments affects SOD1-mediated disease, onset and progression were carefully followed in a cohort of 163 mice. Onset of disease in mice carrying the SOD1G85R transgene mutation was determined as the time when mice were unable to maintain grip when suspended from a metal grid. This loss of grip is preceded by difficulty in extending the hind limbs when suspended from the tail, but the former test was chosen as it provides a simple, unbiased assay of disease progression. Disease onset was scored independently by two observers, who performed the assays without knowing the corresponding genotypes. Comparison of SOD1 mutant animals with and without neurofilaments revealed a neurofilament dose-dependent acceleration in SOD1G85R disease onset (Fig. 2A): mice with normal neurofilament levels developed disease more rapidly [371 ± 10 days (n = 22) for SOD1G85R, NF-L +/+ mice; Table 1] than did neurofilament-free animals [416 ± 12 days (n = 14)]. This difference was highly statistically significant (P < 0.007, t = 2.9; Student’s t test). SOD1G85R mice heterozygous for the NF-L gene showed an apparent intermediate age of onset [mean onset 382 ± 8 days (n = 28)], although this did not reach statistical significance from SOD1G85R mice with normal neurofilament content (P > 0.25, t = 0.17).
Figure 2.
Delayed disease onset and increased survival in SOD1G85R mice lacking neurofilaments. Kaplan–Meier curves showing age of onset (A) and survival (B) of SOD1G85R mice in the presence or absence of neurofilaments.
Table 1.
Absence of neurofilaments slows onset of SOD1-mediated ALS in mice
| Age, days
|
|||
|---|---|---|---|
| NF-L +/+, SOD1G85R | NF-L +/−, SOD1G85R | NF-L −/−, SOD1G85R | |
| Onset | 372 ± 10 (n = 22) | 382 ± 9 (n = 28) | 416 ± 12 (n = 14) |
| Survival | 388 ± 11 (n = 24) | 397 ± 11 (n = 33) | 433 ± 14 (n = 14) |
| Duration | 21 ± 2 (n = 23) | 22 ± 2 (n = 30) | 17 ± 3 (n = 14) |
Results are mean ± SEM.
The absence of neurofilaments also produced a significantly increased lifespan (Fig. 2B; Table 1), with the average survival of 388 ± 11 (n = 24) days for SOD1G85R mice with a normal level of neurofilaments compared with 433 ± 14 (n = 14; P < 0.013, t = 2.6) days for SOD1G85R mice lacking neurofilaments. Acceleration of SOD1-mediated disease by neurofilaments is not due to any changes in SOD1 protein levels, since immunoblotting also revealed that the levels of wild-type and SOD1G85R (which migrates more slowly than the endogenous mouse SOD1; Fig. 1B) were unaffected by neurofilament content.
Neurofilaments Are Important for Efficient Survival of Motor Neurons During Development but Increase the Vulnerability of Motor Neurons to SOD1G85R Toxicity.
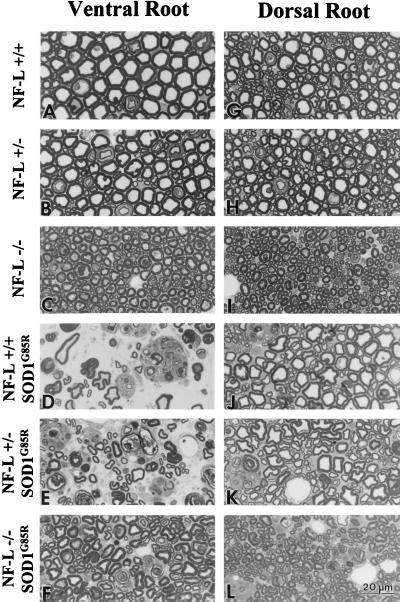
Because neurofilaments are normally an abundant structural feature of axons, a key question is whether the mechanism and the extent of degeneration are the same in SOD1G85R mice with or without neurofilaments. Examination of L5 ventral roots from mice with (Fig. 3A) and without (Fig. 3C) neurofilaments revealed the anticipated neurofilament-dependent growth in axonal caliber. Beyond this result, end-stage SOD1G85R mice displayed swollen axons, Wallerian degeneration, and loss of large motor axons in mice with normal levels (Fig. 3D), half the normal levels (Fig. 3E), or completely lacking neurofilaments (Fig. 3F). At 12 months of age (the mean age of onset for NF-L +/+, SOD1G85R mice), the NF-L −/−, SOD1G85R mice showed no axon loss or axonal degeneration (Fig. 4 A and B), indicating that the delay in onset of disease is really a delay in onset of degeneration rather than a slowing of disease progression.
Figure 3.
Reduced motor axon loss, but increased sensory loss, in SOD1G85R mice lacking neurofilaments. Ventral (A–C) and dorsal (G–I) L5 roots from wild-type, NF-L +/−, and NF-L −/− mice, respectively. Ventral (D–F) and dorsal (J–L) L5 roots from end-stage NF-L +/+, NF-L +/−, or NF-L −/− mice expressing the SOD1G85R transgene. (Scale bar, 20 μm.)
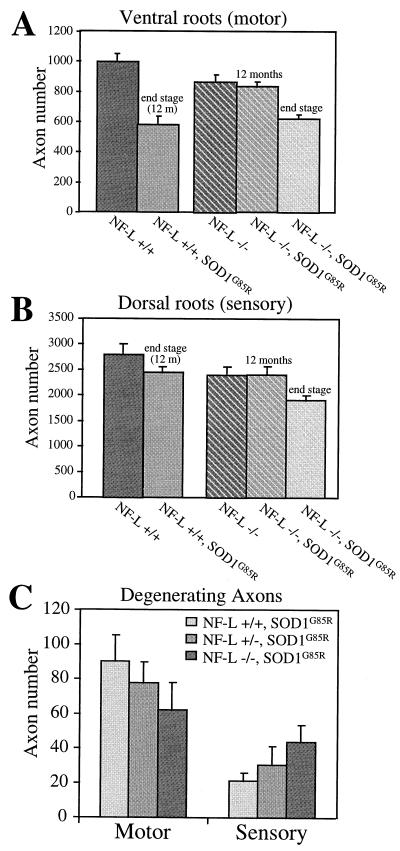
Figure 4.
Histograms to show the reduced motor axon loss (A and C), but increased sensory loss (B and C) in SOD1G85R mice lacking neurofilaments. The total number of axons (A and B) or the number of degenerating axonal profiles (C) was counted in the L5 ventral and dorsal roots of five mice of each genotype. Control mice were age matched with SOD1G85R mice with the same neurofilament content (NF-L +/+, 12 months; NF-L −/−, 13.5 months). Error bars are standard errors.
Counting of axons from mice as young as 3 months revealed that independent of the SOD1G85R transgene, the NF-L −/− mice lose 13% of their lower motor neurons early in life [as initially reported (30)]. The delay in disease onset and extension of lifespan arises in NF-L −/−, SOD1G85R mice despite this significant initial disadvantage in motor neuron number. Consequently, the NF-L +/+, SOD1G85R mice lose 42% of their initial motor axons from disease onset to end stage, whereas the NF-L −/−, SOD1G85R mice lose only 28% of their axons to reach the same end stage number of axons (mean axon loss of 416 and 243 axons respectively; Fig. 4A; Table 2). Comparison of the number of axons lost to the length of disease progression indicates a 35% increase in the rate of axon loss in the mice with neurofilaments compared with those without [19 axons per day (n = 23) versus 14 (n = 14), respectively]. The acceleration of degeneration in the presence of neurofilaments is further supported by the fact that the number of actively degenerating motor axons at end stage is higher in mice with neurofilaments than in NF-L-deleted SOD1G85R mice [90 ± 15 axons (n = 5) per L5 ventral root compared with 62 ± 10 axons (n = 5); Fig. 4C and Table 2]. However, the total number of motor axons at end stage in NF-L −/−, SOD1G85R mice is not significantly different from that of NF-L +/+, SOD1G85R mice [622 ± 29 axons for NF-L −/−, SOD1G85R (n = 5) compared with 582 ± 57 axons for NF-L +/+, SOD1G85R mice (n = 5); Table 2]. This suggests that a critical number of motor neurons are needed for the mice to survive and this number is comparable in the presence or absence of neurofilaments.
Table 2.
Reduced motor neuron degeneration in NF-L −/−, SOD1G85R mice
| Axons | Axon number
|
||||
|---|---|---|---|---|---|
| NF-L +/+ | NF-L −/− | NF-L +/+, SOD1G85R end stage | NF-L −/−, SOD1G85R 12 months | NF-L −/−, SOD1G85R end stage | |
| Ventral | 998 ± 55 | 866 ± 47 | 582 ± 57 | 835 ± 32 | 622 ± 29 |
| Dorsal | 2,791 ± 216 | 2,397 ± 164 | 2,443 ± 119 | 2,403 ± 175 | 1,910 ± 92 |
| Degenerating ventral | 0 | 0 | 90 ± 15 | 0 | 62 ± 9 |
| Degenerating dorsal | 0 | 0 | 21 ± 5 | 0 | 44 ± 2 |
Results are mean ± SEM. For all genotypes n = 5; except n = 2 for NF-L −/−, SOD1G85R at 12 months. NF-L +/+ and NF-L −/− mice are littermate controls for SOD1 mice.
Examination of the pathology arising in SOD1G85R mice with and without neurofilaments by using standard histological stains and antibodies to glial fibrillary acidic protein (GFAP) and SOD1 revealed very similar abnormalities, including reactive gliosis and SOD1 immunoreactive aggregates in astrocytes and motor neurons. Onset of both pathology and clinical disease was delayed to the same extent in the neurofilament-free mice. However, the NF-L −/−, SOD1G85R mice exhibited ½ to  as many SOD1-containing inclusions at end stage (not shown). These results suggest that inclusions may be reflecting neuronal dysfunction rather than being an obligate cause of motor neuron degeneration and death.
as many SOD1-containing inclusions at end stage (not shown). These results suggest that inclusions may be reflecting neuronal dysfunction rather than being an obligate cause of motor neuron degeneration and death.
Neurofilaments Contribute to Selective Vulnerability of Motor Neurons to SOD1G85R.
An unexplained feature of SOD1-mediated disease is the molecular basis for the selective toxicity to motor neurons. Of special note are the peripheral sensory neurons, whose large, neurofilament-rich axons share common structural features with the motor neurons. To determine the effect of neurofilament content on SOD1 mutant-mediated degeneration in these sensory axons, the L5 dorsal roots were examined at disease end stage. SOD1G85R mice with a normal complement of neurofilaments lose a small percentage of sensory neurons (Table 2; Fig. 4B). In contrast to their reduced motor neuron degeneration, SOD1G85R mice lacking neurofilaments show an increased loss of sensory axons (Fig. 4B) and an increased number of degenerating axonal profiles (Fig. 4C). At end stage, NF-L +/+, SOD1G85R mice have 21 ± 4 (n = 5) degenerating sensory axons, compared with 44 ± 2 (n = 5) in the NF-L −/−, SOD1G85R mice (Fig. 4C and Table 2; compare Fig. 3 J and L). Independent of the SOD1G85R transgene, as in the case of motor neurons, the absence of neurofilaments yields an initial sensory loss of ≈14% in the NF-L −/− mice compared with normal mice. In addition, 40% more sensory axons are lost from L5 roots of end-stage SOD1G85R mice without neurofilaments (Fig. 4B and Table 2), resulting in an overall loss of 20% of the initial sensory neurons. Thus, the high selectivity of SOD1G85R mutant toxicity on motor neurons is itself dependent on the presence of neurofilaments.
To examine the possibility that compensatory changes in other neuronal intermediate filament subunits could at least partially underlie the selective SOD1G85R toxicity to motor neurons, extracts of dorsal and ventral roots from wild-type and NF-L −/− mice were immunoblotted to determine levels of peripherin, which is normally expressed by small sensory neurons and some large motor neurons (35–37). This revealed that in normal mice peripherin is 5 times as abundant in sensory axons compared with motor axons (Fig. 5A). However, loss of neurofilaments results in reduced peripherin levels in both ventral and dorsal roots of NF-L −/− mice. Rather than compensating for absence of neurofilaments, axonal peripherin abundance is thus dependent on neurofilament accumulation and cannot contribute significantly to the selective vulnerability of motor neurons to SOD1G85R.
Figure 5.

Peripherin neither compensates for absence of neurofilaments nor contributes significantly to selective vulnerability of motor neurons to SOD1G85R toxicity. (A) Levels of peripherin in sensory and motor axons determined by immunoblotting extracts (20 μg) of ventral or dorsal root tissue from 6-month-old NF-L −/− and NF-L +/+ mice. (B) Coomassie blue stain of a parallel gel.
Slowing of SOD1G85R-Mediated Disease in NF-L Null Mice Correlates with Increased Levels of NF-M and NF-H Subunits in Cell Bodies.
NF-M and NF-H subunits cannot assemble into neurofilaments in the absence of NF-L and do not accumulate to measurable axonal levels in the absence of NF-L (Fig. 1E). To examine the fate of the NF-M and NF-H subunits in motor neuron cell bodies, antibodies were used to identify NF-M and NF-H in spinal cord sections from wild-type mice or mice heterozygous or homozygous for disruption of the NF-L gene. Although immunocytochemistry is not a highly quantitative method, in multiple sections processed in parallel, an antibody (SMI-32) that detects unphosphorylated NF-H revealed an increase in NF-H levels in motor neuron cell bodies in homozygous mice (Fig. 6B) compared with wild-type mice (Fig. 6A). Similar changes in staining intensity (not shown) were seen with NF-M antibodies (Boehringer Mannheim). Thus, reduction or elimination of NF-L is accompanied by an increased level of perikaryal NF-M and NF-H. The barely detectable levels of NF-H in spinal cord, but not nerves, of NF-L −/− mice (Fig. 1E) confirms that the NF-H is accumulating in the motor neuron cell bodies, which represent a relatively small proportion of the spinal cord protein.
Figure 6.

Higher accumulated levels of NF-H subunits in motor neuron cell bodies in mice homozygously deleted for NF-L. Unphosphorylated NF-H was detected with antibody SMI-32 in motor neuron cell bodies in frozen spinal cord sections from normal mice (A) or mice homozygous for disruption of the NF-L gene (B). (Bar, 30 μm.)
DISCUSSION
We have shown that the absence of neurofilaments results in a significant delay in disease onset and extension of lifespan in mice expressing SOD1G85R. This slowing of disease onset clearly arises from delayed axonal degeneration, despite an initial loss of 13% of the motor neurons early in life and the chronic absence of the flexible structural framework normally provided by the axonal neurofilament array. While these findings prove that neither axonal or perikaryal neurofilaments are necessary for SOD1 mutant-mediated motor neuron disease, the absence of neurofilaments does slow motor neuron degeneration and death, even in this example of SOD1 mutant-mediated motor neuron disease in which neurofilament aggregates are not prominent pathological features. Moreover, to the key question of why motor neurons are selectively vulnerable to toxicity from SOD1 mutations despite the ubiquitous expression of SOD1, the current evidence demonstrates that total, chronic absence of neurofilaments significantly reduces the high degree of selectivity of this FALS-causing SOD1 mutant for motor neurons, yielding comparable losses of motor and sensory neurons.
As to what mechanism underlies the toxicity of the SOD1 mutants and how neurofilaments contribute to it, the findings of two additional mating experiments are relevant: First, consistent with the present results is the finding that SOD1G37R can promote disease in the absence of axonal neurofilaments in the NF-H β-galactosidase mice, in which neurofilaments are trapped in the neuron cell bodies (32). However, relative to mice with normal neurofilament levels, the NF-H β-galactosidase mice are at a significant disadvantage: chronic neurofilamentous aggregates in their perikarya and a 25% loss of motor axons during the first 14 months of life (31). Thus, the comparable lifespans of mice expressing the SOD1G37R mutant alone and SOD1G37R, NF-H β-galactosidase doubly transgenic mice (32) firmly supports a conclusion that the absence of axonal neurofilaments retards motor neuron loss from the SOD1G37R mutant, similar to what we report here for SOD1G85R. In the SOD1G37R example, the simplest view is that the axonal dropout throughout development and aging because of the presence of perikaryal aggregates and absence of axonal neurofilaments is compensated for by decreased SOD1 mutant-mediated damage later in life.
Second, an additional mating experiment has tested how elevating synthesis of NF-H affects disease mediated by FALS-mutant SOD1G37R (38). Despite the prior finding that mice expressing high levels of human NF-H develop a late-onset motor neuropathy (23), expression of human NF-H at lower levels reduced SOD1G37R toxicity very significantly, increasing the mean lifespan of SOD1G37R mice by up to 65% (6 months). The increased synthesis of NF-H traps an increased proportion of neurofilaments in the perikarya of motor neurons with a corresponding reduction in axonal neurofilaments, so that maximal slowing of disease onset was achieved with marked reduction in axonal neurofilaments and increased perikaryal levels of both neurofilaments and all three subunits, especially NF-H.
Taken together with the current evidence, amelioration of SOD1 mutant toxicity correlates with reduction in axonal neurofilaments and increased perikaryal neurofilament subunits. One pleasing mechanism would be if neurofilaments, especially axonal ones, were targets for SOD-1 mediated damage. Indeed, one highly visible model has proposed that the aberrant property of mutant SOD1 subunits is that they unfold slightly, thereby allowing increased access of alternative substrates hydrogen peroxide (39) or peroxynitrite (40) to the enzyme-bound copper. While examination of patient samples (8, 41) or SOD1 mutant-expressing mice (42) has provided no in vivo evidence to support mutant-mediated use of hydrogen peroxide, evidence offering some support for accelerated use of peroxynitrite by the mutant enzymes includes the finding of elevated nitrotyrosine in mice expressing SOD1G37R mutant (42) and focally increased reactivity to a nitrotyrosine antibody reported in SOD1-mediated familial (41, 43) and sporadic (43, 44) ALS patients. Further, in vitro studies have shown that NF-L can be a preferential target for SOD1-mediated nitration and that nitration affects NF-L assembly properties (45). All of these findings would support a model of nitrated axonal neurofilaments as one of the toxic species arising from these mutants were it not for the absence of support for neurofilaments as targets for nitration in vivo (42).
It is also possible that the marked beneficial effect in both the NF-H-overexpressing mice and in mice devoid of neurofilaments arises from an increase in perikaryal levels of NF-H (and perhaps NF-M) and a reduction of axonal neurofilaments. A portion of this protective effect may arise from enhancing axonal transport by removing a major cargo whose incorporation is known to correlate with the slowing of slow axonal transport (46, 47). Another appealing explanation for the protective effect of NF-H overexpression is based on previous reports indicating that neurofilament proteins have multiple calcium-binding sites, including high-affinity sites (48, 49), suggesting neurofilament involvement in calcium homeostasis. From these studies, it is therefore conceivable that increasing NF-H protein levels might confer protection in perikarya against rises in intracellular calcium that otherwise result in mutant-mediated damage, particularly as reflected in the mitochondrial damage reported in some mutant SOD1 transgenic mice (10, 50). Direct support for involvement of calcium in SOD1-mediated disease has also come from overexpression of the calcium-binding protein calbindin D28K. This protein was recently reported to confer protection against mutant SOD1-mediated death of PC12 cells (51) and of cultured motor neurons expressing mutant SOD1G93A (Heather Durham, Montreal Neurological Institute, personal communication). A calcium involvement in ALS has been supported by the selective vulnerability of motor neurons lacking typical calcium-binding proteins parvalbumin and calbindin, as shown in studies on ALS patients and monkeys (52–55) as well as in a line of SOD1 transgenic mice (56). Indeed, if neurofilament proteins act as calcium chelators, the dramatic declines in neurofilament mRNA levels that have been reported during aging (57, 58) and to a greater extent in neurodegenerative diseases, including ALS (59), may contribute to increase the susceptibility of specific neuronal populations to calcium-mediated death.
Acknowledgments
This work has been supported by Grant NS 27036 from the National Institutes of Health and a grant from the Muscular Dystrophy Association to D.W.C. T.L.W. and L.I.B. were supported by postdoctoral fellowships from the Muscular Dystrophy Foundation. Salary support for D.W.C. is provided by the Ludwig Institute for Cancer Research.
ABBREVIATIONS
- ALS
amyotrophic lateral sclerosis
- FALS
familial ALS
- SOD1
superoxide dismutase 1
References
- 1.Rosen D R, Siddique T, Patterson D, Figlewicz D A, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan J P, Deng H X, et al. Nature (London) 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.Deng H-X, Hentati A, Tainer J A, Iqbal Z, Cayabyab A, Hung W-Y, Getzoff E D, Hu P, Herzfeldt B, Roos R P, et al. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 3.Siddique, T. & Deng, H. X. (1996) Hum. Mol. Genet. 5, Suppl. 1465–1470. [DOI] [PubMed]
- 4.Radunovic A, Leigh P N. J Neurol Neurosurg Psychiatry. 1996;61:565–572. doi: 10.1136/jnnp.61.6.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fridovich I. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 6.Halliwell B. Lancet. 1994;344:721–724. doi: 10.1016/s0140-6736(94)92211-x. [DOI] [PubMed] [Google Scholar]
- 7.Peter F, Plutner H, Zhu H, Kreis T E, Balch W E. J Cell Biol. 1993;122:1155–1167. doi: 10.1083/jcb.122.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowling A C, Schulz J B, Brown R H, Jr, Beal M F. J Neurochem. 1993;61:2322–2325. doi: 10.1111/j.1471-4159.1993.tb07478.x. [DOI] [PubMed] [Google Scholar]
- 9.Gurney M E, Pu H, Chiu A Y, Dal Canto M C, Polchow C Y, Alexander D D, Caliendo J, Hentati A, Kwon Y W, Deng H X. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 10.Wong P C, Pardo C A, Borchelt D R, Lee M K, Copeland N G, Jenkins N A, Sisodia S S, Cleveland D W, Price D L. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 11.Ripps M E, Huntley G W, Hof P R, Morrison J H, Gordon J W. Proc Natl Acad Sci USA. 1995;92:689–693. doi: 10.1073/pnas.92.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallee R B, Shpetner H S. Nature (London) 1993;365:107–108. doi: 10.1038/365107a0. [DOI] [PubMed] [Google Scholar]
- 13.Bruijn L I, Becher M W, Lee M K, Anderson K L, Jenkins N A, Copeland N G, Sisodia S S, Rothstein J D, Borchelt D R, Price D L, et al. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 14.Dal Canto M C, Gurney M E. Am J Pathol. 1994;145:1271–1279. [PMC free article] [PubMed] [Google Scholar]
- 15.Bruijn L I, Cleveland D W. Neuropathol Appl Neurobiol. 1996;22:373–387. doi: 10.1111/j.1365-2990.1996.tb00907.x. [DOI] [PubMed] [Google Scholar]
- 16.Tu P H, Raju P, Robinson K A, Gurney M E, Trojanowski J Q, Lee V M-Y. Proc Natl Acad Sci USA. 1996;93:3155–3160. doi: 10.1073/pnas.93.7.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpenter S. Neurology. 1968;18:841–851. doi: 10.1212/wnl.18.9.841. [DOI] [PubMed] [Google Scholar]
- 18.Chou S M, Fakadej A V. J Neuropathol Exp Neurol. 1971;30:42–55. doi: 10.1097/00005072-197107000-00005. [DOI] [PubMed] [Google Scholar]
- 19.Hirano A, Nakano I, Kurland L T, Mulder D W, Holley P W, Saccomanno G. J Neuropathol Exp Neurol. 1984;43:471–480. doi: 10.1097/00005072-198409000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Hirano A, Donnenfeld H, Sasaki S, Nakano I. J Neuropathol Exp Neurol. 1984;43:461–470. doi: 10.1097/00005072-198409000-00001. [DOI] [PubMed] [Google Scholar]
- 21.Hirano A. Adv Neurol. 1991;56:91–101. [PubMed] [Google Scholar]
- 22.Xu Z, Cork L C, Griffin J W, Cleveland D W. Cell. 1993;73:23–33. doi: 10.1016/0092-8674(93)90157-l. [DOI] [PubMed] [Google Scholar]
- 23.Cote F, Collard J F, Julien J P. Cell. 1993;73:35–46. doi: 10.1016/0092-8674(93)90158-m. [DOI] [PubMed] [Google Scholar]
- 24.Collard J F, Cote F, Julien J P. Nature (London) 1995;375:61–64. doi: 10.1038/375061a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee M K, Marszalek J R, Cleveland D W. Neuron. 1994;13:975–988. doi: 10.1016/0896-6273(94)90263-1. [DOI] [PubMed] [Google Scholar]
- 26.Kawamura Y, Dyck P J, Shimono M, Okazaki H, Tateishi J, Doi H. J Neuropathol Exp Neurol. 1981;40:667–675. doi: 10.1097/00005072-198111000-00008. [DOI] [PubMed] [Google Scholar]
- 27.Lee M K, Xu Z, Wong P C, Cleveland D W. J Cell Biol. 1993;122:1337–1350. doi: 10.1083/jcb.122.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ching G Y, Liem R K. J Cell Biol. 1993;122:1323–1335. doi: 10.1083/jcb.122.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohara O, Gahara Y, Miyake T, Teraoka H, Kitamura T. J Cell Biol. 1993;121:387–395. doi: 10.1083/jcb.121.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- 31.Eyer J, Peterson A. Neuron. 1994;12:389–405. doi: 10.1016/0896-6273(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 32.Eyer J, Cleveland D W, Wong P, Peterson A C. Nature (London) 1998;391:584–587. doi: 10.1038/35378. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Pardo C A, Xu Z, Borchelt D R, Price D L, Sisodia S S, Cleveland D W. Proc Natl Acad Sci USA. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonard D G B, Gorham J D, Cole P, Green L A, Ziff E B. J Cell Biol. 1988;106:181–193. doi: 10.1083/jcb.106.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parysek L M, Goldman R D. J Neurosci. 1988;8:555–563. doi: 10.1523/JNEUROSCI.08-02-00555.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brody B A, Ley C A, Parysek L M. J Neurosci. 1989;9:2391–2401. doi: 10.1523/JNEUROSCI.09-07-02391.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Couillard-Depres S, Zhu Q, Wong P, Price D L, Cleveland D W, Julien J-P. Proc Natl Acad Sci USA. 1998;95:9626–9630. doi: 10.1073/pnas.95.16.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiedau-Pazos M, Goto J J, Rabizadeh S, Gralla E D, Roe J A, Valentine J S, Bredesen D E. Science. 1996;271:515–518. doi: 10.1126/science.271.5248.515. [DOI] [PubMed] [Google Scholar]
- 40.Beckman J S, Carson M, Smith C D, Koppenol W H. Nature (London) 1993;364:584. doi: 10.1038/364584a0. [DOI] [PubMed] [Google Scholar]
- 41.Beal M F, Ferrante R J, Browne S E, Matthews R T, Kowall N W, Brown R H. Ann Neurol. 1997;42:644–654. doi: 10.1002/ana.410420416. [DOI] [PubMed] [Google Scholar]
- 42.VanRenterghem B, Gibbs J B, Maller J L. J Biol Chem. 1993;268:19935–19938. [PubMed] [Google Scholar]
- 43.Bruijn L I, Beal M F, Becher M W, Schulz J B, Wong P C, Price D L, Cleveland D W. Proc Natl Acad Sci USA. 1997;94:7606–7611. doi: 10.1073/pnas.94.14.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abe K, Pan L H, Watanabe M, Kato T, Itoyama Y. Neurosci Lett. 1995;199:152–154. doi: 10.1016/0304-3940(95)12039-7. [DOI] [PubMed] [Google Scholar]
- 45.Crow J P, Ye Z Y, Strong M, Kirk M, Barnes S, Beckman J S. J Neurochem. 1997;69:1945–1953. doi: 10.1046/j.1471-4159.1997.69051945.x. [DOI] [PubMed] [Google Scholar]
- 46.Hoffman P N, Lasek R J, Griffin J W, Price D L. J Neurosci. 1983;3:1694–1700. doi: 10.1523/JNEUROSCI.03-08-01694.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willard M, Simon C. Cell. 1983;35:551–559. doi: 10.1016/0092-8674(83)90189-7. [DOI] [PubMed] [Google Scholar]
- 48.Lefebvre S, Mushynski W E. Biochem Biophys Res Commun. 1987;145:1006–1011. doi: 10.1016/0006-291x(87)91535-x. [DOI] [PubMed] [Google Scholar]
- 49.Lefebvre S, Mushynski W E. Biochemistry. 1988;27:8503–8508. doi: 10.1021/bi00422a031. [DOI] [PubMed] [Google Scholar]
- 50.Dal Canto M C, Gurney M E. Brain Res. 1995;676:25–40. doi: 10.1016/0006-8993(95)00063-v. [DOI] [PubMed] [Google Scholar]
- 51.Ghadge G D, Lee J P, Bindokas V P, Jordan J, Ma L, Miller R J, Roos R P. J Neurosci. 1997;17:8756–8766. doi: 10.1523/JNEUROSCI.17-22-08756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiner A, Medina L, Figueredo-Cardenas G, Anfinson S. Exp Neurol. 1995;131:239–250. doi: 10.1016/0014-4886(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 53.Elliott J L, Snider W D. NeuroReport. 1995;6:449–452. doi: 10.1097/00001756-199502000-00011. [DOI] [PubMed] [Google Scholar]
- 54.Ince P, Stout N, Shaw P, Slade J, Hunziker W, Neizmann C W, Baimbridge K G. Neuropathol Appl Neurobiol. 1993;19:291–299. doi: 10.1111/j.1365-2990.1993.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 55.Ho B K, Alexianu M E, Colom L V, Mohamed A H, Serrano F, Appel S. Proc Natl Acad Sci USA. 1996;93:6796–6801. doi: 10.1073/pnas.93.13.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison B M, Gordon J W, Ripps M E, Morrison J H. J Comp Neurol. 1996;373:619–631. doi: 10.1002/(SICI)1096-9861(19960930)373:4<619::AID-CNE9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 57.Parhad I M, Scott J N, Cellars L A, Bains J S, Krekowski C A, Clark A. J Neurosci Res. 1995;41:355–366. doi: 10.1002/jnr.490410308. [DOI] [PubMed] [Google Scholar]
- 58.Kuchel G A, Poon T, Irshad K, Richard C, Julien J P, Cowen T. NeuroReport. 1996;7:1353–1359. doi: 10.1097/00001756-199605310-00004. [DOI] [PubMed] [Google Scholar]
- 59.Bergeron C, Beric-Maskarel K, Muntasser S, Weyer L, Somerville M J, Percy M E. J Neuropathol Exp Neurol. 1994;53:221–230. doi: 10.1097/00005072-199405000-00002. [DOI] [PubMed] [Google Scholar]
- 60.Zhang B, Tu P, Abtahian F, Trojanowski J Q, Lee V M. J Cell Biol. 1997;139:1307–1315. doi: 10.1083/jcb.139.5.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elder G A, Friedrich U L, Bosco P, Kang C, Gourov A, Tu P H, Lee V M, Lazzarini R A. J Cell Biol. 1998;141:727–737. doi: 10.1083/jcb.141.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]