Abstract
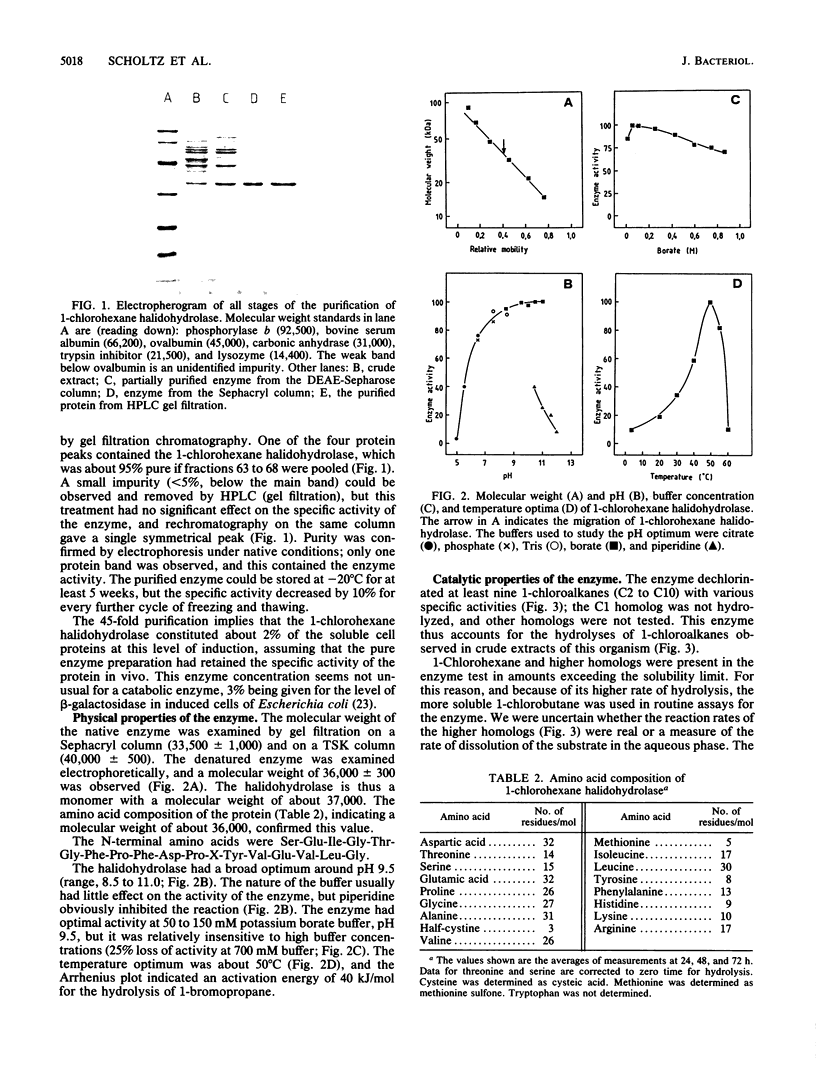
1-Chlorohexane halidohydrolase from Arthrobacter sp. strain HA1 was purified to homogeneity by fractional precipitation, ion-exchange chromatography, gel filtration, and high-performance liquid chromatography gel filtration. The enzyme was a monomer with a molecular weight of about 37,000; its amino acid composition and N-terminal sequence were determined. The enzyme had a broad optimum around pH 9.5, a temperature optimum near 50 degrees C, an activation energy of 40 kJ/mol, and a molecular activity of 0.9 kat/mol. The substrate range of the enzyme included at least 50 halogenated compounds. 1-Chloroalkanes (C3 to C10), 1-bromoalkanes (C1 to C9), and 1-iodoalkanes (C1 to C7), but no 1-fluoroalkane, were substrates. Subterminally substituted, branched-chain, and nonsaturated haloalkanes were dehalogenated. Some halogenated aromatic substrates, e.g., bromobenzene and benzyl bromide, were hydrolyzed. Several alpha,omega-dihaloalkanes were subject to double dehalogenation. Thus, 1,2-dibromoethane was hydrolyzed first to 2-bromoethanol and then to 1,2-dihydroxyethane. Crude extracts of strain HA1 were found to contain a debrominase that cleaved bromoalkanes with long alkyl chains.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. The gel-filtration behaviour of proteins related to their molecular weights over a wide range. Biochem J. 1965 Sep;96(3):595–606. doi: 10.1042/bj0960595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Eisenthal R., Cornish-Bowden A. The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. Biochem J. 1974 Jun;139(3):715–720. doi: 10.1042/bj1390715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank G., Zuber H. The complete amino acid sequences of both subunits of the sweet protein monellin. Hoppe Seylers Z Physiol Chem. 1976 Apr;357(4):585–592. doi: 10.1515/bchm2.1976.357.1.585. [DOI] [PubMed] [Google Scholar]
- GOLDMAN P. THE ENZYMATIC CLEAVAGE OF THE CARBON-FLUORINE BOND IN FLUOROACETATE. J Biol Chem. 1965 Aug;240:3434–3438. [PubMed] [Google Scholar]
- Janssen D. B., Jager D., Witholt B. Degradation of n-haloalkanes and alpha, omega-dihaloalkanes by wild-type and mutants of Acinetobacter sp. strain GJ70. Appl Environ Microbiol. 1987 Mar;53(3):561–566. doi: 10.1128/aem.53.3.561-566.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuning S., Janssen D. B., Witholt B. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J Bacteriol. 1985 Aug;163(2):635–639. doi: 10.1128/jb.163.2.635-639.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler-Staub D., Leisinger T. Dichloromethane dehalogenase of Hyphomicrobium sp. strain DM2. J Bacteriol. 1985 May;162(2):676–681. doi: 10.1128/jb.162.2.676-681.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Motosugi K., Soda K. Microbial degradation of synthetic organochlorine compounds. Experientia. 1983 Nov 15;39(11):1214–1220. doi: 10.1007/BF01990358. [DOI] [PubMed] [Google Scholar]
- Nelson M. J., Montgomery S. O., O'neill E. J., Pritchard P. H. Aerobic metabolism of trichloroethylene by a bacterial isolate. Appl Environ Microbiol. 1986 Aug;52(2):383–384. doi: 10.1128/aem.52.2.383-384.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. N., Hou C. T., Laskin A. I., Felix A. Microbial Oxidation of Hydrocarbons: Properties of a Soluble Methane Monooxygenase from a Facultative Methane-Utilizing Organism, Methylobacterium sp. Strain CRL-26. Appl Environ Microbiol. 1982 Nov;44(5):1130–1137. doi: 10.1128/aem.44.5.1130-1137.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]