Abstract
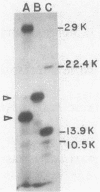
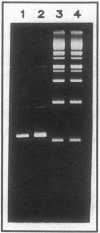
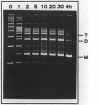
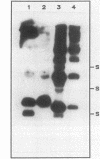
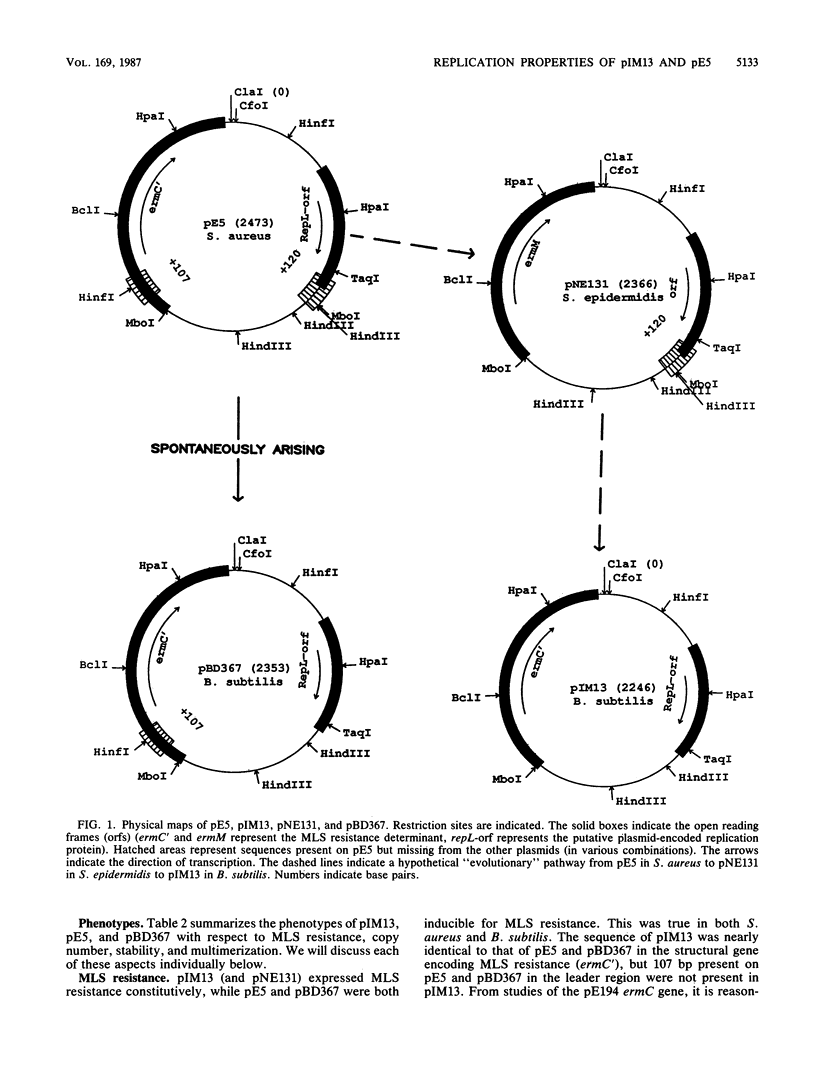
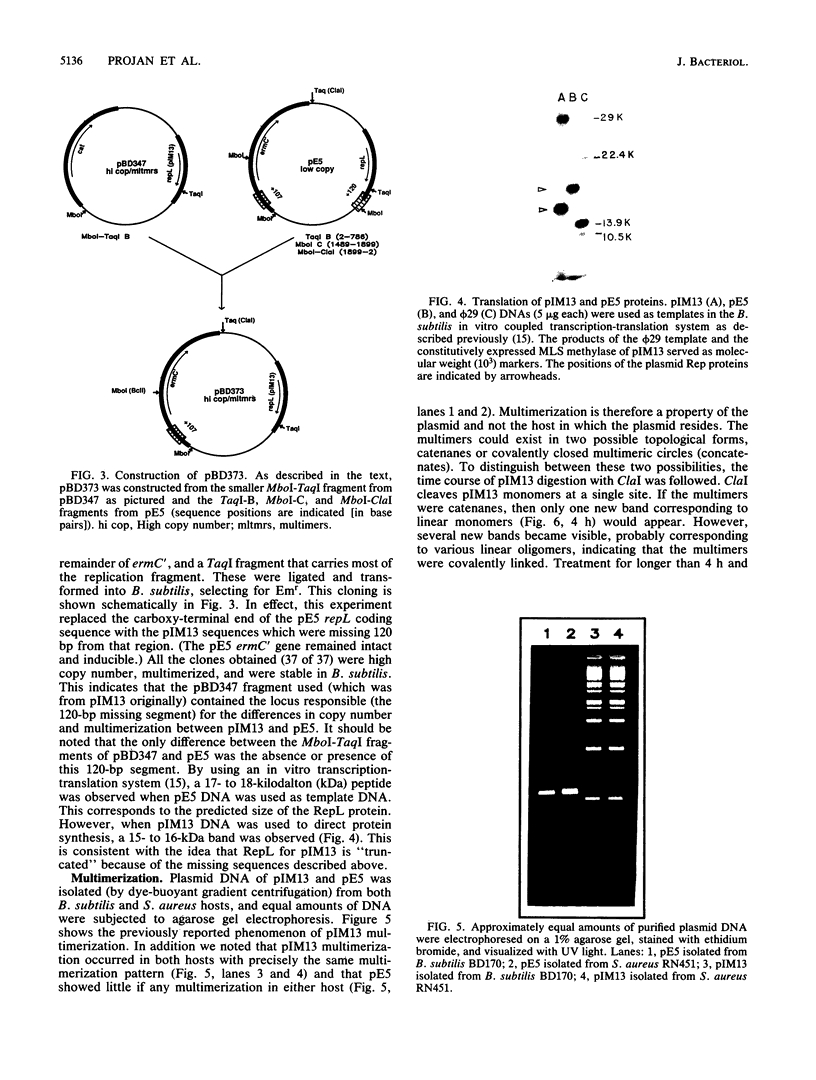
A naturally occurring plasmid from Bacillus subtilis, pIM13, codes for constitutively expressed macrolide-lincosamide-streptogramin B (MLS) resistance, is stably maintained at a high copy number, and exists as a series of covalent multimers. The complete sequence of pIM13 has been reported (M. Monod, C. Denoya, and D. Dubnau, J. Bacteriol. 167:138-147, 1986) and two long open reading frames have been identified, one of which (ermC') is greater than 90% homologous to the ermC MLS resistance determinant of the Staphylococcus aureus plasmid pE194. The second reading frame (repL) shares homology with the only long open reading frame of the cryptic S. aureus plasmid pSN2 and is probably involved in plasmid replication. The map of pIM13 is almost a precise match with that of pE5, a naturally occurring, stable, low-copy-number, inducible MLS resistance plasmid found in S. aureus. pIM13 is unstable in S. aureus but still multimerizes in that host, while pE5 is unstable in B. subtilis and does not form multimers in either host. The complete sequence of pE5 is presented, and comparison between pIM13 and pE5 revealed two stretches of sequence present in pE5 that were missing from pIM13. It is likely that a 107-base-pair segment in the ermC' leader region missing from pIM13 accounts for the constitutive nature of the pIM13 MLS resistance and that the lack of an additional 120-base-pair segment in pIM13 that is present on pE5 gives rise to the high copy number, stability, and multimerization in B. subtilis. The missing 120 base pairs occur at the carboxyl-terminal end of the putative replication protein coding sequence and results in truncation of that protein. It is suggested either that the missing segment contains a site involved in resolution of multimers into monomers or that the smaller replication protein causes defective termination of replication. It is concluded that pIM13 and pE5 are coancestral plasmids and it is probable that pIM13 arose from pE5.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakour R., Laroche Y., Cornelis G. Study of the incompatibility and replication of the 70-kb virulence plasmid of Yersinia. Plasmid. 1983 Nov;10(3):279–289. doi: 10.1016/0147-619x(83)90042-2. [DOI] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Contente S., Dubnau D. Characterization of plasmid transformation in Bacillus subtilis: kinetic properties and the effect of DNA conformation. Mol Gen Genet. 1979 Jan 2;167(3):251–258. doi: 10.1007/BF00267416. [DOI] [PubMed] [Google Scholar]
- Docherty A., Grandi G., Grandi R., Gryczan T. J., Shivakumar A. G., Dubnau D. Naturally occurring macrolide-lincosamide-streptogramin B resistance in Bacillus licheniformis. J Bacteriol. 1981 Jan;145(1):129–137. doi: 10.1128/jb.145.1.129-137.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A. D., Ross H. F., Novick R. P. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2165–2169. doi: 10.1073/pnas.84.8.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J., Dubnau D. Analysis of plasmid deletional instability in Bacillus subtilis. J Bacteriol. 1985 Jun;162(3):1014–1023. doi: 10.1128/jb.162.3.1014-1023.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Structural analysis of plasmid pSN2 in Staphylococcus aureus: no involvement in enterotoxin B production. J Bacteriol. 1982 Feb;149(2):642–649. doi: 10.1128/jb.149.2.642-649.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson B. C., Parisi J. T. Nucleotide sequence of the constitutive macrolide-lincosamide-streptogramin B resistance plasmid pNE131 from Staphylococcus epidermidis and homologies with Staphylococcus aureus plasmids pE194 and pSN2. J Bacteriol. 1986 Sep;167(3):888–892. doi: 10.1128/jb.167.3.888-892.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler I., Halvorson H. O. Two erythromycin-resistance plasmids of diverse origin and their effect on sporulation in Bacillus subtilis. J Gen Microbiol. 1980 Sep;120(1):259–263. doi: 10.1099/00221287-120-1-259. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Monod M., Denoya C., Dubnau D. Sequence and properties of pIM13, a macrolide-lincosamide-streptogramin B resistance plasmid from Bacillus subtilis. J Bacteriol. 1986 Jul;167(1):138–147. doi: 10.1128/jb.167.1.138-147.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R. E., Ano T., Imanaka T., Aiba S. Complete nucleotide sequences of Bacillus plasmids pUB110dB, pRBH1 and its copy mutants. Mol Gen Genet. 1986 Jan;202(1):169–171. doi: 10.1007/BF00330534. [DOI] [PubMed] [Google Scholar]
- Murphy E. Inhibition of Tn554 transposition: deletion analysis. Plasmid. 1983 Nov;10(3):260–269. doi: 10.1016/0147-619x(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Narayanan C. S., Dubnau D. An in vitro study of the translational attenuation model of ermC regulation. J Biol Chem. 1987 Feb 5;262(4):1756–1765. [PubMed] [Google Scholar]
- Novick R. P., Brodsky R. Studies on plasmid replication. I. Plasmid incompatibility and establishment in Staphylococcus aureus. J Mol Biol. 1972 Jul 21;68(2):285–302. doi: 10.1016/0022-2836(72)90214-8. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Murphy E., Gryczan T. J., Baron E., Edelman I. Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid. 1979 Jan;2(1):109–129. doi: 10.1016/0147-619x(79)90010-6. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Projan S. J., Kumar C. C., Carleton S., Gruss A., Highlander S. K., Kornblum J. Replication control for pT181, an indirectly regulated plasmid. Basic Life Sci. 1985;30:299–320. doi: 10.1007/978-1-4613-2447-8_24. [DOI] [PubMed] [Google Scholar]
- Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967 Sep;33(1):155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Carleton S., Novick R. P. Determination of plasmid copy number by fluorescence densitometry. Plasmid. 1983 Mar;9(2):182–190. doi: 10.1016/0147-619x(83)90019-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer-Abramowitz J., Gryczan T. J., Dubnau D. Origin and mode of replication of plasmids pE194 and pUB110. Plasmid. 1981 Jul;6(1):67–77. doi: 10.1016/0147-619x(81)90054-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Summers D. K., Sherratt D. J. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984 Apr;36(4):1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]