Abstract
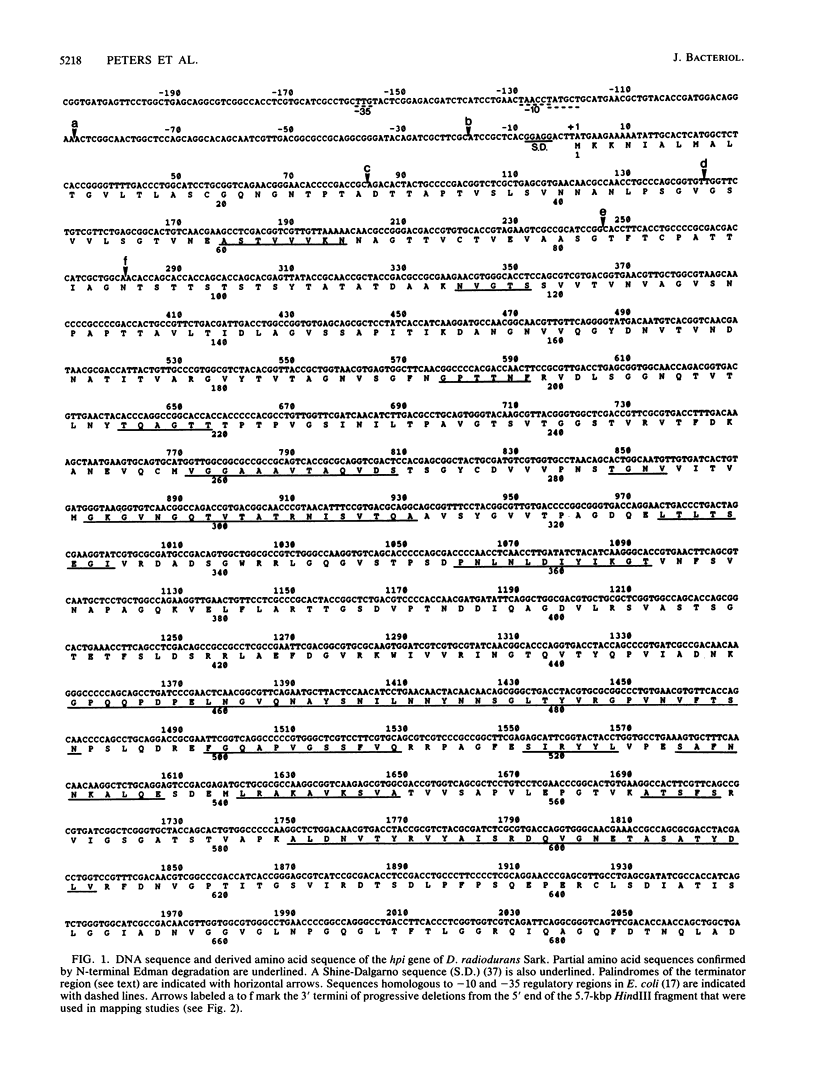
The complete nucleotide sequence of the gene encoding the surface (hexagonally packed intermediate [HPI])-layer polypeptide of Deinococcus radiodurans Sark was determined and found to encode a polypeptide of 1,036 amino acids. Amino acid sequence analysis of about 30% of the residues revealed that the mature polypeptide consists of at least 978 amino acids. The N terminus was blocked to Edman degradation. The results of proteolytic modification of the HPI layer in situ and Mr estimations of the HPI polypeptide expressed in Escherichia coli indicated that there is a leader sequence. The N-terminal region contained a very high percentage (29%) of threonine and serine, including a cluster of nine consecutive serine or threonine residues, whereas a stretch near the C terminus was extremely rich in aromatic amino acids (29%). The protein contained at least two disulfide bridges, as well as tightly bound reducing sugars and fatty acids.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baumeister W., Barth M., Hegerl R., Guckenberger R., Hahn M., Saxton W. O. Three-dimensional structure of the regular surface layer (HPI layer) of Deinococcus radiodurans. J Mol Biol. 1986 Jan 20;187(2):241–250. doi: 10.1016/0022-2836(86)90231-7. [DOI] [PubMed] [Google Scholar]
- Baumeister W., Karrenberg F., Rachel R., Engel A., ten Heggeler B., Saxton W. O. The major cell envelope protein of Micrococcus radiodurans (R1). Structural and chemical characterization. Eur J Biochem. 1982 Jul;125(3):535–544. doi: 10.1111/j.1432-1033.1982.tb06715.x. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein P., Balian G. Cleavage at Asn-Gly bonds with hydroxylamine. Methods Enzymol. 1977;47:132–145. doi: 10.1016/0076-6879(77)47016-2. [DOI] [PubMed] [Google Scholar]
- Brendel V., Trifonov E. N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984 May 25;12(10):4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Edman P., Begg G. A protein sequenator. Eur J Biochem. 1967 Mar;1(1):80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Devereux J., Burgess R. R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry R. J., Blakeney A. B., Harris P. J., Stone B. A. Detection of neutral and aminosugars from glycoproteins and polysaccharides as their alditol acetates. J Chromatogr. 1983 Feb 18;256(3):419–427. doi: 10.1016/s0021-9673(01)88259-5. [DOI] [PubMed] [Google Scholar]
- Inoyye S., Takeishi K., Lee N., DeMartini M., Hirashima A., Inouye M. Lipoprotein from the outer membrane of Escherichia coli: purification, paracrystallization, and some properties of its free form. J Bacteriol. 1976 Jul;127(1):555–563. doi: 10.1128/jb.127.1.555-563.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa Y., Nojima H., Nukiwa N., Ishizuka M., Nakajima T., Yasuhara T., Tanaka T., Oshima T. High guanine plus cytosine content in the third letter of codons of an extreme thermophile. DNA sequence of the isopropylmalate dehydrogenase of Thermus thermophilus. J Biol Chem. 1984 Mar 10;259(5):2956–2960. [PubMed] [Google Scholar]
- Koval S. F., Murray R. G. The isolation of surface array proteins from bacteria. Can J Biochem Cell Biol. 1984 Nov;62(11):1181–1189. doi: 10.1139/o84-152. [DOI] [PubMed] [Google Scholar]
- Koval S. F., Murray R. The superficial protein arrays on bacteria. Microbiol Sci. 1986 Dec;3(12):357–361. [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lottspeich F., Kellermann J., Henschen A., Rauth G., Müller-Esterl W. Human low-molecular-mass kininogen. Amino-acid sequence of the light chain; homology with other protein sequences. Eur J Biochem. 1984 Jul 16;142(2):227–232. doi: 10.1111/j.1432-1033.1984.tb08275.x. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Michel H., Weyer K. A., Gruenberg H., Dunger I., Oesterhelt D., Lottspeich F. The 'light' and 'medium' subunits of the photosynthetic reaction centre from Rhodopseudomonas viridis: isolation of the genes, nucleotide and amino acid sequence. EMBO J. 1986 Jun;5(6):1149–1158. doi: 10.1002/j.1460-2075.1986.tb04340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K., Mundy A. R., Blackford H. N., Stephenson T. P. Transvesical phenolisation of the pelvic plexuses: a simple technique for the treatment of refractory detrusor instability and hyperreflexia. Urol Int. 1986;41(3):202–206. doi: 10.1159/000281198. [DOI] [PubMed] [Google Scholar]
- Nakayama N., Arai N., Bond M. W., Kaziro Y., Arai K. Nucleotide sequence of dnaB and the primary structure of the dnaB protein from Escherichia coli. J Biol Chem. 1984 Jan 10;259(1):97–101. [PubMed] [Google Scholar]
- Peters J., Baumeister W. Molecular cloning, expression, and characterization of the gene for the surface (HPI)-layer protein of Deinococcus radiodurans in Escherichia coli. J Bacteriol. 1986 Sep;167(3):1048–1054. doi: 10.1128/jb.167.3.1048-1054.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer G. F., Walkinshaw M. D., Arnott S., Morré D. J. The ribosome binding sites recognized by E. coli ribosomes have regions with signal character in both the leader and protein coding segments. Nucleic Acids Res. 1980 Sep 11;8(17):3895–3907. doi: 10.1093/nar/8.17.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleytr U. B., Messner P. Crystalline surface layers on bacteria. Annu Rev Microbiol. 1983;37:311–339. doi: 10.1146/annurev.mi.37.100183.001523. [DOI] [PubMed] [Google Scholar]
- Sleytr U. B., Sára M., Küpcü Z., Messner P. Structural and chemical characterization of S-layers of selected strains of Bacillus stearothermophilus and Desulfotomaculum nigrificans. Arch Microbiol. 1986 Oct;146(1):19–24. doi: 10.1007/BF00690152. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi A., Uchihi R., Tabata R., Takahashi Y., Hashiba H., Sasaki T., Yamagata H., Tsukagoshi N., Udaka S. Characterization of the genes coding for two major cell wall proteins from protein-producing Bacillus brevis 47: complete nucleotide sequence of the outer wall protein gene. J Bacteriol. 1986 Oct;168(1):365–373. doi: 10.1128/jb.168.1.365-373.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildhaber I., Baumeister W. The cell envelope of Thermoproteus tenax: three-dimensional structure of the surface layer and its role in shape maintenance. EMBO J. 1987 May;6(5):1475–1480. doi: 10.1002/j.1460-2075.1987.tb02389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]




