Abstract
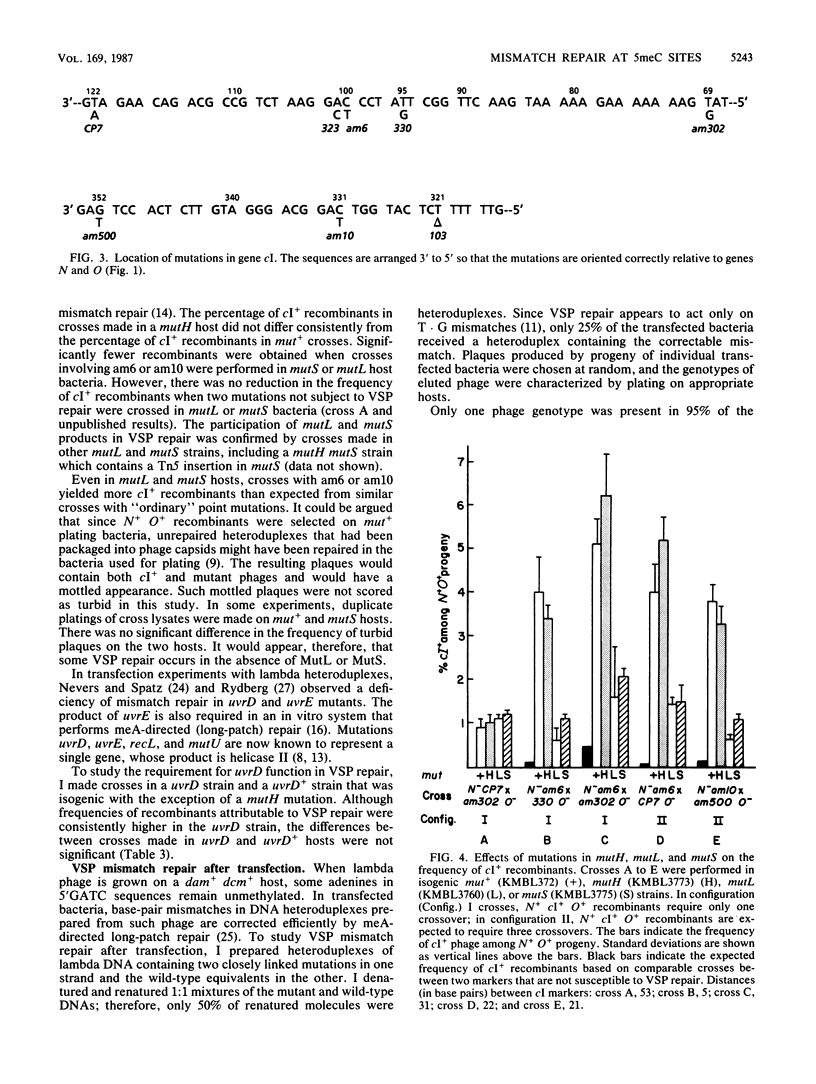
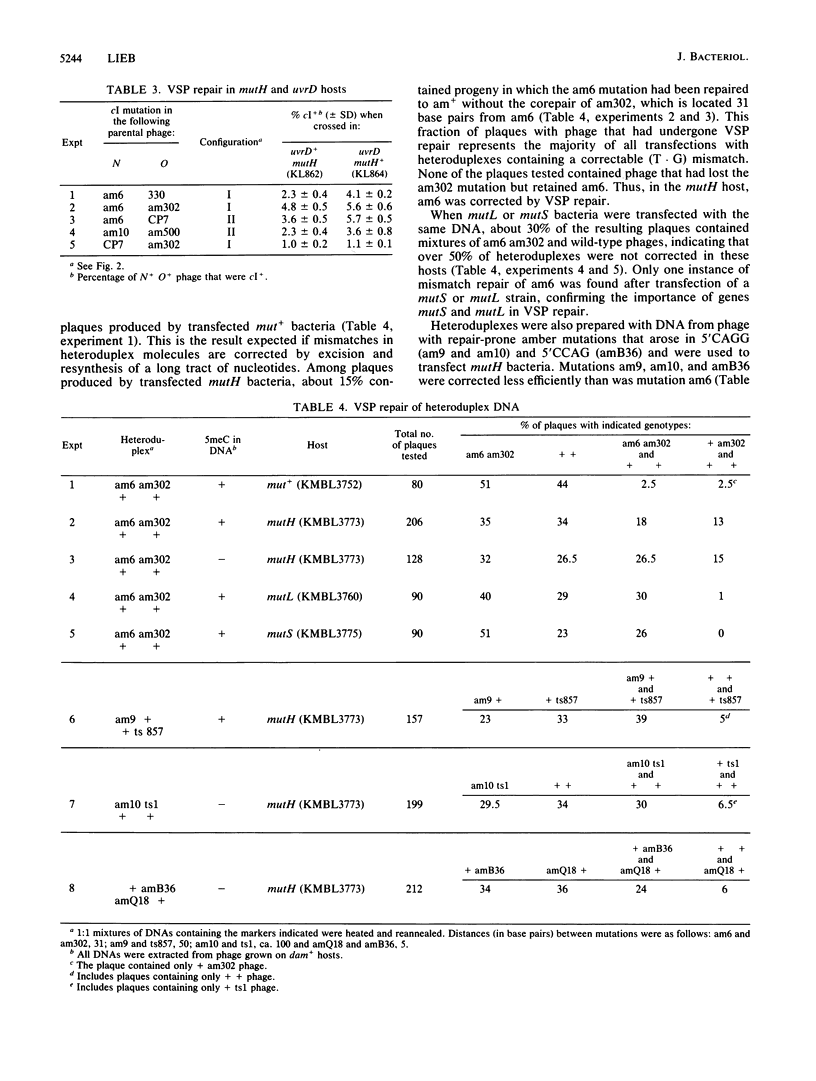
Certain amber mutations in the cI gene of bacteriophage lambda appear to recombine very frequently with nearby mutations. The aberrant mutations included C-to-T transitions at the second cytosine in 5'CC(A/T)GG sequences (which are subject to methylation by bacterial cytosine methylase) and in 5'CCAG and 5'CAGG sequences. Excess cI+ recombinants arising in crosses that utilize these mutations are attributable to the correction of mismatches by a bacterial very-short-patch (VSP) mismatch repair system. In the present study I found that two genes required for methyladenine-directed (long-patch) mismatch repair, mutL and mutS, also functioned in VSP mismatch repair; mutH and mutU (uvrD) were dispensable. VSP mismatch repair was greatly reduced in a dcm Escherichia coli mutant, in which 5-methylcytosine was not methylated. However, mismatches in heteroduplexes prepared from lambda DNA lacking 5-methylcytosine were repaired in dcm+ bacteria. These results indicate that the product of gene dcm has a repair function in addition to its methylase activity.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat A. S., Sohail A., Roberts R. J. Cloning and characterization of the dcm locus of Escherichia coli K-12. J Bacteriol. 1986 Jun;166(3):751–755. doi: 10.1128/jb.166.3.751-755.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol. 1977 Dec 15;117(3):525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein S. I., Low K. B. Hyper-recombining recipient strains in bacterial conjugation. Genetics. 1986 May;113(1):13–33. doi: 10.1093/genetics/113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B. W., Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I. D., Arthur H. M., Bramhill D., Emmerson P. T. The E. coli uvrD gene product is DNA helicase II. Mol Gen Genet. 1983;190(2):265–270. doi: 10.1007/BF00330649. [DOI] [PubMed] [Google Scholar]
- Huisman O., Fox M. S. A genetic analysis of primary products of bacteriophage lambda recombination. Genetics. 1986 Mar;112(3):409–420. doi: 10.1093/genetics/112.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M., Wagner R., Radman M. Mismatch repair of deaminated 5-methyl-cytosine. J Mol Biol. 1987 Mar 5;194(1):155–159. doi: 10.1016/0022-2836(87)90724-8. [DOI] [PubMed] [Google Scholar]
- Jones M., Wagner R., Radman M. Repair of a mismatch is influenced by the base composition of the surrounding nucleotide sequence. Genetics. 1987 Apr;115(4):605–610. doi: 10.1093/genetics/115.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Lieb M., Allen E., Read D. Very short patch mismatch repair in phage lambda: repair sites and length of repair tracts. Genetics. 1986 Dec;114(4):1041–1060. doi: 10.1093/genetics/114.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb M. Recombination in the lambda repressor gene: evidence that very short patch (VSP) mismatch correction restores a specific sequence. Mol Gen Genet. 1985;199(3):465–470. doi: 10.1007/BF00330759. [DOI] [PubMed] [Google Scholar]
- Lieb M. Specific mismatch correction in bacteriophage lambda crosses by very short patch repair. Mol Gen Genet. 1983;191(1):118–125. doi: 10.1007/BF00330898. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol Gen Genet. 1973 Dec 14;127(1):47–55. doi: 10.1007/BF00267782. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Poteete A., Arraj J. A. Correlation of DNA adenine methylase activity with spontaneous mutability in Escherichia coli K-12. Gene. 1984 Apr;28(1):123–125. doi: 10.1016/0378-1119(84)90095-7. [DOI] [PubMed] [Google Scholar]
- May M. S., Hattaman S. Deoxyribonucleic acid-cytosine methylation by host- and plasmid-controlled enzymes. J Bacteriol. 1975 Apr;122(1):129–138. doi: 10.1128/jb.122.1.129-138.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Nevers P., Spatz H. C. Escherichia coli mutants uvr D and uvr E deficient in gene conversion of lambda-heteroduplexes. Mol Gen Genet. 1975 Aug 27;139(3):233–243. doi: 10.1007/BF00268974. [DOI] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- Rydberg B. Bromouracil mutagenesis and mismatch repair in mutator strains of Escherichia coli. Mutat Res. 1978 Oct;52(1):11–24. doi: 10.1016/0027-5107(78)90091-x. [DOI] [PubMed] [Google Scholar]
- Som S., Bhagwat A. S., Friedman S. Nucleotide sequence and expression of the gene encoding the EcoRII modification enzyme. Nucleic Acids Res. 1987 Jan 12;15(1):313–332. doi: 10.1093/nar/15.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S. S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R., Fritz H. J. DNA mismatch-repair in Escherichia coli counteracting the hydrolytic deamination of 5-methyl-cytosine residues. EMBO J. 1987 Jun;6(6):1809–1815. doi: 10.1002/j.1460-2075.1987.tb02435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



