Abstract
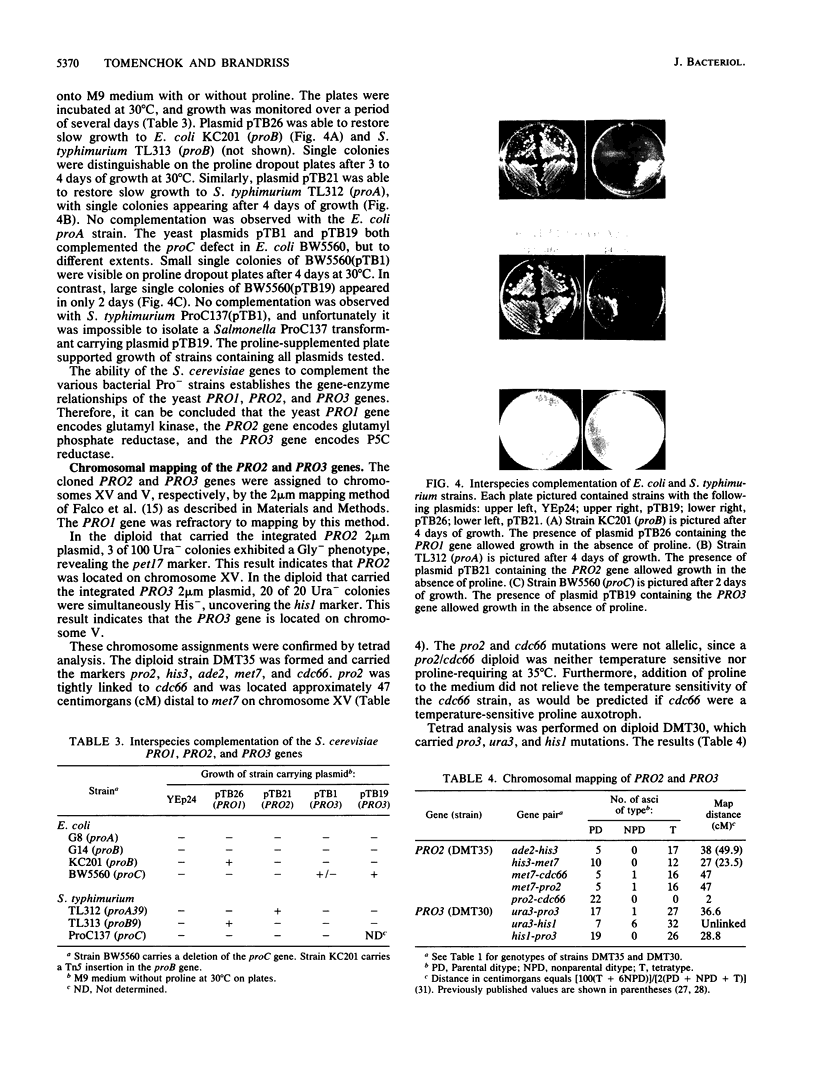
The PRO1, PRO2, and PRO3 genes were isolated by functional complementation of pro1, pro2, and pro3 (proline-requiring) strains of Saccharomyces cerevisiae. Independent clones with overlapping inserts were isolated from S. cerevisiae genomic libraries in YEp24 (2 microns) and YCp50 (CEN) plasmids. The identity of each gene was determined by gene disruption, and Southern hybridization and genetic analyses confirmed that the bona fide genes had been cloned. Plasmids containing each gene were introduced into known bacterial proline auxotrophs, and the ability to restore proline prototrophy was assessed. Interspecies complementation demonstrated that the S. cerevisiae PRO1 gene encoded gamma-glutamyl kinase, PRO2 encoded gamma-glutamyl phosphate reductase, and PRO3 encoded delta 1-pyrroline-5-carboxylate reductase. The presence of the PRO3 gene on a high-copy-number plasmid in S. cerevisiae caused a 20-fold overproduction of delta 1-pyrroline-5-carboxylate reductase. The PRO2 gene mapped on chromosome XV tightly linked to cdc66, and the PRO3 gene was located on the right arm of chromosome V between HIS1 and the centromere.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkinson K. D. Two recessive suppressors of Saccharomyces cerevisiae cho1 that are unlinked but fall in the same complementation group. Genetics. 1985 Sep;111(1):1–6. doi: 10.1093/genetics/111.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baich A. Proline synthesis in Escherichia coli. A proline-inhibitable glutamic acid kinase. Biochim Biophys Acta. 1969 Dec 30;192(3):462–467. doi: 10.1016/0304-4165(69)90395-x. [DOI] [PubMed] [Google Scholar]
- Baich A. The biosynthesis of proline in Escherichia coli: phosphate-dependent glutamate -semialdehyde dehydrogenase (NADP), the second enzyme in the pathway. Biochim Biophys Acta. 1971 Jul 20;244(1):129–134. doi: 10.1016/0304-4165(71)90129-2. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brandriss M. C. Isolation and preliminary characterization of Saccharomyces cerevisiae proline auxotrophs. J Bacteriol. 1979 Jun;138(3):816–822. doi: 10.1128/jb.138.3.816-822.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Magasanik B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: enzyme induction by proline. J Bacteriol. 1979 Nov;140(2):498–503. doi: 10.1128/jb.140.2.498-503.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Magasanik B. Subcellular compartmentation in control of converging pathways for proline and arginine metabolism in Saccharomyces cerevisiae. J Bacteriol. 1981 Mar;145(3):1359–1364. doi: 10.1128/jb.145.3.1359-1364.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT2 gene. Mol Cell Biol. 1983 Oct;3(10):1846–1856. doi: 10.1128/mcb.3.10.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutch A. H., Rushlow K. E., Smith C. J. Analysis of the Escherichia coli proBA locus by DNA and protein sequencing. Nucleic Acids Res. 1984 Aug 10;12(15):6337–6355. doi: 10.1093/nar/12.15.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutch A. H., Smith C. J., Rushlow K. E., Kretschmer P. J. Escherichia coli delta 1-pyrroline-5-carboxylate reductase: gene sequence, protein overproduction and purification. Nucleic Acids Res. 1982 Dec 11;10(23):7701–7714. doi: 10.1093/nar/10.23.7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S. C., Li Y., Broach J. R., Botstein D. Genetic properties of chromosomally integrated 2 mu plasmid DNA in yeast. Cell. 1982 Jun;29(2):573–584. doi: 10.1016/0092-8674(82)90173-8. [DOI] [PubMed] [Google Scholar]
- Hayzer D. J., Leisinger T. Proline biosynthesis in Escherichia coli. Stoichiometry and end-product identification of the reaction catalysed by glutamate semialdehyde dehydrogenase. Biochem J. 1981 Aug 1;197(2):269–274. doi: 10.1042/bj1970269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayzer D. J., Moses V. Proline biosynthesis by cell-free extracts of Escherichia coli and potential errors arising from the use of a bioradiological assay procedure. Biochem J. 1978 Jul 1;173(1):207–217. doi: 10.1042/bj1730207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C., Meeks-Wagner D. W., Fangman W. L., Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42(2):169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E. A new mapping method employing a meiotic rec-mutant of yeast. Genetics. 1982 Mar;100(3):387–412. doi: 10.1093/genetics/100.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake T, Demerec M. Proline Mutants of Salmonella Typhimurium. Genetics. 1960 Jun;45(6):755–762. doi: 10.1093/genetics/45.6.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae. Microbiol Rev. 1980 Dec;44(4):519–571. doi: 10.1128/mr.44.4.519-571.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederberger P., Miozzari G., Hütter R. Biological role of the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Jul;1(7):584–593. doi: 10.1128/mcb.1.7.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent S. A., Fenimore C. M., Bostian K. A. Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast. 1985 Dec;1(2):83–138. doi: 10.1002/yea.320010202. [DOI] [PubMed] [Google Scholar]
- Perkins D. D. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949 Sep;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzkin B., Carbon J. Functional expression of cloned yeast DNA in Escherichia coli. Proc Natl Acad Sci U S A. 1977 Feb;74(2):487–491. doi: 10.1073/pnas.74.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Vogel H. J., Bonner D. M. ON THE GLUTAMATE-PROLINE-ORNITHINE INTERRELATION IN NEUROSPORA CRASSA. Proc Natl Acad Sci U S A. 1954 Aug;40(8):688–694. doi: 10.1073/pnas.40.8.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams I., Frank L. Improved chemical synthesis and enzymatic assay of delta-1-pyrroline-5-carboxylic acid. Anal Biochem. 1975 Mar;64(1):85–97. doi: 10.1016/0003-2697(75)90408-x. [DOI] [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]
- Yura T. GENETIC ALTERATION OF PYRROLINE-5-CARBOXYLATE REDUCTASE IN NEUROSPORA CRASSA. Proc Natl Acad Sci U S A. 1959 Feb;45(2):197–204. doi: 10.1073/pnas.45.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]