Abstract
Rho-like GTPases, including Cdc42, Rac, and Rho, regulate signaling pathways that control actin cytoskeletal structures and transcriptional activation. The Tiam1 gene encodes an activator of Rac1, and similarly to constitutively activated (V12)Rac1, overexpression of Tiam1 in fibroblasts induces the formation of membrane ruffles. Tiam1 contains a Dbl homology (DH) domain and adjacent pleckstrin homology (PH) domain, hallmarks for activators of Rho-like GTPases. Unique for Tiam1 are an additional PH domain and a Discs-large homology region in the NH2-terminal part of the protein. Here we show that both in fibroblasts and COS cells, membrane localization of Tiam1 is required for the induction of membrane ruffling. A detailed mutational analysis, in combination with confocal laser scanning microscopy and immunoelectron microscopy, demonstrates that the NH2-terminal PH domain of Tiam1, but not the DH-adjacent PH domain, is essential for membrane association. This NH2-terminal PH domain of Tiam1 can be functionally replaced by the myristoylated membrane localization domain of c-Src, indicating that the primary function of this PH domain is to localize the protein at the membrane. After serum starvation, both membrane association of Tiam1 and ruffling can be induced by serum, suggesting that receptor stimulation induces membrane translocation of Tiam1. Similar to V12Rac1, Tiam1 stimulates the activity of the c-Jun NH2-terminal kinase (JNK). This Rac-dependent stimulation of JNK also requires membrane association of Tiam1. We conclude that the regulated membrane localization of Tiam1 through its NH2-terminal PH domain determines the activation of distinct Rac-mediated signaling pathways.
Rho-like GTPases, which include Cdc42, Rac1, and RhoA, control distinct signal transduction pathways that determine the organization of the actin cytoskeleton in response to various extracellular stimuli. In fibroblast cells, activation of Cdc42 and Rac results in the formation of focal complexes and filopodia or lamellipodia, respectively, while Rho is involved in the formation of stress fibers and focal contacts (Ridley and Hall, 1992; Ridley et al., 1992; Kozma et al., 1995; Nobes and Hall, 1995a ). Each of these GTPases can be activated by specific growth factors, and they may also constitute a hierarchical signaling cascade (Kozma et al., 1995; Nobes and Hall, 1995a ). The resulting cytoskeletal rearrangements influence both adhesion and motility of cells, implicating Rholike GTPases as key regulators of these processes (Nobes and Hall, 1995b ).
Apart from their effects on the cytoskeleton, both Cdc42 and Rac1 can activate the c-Jun NH2-terminal kinase (JNK)1 pathway, while all three Rho-like GTPases activate serum response factor (Coso et al., 1995; Hill et al., 1995; Minden et al., 1995; Olson et al., 1995). Rho-like proteins thus may also play a role in transcriptional activation of genes. Moreover, both Rac1 and RhoA are involved in the transformation of cells by Ras (Khosravi-Far et al., 1995; Qiu et al., 1995a ,b), and constitutively active mutants of Rac1 (V12Rac1) and RhoA (V14RhoA) can induce an oncogenic phenotype in NIH3T3 cells (Perona et al., 1993; Qiu et al., 1995a ; Van Leeuwen et al., 1995).
Rho-like GTPases are activated by guanine nucleotide dissociation stimulators (GDSs), also known as guanine nucleotide exchange factors. Inactivation occurs by hydrolysis of bound GTP, which is stimulated by interaction with GTPase activating proteins, while guanine nucleotide dissociation inhibitors (GDIs) lock the GTPases either in the active or in the inactive state (Boguski and McCormick, 1993). The role of these regulators in the diverse signaling pathways mediated by Rho-like proteins is largely unknown.
Tiam1 was originally identified in T-lymphoma cells as an invasion- and metastasis-inducing gene (Habets et al., 1994). The predicted Tiam1 protein consists of 1,591 amino acids and contains a Dbl homology (DH) domain. This domain is present in GDS proteins for Rho-like GTPases and is considered to be the catalytic domain (Hart et al., 1994). DH domains are found in a rapidly expanding group of proteins, many of which are encoded by transforming genes that were identified after transfection of genomic DNA or cDNA libraries into NIH3T3 cells (Cerione and Zheng, 1996; Collard, 1996). A number of these proteins, like Dbl, Ost, Cdc24, Lbc, and Vav were shown to activate Rho-like GTPases (Hart et al., 1991; Horii et al., 1994; Zheng et al., 1994, 1995; Olson et al., 1996). Tiam1 is the only GDS protein identified that specifically activates Rac in vitro as well as in vivo (Michiels et al., 1995). In fibroblasts, Tiam1 induces the formation of membrane ruffles, similarly to constitutively activated V12Rac1 (Michiels et al., 1995). The Tiam1-induced phenotype is inhibited by coexpression of dominant-negative mutant (N17)Rac1, but not by coexpression of (N17)Cdc42 or by inactivation of RhoA with C3 transferase (Michiels et al., 1995; Collard, 1996). This indicates that the Tiam1-induced membrane ruffling is primarily caused by activation of Rac and that other Rho-like GTPases are not involved. Moreover, V12Rac1, but not V14RhoA, induces an invasive phenotype in T-lymphoma cells, similar to Tiam1, suggesting that Tiam1-induced invasiveness is also caused by activation of Rac (Michiels et al., 1995).
Apart from the DH domain, Tiam1 contains several other conserved motifs, including a consensus myristoylation sequence at the NH2 terminus, a Discs-large homology region (DHR), and two pleckstrin homology (PH) domains (Habets et al., 1994; Collard, 1996; see Fig. 1 A). One of the PH domains is present COOH-terminally adjacent to the DH domain, as found in all other GDS proteins for Rho-like GTPases (Collard, 1996), while a second PH domain is located in the NH2-terminal part of Tiam1. DHR domains have been implicated as protein–protein interaction motifs (Ponting and Phillips, 1995). PH domains, primarily found in signaling molecules, are thought to determine the cellular localization and/or activity of these proteins by interacting with proteins or phospholipids (for review see Lemmon et al., 1996).
Figure 1.
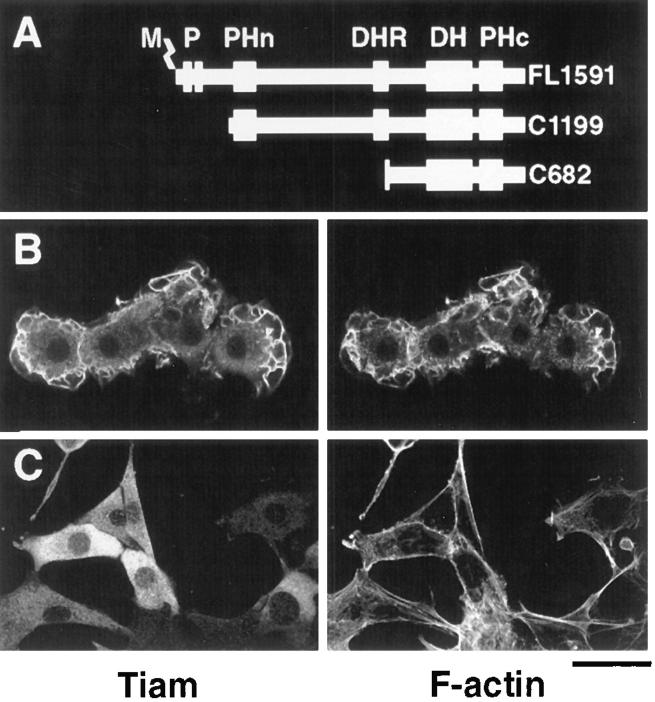
Confocal images of NIH3T3 cells expressing truncated Tiam1 proteins. A depicts the Tiam1 constructs encoding the COOH-terminal 1,199 amino acids (C1199) or 682 amino acids (C682) that were used for transfection of NIH3T3 cells, in comparison to the full-length Tiam1 protein (FL1591). Established cell lines that express the C1199 Tiam1 protein (B) or the C682 Tiam1 protein (C) were fixed and stained with anti-Tiam1 antibodies (Habets et al., 1994), followed by FITC-coupled secondary antibodies (left) and by TRITC-labeled phalloidin for F-actin (right). Note the prominent membrane ruffling in cells that express the C1199 Tiam1 protein. Stress fibers are hardly visible in these cells, since the optical sections were made through the upper half of the cells to reveal the presence or absence of membrane ruffling. No significant differences were found in the amount or appearance of stress fibers between Tiam1-expressing cells and controls. M, myristoylation signal; P, PEST regions; PHn and PHc, NH2-terminal and COOH-terminal pleckstrin homology domains. Magnification is 250×. Bar, 40 μm.
Since little is known of the intracellular localization and requirements of GDS proteins to stimulate Rho-like GTPases, we analyzed the conditions for Tiam1 to stimulate Racdependent membrane ruffling and JNK activity. For this, truncated Tiam1 proteins, or proteins that contain small deletions in each of the conserved domains, were expressed in fibroblast cells and COS cells, and their intracellular localization and biological activity were analyzed by confocal microscopy, immunoelectron microscopy, and analysis of JNK activity. We demonstrate that membrane association of Tiam1 is absolutely required for Rac-dependent induction of membrane ruffling and stimulation of JNK activity. Membrane association of Tiam1 is accomplished through the NH2-terminal PH domain, but not the COOH-terminal PH domain or the DHR domain. Moreover, PH domain– dependent membrane association of Tiam1 could be induced by serum. These results suggest that the controlled membrane association of Tiam1 is an important factor in the activation of distinct Rac-mediated signaling pathways.
Materials and Methods
Transfection Constructs
Full-length mouse Tiam1 (FL1591) and the NH2-terminally truncated C1199 and C580 Tiam1 constructs (nomenclature refers to the number of COOH-terminal amino acids [aa] of the encoded Tiam1 proteins) contain artificial SpeI and SalI restriction sites in front of the first ATG and a hemagglutinin tag (HA-tag), followed by a SalI restriction site, at their 3′ ends. They were cloned as SpeI-SalI fragments into the eukaryotic expression vector pUTSV1 (Eurogentec S.A., Belgium), which was linearized by SpeIXhoI. C1199-ΔPHc carries a small internal PstI (aa 1377) to BsmI (aa 1390) deletion in the conserved COOH-terminal part of the DH-adjacent PH domain. C1199-ΔDH1 carries a BamHI (aa 1103) to XbaI (aa 1130) deletion in a nonconserved part of the DH domain. C1199-ΔDHR carries a large NotI (aa 853) to Asp718 (aa 905) deletion in the DHR region that eliminates most of the DHR region. C1199-ΔPHn carries a small internal Asp700 (aa 513) to EarI (aa 528) deletion in the conserved COOH-terminal part of the NH2-terminal PH domain, which is quite similar to the deletion made in the COOH-terminal PH domain (C1199-ΔPHc). Overhangs were blunted by either Klenow end filling reactions (Boehringer Mannheim Corp., Indianapolis, IN) or with T4 DNA polymerase (Boehringer Mannheim Corp.) before ligation. The myristoylation sequence of c-Src, encoding the NH2-terminal 20 amino acids, was amplified with SpeISalI adaptors by PCR with Pfu polymerase (Stratagene, La Jolla, CA) and cloned between the SpeI and SalI sites at the 5′ ends of the FL1591-ΔPHn, C1191-ΔPHn, and C580 Tiam1 constructs. Fragments containing the COOHterminal PH domain of Tiam1, the PH domain of Dbl, or the PH domain of β-Ark were amplified by PCR with SmaI and PvuII adapters and used to replace the region containing the NH2-terminal PH domain of Tiam1 in the C1199 construct. The exchanged regions contain 17 additional aa at the NH2-terminal end and 36 additional aa (33 aa for β-Ark) at the COOHterminal end, in comparison to the consensus PH domain (Gibson et al., 1994). To analyze whether the NH2-terminal PH domain is sufficient for membrane targeting, a region containing the NH2-terminal PH domain, from SalI (aa 393) to BstX1 (aa 647), was fused to the Bsu36I site (aa 1022) at the NH2 terminus of the C580 Tiam1 protein. All amplified fragments were sequenced with the T7 polymerase sequencing kit (Pharmacia LKB Biotechnology Inc., Uppsala, Sweden).
Cells and Transfection Experiments
NIH3T3 cells, stably expressing the C1199 Tiam1 protein and C682 Tiam1 protein, have been described previously (Van Leeuwen et al., 1995). NIH3T3 cells and COS-7 cells were cultured in a humidified CO2 incubator in DME supplemented with 10% newborn calf serum or 10% fetal calf serum, respectively. For transient expression assays, NIH3T3 cells were seeded in dishes with or without coverslips (8,000 cells/cm2). 18 h after seeding, cells were transfected with lipofectamin (Life Technologies, Inc., Grand Island, NY). COS-7 cells were seeded in dishes with or without coverslips and transfected using DEAE dextran (Seed and Aruffo, 1987). 16 h after transfection, the medium was either replaced and cells were incubated for an additional 30 h before washing and fixation, or cells were incubated for 24 h in serum-free medium, followed by the addition of serum for 2 h. Western blot analyses were performed to monitor the size and expression levels of the mutant proteins, using either polyclonal antibodies directed against the COOH-terminal part of Tiam1 (Habets et al., 1994) or monoclonal antibodies against the HA-tag (12CA5; Boehringer Mannheim Corp.) at the COOH terminus of all mutant Tiam1 proteins. Proteins were visualized using the enhanced chemiluminescence protocol (Amersham Corp., Arlingtin Heights, IL).
Immunocytochemistry
Cells on coverslips were washed and fixed in 3.7% formaldehyde in PBS. Cells were subsequently stained as described (Michiels et al., 1995) with a polyclonal rabbit antiserum against Tiam1 (Habets et al., 1994). Antibodies were visualized with FITC-labeled secondary antibodies (Zymed Labs Inc., S. San Fransisco, CA). Cells were stained simultaneously for F-actin with TRITC-labeled phalloidin (Molecular Probes, Eugene, OR) at a concentration of 2 U/ml. At least 100 positive cells were analyzed by fluorescence microscopy and confocal laser scanning microscopy. Optical sections were taken through the apical part of the cells to show the presence or absence of membrane ruffles. Confocal images were recorded separately and subsequently merged. Colocalization of green (FITC) and red (TRITC) fluorescence appears in yellow in the merged images.
Immunoelectron Microscopy
48 h after transfection, cells were fixed for 1 h on the dish with a mixture of 4% paraformaldehyde and 0.5% glutaraldehyde in 0.1 M phosphate buffer. Fixed cells were scraped with a rubber policeman, washed, embedded in 10% gelatin, and infiltrated overnight with 1 M sucrose. After a further infiltration (2 h) with 1.8 M sucrose with 20% polyvinylpyrolidone, the samples were frozen in liquid nitrogen. Frozen thin sections were collected on formvar/carbon-coated grids, preincubated with 0.15% glycine in PBS, followed by immunoincubations with antibodies for 45 min at 25°C. Subsequently, the sections were incubated with 10-nm gold-labeled secondary antibodies for 30 min at 25°C. Antibody dilutions were made in phosphate buffer with 1% BSA and sections were washed in the same buffer, before and in between the incubation steps. Finally, sections were embedded and stained with 1.5% methylcellulose containing 0.3% uranyl acetate and examined in an electron microscope (model CM10; Philips Electronic Instrs., Mahwah, NJ). Where appropriate, the distribution of immunogold particles in at least 40 cross sections of cells was analyzed.
JNK Assay
Semiconfluent COS-7 cells or NIH3T3 cells were transfected with pCMVFlag-JNK1 and pcDNA-neo-Rac1– or Tiam1-expression vectors as described above. 48 h after transfection, cells were washed with ice-cold PBS and lysed on ice in a buffer containing 50 mM Tris-HCl, pH 7.5, 1% NP40, 100 mM NaCl, 5 mM EDTA, 100 μM sodium orthovanadate, 1 mM sodium fluoride, 20 μM aprotonin, 20 μM leupeptin, and 1 mM Pefabloc. After centrifugation (10 min, 10,000 g), flag-tagged c-Jun kinase was immunoprecipitated using 2 μg/ml of M2 monoclonal antibody (Kodak, Rochester, NY) for 1 h (4°C). Immune complexes were captured using protein A–Sepharose beads (Pharmacia LKB Biotechnology Inc.) coated with rabbit anti–mouse immunoglobulins (Nordic Immunological Labs, Capistrano Beach, CA). Beads were washed three times in lysis buffer and once in kinase buffer (20 mM Hepes, 10 mM MgCl2). Dried beads were incubated for 20 min at 30°C with 5 μg bacterially expressed glutathione S–transferase (GST)-c-Jun (amino acids 1–135) in 40 μl kinase buffer containing 1 mM DTT, 20 μM ATP, 5 μCi [γ-32P]ATP (5,000 Ci/mmol), and 0.5 mM para-nitrophenylphosphate. Reactions were stopped with SDSsample buffer, and the amount of radioactive GST-c-Jun was quantified after SDS-PAGE by phosphorimaging. The level of expression of Tiam1, mutant Rac, and tagged JNK in the NIH3T3 or COS-7 cells was monitored by Western blotting.
Results
Membrane Association of Tiam1 Is Required for Membrane Ruffling
We have shown previously that full-length Tiam1 (FL1591) or a large COOH-terminal fragment of the Tiam1 protein (C1199) causes Rac1-dependent induction of membrane ruffling in NIH3T3 fibroblast cells (Michiels et al., 1995). Established NIH3T3 cell lines expressing either of these proteins are flat and epithelial-like and contain many membrane ruffles (Fig. 1 B), a phenotype that is also induced by transfection or microinjection of constitutively activated (V12)Rac1 (Ridley et al., 1992; Michiels et al., 1995; Van Leeuwen et al., 1995). In contrast, a smaller COOH-terminal part of the Tiam1 protein (C682), which contains only the DH domain and the adjacent PH domain, did not induce this phenotype (Fig. 1 C). As shown by confocal laser scanning immunofluorescence microscopy, the truncated large C1199 Tiam1 protein was present in the cytoplasm and colocalized with F-actin in membrane ruffles (Fig. 1 B). In contrast, the short C682 Tiam1 protein seemed to be restricted to the cytoplasm (Fig. 1 C). Western blot analyses (see Fig. 4, lanes 1–3) indicated that both proteins were intact and equally expressed. This suggested that the difference in phenotypes induced by these truncated proteins is probably caused by a different intracellular localization, and not by differences in stability. Immunoelectron microscopy (immuno-EM) indeed confirmed that the C1199 Tiam1 protein is present in the cytoplasm as well as at the cell membrane and particularly in the membrane ruffles, whereas the C682 Tiam1 protein is almost exclusively located in the cytoplasm (Fig. 2). We hypothesized, therefore, that membrane localization of Tiam1 is required for morphological transformation of NIH3T3 cells, including the formation of membrane ruffles.
Figure 4.
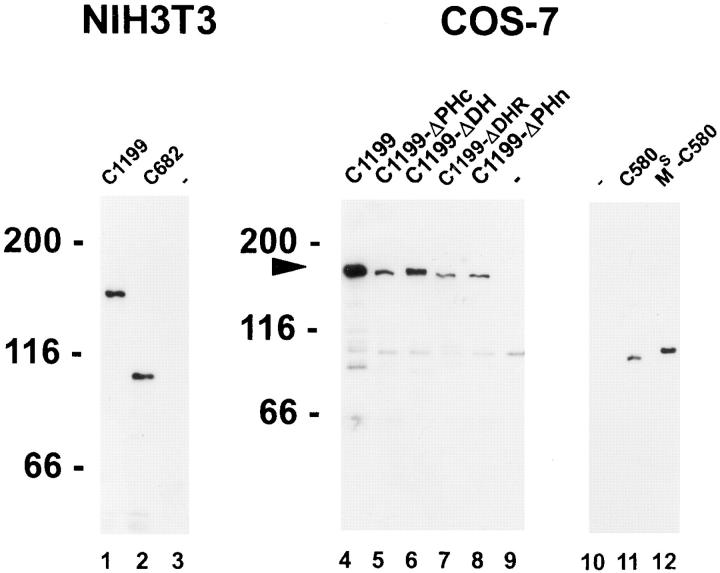
Immunoblotting of mutant Tiam1 proteins. Cell lysates were separated on a 7.5% SDS-polyacrylamide gel and blotted to nitrocellulose. All mutant C1199 and C682 Tiam1 proteins were detected with a polyclonal anti-Tiam1 antibody (Habets et al., 1994). Mutant C580 Tiam1 proteins were detected with a monoclonal antibody against the HA-tag (12CA5). Proteins were visualized using enhanced chemiluminescence (Amersham Corp.). Molecular mass markers (in kD) are shown. The arrow indicates the position of the mutant C1199 Tiam1 proteins.
Figure 2.

Immunoelectron microscopic localization of truncated Tiam1 proteins. Frozen sections of established NIH3T3 cell lines expressing either C1199 Tiam1 (A) or C682 Tiam1 (B) were stained with anti-Tiam1 antibodies followed by gold-conjugated secondary antibodies. The C682 Tiam1 protein is almost exclusively located in the cytoplasm, whereas the C1199 Tiam1 protein is also present at the plasma membrane and in the membrane ruffles. Bar, 400 nm.
The NH2-terminal PH Domain Is Essential for Membrane Localization of Tiam1
To investigate which of the conserved domains in Tiam1 determines the intracellular localization, small deletions were made in each of these domains within the C1199 construct (see Fig. 3 and Materials and Methods). These mutant proteins were transiently expressed in COS-7 cells to analyze their intracellular localization and ability to induce membrane ruffling. Transfection of these mutant Tiam1 constructs in NIH3T3 cells resulted in the same phenotypic changes (not shown). Although Tiam1 is endogenously expressed in COS-7 cells and NIH3T3 cells, the levels are too low to be visualized by immunocytochemistry or Western blotting. If phenotypic changes were induced by the transfection of Tiam1 mutants, they were found in >80% of the transfected cells.
Figure 3.



Confocal images of COS-7 cells expressing mutated C1199 Tiam1 proteins. In this figure, the first column depicts the mutant Tiam1 proteins that were used. The second column shows the FITC-staining for Tiam1 expression. The third column shows the TRITClabeled phalloidin staining for F-actin, while the merged images of Tiam1 (green) and F-actin (red) are shown in the fourth column. Colocalization appears in yellow. Deletions within the conserved domains are indicated by arrowheads. See Materials and Methods section for a description of the mutations. For abbreviations, see legend to Fig. 1. Magnification is 200×. Bar, 40 μm.
As shown in Fig. 3 B, the C1199 Tiam1 protein carrying a small internal deletion in the conserved COOH-terminal part of the DH-adjacent PH domain (C1199-ΔPHc) was still able to induce membrane ruffling. This was an unexpected finding since deletions in the DH-adjacent PH domain of other GDS proteins interfered with their proper functioning (Hart et al., 1994; Whitehead et al., 1995b ), and mutations in this region of the PH domain of β-Ark abolished interactions with both Gβγ proteins and phospholipids (Touhara et al., 1995). In contrast, a C1199 Tiam1 protein with a deletion in the DH domain (C1199ΔDH) did not induce membrane ruffling (Fig. 3 C), corroborating a previously proposed catalytic function for the DH domain (Hart et al., 1994). Deletion of most of the DHR region (C1199-ΔDHR) did not affect ruffling (Fig. 3 D), indicating that the DHR domain is not required for the induction of membrane ruffles. However, a small deletion in the NH2-terminal PH domain (C1199-ΔPHn), comparable to the deletion made in the COOH-terminal PH domain, completely abolished membrane ruffling (Fig. 3 E). Western blot analyses showed that all mutant proteins were of the predicted size and expressed at similar levels (see Fig. 4, lanes 4–9), although somewhat less than the C1199 Tiam1 protein. This excludes the possibility that these results were due to major differences in protein stability. We conclude that both the catalytic DH domain and the NH2-terminal PH domain are essential for Tiam1-induced membrane ruffling.
Immuno-EM analyses of transfected COS cells showed that all mutant proteins were equally present in the cytoplasm and at the plasma membrane (Fig. 5), except for mutant C1199 Tiam1, which contains a deletion in the NH2terminal PH domain (C1199-ΔPHn). This protein was almost exclusively localized in the cytoplasm (Fig. 5 D), similar to the C682 Tiam1 protein (Fig. 2 B). The C1199-ΔDH Tiam1 protein, which carries a deletion in the catalytic DH domain and does not induce membrane ruffling, was still capable of associating with the plasma membrane (Fig. 5 B). This clearly shows that membrane localization of Tiam1 is not a consequence of the induction of membrane ruffling. Therefore, it can be concluded that the NH2-terminal PH domain is essential for the membrane localization of C1199 Tiam1 and that this membrane localization is required for the formation of membrane ruffles.
Figure 5.

Ultrastructural localization of mutant C1199 Tiam1 proteins in COS-7 cells. Immunogold labeling of mutant C1199 Tiam1 proteins with a deletion in the COOH-terminal PH domain (A), a deletion in the DH domain (B), a deletion in the DHR domain (C), and a deletion in the NH2-terminal PH domain (D). C1199-ΔPHn Tiam1 is almost exclusively localized in the cytoplasm, while other mutant Tiam1 proteins also associate with the plasma membrane. Bar, 400 nm.
The NH2-terminal PH Domain of Tiam1 Can Be Functionally Replaced by the c-Src Membrane Localization Domain, but Not by Other PH Domains
To determine whether membrane localization is the only function of the NH2-terminal PH domain, we fused the NH2terminal 20 amino acids of c-Src, containing the myristoylation site and a basic region, in front of a C580 Tiam1 protein, resulting in MS-C580 Tiam1 (see Materials and Methods). C580 Tiam1 encompasses the COOH-terminal 580 amino acids and encodes the DH domain and adjacent PH domain (Fig. 6 A). Similar to the C682 Tiam1 protein (see Fig. 2 B), the C580 Tiam1 protein did not induce membrane ruffling and was not localized at the plasma membrane (Figs. 6 A and 7 A). In contrast, the MS-C580 Tiam1 protein induced ruffling in both NIH3T3 cells and COS-7 cells (Fig. 6 B), albeit less extensively than C1199 Tiam1 (Fig. 3 A). No differences were observed in the expression levels of these proteins (Fig. 4, lanes 10–12). Immuno-EM showed that MS-C580 Tiam1 was able to associate with the plasma membrane (Fig. 7 B), although this protein seemed to be more interiorly located than the mutant C1199 Tiam1 proteins (compare Figs. 5 and 7 B). Furthermore, part of the expressed MS-C580 Tiam1 protein was present around vesicle-like structures in the cytoplasm, similar to other MS-containing Tiam1 fusion proteins (see for example Fig. 6 E, inset). A fusion protein containing the NH2-terminal Src domain in front of the C1199-ΔPHn Tiam1 protein was also able to localize at the plasma membrane and to cause ruffling (data not shown). The Src membrane localization domain thus enables C580 Tiam1 and C1199-ΔPHn to induce membrane ruffling by tethering these proteins to the plasma membrane.
Figure 6.



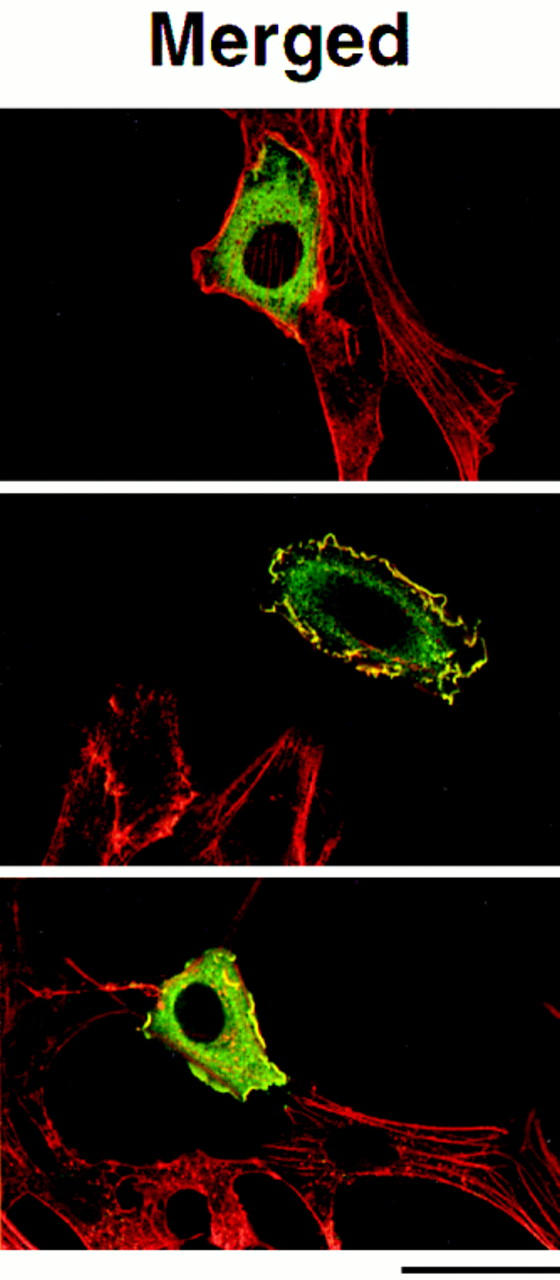
Confocal images of COS-7 cells expressing mutant Tiam1 proteins. Confocal laser scanning microscopic images of COS-7 cells expressing C580 Tiam1 (A), MS-C580 Tiam1 (B), full-length Tiam1 (FL1591) (C), full-length Tiam1 with a deletion in the NH2-terminal PH domain (FL-ΔPHn) (D), and FL-ΔPHn containing a Src myristoylation signal at the NH2 terminus (MS-FL-ΔPHn) (E). The insert in E is a higher magnification of the framed region showing the vesicle-like structures that are surrounded by the MS-FL-ΔPHn Tiam1 protein. Indicated in blue is the myristoylation signal from c-Src (MS), containing the NH2-terminal 20 amino acids. In the merged images, Tiam1 is shown in green and F-actin in red. Magnification is 200×. Bar, 40 μm.
Figure 7.
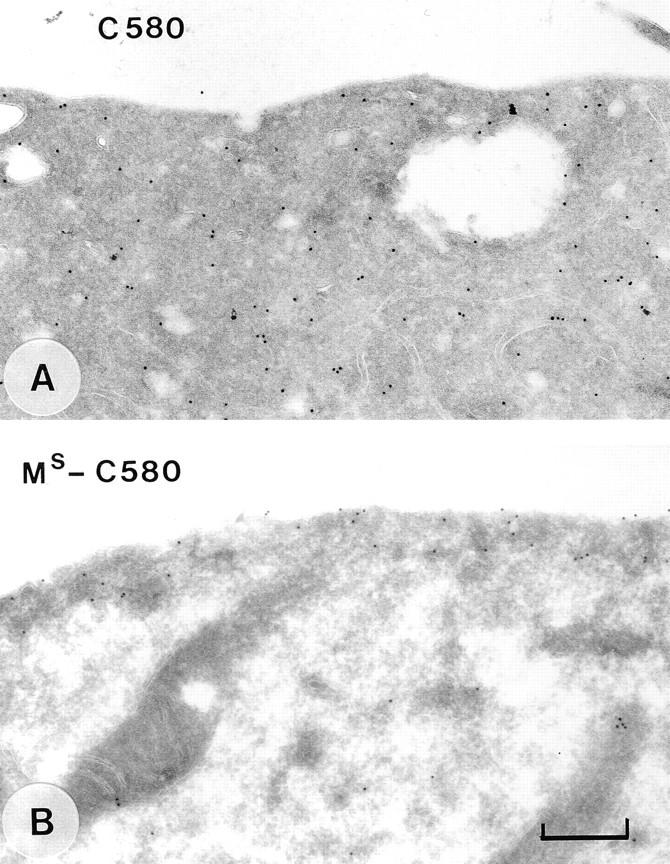
Immunoelectron microscopic localization of mutant C580 Tiam1 proteins. Immunogold labeling of transiently transfected COS cells expressing C580 Tiam1 (A) or MS-C580 Tiam1 (B). Note that the MS-C580 Tiam1 protein is located at and underneath the plasma membrane. Bar, 400 nm.
To test whether the NH2-terminal PH domain of Tiam1 is sufficient for membrane localization, we fused a region containing this PH domain to the C580 Tiam1 protein (see Materials and Methods). The resulting protein, however, did not induce membrane ruffling and was not associated with the plasma membrane (Michiels, F., and J.C. Stam, unpublished results). This might argue that additional sequences are required to ensure membrane localization. Alternatively, the juxtaposition of the PH domain to the DH domain might interfere with the proper folding of these domains. A construct expressing solely the region containing the NH2-terminal PH domain did not function as a dominant-negative Tiam1 mutant. However, C1199-ΔDH, which contains a deletion in the catalytic DH domain, also does not interfere with the induction of membrane ruffling by Tiam1. The reason for this is not clear at the moment.
To analyze the specificity of the NH2-terminal PH domain of Tiam1, a region containing this PH domain was exchanged for the corresponding regions containing the PH domain of DbL, the PH domain of β-ARK, or the COOHterminal PH domain of Tiam1 (see Materials and Methods). However, none of these mutant Tiam1 proteins localized at the plasma membrane and induced membrane ruffling (data not shown). This means either that additional sequences are required for correct functioning of these PH domains or that the primary function of the NH2terminal PH domain, namely to localize Tiam1 through specific interactions with components of the plasma membrane, cannot be substituted for by other PH domains.
The NH2-terminal PH Domain Is Required for the Induction of Membrane Ruffles by the Full-Length Tiam1 Protein
The full-length Tiam1 protein (FL1591) carries a consensus myristoylation sequence at the NH2 terminus. By labeling with [3H]myristate, we have confirmed that the NH2 terminus of Tiam1 is myristoylated (not shown). To test whether the myristoylation is sufficient for the membrane association of full-length Tiam1, a small deletion was also made in the NH2-terminal PH domain of full-length FL1591 Tiam1. However, similar to C1199-ΔPHn, mutant fulllength Tiam1 (FL1591-ΔPHn) was unable to induce membrane ruffling in NIH3T3 cells or COS cells (Fig. 6, C and D), and immuno-EM showed that the protein was not localized at the plasma membrane (data not shown). Apparently, the NH2-terminal myristoylation of full-length Tiam1 alone is not sufficient for membrane localization. To further substantiate this, the NH2-terminal c-Src sequences were fused in front of the FL1591-ΔPHn1 Tiam1 protein. This region of Src contains a basic region that is required for optimal membrane translocation in addition to the myristoylation sequence, whereas other Src-like tyrosine kinases contain a palmitoylated cysteine in this region (Superti-Furga and Courtneidge, 1995). Both additional sequences are absent in Tiam1. Cells expressing the MS-FL1591-ΔPHn1 Tiam1 protein again showed membrane ruffling, although less extensive than cells expressing FL1591 Tiam1 (Fig. 6, C–E). Again, no main differences in expression levels were observed between these proteins (data not shown). Immuno-EM confirmed that the MS-FL1591-ΔPHn1 Tiam1 protein was present at and close to the plasma membrane, as found for MS-C580 Tiam1 (data not shown). Apparently, the accessory basic region of c-Src also functions to properly localize MS-FL1591ΔPHn1 Tiam1. Similar to MS-C580, the expression of MSFL1591-ΔPHn Tiam1 led to the formation of small cytoplasmic vesicle-like structures, and the protein was also located around these structures (Fig. 6 E). These observations indicate that the NH2-terminal PH domain also serves as an essential membrane tag, or perhaps membrane anchor, for full-length Tiam1.
Controlled Cellular Localization of Tiam1
PH domains have been identified in other signaling molecules as protein–protein and/or protein–phospholipid interaction motifs that are required for the controlled targeting of these proteins to the plasma membrane (Lemmon et al., 1996). In tissue sections of skin and certain carcinomas, endogenous Tiam1 is predominantly present in the cytoplasm. Also in some cell lines, where we can envision endogenous Tiam1 by Western blotting, including T-lymphoma cells and neuronal cells, it is mainly localized in the cytoplasmic fraction (data not shown), suggesting that Tiam1 may translocate to the plasma membrane after receptor stimulation. So far, however, we have not been able to identify a receptor-mediated signaling pathway involving Tiam1 activation. To investigate whether membrane translocation of exogenous Tiam1 could be visualized in NIH3T3 cells, we analyzed whether serum could affect the localization and capacity of Tiam1 to induce membrane ruffling. As shown in Fig. 8 A, NIH3T3 cells transiently expressing the C1199 Tiam1 protein showed no membrane ruffling after serum starvation for 24 h. Almost no Tiam1 protein was present at the plasma membrane, and F-actin was mostly concentrated in lamellipodia in the Tiam1-expressing cells. Since these optical sections were taken at the basal site of the cells to illustrate the lamellipodia, stress fibers are also visible. Note that after serum starvation for 24 h, NIH3T3 cells still contain some stress fibers, in contrast to Swiss 3T3 cells. Immuno-EM confirmed that most of the cells contained significantly less Tiam1 at the plasma membrane after serum starvation (Fig. 8, D and F). The residual membrane-associated Tiam1 might be sufficient for the presence of lamellipodia under these conditions. Addition of serum induced membrane localization and subsequent ruffling of C1199 Tiam1-expressing cells after 2 h (Fig. 8, B and E), at which time point seruminduced stress fibers have already decreased. A quantification of these results is presented in Fig. 8 F. Similar results were obtained with FL1591 Tiam1, C1199-ΔPHc, and C1199ΔDHR Tiam1 (data not shown). In contrast, after 24 h of serum starvation the MS-C580 Tiam1 protein remained at the plasma membrane and still induced membrane ruffling (Fig. 8 C). This suggests that the serum-induced membrane translocation of Tiam1 is mediated by the NH2-terminal PH domain.
Figure 8.


Serum-dependent localization of Tiam1. Confocal images of NIH3T3 cells expressing C1199 Tiam1 (A and B) or MS-C580 Tiam1 (C) proteins. Cells were serum starved for 24 h (A and C), followed by stimulation with serum for 2 h (B). In the merged pictures, Tiam1 proteins are shown in green and F-actin in red. These optical sections were taken at the basal sites of the cells and reveal the presence of stress fibers in serum-starved NIH3T3 cells. Magnification is 300×. (D and E) Immuno-EM localization of C1199 Tiam1 after serum starvation (D) or after readdition of serum for 2 h (E). In F, the distribution of gold particles in at least 40 cross sections of cells were scored and presented as the percentage of cross sections with gold particles being predominantly at the membrane (M) or in the cytoplasm (C). Bars: (A–C) 40 μm; (D and E) 400 nm.
Unexpectedly, neither PDGF nor insulin could substitute for serum in the induction of membrane ruffling (data not shown). Both growth factors induce lamellipodia formation in NIH3T3 cells within 5 min, which can easily be discriminated from the Tiam1-induced ruffling. This might indicate that Tiam1 is not activated by PDGF or insulin.
Membrane-associated Tiam1 Activates JNK
To investigate whether membrane association of Tiam1 is also required for the induction of other Rac-mediated signaling pathways besides membrane ruffling, the activity of JNK was determined after cotransfection with different Tiam1 mutants (see Materials and Methods). Both in COS-7 cells and NIH3T3 cells, cotransfection of fulllength Tiam1 led to an almost sevenfold stimulation of JNK activity, similar to constitutively activated (V12)Rac1 (Fig. 9 A). Cotransfection of tagged mitogen-activated protein kinase (MAPK) with full-length or mutant Tiam1 or (V12)Rac did not result in activation of MAPK (not shown). The stimulation of JNK by Tiam1 was dependent on the activation of Rac, since cotransfection of dominantnegative (N17)Rac1 partly blocked the activation of JNK by Tiam1 (Fig. 9 A). Higher amounts of (N17)Rac interfered with the expression of JNK. Furthermore, a deletion in the catalytic DH domain of Tiam1 prevented JNK activation (not shown). Cotransfection of dominant-negative (N17)Cdc42 did not interfere with the activation of JNK by Tiam1 (data not shown). Transfection of C1199 Tiam1 stimulated JNK activity to a similar extent as FL1591 (Fig. 9 B). However, shorter COOH-terminal Tiam1 fragments such as C682 and C580 Tiam1, which did not localize to the plasma membrane, were not able to activate JNK above background levels. Also, mutant C1199-ΔPHn Tiam1, which did not localize to the plasma membrane because of the deletion in the NH2-terminal PH domain, was unable to activate JNK (Fig. 9 B). MS-C580 Tiam1 caused no stimulation of JNK activity (Fig. 9 B), and neither did MS-C1199-ΔPHn or MS-FL1591-ΔPHn Tiam1 proteins (data not shown). These proteins, however, induced membrane ruffling and were present at the plasma membrane (Fig. 6). A possible explanation for this somewhat unexpected result is that activation of endogenous Rac by MS-containing Tiam1 proteins is sufficient for induction of membrane ruffling but not for stimulation of JNK. Alternatively, the lack of induction of JNK activity is caused by the different ultrastructural localization of the MS-C580 Tiam1 protein (compare Figs. 5 and 7) or may reflect the need for an intact NH2-terminal PH domain. Interestingly, similar results were obtained with membrane-targeted p110 phosphoinositide-3-kinase which was able to stimulate membrane ruffling but not gene transcription from the Jun-inducible Fos promoter (Reif et al., 1996). Whatever the mechanism, these results indicate that the localization of Tiam1 at the plasma membrane is also required for Rac-mediated stimulation of JNK activity.
Figure 9.

Stimulation of JNK activity by Tiam1. (A) COS-7 cells were transfected with pCMV-Flag-JNK1 and empty vector (pcDNA-neo), full-length Tiam1 (FL1591), or constitutively activated (V12)Rac1. Cotransfection with dominant-negative (N17)Rac1 was performed as indicated. The activity of immunoprecipitated Flag-JNK was determined 48 h after transfection by an in vitro kinase assay using bacterially expressed GST-Jun as a substrate. The expression of Tiam1, Myc-tagged (N17)Rac, and Flag-tagged JNK was analyzed by Western blotting and is shown underneath the graph. (B) NIH3T3 cells were transfected with pCMV-Flag-JNK1 and mutant Tiam1 constructs. JNK activity was determined as described. All data are presented as fold stimulation of empty vector controls. Equal amounts of expressed Flag-JNK1 and Tiam1 proteins, as determined by Western blotting, were used. Data represent mean ± standard deviation of at least two independent experiments.
Discussion
The data presented here demonstrate that Tiam1 is a cytoplasmic protein that is capable of associating with the plasma membrane. The NH2-terminal PH domain is essential for membrane association of Tiam1, and this membrane association is required for Rac-mediated induction of membrane ruffling and stimulation of JNK activity.
A short COOH-terminal fragment of Tiam1, C682 Tiam1, which lacks the NH2-terminal PH domain, did not associate with the plasma membrane in NIH3T3 cells or COS-7 cells and was unable to induce membrane ruffling (see Figs. 1 and 2). Similar to C1199 Tiam1 and (V12)Rac1, the C682 Tiam1 protein induces a tumorigenic phenotype in NIH3T3 cells (Van Leeuwen et al., 1995). This indicates that the Tiam1-induced formation of membrane ruffles and induction of a tumorigenic phenotype are triggered by two distinct signaling pathways. Since JNK activity could not be stimulated by C682 Tiam1 and none of the truncated or mutant Tiam1 proteins activated MAPK, it is unlikely that the induction of a tumorigenic phenotype is due to activation of JNK or MAPK. An explanation could be that tumorigenicity is caused by activation of cytoplasmic Rac, although we cannot exclude that cytoplasmic Tiam1 activates another, yet unknown GTPase. Both the C1199 and C682 Tiam1 proteins were also able to induce an invasive phenotype in T-lymphoma cells (Habets et al., 1994). As the localization of Tiam1 might be differently regulated in lymphoid cells compared to adherent COS and fibroblast cells, we are currently testing whether or not JNK is involved in the Tiam1/Rac-mediated induction of an invasive phenotype in T-lymphoma cells.
A detailed structural analysis showed that a small deletion in the NH2-terminal PH domain, but not the DHadjacent COOH-terminal PH domain, abolished the membrane localization of Tiam1 and the subsequent induction of membrane ruffling. In all DH domain–containing GDS proteins analyzed thus far, the DH domain is flanked by a PH domain (Cerione and Zheng, 1996; Collard, 1996), suggesting that the DH–PH combination is a functional unit. Indeed, deletions in either the DH domain or the flanking PH domain of the protooncogenes Dbl, Ost, Dbs, and Lfc result in loss of transforming capacity (Ron et al., 1991; Horii et al., 1994; Whitehead et al., 1995a ,b; Zheng et al., 1996). The transforming capacity of a mutant of Lfc, which carried a deletion in the PH domain, could be restored by the addition of a COOH-terminal Ras CAAX motif, suggesting that the localization of Lfc at the plasma membrane is required for its transforming capacity (Whitehead et al., 1995b ). In contrast, the Ras-CAAX motif was not sufficient to replace the PH domain of Dbl, which is required for the presence of Dbl in the particulate fraction of cells (Zheng et al., 1996). Corresponding results were obtained for the NH2-terminal PH domain of Ras-guanine nucleotide release factor (GRF), suggesting that the association of Ras-GRF and Dbl in the particulate fraction is a prerequisite for their activity (Buchsbaum et al., 1996; Zheng et al., 1996). Together with pleckstrin and RasGRF, Tiam1 is one of a few proteins that contains two PH domains (Habets et al., 1994). Similarly to Ras-GRF, only the NH2-terminal PH domain of Tiam1 is involved in the localization of the protein. The COOH-terminal PH domain of Tiam1 has only moderate homology with other PH domains and lacks a highly conserved tryptophan residue in the COOH-terminal α-helix (Habets et al., 1994). Although a complete deletion of the DH-adjacent PH domain of Tiam1 abolished membrane ruffling, it did not interfere with the membrane localization of the protein (data not shown). What other functions can be attributed to this PH domain remains to be elucidated.
Several reports have demonstrated that isolated PH domains can bind in vitro to Gβγ subunits and phospholipids like phosphatidylinositol (4,5)bisphosphate (PIP2) and inositol (3,4,5)trisphosphate (IP3) (Harlan et al., 1994; Ferguson et al., 1995; Mahadevan et al., 1995; Touhara et al., 1995). Phosphoinositide-3-kinase, which stimulates the synthesis of phosphatidylinositol (3,4,5)trisphosphate (PIP3) from PIP2 (Hiles et al., 1992), has been implicated in the activation of Rac by PDGF and insulin (Wennstrom et al., 1994; Hawkins et al., 1995) and might be required for the activation of Rac by Ras (Rodriguez-Viciana et al., 1994). Although we have shown that the association of Tiam1 with the plasma membrane is induced by serum, neither PDGF nor insulin could substitute for serum in NIH3T3 cells. Gβγ subunits and phospholipids represent potential downstream effectors of G-protein–coupled receptors and/or activated tyrosine kinase receptors. Based on the in vitro binding data of PH domains, it is tempting to speculate that Gβγ subunits and/or specific phospholipids regulate the association of Tiam1 with the plasma membrane, as depicted in Fig. 10. The fact that we were not able to functionally exchange the NH2-terminal PH domain of Tiam1 for other PH domains argues for a specific targeting function for this PH domain, and similar results have been reported for PH domains in other signaling proteins (Buchsbaum et al., 1996; Kolanus et al., 1996). However, at the moment we cannot determine whether PH domains can function as independent modules or whether additional sequences are required for their activity.
Figure 10.
Model for activation of Rac by Tiam1. Activation of Rac requires membrane translocation of Tiam1 and might be analogous to activation of Ras by receptor-induced translocation of SOS. Membrane association of Tiam1 could be mediated by interactions between the NH2-terminal PH domain and putative effectors of activated G-protein coupled receptors (GPCR), tyrosine kinase receptors (TKR), or Ras, which may include Gβγ subunits, PIP2, PIP3, IP3, and perhaps Gα subunits. Activation of Rac by membrane-associated Tiam1 leads to membrane ruffling and JNK activation. It is unlikely that JNK is involved in the induction of an oncogenic phenotype in NIH3T3 cells, since mutant C682 Tiam1 induces an oncogenic phenotype without activation of JNK. Which Tiam1/Rac-mediated signaling pathways are involved in the induction of an invasive phenotype in T-lymphoma cells remains to be established.
Since Rac-dependent membrane ruffling and activation of JNK are induced by membrane-associated Tiam1, the question of how Rac is activated arises. The current hypothesis is that inactive Rac is complexed to RhoGDI and localized in the cytoplasm, as suggested for endogenous Rac in neutrophils (Bokoch, 1994; Hawkins et al., 1995) and for exogenous Rac and Rho proteins in other cell types (Adamson et al., 1992; Ridley et al., 1992; Takaishi et al., 1995; Robertson et al., 1995). According to this model, activation involves the dissociation of the Rac-GDI complex, the translocation of GDP-bound Rac to the plasma membrane, followed by the exchange of GDP for GTP by interaction with a membrane-bound GDS (Bokoch, 1994; Hawkins et al., 1995). Our findings, however, also indicate that Tiam1 translocates from the cytoplasm to the plasma membrane after induction by serum. The most straightforward explanation for the fact that membrane-associated C1199 Tiam1, but not cytoplasmic C580 Tiam1, induces ruffling and activates JNK would be that GDP-bound Rac is partly present at the plasma membrane in NIH3T3 and COS-7 cells. The activation of this pool of membranebound, inactive Rac by Tiam1 is then sufficient for the induction of membrane ruffles and activation of JNK. Alternative explanations would be that the activity of Tiam1 is enhanced at the plasma membrane, or that Tiam1 is also involved in the translocation of Rac from the cytoplasm to the plasma membrane. In all these models, the controlled membrane association of Tiam1 is the only determinant for the induction of membrane ruffling by Rac. Since MSC580 Tiam1, but not C580 Tiam1, induces membrane ruffling, it is unlikely that the activation of cytoplasmic Rac by Tiam1 leads to the membrane translocation of GTPbound Rac. As discussed before, however, cytoplasmic GTPbound Rac might be responsible for the induction of a tumorigenic phenotype by C682 Tiam1. Further exploration of these models will require the generation of antibodies that efficiently detect endogenous Tiam1 and Rac and tools to discriminate between active and inactive endogenous Rac.
Given the importance of PH domains in Tiam1 and other GDS proteins, it is conceivable that phospholipids and/or Gβγ subunits are involved in their intracellular localization and/or activation. The next challenge will be to identify the upstream signaling pathways and molecules that translocate and activate these GDS proteins.
Acknowledgments
We thank Drs. M. Karin, A. Eva, and T. van Biezen for their gift of constructs; G. Habets and F. van Leeuwen for their help in the initial studies; L. Oomen for his assistance with the confocal microscope; L. Oomen and N. Ong for preparation of some of the figures; and F. van Leeuwen, E. Roos, and R. Engers for critically reading the manuscript.
This work was supported by grants from the Netherlands Organization for Scientific Research and the Dutch Cancer Society to J.G. Collard.
Footnotes
1. Abbreviations used in this paper: aa, amino acids; DH, Dbl homology; DHR, Discs-large homology region; GDI, guanine nucleotide dissociation inhibitors; GDS, guanine nucleotide dissociation stimulator; GRF, guanine nucleotide release factor; GST, glutathione S-transferase; HA, hemagglutinin; IP3, inositol (3,4,5)trisphosphate; JNK, c-Jun NH2-terminal kinase; MAPK, mitogen-activated protein kinase; PH, pleckstrin homology; PIP2 and PIP3, phosphatidylinositol (4,5)bisphosphate and (3,4,5)trisphosphate.
Address all correspondence to Dr. J.G. Collard, The Netherlands Cancer Institute, Division of Cell Biology (H1), 121 Plesmanlaan, 1066 CX Amsterdam, The Netherlands. Fax: (31) 20 5121944. E-mail: JCOLL@NKI.NL
References
- Adamson P, Paterson HF, Hall A. Intracellular localization of the p21-Rho proteins. J Cell Biol. 1992;119:617–627. doi: 10.1083/jcb.119.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature (Lond) 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Bokoch GM. Regulation of the human neutrophil NADPH oxidase by the Rac GTP-binding proteins. Curr Opin Cell Biol. 1994;6:212–218. doi: 10.1016/0955-0674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Buchsbaum R, Telliez J-P, Goonesekera S, Feig LA. The NH2terminal pleckstrin, coiled-coil, and IQ domains of the exchange factor RasGRF act cooperatively to facilitate activation by calcium. Mol Cell Biol. 1996;16:4888–4896. doi: 10.1128/mcb.16.9.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerione RA, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- Collard JG. Signaling pathways regulated by Rho-like proteins: a possible role in tumor formation and metastasis. Int J Oncol. 1996;8:131–138. [PubMed] [Google Scholar]
- Coso OA, Chiariello M, Yu JC, Teramoto H, Crespo P, Xu NG, Miki T, Gutkind JS. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- Gibson TJ, Hyvonen M, Masacchio A, Saraste M, Birney E. PH domains: the first anniversary. Trends Biochem Sci. 1994;19:349–353. doi: 10.1016/0968-0004(94)90108-2. [DOI] [PubMed] [Google Scholar]
- Habets GGM, Scholtes EHM, Zuydgeest D, Van der Kammen RA, Stam JC, Berns A, Collard JG. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for rho-like proteins. Cell. 1994;77:537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- Harlan JE, Hajduk PJ, Yoon HS, Fesik SW. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature (Lond) 1994;371:168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- Hart MJ, Eva A, Evans T, Aaronson AA, Cerione RA. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the Dbl oncogene product. Nature (Lond) 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- Hart MJ, Eva A, Zangrilli D, Aaronson SA, Evans T, Cerione RA, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the Dbl oncogene product. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- Hawkins PT, Eguinoa A, Qiu RG, Stokoe D, Cooke FT, Walters R, Wennstrom S, Claessonwelsh L, Evans T, Symons M, Stephens L. PDGF stimulates an increase in GTP-Rac via activation of phosphoinositide 3-kinase. Curr Biol. 1995;5:393–403. doi: 10.1016/s0960-9822(95)00080-7. [DOI] [PubMed] [Google Scholar]
- Hiles ID, Otsu M, Volinia S, Fry MJ, Gout I, Dhand R, Panayotou G, Ruiz-Larrea F, Thompson A, Totty NF, et al. Phosphatidylinositol 3-kinase: structure and expression of the 110 kd catalytic subunit. Cell. 1992;70:419–429. doi: 10.1016/0092-8674(92)90166-a. [DOI] [PubMed] [Google Scholar]
- Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- Horii Y, Beeler JF, Sakaguchi K, Tachibana M, Miki T. A novel oncogene, Ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signaling pathways. EMBO (Eur Mol Biol Organ) J. 1994;13:4776–4786. doi: 10.1002/j.1460-2075.1994.tb06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi-Far R, Solski PA, Clark GJ, Kinch MS, Der CJ. Activation of Rac and Rho, and mitogen activated protein kinases, are required for Ras transformation. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolanus W, Nagel W, Schiller B, Zeitlmann L, Godar S, Stockinger H, Seed B. aLb2 integrin/LFA-1 binding to ICAM-1 induced by cytohesin-1, a cytoplasmic regulatory molecule. Cell. 1996;86:233–242. doi: 10.1016/s0092-8674(00)80095-1. [DOI] [PubMed] [Google Scholar]
- Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Schlessinger J. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- Mahadevan D, Thanki N, Singh J, McPhie P, Zangrilli D, Wang LM, Guerrero C, Levine H, Humblet C, Saldanha J, et al. Structural studies on the PH domains of Dbl, Sos1, IRS-1, and beta-ARK1 and their differential binding to Gβγ subunits. Biochemistry. 1995;34:9111–9117. doi: 10.1021/bi00028a021. [DOI] [PubMed] [Google Scholar]
- Michiels F, Habets GGM, Stam JC, Van der Kammen RA, Collard JG. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature (Lond) 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- Minden A, Lin AN, Claret FX, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995a;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, Rac and Cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem Soc Trans. 1995b;23:456–459. doi: 10.1042/bst0230456. [DOI] [PubMed] [Google Scholar]
- Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science (Wash DC) 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- Olson MF, Pasteris NG, Gorski JL, Hall A. Faciogenital dysplasia protein (FGD1) and Vav, two related proteins required for normal embryonic development, are upstream regulators of Rho-like GTPases. Curr Biol. 1996;6:1629–1633. doi: 10.1016/s0960-9822(02)70786-0. [DOI] [PubMed] [Google Scholar]
- Perona R, Esteve P, Jimenez B, Ballestero RP, Cajal SR. Tumorigenic activity of Rho genes from Aplysia-Californica. Oncogene. 1993;8:1285–1292. [PubMed] [Google Scholar]
- Ponting CP, Phillips C. DHR domains in synthropins, neuronal NO synthases and other intracellular proteins. Trends Biochem Sci. 1995;20:102–103. doi: 10.1016/s0968-0004(00)88973-2. [DOI] [PubMed] [Google Scholar]
- Qiu RG, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature (Lond) 1995a;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- Qiu RG, Chen J, McCormick F, Symons M. A role for Rho in Ras transformation. Proc Natl Acad Sci USA. 1995b;92:11781–11785. doi: 10.1073/pnas.92.25.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif K, Nobes CD, Thomas G, Hall A, Cantrell DA. Phosphatidylinositol 3-kinase signals activate a selective subset of Rac/Rho-dependent effector pathways. Curr Biol. 1996;6:1445–1455. doi: 10.1016/s0960-9822(96)00749-x. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Robertson D, Paterson HF, Adamson P, Hall A, Monaghan P. Ultrastructural localization of Ras-related proteins using epitope-tagged plasmids. J Histochem Cytochem. 1995;43:471–480. doi: 10.1177/43.5.7537292. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warna PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. Phosphatidylinositol3-OH kinase as a direct target of Ras. Nature (Lond) 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- Ron D, Zannini M, Lewis M, Wickner RB, Hunt LT, Graziani G, Tronick SR, Aaronson SA, Eva A. A region of proto-dbl essential for its transforming activity shows sequence similarity to a yeast cell cycle gene, CDC24, and the human breakpoint cluster gene, bcr. New Biol. 1991;3:372–379. [PubMed] [Google Scholar]
- Seed B, Aruffo A. Molecular cloning of the CD2 antigen, the T-cell erythrocyte receptor, by a rapid immunoselection procedure. Proc Natl Acad Sci USA. 1987;84:3365–3369. doi: 10.1073/pnas.84.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Superti-Furga G, Courtneidge SA. Structure-function relationships in Src family and related protein tyrosine kinases. BioEssays. 1995;17:321–330. doi: 10.1002/bies.950170408. [DOI] [PubMed] [Google Scholar]
- Takaishi K, Sasaki T, Kameyama T, Tsukita S, Tsukita S, Takai Y. Translocation of activated Rho from the cytoplasm to membrane ruffling area, cell-cell adhesion sites and cleavage furrow. Oncogene. 1995;11:39–48. [PubMed] [Google Scholar]
- Touhara K, Koch WJ, Hawes BE, Lefkowitz RJ. Mutational analysis of the pleckstrin homology domain of the beta-adrenergic receptor kinase. Differential effects on Gβγ and phosphatidylinositol 4,5-bisphosphate binding. J Biol Chem. 1995;270:17000–17005. doi: 10.1074/jbc.270.28.17000. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen FN, Van der Kammen RA, Habets GGM, Collard JG. Oncogenic activity of Tiam1 and Rac1 in NIH3T3 cells. Oncogene. 1995;11:2215–2221. [PubMed] [Google Scholar]
- Wennstrom S, Hawkins P, Cooke F, Hara K, Yonezawa K, Kasuga M, Jackson T, Claesson-Welsh L, Stephens L. Activation of phosphoinositide 3-kinase is required for PDGF-stimulated membrane ruffling. Curr Biol. 1994;4:385–393. doi: 10.1016/s0960-9822(00)00087-7. [DOI] [PubMed] [Google Scholar]
- Whitehead I, Kirk H, Kay R. Retroviral transduction and oncogenic selection of a cDNA encoding Dbs, a homolog of the Dbl guanine nucleotide exchange factor. Oncogene. 1995a;10:713–725. [PubMed] [Google Scholar]
- Whitehead I, Kirk H, Tognon C, Trigogonzalez G, Kay R. Expression cloning of Lfc, a novel oncogene with structural similarities to guanine nucleotide exchange factors and to the regulatory region of protein kinase C. J Biol Chem. 1995b;270:18388–18395. doi: 10.1074/jbc.270.31.18388. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Cerione R, Bender A. Control of the yeast bud-site assembly GTPase Cdc42—catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994;269:2369–2372. [PubMed] [Google Scholar]
- Zheng Y, Olson MF, Hall A, Cerione RA, Toksoz D. Direct involvement of the small GTP-binding protein Rho in Lbc oncogene function. J Biol Chem. 1995;270:9031–9034. doi: 10.1074/jbc.270.16.9031. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Zangrilli D, Cerione RA, Eva A. The pleckstrin homology domain mediates transformation by oncogenic Dbl through specific intracellular targeting. J Biol Chem. 1996;271:19017–19020. doi: 10.1074/jbc.271.32.19017. [DOI] [PubMed] [Google Scholar]