Abstract
The Drosophila protein Hrb57A has sequence homology to mammalian heterogenous nuclear ribonucleoprotein (hnRNP) K proteins. Its in vivo distribution has been studied at high resolution by confocal laser scanning microscopy (CLSM) in embryos injected with fluorescently labeled monoclonal antibody. Injection of antibody into living embryos had no apparent deleterious effects on further development. Furthermore, the antibody-protein complex could be observed for more than 7 cell cycles in vivo, revealing a dynamic redistribution from the nucleus to cytoplasm at each mitosis from blastoderm until hatching. The evaluation of two- and three-dimensional CLSM data sets demonstrated important differences in the localization of the protein in the nuclei of living compared to fixed embryos. The Hrb57A protein was recruited to the 93D locus upon heat shock and thus serves as an in vivo probe for the activity of the gene in diploid cells of the embryo. Observations during heat shock revealed considerable mobility within interphase nuclei of this transcription site. Furthermore, the reinitiation as well as the down regulation of transcriptional loci in vivo during the recovery from heat shock could be followed by the rapid redistribution of the hnRNP K during stress recovery. These data are incompatible with a model of the interphase nucleus in which transcription complexes are associated with a rigid nuclear matrix.
Chromatin structure has been resolved at the nucleosomal level, yet the structural and compositional features defining the higher levels of organization of the interphase chromosome are hotly debated issues. The chromosome constitutes the structural basis for transcription and replication and may play a critical role in the organization of pre-mRNA processing as well. These processes have to be regulated and coordinated in an efficient way according to the specific requirements of the cell. The efficiency of in vitro transcription and processing systems is significantly lower than those in vivo. This difference may be explained by the reduced local concentrations of these factors as well as a lack of long range chromosomal order in these soluble systems. According to present knowledge, we assume that some ordered structure exists at the chromosomal level within the interphase nucleus. In early developing Drosophila embryos the chromosomes are positioned inside the nucleus with a defined centromere-telomere polarity following a rule first described by Rabl (1885; Swedlow et al., 1993). However, during gastrulation this orientation largely disappears, and homologous associations are formed (Foe and Alberts, 1983; Campos-Ortega and Hartenstein, 1985; Hiraoka et al., 1993; Dernburg et al., 1996; Gemkow et al., 1996). In many other species or cell types one can observe only a territorial delineation with no defined polarity or homologous pairing of the chromosomes (Cremer et al., 1994).
The functional organization of the nucleus is under investigation in a number of laboratories (for review see van Driel et al., 1995; Strouboulis and Wolffe, 1996). Certain biochemical procedures lead to the isolation of a nuclear scaffold or nuclear matrix (Lewis et al., 1984). Experiments demonstrating and characterizing the components of such scaffolds have led to ambiguous results (Dworetzky et al., 1992; Stuurman et al., 1992; Kallajoki and Osborn, 1994; He et al., 1995; Mattern et al., 1996). Unfortunately, existing data regarding the organization of transcriptional complexes within the nucleus are conflicting, some data indicating preferential activity towards the nuclear periphery (Blobel, 1985; Hutchison and Weintraub, 1985) but others showing a random distribution of sites throughout the nucleus (Wansink et al., 1993, 1994; Xing et al., 1993). As we have discussed previously (Buchenau et al., 1993 a), many conflicts in the results concerning nuclear organization may be attributed to the use of different protocols for staining in situ preparations of fixed cells. It is obvious that it is not possible to make a linear extrapolation of data derived from the fixed state directly to the living organism. An experimental strategy to study protein macromolecular assemblies in vivo may afford more direct insight into nuclear processes. Just such an approach has been taken in this investigation of the 93D transcriptional locus.
Nascent transcripts within the nucleus are complexed with proteins as heterogeneous nuclear (hn)1 RNP particles. It has been proposed that these particles are the true substrates for the assembly of splicing complexes by the addition of small nuclear (sn)RNP particles and non–snRNP splicing factors. The hnRNP particles presumably constitute the vehicle for mRNA transport to the cytoplasm as well (Mayeda and Krainer, 1992). The major hnRNPs of Drosophila have been isolated and characterized (Matunis et al., 1992a ). On polytene chromosomes of larval salivary glands (Matunis et al., 1993) some hnRNPs are abundant molecules, present in many copies on most transcribed loci where they possibly serve in some hnRNA packaging function. In addition, specific minor components have been identified by fractionation of Drosophila hnRNP particles (Saumweber et al., 1980; Risau et al., 1983). These proteins are also present in most of the transcriptionally active regions of polytene chromosomes but in an amount estimated at only one to five protein molecules per transcript. One of these proteins, a 55-kD protein that is specifically recognized by the monoclonal antibody Q18 (Saumweber et al., 1980), has a strong sequence homology to the mammalian hnRNP K family of proteins, and its gene has been mapped on the 2R polytene chromosome to the 57A region (B. Hovemann, personal communication). Following a nomenclature introduced by Haynes et al. (1990), we refer to the protein as hnRNA binding protein at region 57A or Hrb57A. This protein has been shown to be present in some 100 transcriptionally active loci on larval salivary gland polytene chromosomes (Saumweber et al., 1980; Kabisch and Bautz, 1983; Risau et al., 1983) and similarly, at active loci on chromosomes in other larval tissues with a lower degree of polytenization (H. Saumweber, unpublished observations). We describe in this paper the distribution of this protein both in vivo and in situ at high resolution in developing Drosophila embryos.
Hrb57A is seen to be almost completely restricted to puff 93D on salivary gland polytene chromosomes following heat shock treatment (Dangli and Bautz, 1983; Dangli et al., 1983). The heat shock locus 93D of Drosophila melanogaster contains the transcription unit heat shock RNA omega (hsr-ω; Garbe and Pardue, 1986; Hovemann et al., 1986; Hogan et al., 1994; Lakhotia and Sharma, 1995). Two polyadenylated transcripts are produced by alternate termination. The shorter ω-c RNA is spliced and exported from the nucleus. The ω-n RNA is 8–18 kb long with no known coding capacity and remains in the nucleus. The 93D locus is constitutively expressed in all cells and is strongly induced upon heat shock (Bonner and Pardue, 1976; Bendena et al., 1991) or the application of certain chemicals (Lakhotia and Sharma, 1995). It has been postulated that the 93D transcript serves as a docking site in the assembly and stabilization of a number of important nuclear proteins for use immediately following recovery from heat shock. Although we lack knowledge about a definitive biological function for the transcripts from the 93D locus, their sequences provide us with an ideal system to identify a specific transcriptionally active site in embryonic nuclei. Similarly, we are able to take advantage of the redistribution of Hrb57A during heat shock so as to distinguish this transcriptional locus in all of the nuclei of the embryo and follow its behavior in vivo. These studies illuminate facets of the transcription process and nuclear structure in the Drosophila embryo and have implications about the temporal and spatial constraints within interphase nuclei. To our knowledge, the results reported here for the Hrb57A protein constitute the first direct in vivo observation of gene activation and repression brought about by heat shock. We are able to follow the kinetics by which the hnRNP K protein is recruited to the activated 93D locus in diploid cells from other chromosomal sites after heat shock. Finally, we demonstrate that active transcription sites are mobile within nuclei, and that the formation and dissolution of these sites occurs in a time domain compatible with in vivo observations by fast confocal scanning microscopy. These dynamic observations corroborate the mobility of transcription sites described in an earlier study (Buchenau et al., 1993 a) of the in vivo imaging of NonA, a protein component of active chromatin.
Materials and Methods
Drosophila Strain
Drosophila melanogaster wild-type strain Oregon R-P2 (Allis et al., 1977) was used throughout. Flies were raised at room temperature on a medium of cornmeal, agar, soy bean meal, malt extract, molasses, yeast, 0.5% (vol/ vol) propionic acid, and the mold inhibitor methyl p-hydroxybenzoate (Caesar and Lorentz, Hilden, Germany).
Antibodies
The generation and maintenance of the hybridoma cell line, Q18, secreting antibodies, mAb Q18, against the Hrb57A protein have been described (Saumweber et al., 1980). The antibody was purified from cell culture supernatants by chromatography on a protein A–Sepharose column (Pharmacia, Freiburg, Germany) before use. For microinjection experiments the antibody was labeled for 2 h at a protein concentration of 0.6 mg/ml with a 15-fold molar excess of carboxytetramethylrhodamine-succinimidylester (TMrhodamine; Molecular Probes Inc., Eugene, OR) in carbonate buffer (pH 8.5) at room temperature. The reaction was quenched with hydroxlamine, and the labeled antibodies were freed from unbound dye by chromatography over Sephadex G25 in PBS. The antibodies were concentrated by centrifugation in a microporus membrane, Centricon-10 (Amicon Corp., Danvers, MA). An absorbance spectrum of the final solution was taken, and the protein concentration and dye/protein labeling ratio were determined to be 0.54 mg/ml and 1.86, respectively. For indirect immunofluorescence with the Q18 antibody we used a Cy3-coupled (Fab)2 goat anti–mouse IgG (Jackson ImmunoResearch Labs, Inc., West Grove, PA).
Fluorescent Deoxyribonucleotide Probe to the ω-n Transcript
The NH2-linked antisense 29-mer, 5′-CA GTT GTT AAG ATA TAC CAT ATA AAA AA (NH2)-3′, was synthesized on a DNA synthesizer (model 382A; Applied Biosystems, Foster City, CA) and purified by HPLC chromatography (Waters Millipore Corp., Milford, MA). The oligonucleotide was fluorescently labeled at a concentration of 1.5 mM with an eightfold excess of carboxyfluorescein–succinimidylester (Molecular Probes Inc.) in 150 mM carbonate buffer (pH 9.3) at room temperature. After shaking overnight, the reaction was stopped by addition of 100 mM Tris buffer and the probe freed of unbound dye by repeated centrifugation over Centricon-3 (Amicon Corp.). The labeling ratio was 0.58 dye molecules per oligonucleotide.
Squashed Polytene Chromosomes and Hybridization
The preparation and fixation of polytene chromosomes from larval salivary glands was described previously (Saumweber et al., 1980).
DNA–RNA hybridization.
Chromosomes were washed in 2× SSC and stabilized for 30 min at 80°C. Hybridization of the oligonucleotide to chromosomal RNA was carried out for 2 h without prior denaturation and using a probe concentration of 7 ng/μl in 2× SSC containing 0.1% Tween 20. The chromosomes were washed in 2× SSC/Tween, stained with 5 μM DAPI (diamidino-phenylindol) or 3 μM Hoechst 33342, and mounted in glycerol-Mowiol 4-88 (Osborn and Weber, 1982).
DNA–DNA hybridization.
Denaturation of the DNA as well as the hybridization of the labeled oligonucleotides to DNA was performed as described by Schmidt (1992). Stabilized chromosomes were denatured in 0.1 N NaOH (90 s at room temperature), washed in 2× SSC, dehydrated via increasing ethanol concentrations, and air dried. Hybridizations were performed using 7.5 ng labeled oligonucleotide in 5 μl 5× SSC under a sealed coverslip at 50°C. After washing (5 min, 2× SSC, 2 min, PBS) the preparations were stained for DNA and mounted as above.
Whole Mount In Situ Fixation
Fixation of Drosophila embryos was carried out with slight modifications of the method described by Mitchison and Sedat (1983). Collected embryos of the appropriate stages were bleach dechlorionated in 50% hypochloride for 90 s and fixed in a heptane/buffer A/37% paraformaldehyde mixture (9: 0.9: 0.1, by vol) for 20–40 min. Buffer A is 60 mM KCl, 15 mM NaCl, 0.5 mM spermidine, 0.15 mM spermine, 2 mM EDTA, 0.5 mM EGTA, 15 mM Pipes, pH 7.4. After removal of the water layer the embryos were devitellinized in a mixture of heptane/methanol (1:1) by vigorous shaking for 60 s and rehydrated through a methanol/buffer A series (90, 75, 50, and 25% methanol) followed by extensive washing in buffer A containing 0.05% Tween 20 in vol of 1 ml on a rotating wheel.
Heat Shock of Embryos
Flies in population cages were induced to lay eggs onto an agar plate covered with a gauze net. After appropriate staging of the embryos at room temperature the gauze net was transferred to a plate in an oven at 37°C for 20–60 min. The embryos were then fixed as described above with all solutions warmed to 37°C.
A thermal microscope stage (Sensortek, Clifton, NJ) was used to heat shock embryos during in vivo observations.
Immuno and DNA Staining of Fixed Embryos
All incubations and washing steps were done in conical tubes on a rotating wheel. Antibodies were diluted in PBS containing 0.05% Tween 20 or, after the RNA hybridization (see below), in 2× SSC containing 0.1% Tween 20. Fixed in situ embryos were incubated with the Q18 antibody using a concentration of 1–4 μg/ml and with the Cy3-coupled secondary antibody at 5 μg/ml, either at room temperature for 3–4 h or at 4°C overnight. Embryos were washed at least three times for 15 min between each antibody in a buffer vol of 1 ml.
DNA was stained for 15–30 min with 5 μm DAPI or 3 μm Hoechst 33342 without prior RNA digestion. Embryos were mounted in glycerol Mowiol 4-88.
RNA Hybridization in Fixed Embryos
Fixed embryos were first exchanged into 2× SSC buffer containing 0.1% Tween 20. Hybridization was performed for 3 h at room temperature in 500 μl using the fluorescently labeled oligonucleotides at a concentration of 500 ng/ml. Embryos were freed from unhybridized oligonucleotide by washing successively in 5× SSC/Tween and 2× SSC/Tween followed by immunostaining in the same buffer. Identical hybridization signals to the ω-n RNA were achieved with more stringent washing conditions as well (0.5× SSC/Tween at 37°C).
Microinjection, Microscopy, and Image Analysis
Microinjection of antibodies into living Drosophila embryos was performed as described previously (Buchenau et al., 1993 a,b) using siliconized injection needles. After injection of 200–500 pl of the TMrhodamine-labeled mAb Q18 (0.54 mg/ml in the needle) the embryos were oxygenated and incubated in a humid chamber before microscopic analysis. Digital images of optical sections through living as well as fixed embryos were collected using a confocal laser scanning microscope (LSM 310; Zeiss, Inc., Jena, Germany) and either a 40× NA 0.9 Plan-Neofluar immersion objective equipped with a refractive index correction collar or a 63× NA 1.4 Plan-Neofluar oil immersion objective. Details about this system and the image acquisition have been described previously (Buchenau et al., 1993 a).
Dyes were excited using the following laser lines of the microscope system: fluorescein at 488 nm and tetramethylrhodamine and Cy3 at 514 nm. Depending upon the combination of dyes within the preparation, the following emission filters were used: LP515 (Schott, Mainz, Germany) or BP530 DF30 (Omega Optical Inc., Brattleboro, VT) for fluorescein; LP550, LP575, LP610 (Schott), or BP590 DF35 (Omega Optical Inc.) for tetramethylrhodamine and Cy3. For every preparation containing two dyes, the respective single-labeled controls were prepared under identical conditions and used for selecting the appropriate excitation wavelength and emission filter combinations.
Images (8 bit) were acquired with an appropriate scanning time and frame averaging. For double staining, the images of the two fluorophore distributions were recorded separately and saved to separate channels of an RGB image. Reconstructions of stereo images were performed using the projection functions of the LSM310 software or NIH-Image (National Institutes of Health, Bethesda, Maryland). Additional image processing was performed on some images which included contrast stretching, uniform filtering, bit plane masking, and intensity quantitation using Scil Image (Technical University, Delft, The Netherlands), NIH-Image (National Institutes of Health), Photoshop 3.0 (Adobe Systems, Mountain View, CA), and Imaris 2.2.6 (Bitplane AG, Zürich, Switzerland). Quantitative image processing was performed on confocal laser scanning microscopy (CLSM) data on a Silicon Graphics (Mountain View, CA) workstation using Scil Image or the depth analyzer module of Imaris 2.2.6. The latter program permits the interactive definition of polygons in three dimensional (3-D) stacks of sequential sections in up to three different fluorescence channels and calculates volume, mean, and integrated greyvalues for each of the channels in three dimensions. Nuclear/chromosomal and cellular volumes were outlined by such polygons for the determination of the mean pixel intensities as calculated in Fig. 2 B. Masks for the 93D subnuclear region were generated from the Fl-P2 oligonucleotide fluorescence image stacks by intensity thresholds and used to calculate the distribution of the Hrb57A protein coincident signal in comparison to the protein signal in the total nucleus. Minimal translational distance measurements such as those shown in Fig. 7, D and E were calculated from 2-D maximal intensity 3-D projection images using NIH-Image on a Macintosh Power PC.
Figure 2.
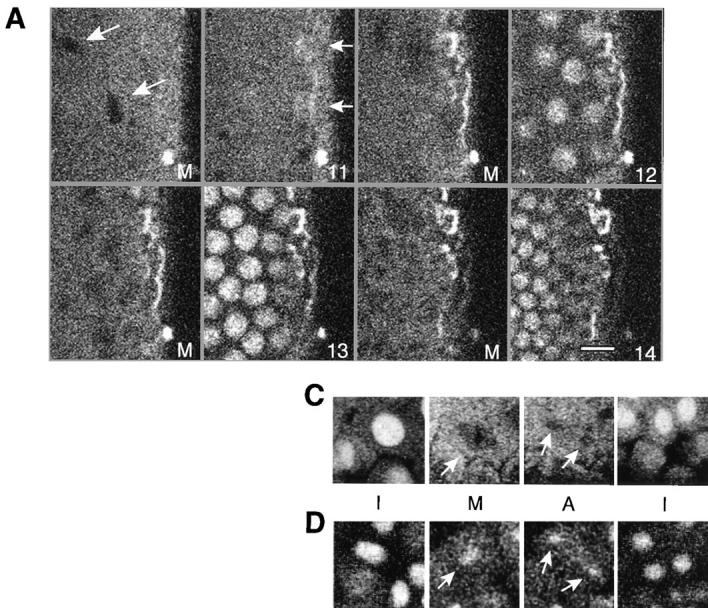
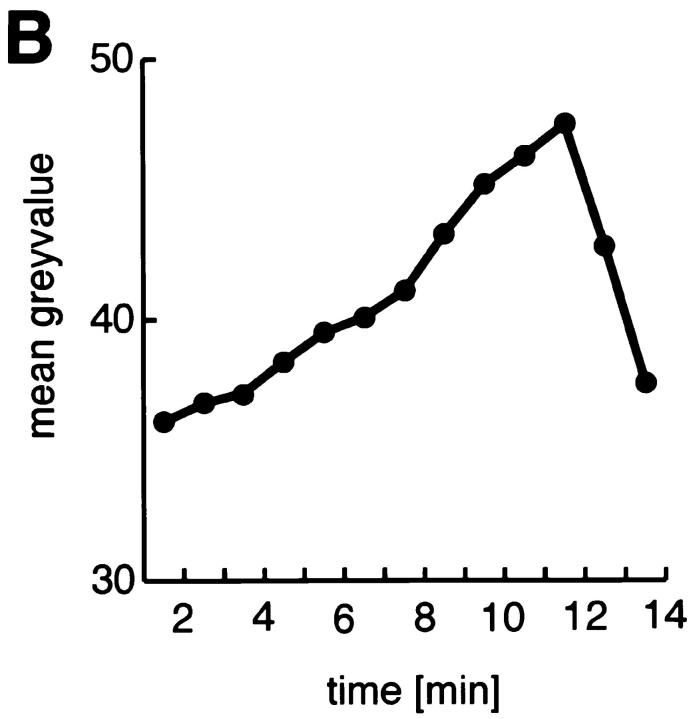
Nuclear import and mitotic distribution of Hrb57A in living embryos. A shows selected frames from a time series of confocal images from a single blastoderm embryo during the nuclear cycles 11–14. The cycle number is given at the bottom of each panel. M denotes mitosis. Arrows indicate the nonfluorescent mitotic chromatin (first panel) and a weak accumulation of Hrb57A in cycle 11 interphase nuclei (second panel). (B) Plot of the mean nuclear fluorescence intensity measured in a single focal plane for a series of images measured at 1 min intervals during cycle 13. An import of Hrb57A is evident for the 10 min during interphase while the sharp decline of fluorescence falls together with nuclear division. C and D are time series of confocal images showing nuclear division in blastoderm (C) and after gastrulation (D). I, interphase; M, metaphase; A, anaphase. Arrows denote the location of the dividing chromatin which contains a detectable amount of Hrb57A after (D) but not before (C) gastrulation. Bar, 10 μm.
Figure 7.
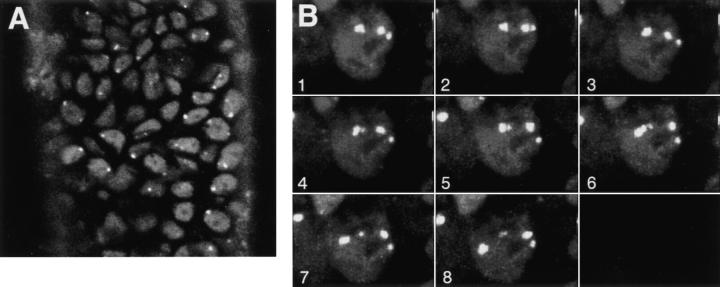
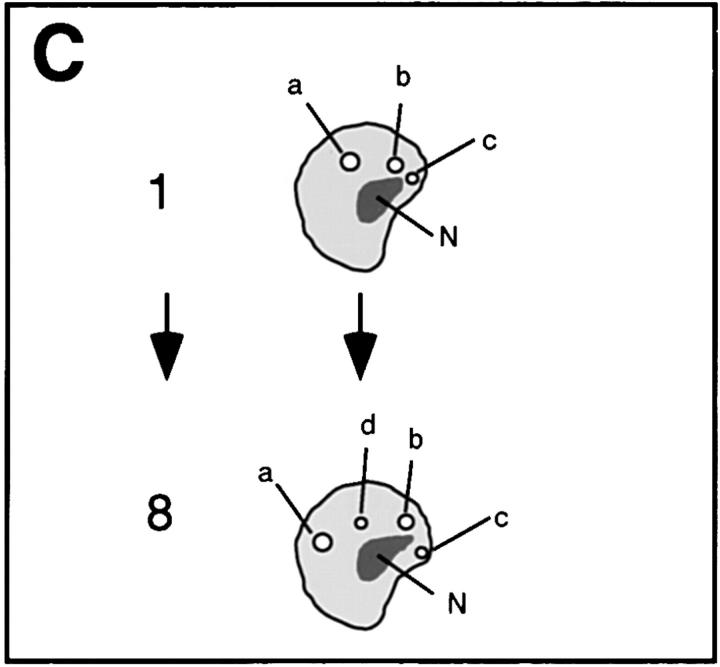
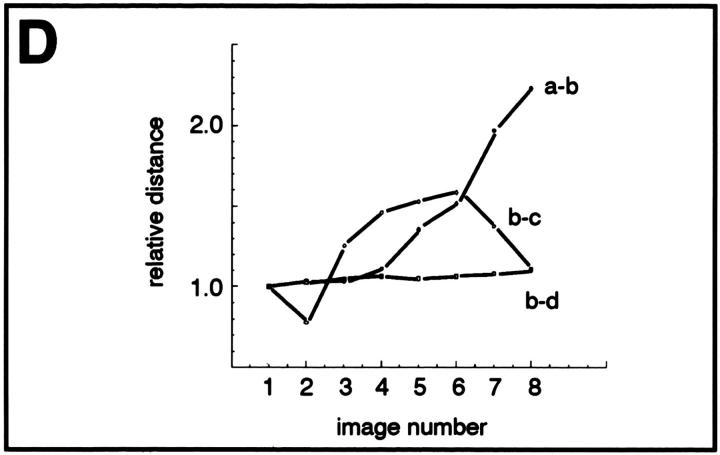
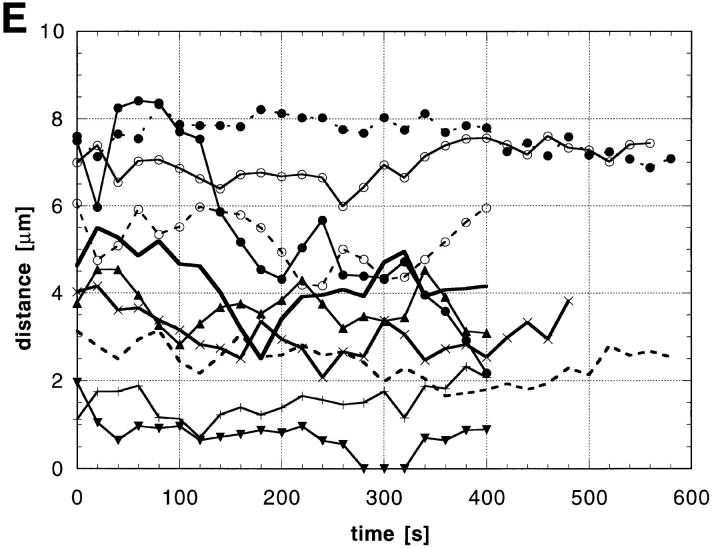
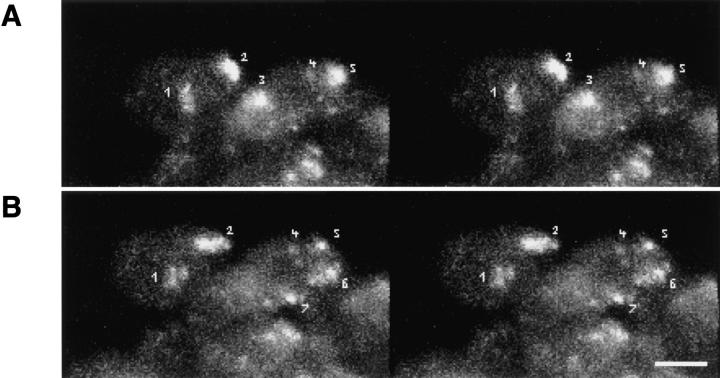
Detection and mobility of the 93D locus in vivo. Living embryos were injected with rhodaminecoupled mAb Q18. After development at room temperature, embryos were heat shocked at 37°C directly on the stage of the microscope to mark the 93D locus. (A) A field of epidermal interphase nuclei from an embryo 75 min after the beginning of heat shock. (B) Time series of 2-D projections of a single amnioserosa nucleus from a heat-shocked embryo during germ band elongation. The original data set consisted of seven 1-μm Z-axis sections per time point. The schematic drawing in C shows the location of the nucleolus (N) and the 93D loci (a–d) within the nucleus in B. (D) A plot of the relative minimal distances between the loci of the nucleus in B against time. The distances are given relative to the first image of the series. (E) 10 two-body distance plots against time demonstrating the time-dependent changes in the distances between 93D loci during heat shock. The distances were measured from 2-D projections of image stacks from seven different interphase nuclei in several embryos.
Results
Dynamics of the Hrb57A Distribution in Living Embryos: Nuclear Import and Mitotic Distribution of the Hrb57A Protein
Fixation may introduce artifacts through the aggregation of structural components, the destruction of sensitive biological structures, or by selective extraction of components. In addition, the temporal dimension in biological processes is lost or often obscured by fixation. Therefore, we concentrated on in vivo observations of the RNA binding protein Hrb57A to track the disposition of zygotic transcription and RNA processing loci in early Drosophila embryos. The monoclonal antibody Q18, specific for Hrb57A, was fluorescently labeled and microinjected into living embryos several nuclear cycles before the formation of the cell membranes (nuclear cycle 14; Foe and Alberts, 1983) so as to achieve a uniform distribution of the antibody within the embryos. The in vivo distribution of the labeled mAb Q18 was followed by CLSM, and the immunofluorescence of fixed embryos at similar time points was monitored as a reference. The antibody injections as well as the irradiation of embryos during the observation had no discernible detrimental effect on the further progress of embryogenesis. Similar viability between antibody injected and control embryos has been documented in a previous study (Buchenau et al., 1993 a). Not only were microinjected embryos viable, but the antibody–antigen complex remained stable and shuttled between the nucleus and the cytoplasm faithfully at each nuclear division following the midblastoderm, permitting us to follow the complex by CLSM for at least seven cell cycles. Thus, the Hrb57A protein/Q18 mAb complex constitutes a tool for the direct in vivo observation of transcriptionally active loci during early Drosophila development.
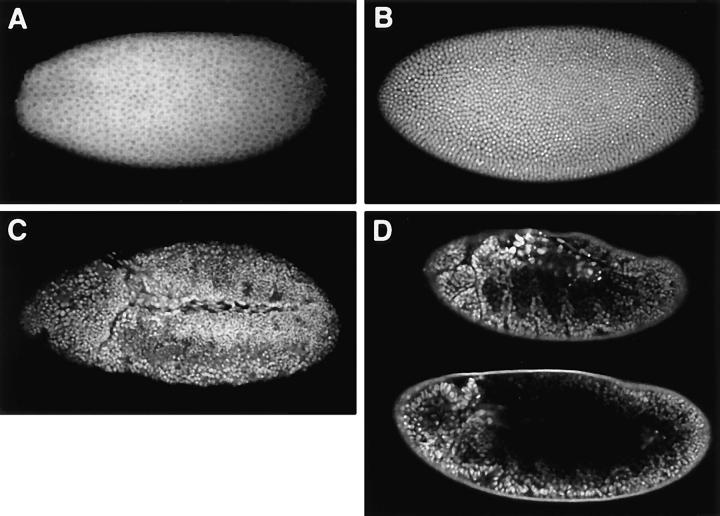
Hrb57A showed a dynamic redistribution from the cytoplasm to the nuclei at the time of the initiation of massive zygotic transcription in early embryos. Fixed embryos of the nuclear cycles 10–12 were characterized by strong cytoplasmic staining with very few accumulations of Hrb57A within the interphase nuclei (Fig. 1 A). In contrast, embryos of the cycle 14 showed an exclusively nuclear Hrb57A signal (Fig. 1 B) whereas cycle 13 embryos were clearly in transition, demonstrating examples of both patterns (data not shown). From cycle 14 throughout the rest of embryogenesis, the protein was found in all interphase nuclei of fixed embryos without any preferential tissue distribution (Fig. 1 C). In living embryos we observed similar cytoplasmic antibody fluorescence in the early nuclear cycles and an almost exclusively nuclear staining at later stages. Fig. 1 D shows an example of two optical sections through a living embryo during germ band retraction. By this time of development, about 12 h after microinjection, most of the nuclei had divided two to three times after cellularization and have gone through six to eight cell cycles since microinjection.
Figure 1.
The distribution of Hrb57A in fixed and living embryos. The overall distribution of Hrb57A during embryogenesis was observed in fixed embryos (A–C) using indirect antibody staining or followed directly in vivo by microinjection of rhodaminecoupled mAb Q18 into living embryos (D). (A) Preferential cytoplasmic localization of Hrb57A, syncytial blastoderm, nuclear cycle 13. (B) Nuclear localization, cellular blastoderm, cycle 14. (C) Hrb57A is nuclear in all tissues during later embryogenesis, as shown here for an embryo of stage 10, at the time of full germ band elongation. (D) The similar complete localization of Hrb57A in late embryogenesis occurs in the microinjected embryos. Two optical sections through a living embryo of stage 12 during germ band retraction, showing the in vivo labeling pattern of Hrb57A. All images are single confocal sections.
The labeling of Hrb57A in living embryos allowed us to monitor continuously and directly cell cycle–dependent changes in the distribution of the protein. Hrb57A was excluded from the nuclei until nuclear cycle 10 (data not shown). Thereafter, following each mitosis we see a time- dependent accumulation of the protein in every nucleus of all of the many embryos injected. This observation is best demonstrated by a montage of representative data shown in Fig. 2. Fig. 2 A is comprised of selected images from a time series of 25 frames recorded from one such living embryo, demonstrating an increasingly nuclear localization of the bulk of Hrb57A during the interphases of the nuclear cycles 11–14. Already in the interphase of cycle 11 the nuclear area contains protein, the concentration of which increases dramatically in the next two cycles until by the end of cycle 14 (Fig. 2 D) most of the protein resides in the nuclei. Computer animations of the recorded time series show this dynamic transport and accumulation after each mitosis. The kinetics of the Hrb57A nuclear import can be represented semiquantitatively as shown in Fig. 2 B by measuring the mean pixel intensities of the nuclear/chromosomal areas of a portion of a cycle 13 embryo, imaged at 1 min intervals. In every nucleus of the living embryos at cycle 13, the import of the protein was almost linear lasting through the entire interphase. In contrast, the dispersal of the protein from the nucleus was achieved within 1–2 min, presumably during the time between breakdown of the nuclear envelope and formation of the metaphase plate. This nuclear localization in vivo can be detected two cycles earlier than that seen by the analysis of fixed preparations (compare Fig. 2 to Fig. 1, A and B). There was complete dissociation of Hrb57A from mitotic chromosomes in early embryos (Fig. 2 C), whereas mitotic nuclei of gastrulating and older embryos continue to show small amounts of protein remaining in the volume defined by the metaphase chromosomes (Fig. 2 D). This phenomenon in later embryos was best visualized by computer animation of time lapse sequences of images. The protein may be associated with the extrachromosomal protein layer surrounding metaphase chromosomes identified by electron microscopy (Hernandez-Verdun and Gautier, 1994).
Comparison of the Distribution of Hrb57A in Fixed and Living Interphase Nuclei of Embryos
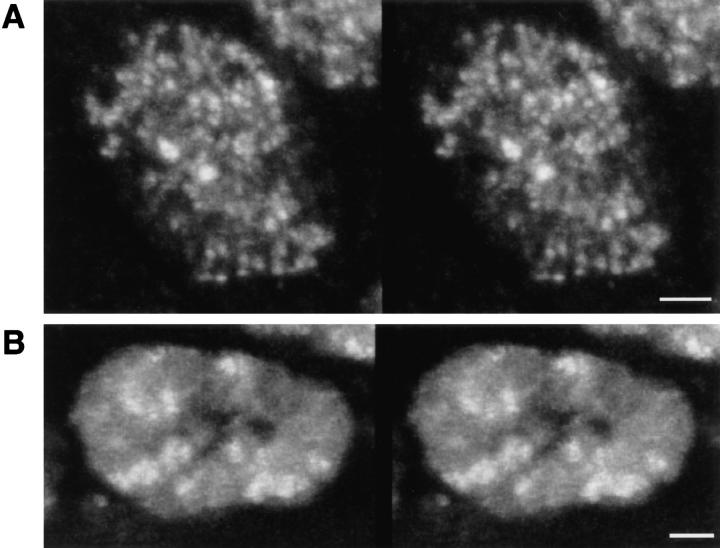
Using the high spatial resolution of the fluorescence CLSM we have analyzed the 3-D distribution of Hrb57A in the nuclei of numerous fixed and living embryos. Fig. 3 A shows a stereo image of the protein distribution in a nucleus from the amnioserosa of a fixed embryo. Hrb57A was localized in a number of optically resolvable spots throughout the nuclear volume excluding the nucleoli, which can be identified as unstained domains. The number of discrete protein loci in such interphase nuclei was determined by intensity segmentation of the 3-D data sets and generation of labeled binary objects. The representative nucleus of Fig. 3 A contains about 200 Hrb57A spots. This number is close to the number of Hrb57A-bound transcriptional loci on polytene chromosomes and confirms our supposition that the protein denotes gene loci sensitive to activation and binding of the hnRNP K in the embryonic nuclei.
Figure 3.
The 3-D intranuclear distribution of Hrb57A in situ and in vivo. (A) Stereo image showing the distribution in discrete loci of the Hrb57A protein in a fixed interphase nucleus of a wholemount embryo reconstructed from 25 optical sections with a Z axis distance of 0.25 μm. (B) Stereo image of a part of a similar nucleus in a living embryo, reconstructed from 6 optical sections separated by 0.4 μm. Both image stacks have been recorded with a Plan-neofluar oil immersion objective 63×, NA 1.4, with a 0.14-mm coverslip. Bars, 2 μm.
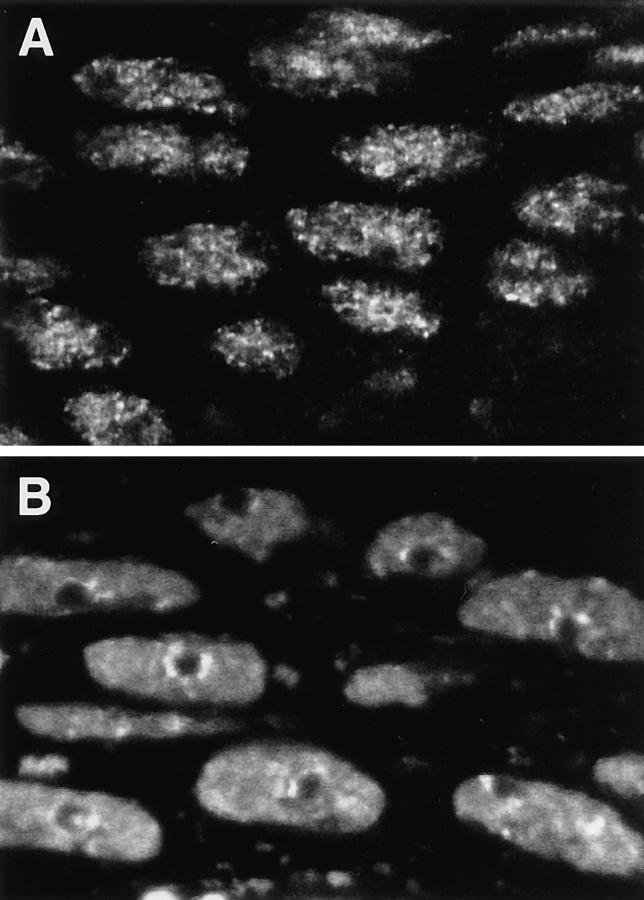
A nucleus from the same tissue of a living embryo 5 h after the injection of rhodamine-labeled mAb Q18 is shown in Fig. 3 B. In this case the unstained nucleoli were surrounded by a positively fluorescent nuclear volume punctated by discrete loci with higher concentrations of Hrb57A. These loci are somewhat larger and fewer in number than those resolved in the fixed embryos. Both stereo images in Fig. 3 have been reconstructed from confocal sections acquired under optical conditions as nearly identical as possible. Fig. 4 shows overviews of a field of amnioserosa cells from another fixed (A) and another living (B) embryo which demonstrate that these differences in the distribution of Hrb57A between fixed and living embryos are highly reproducible and visible already in single confocal sections. We attribute the differences to a differential extraction of the protein as well as to changes in the chromatin structure introduced by the fixation procedure and/or to the dynamic movement of transcription sites in living embryos (see Discussion).
Figure 4.
Fields of interphase nuclei in the amnioserosa of a fixed (A) and a living (B) embryo during stage 11 (elongated germ band) stained for Hrb57A. Both images are single confocal sections from stacks recorded as in Fig. 3. Width of the total field is 40 μm.
Activation of a Transcriptional Locus
It has been observed previously that following heat shock, the Hrb57A protein is concentrated exclusively on the heat shock locus 93D of chromosome 3R in salivary gland polytene chromosomes of third instar larvae (Dangli and Bautz, 1983). This locus is constitutively transcribed but can be further induced by heat shock, both in salivary glands (Bonner and Pardue, 1976) and in embryos (Bendena et al., 1989). We were interested to determine how much of the Hrb57A is associated with 93D in normal embryos and after heat shock.
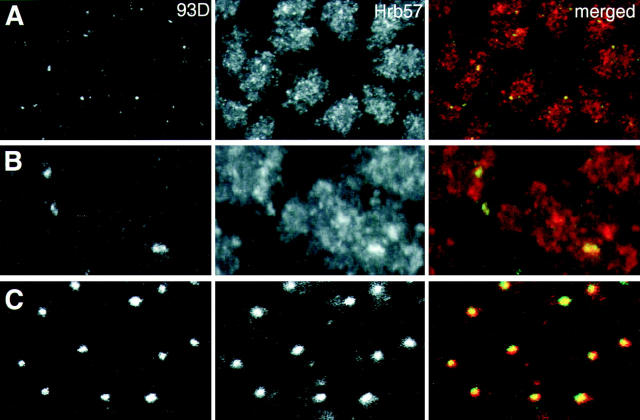
A 29-base oligonucleotide with a sequence complementary to a repeated region in the 3′-end of the 93D transcript ω-n (Hovemann et al., 1986; Hogan et al., 1994) was synthesized and covalently labeled with fluorescein (Fig. 5 A). In squashed salivary gland chromosomes the labeled oligonucleotide hybridized exclusively to the 93D region both under conditions for DNA–DNA (data not shown) and for DNA–RNA hybridization (Fig. 5, B and C), demonstrating the specificity of the probe and the constitutive nature of the transcription from this locus. When formaldehyde fixed, whole-mount embryos were hybridized with the probe to RNA, i.e., under nondenaturing conditions, we detected one or two prominent spots per diploid nucleus with almost no labeling of the residual nuclear volume (Fig. 6 A). The polyploid nuclei of the embryo very often contained more than two hybridization spots, indicating that in these cases the loci were not synapsed. The fact that in all larval and embryonic tissues observed, the number of transcript accumulations as revealed by the hybridization parallels the number of expected chromosomal 93D loci leads us to conclude that within the resolution of the CLSM, our oligonucleotide probe reflects the location of nascent transcripts at the 93D locus. Therefore it can be used as a marker for the spatial location of this transcriptionally active gene in developing embryos. Fig. 6 A shows a projection image of normal diploid nuclei from a region of a fixed embryo hybridized to RNA (nondenaturing conditions) with the ω-n probe and stained with mAb Q18 and the merged multicolor image. These images have been reconstructed from the maximal intensities of 15 confocal sections for each fluorescence channel. A comparison of the ω-n/93D signal (Fig. 6 A) and the corresponding Hrb57A protein distribution in the merged panel indicates clearly that the protein was constitutively associated with the transcripts at 93D in these nonheat shocked embryos. Fig. 6 B contains a higher magnification image of two nuclei from the field in Fig. 6 A. The shape and size of the hybridization signal correlated almost precisely with that of an intense Q18 antibody binding region. In many but not in all nuclei of nonstressed embryos, Hrb57A showed the highest concentration in the 93D region. To determine the relative amount of the protein associated with 93D we measured the fluorescence intensities of the anti-Hrb57A antibodies in whole nuclei and in the nuclear subregion defined by the ω-n probe. Scoring many nuclei in several embryos, we found 2–5% of the total nuclear Hrb57A protein was associated with the 93D locus in normally developing embryos.
Figure 5.
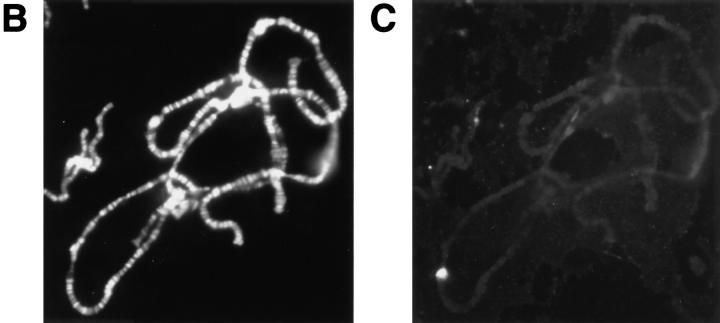
Transcripts from the 93D region and detection of the locus by oligonucleotide hybridization. (A) Schematic representation of the transcripts from the heat shock RNA-ω region in 93D. The gray box indicates the location of the intron which is spliced out of the smaller ω-c RNA. The repeat region of the long ω-n transcript is shown in black. The sequence of the fluorescently labeled oligonucleotide, Fl-P2, used as a probe for ω-n is given below a 29 base transcript sequence which is strongly conserved between the tandem repeats. B and C demonstrate that the probe specifically binds to 93D RNA in squashed polytene chromosomes. (B) DNA stained with DAPI. (C) 93D hybridized with FlP2 under nondenaturing conditions.
Figure 6.
Association of Hrb57A with the 93D locus in situ. The RNA at the 93D locus in the nuclei of fixed embryos was detected by hybridization with the Fl-P2 oligonucleotide (left column) and the Hrb57A protein by mAb Q18 followed by a Cy3-coupled secondary antibody (center column). The superposition of both signals in an RGB image generates a yellow color where 93D and the protein are colocalized (right column). (A) A group of interphase nuclei from a nonstressed embryo. Overlay of 15 optical sections separated by 0.35 μm. (B) Higher magnification of a subregion from the field shown in A demonstrating the precise overlap of the 93D signals with some of the Hrb57A accumulations. (C) A group of interphase nuclei from an embryo that had been heat shocked at 37°C for 60 min before fixation. The restriction of Hrb57A to the 93D locus is evident.
The distribution of Hrb57A in embryos fixed after a heat shock of 20–60 min at 37°C showed dramatic changes. Fewer nuclear loci were positive for Hrb57A and 93D always contained the highest concentration of the protein (Fig. 6 C). Over 50% of the total nuclear Hrb57A protein was coincident with 93D even after relatively short heatshock times. The extent of this heat shock response, i.e., the increased transcription of ω-n and the progressive restriction of the protein to 93D was dependent on the age of the embryos. As expected, embryos of the preblastoderm stage did not show any reaction due to the lack of transcriptional activity. After gastrulation the response to the stress situation was unambiguous but progressively stronger in older embryos. The disappearance of Hrb57A from most of the nuclear volume and its concentration on the 93D locus under heat shock conditions parallels exactly the redistribution of the protein in polytene nuclei after heat shock (Dangli and Bautz, 1983).
To follow the same process in vivo we injected embryos of the blastoderm stage with rhodamine-coupled mAb Q18. After an incubation time of 2–3 h at room temperature the embryos were heat shocked at 37°C either in an incubator or by a thermo-controlled stage mounted directly onto the stage of the confocal microscope. The first signs of a heat shock response were clearly visible 10–15 min after raising the temperature. At room temperature the embryonic nuclei were characterized by a moderately differential Hrb57A staining (Figs. 2 and 3), whereas after heat shock they developed one, and in polyploid nuclei up to a few, very intensely fluorescing spots (Fig. 7 A).
Dynamic Behavior of Transcriptional Loci in Living Embryos
The redistribution of Hrb57A during heat shock to an almost exclusive localization at 93D (Figs. 6 C and 7 A) provided us with a system amenable for the study of the dynamic behavior of transcriptional loci in living embryos, due to the reduced number of sites as compared with nonstressed embryos. We have observed literally thousands of cells in many tens of living, heat shocked embryos. There is constant movement of loci throughout the embryo, some of which may originate from rotation of whole nuclear contents whereas others are clearly local movements of the transcription sites themselves. The exact nature of the 3-D movement of the single 93D locus in 2N cells is often impossible to unambiguously determine. The types and extents of movement can be more clearly examined and documented in the polyploid cells with multiple 93D loci. Time series of 3-D data sets from heat shocked embryos are acquired at high spatial resolution. Computer animated displays of the series show jitter, slow movements, rapid movements over both large and small distances, and apparently, both random walk and directed movements. An example of one such nucleus in a series is seen in Fig. 7, B–D. B is composed of 2-D image projections of a polyploid amnioserosa nucleus at successive time points over a period of ∼20 min. At the beginning of the observation the nucleus exhibited 3 intensely fluorescing 93D loci (labeled a–c in the first schematic drawing in C) lying adjacent to a nonfluorescent nucleolus. In computer animations of time series, such loci appeared to “jitter” around an average position, probably due to continuous small changes in the nuclear shape in vivo. This jitter has been removed from the traces. With increasing time, one of these loci, a, moved away from its original position, exposing a fourth locus. Presumably, this “new” locus d was not optically resolved from a at the beginning of the recording. While the relative positions of the loci b–d and the nucleolus remained almost constant during the observation period, a moved a distance of >3 μm in <10 min (relative distance trajectories are plotted in Fig. 7 D). Similar movements were observed in many of the polyploid nuclei of all heat shock embryos. Fig. 7 E shows typical traces of two body temporal distance measurements from other nuclei in other embryos.
We were able to observe another type of dynamic behavior of Hrb57A in vivo after the transition from the high (37°C) to normal temperature (25°C), i.e., during recovery of the embryos from heat shock. Onset of recovery begins immediately, as reflected by the change in the disposition of Hrb57A at the 93D locus. The earliest recordings were made at 1 min after the temperature drop and subsequent images at 2 or 4 min intervals. The intensity of the Q18 staining of the 93D locus begins to decrease in almost all loci immediately. Many new weaker loci appear throughout the nuclear volume. There is large variability in the rate of appearance and number of resolvable new loci in individual nuclei. The rate of recovery also is dependent upon the extent and duration of the prior heat shock treatment. An example of recovery behavior is demonstrated in Fig. 8. Two stereo image pairs of the amnioserosa of an embryo are reconstructed from image stacks separated by 4 min in time after return to normal temperature. Local accumulations of mAb Q18 in the two fully visible nuclei in this field have been marked by numbers. The nucleus on the left shows small reorganizations of the Hrb57A loci at 6 and 10 min into heat shock recovery, and the composite loci are labeled 1 and 2 for simplicity. The nucleus on the right displayed dramatic changes in the Hrb57A pattern at these time points. One of the spots disappeared completely (3), another one became smaller (5), and two new accumulations appeared (6 and 7). Computer animations of the complete time series of this recovery show increasingly complex reorganization of the Hrb57A protein. These dynamic changes cannot be attributed to a rotation of the nucleus as a whole or to a simple translational mobility of most of the loci as is documented in Fig. 7, B–D. We have observed a large number of similar and more complicated dynamic redistributions of the Hrb57A protein during heat shock recovery in many embryos. We conclude that the redistribution of Hrb57A is causally related to the reactivation of gene expression that occurs in the cell nucleus during recovery from heat shock.
Figure 8.
Dynamic changes in the intranuclear distribution of Hrb57A during recovery from heat shock. Stereo images of amnioserosa nuclei reconstructed from five optical sections separated by 2 μm. Intranuclear accumulations of Hrb57A are numbered (see text). (A) 6 min after reducing the temperature to 25°C. (B) 10 min into recovery. Bar, 5 μm.
Discussion
In this study we show the dynamic distribution of the hnRNP K-homologous protein Hrb57A in the nuclei of developing Drosophila embryos by in vivo immunostaining with the monoclonal antibody Q18. The protein was originally isolated in hnRNP particles from Kc cells (Risau et al., 1983) and localized to a number of transcription sites on salivary gland polytene chromosomes from third instar larvae (Dangli and Bautz, 1983). A partial cDNA sequence of the gene has been determined and localized to region 57A on squashed polytene chromosomes (Hovemann, B., personal communication). The amino acid sequence derived from this nucleotide sequence reveals several glycine-rich regions, a K-homology domain, as well as other sequence motifs which have been found in hnRNP K proteins of other species and other RNA-binding proteins (Hovemann, B., personal communication). In vitro binding studies with other hnRNP K proteins have suggested that at least one mode of binding for this class of proteins is through polyC-rich sequences in RNA transcripts (Matunis et al., 1992b ; Dreyfuss et al., 1993; Leffers et al., 1995). Recent in vitro experiments demonstrate that hnRNP K acts as a transcription factor, activating genes containing CT elements (Michelotti et al., 1996). Our studies demonstrated a 1:1 correspondence between the shape of the protein immunofluorescence and the fluorescent oligonucleotide in situ hybridization signal on the ω-n transcript at the 93D locus (Fig. 6). The tandem repeats in the ω-n transcript from the 93D locus are, however, very AT rich. The only part of the sequence with long stretches of polyC is in the 5′ end, contained also in the shorter ω-c transcript (Garbe et al., 1986). It is possible that the binding of the Hrb57A protein to the transcript is mediated through this region and that the repeat sequences are primarily involved in secondary or tertiary structure which facilitate the localization or stabilization of the RNA at the site of transcription.
In fixed embryos we were able to distinguish ∼200 discrete loci of the Hrb57A protein distributed throughout the nuclear volume (Figs. 3 A and 4 A). This number of loci closely approximates that which has been observed on spread salivary glands where the protein is always associated with transcriptionally active sites on the chromosomes. The colocalization of the protein with transcription sites can be followed through other larval tissues with lower levels of polyteny like the Malpighian tubules (Saumweber, H., unpublished observations). By extrapolation we presume that the protein accumulation denotes sites of transcription activation of genes and hnRNP K binding in the fixed embryonic nuclei.
There are striking differences in the distribution of the protein in the nuclei of living embryos compared with fixed preparations. We see the protein distributed throughout the entire nucleus except for the nucleoli in vivo. The intensities within the nuclei are less differential than in the fixed embryos, and individual loci are more difficult to resolve or appear to form larger aggregates, as can be seen in the examples shown in Figs. 3 B and 4 B. We consider three possible, not mutually exclusive explanations for the difference between the appearance of fixed and live nuclei stained for Hrb57A. Extrapolating from studies on the distribution of fluorescently labeled histones in live and fixed embryos, we suggest that some of these differences may be attributable to changes in the chromatin structure introduced by the fixation procedure itself (Buchenau, 1996). It is possible that the interphase chromosome structure in vivo is less compacted than after fixation and that a microscopically visible higher order structure of fixed chromatin is partly the result of precipitation artifacts. Support for this view comes from comparing electron microscopic images of cells conventionally prepared by fixation and dehydration with nonfixed, vitrified mammalian cells observed at −160°C. Such rapidly frozen cells with their fully hydrated interphase chromosomes lack any obvious chromatin organization above ∼11 nm (Dubochet, 1995). A second, alternative explanation for the differences observed in our study is the possible existence of a nucleoplasmic fraction of the Q18–antigen complex that is not tightly associated with transcription sites or RNA in vivo. This fraction might be less efficiently cross linked during fixation and therefore differentially extracted by the fixative solutions. After fixation, the remaining protein complex, Q18– Hrb57A-RNP, would then appear as discrete fluorescent loci in a darker, protein-free background. Support for this interpretation is provided by the change in the protein distribution from the “living” to the “fixed” pattern upon fixation of microinjected embryos and by the fact that we observe nuclear localization of Hrb57A in living embryos two cell cycles earlier (cycles 11 and 12) than in fixed preparations (Figs. 1 and 2). Since only a very limited set of genes is transcribed during these early nuclear cycles, it is possible that the bulk of the intranuclear Hrb57A is not yet associated with transcript or transcribing genes and, in line with the argument presented above, would be more easily extracted during fixation. A third explanation for our results relates to the dynamic behavior of transcriptional sites, which we have documented in in vivo temporal studies, examples of which are shown in Figs. 7 and 8. Our observations indicate that the chromatin-associated transcription sites in a living nucleus undergo dynamic rearrangements, suggesting great flexibility in the chromatin. Microscopic movements, in a time domain below or comparable to the temporal resolution of the microscope, could lead to a more homogeneous intensity distribution of fluorescence and to an increase in the apparent size of areas of protein concentration in in vivo images by the convolution of a spatial pattern with dynamic temporal changes. At present it is not possible to determine which of these explanations accounts for the differential distribution of Hrb57A, and presumably other RNA associated proteins, within the living nucleus.
As has been demonstrated for a number of other proteins involved in RNA metabolism in Drosophila (Dequin et al., 1984; Segalat and Lepesant, 1992), Hrb57A is excluded from the nucleus until the onset of zygotic transcription. Our in vivo studies in blastoderm embryos allowed a direct observation of the kinetics of nuclear import of Hrb57A (Fig. 2). During the interphases of these early cycles the intranuclear concentration of the protein increased almost linearly. In contrast, the kinetics of the export of the hnRNP from the nucleus at mitosis was very rapid. The fact that Hrb57A is continuously imported throughout most of the interphase period in early embryogenesis suggests that the protein might require the assembly of active transcription before nuclear transport. However, a similar kinetics of nuclear import was observed for the transcription-associated protein, NonA (Buchenau et al., 1993 a, 1996). This protein is still transported in the presence of the RNA polymerase II inhibitor α-amanitin. We are presently investigating the behavior of Hrb57A under these conditions.
Dangli and Bautz (1983) have shown that the Hrb57A protein is almost exclusively localized in the puff at 93D of heat shocked third instar larvae. The experiments described here clearly demonstrate that this phenomenon also occurs in embryos and therefore indicate that it might reflect an important and basic function of the 93D transcript. Transcription of this locus and its influence on other heat shock loci have been studied in some detail (Garbe and Pardue, 1986; Hovemann et al., 1986; Bendena et al., 1991; Vazquez et al., 1993; Hogan et al., 1994; Sharma and Lakhotia, 1995; Lakhotia and Sharma, 1995). Since a variety of proteins concentrate at the 93D locus upon heat shock, for example, Hrb87F (Haynes et al., 1991; Hovemann et al., 1991) and the heat shock protein HSP90 (Morcillo et al., 1993), it has been proposed that the ω-n transcript fulfills a storage function for the sequestration and protection of transcription factors and other nuclear proteins during heat shock (Hogan et al., 1995). However, the fact that its transcription is modulated in nonheat shock conditions (Bendena et al., 1991) suggests that it could also serve as a sink for these proteins when they are not actively engaged in transcription or mRNA processing. Our in vivo observations support this hypothesis. During heat shock and the termination of transcription at most nuclear loci, we measured Hrb57A redistribution leading to the accumulation of >50% of the protein at the 93D locus, as opposed to the disappearance of the protein from the nucleus under these conditions (Figs. 6 and 7 A). After heat shock, during the early stages of recovery, there is obvious retrograde redistribution and appearance of new protein loci throughout the nuclear volume (Fig. 8), presumably representing reinitiated transcriptional loci.
One of the striking results of our experiments is the demonstration that nuclear components, and transcriptional loci in particular, behave in a very dynamic manner in the living embryo. The few in vivo studies of nuclei in the literature corroborate this plasticity and movement in the nucleus (Buchenau et al., 1993 a; de Boni, 1994). Most of the models for nuclear architecture are derived, on the other hand, from experiments performed on fixed cells or in vitro preparations of nuclear extracts. An interesting model by which transcription might occur in the interchromosomal space on the periphery of the chromosome territories is derived from observations of fluorescence in situ hybridization of chromosome DNA sequence libraries, transcript labeling, and antibody staining for snRNPs (Zirbel et al., 1993; Cremer et al., 1994). The extensive literature on models involving the coupling of transcription to a nuclear matrix whereby the “RNA polymerases are immobilized onto the nuclear matrix structure and DNA reels through the immobilized transcription complexes” has been reviewed recently by van Driel et al. (1995).
We have taken advantage of the fact that the majority of the Hrb57A protein is associated with the locus 93D after heat shock to follow for the first time in vivo a specific transcriptional locus, as well as the formation of new loci. During heat shock, morphogenesis is slowed down, and most metabolic reactions are greatly reduced. Despite this reduction in metabolism, a large variety of movements of the transcription loci can be observed throughout the embryo in all types of cells. We have described the types of movements seen from computer animated displays of time lapse sequences of 2- and 3-D data stacks. Examples of these movements of the 93D locus are documented in Fig. 7. Changes in the distribution of chromosomal domains with the differentiation state of a cell (Manuelidis and Borden, 1988) or of centrosomal regions after induction of gene expression (Park and de Boni, 1992; Sahlas et al., 1993; Spector et al., 1994; Janevski et al., 1995) have been inferred from the analysis of fixed cultured cells. We previously suggested that transcriptional loci could move rapidly about the nucleus from the in vivo distribution of a transcription associated protein (Buchenau et al., 1993 a).
During recovery from heat shock the embryo can rapidly return to constitutive levels of transcription from the 93D locus with the concomitant reinitiation or activation of other gene loci (Hogan et al., 1994). We were able to follow this return to normal transcription by direct observation of the microinjected, heat shocked embryos. The reduced binding of Hrb57A at the 93D locus as well as the appearance of new protein accumulations are obvious. A typical redistribution over a period of only 4 min early during this recovery process is documented in Fig. 8.
We emphasize that the phenomena described in this paper and documented in the figures have been observed in diploid as well as polyploid nuclei of a large number of embryos, that the translocations of the 93D locus are sometimes even more rapid than shown here, and that the formation and dissolution of local concentrations of Hrb57A occurs frequently in normal embryos as well as those recovering from heat shock. The distribution of Hrb57A could not be as well resolved in living and as in fixed embryos, which suggests that many redistributions or local movements of the transcription apparatus are occurring in a time scale faster than our present technical imaging resolution. These in vivo results directly support the idea that specific repositioning of transcription sites, and thus, chromosomal loci, within the interphase nucleus is a part of the functional repertoire of the cell. Our data are not easily reconciled with a model by which DNA and the transcriptional machinery are anchored to a relatively rigid nuclear scaffold or matrix during interphase. In addition, the observation from our fixed embryos of transcriptional loci throughout the nuclear volume supports models of chromatin arrangement according to which transcription is not restricted to the periphery of the nucleus or to a limited set of regions within the nucleus but rather occurs at a large number of sites, perhaps close to the number of transcribed genes themselves, where simultaneous transcription and processing take place. From these sites the packaged mRNAs would then disperse throughout the nucleoplasm and exit the nuclear membrane at unrestricted pores.
Finally, these in vivo studies in whole embryos indicate that one can observe metabolic processes at specific gene loci with specific probes and fast scanning CLSM. Further studies could include dynamic inhibition of specific transcription, the observation of embryos with mutant enzymes for transcription or replication, and further studies of the heat shock phenomenon.
Acknowledgments
We thank Dr. B. Hovemann for making unpublished data on the sequence and localization of the partial cDNA clone for Hrb57A available to us.
Abbreviations used in this paper
- CLSM
confocal laser scanning microscopy
- hn
heterogeneous nuclear
- 3-D
three dimensional
- sn
small nuclear
Footnotes
Please address all correspondence to Donna Arndt-Jovin, Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, 370170 Gottingen, Germany. Tel.: (49) 551-2011-393; Fax.: (49) 551-2011467; E-mail: djovin@mpc186.mpibpc.gwdg.de
P. Buchenau's present address is Botanical Institute, University of Bonn, 53115 Bonn, Germany.
References
- Allis CD, Waring GL, Mahowald AP. Mass isolation of pole cells from Drosophila melanogaster. . Dev Biol. 1977;56:372–381. doi: 10.1016/0012-1606(77)90277-9. [DOI] [PubMed] [Google Scholar]
- Bendena WG, Garbe JC, Traverse KL, Lakhotia SC, Pardue ML. Multiple inducers of the Drosophila heat shock locus 93D (hsrω): Inducer-specific patterns of the three transcripts. J Cell Biol. 1989;108:2017–2028. doi: 10.1083/jcb.108.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendena WG, Ayme-Southgate A, Garbe JC, Pardue ML. Expression of heat-shock locus hsr-omega in nonstressed cells during development in Drosophila melanogaster. . Dev Biol. 1991;144:65–77. doi: 10.1016/0012-1606(91)90479-m. [DOI] [PubMed] [Google Scholar]
- Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JJ, Pardue ML. The effect of heat shock on RNA synthesis in Drosophilatissues. Cell. 1976;8:43–50. doi: 10.1016/0092-8674(76)90183-5. [DOI] [PubMed] [Google Scholar]
- Buchenau, P. 1996. Verteilung, Funktion und in vivo-Dynamik chromosomaler Proteine in Drosophila melanogaster. Ph.D. thesis. Humboldt University of Berlin, Berlin, Germany.
- Buchenau P, Arndt-Jovin DJ, Saumweber H. a. In vivo observation of the puff-specific protein no–on transient A (NONA) in nuclei of Drosophilaembryos. J Cell Sci. 1993;106:189–199. doi: 10.1242/jcs.106.1.189. [DOI] [PubMed] [Google Scholar]
- Buchenau P, Saumweber H, Arndt-Jovin DJ. Consequences of topoisomerase II inhibition in early embryogenesis of Drosophila revealed by in vivoconfocal laser scanning microscopy. J Cell Sci. 1993b;104:1175–1185. doi: 10.1242/jcs.104.4.1175. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega, J., and V. Hartenstein. 1985. The Embryonic Development of Drosophila melanogaster. Springer-Verlag Inc., Heidelberg, Germany. 227 pp.
- Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schröck E, Speicher M, Mathieu U, Jauch A, Emmerich P, et al. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harb Symp Quant Biol. 1994;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- Dangli A, Bautz EKF. Differential distribution of nonhistone proteins from polytene chromosomes of Drosophila melanogasterafter heat shock. Chromosoma (Berlin) 1983;88:201–207. doi: 10.1007/BF00285621. [DOI] [PubMed] [Google Scholar]
- Dangli A, Grond C, Kloetzel P, Bautz EKF. Heat-shock puff 93D from Drosophila melanogaster: accumulation of an RNP-specific antigen associated with giant particles of possible storage function. EMBO (Eur Mol Biol Organ) J. 1983;2:1747–1751. doi: 10.1002/j.1460-2075.1983.tb01652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boni U. The interphase nucleus as a dynamic structure. Int Rev Cytol. 1994;150:149–171. doi: 10.1016/s0074-7696(08)61541-7. [DOI] [PubMed] [Google Scholar]
- Dequin R, Saumweber H, Sedat JW. Proteins shifting from the cytoplasm into the nuclei during early embryogenesis of Drosophila melanogaster. . Dev Biol. 1984;104:37–48. doi: 10.1016/0012-1606(84)90034-4. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW. Perturbation of nuclear architecture by longdistance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Dubochet J. High-pressure freezing for cryoelectron microscopy. Trends Cell Biol. 1995;5:366–368. doi: 10.1016/s0962-8924(00)89071-6. [DOI] [PubMed] [Google Scholar]
- Dworetzky SI, Wright KL, Fey EG, Penman S, Lian JB, Stein JL, Stein GS. Sequence-specific DNA-binding proteins are components of a nuclear matrix-attachment site. Proc Natl Acad Sci USA. 1992;89:4178–4182. doi: 10.1073/pnas.89.9.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V, Alberts B. Studies of nuclear and cytoplasmic behavior during the five mitotic cycles that precede gastrulation in Drosophilaembryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Garbe JC, Pardue ML. Heat shock locus 93D of Drosophila melanogaster: A spliced RNA most strongly conserved in the intron sequence. Proc Natl Acad Sci USA. 1986;83:1812–1816. doi: 10.1073/pnas.83.6.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe JC, Bendena WG, Alfano M, Pardue ML. A Drosophilaheat shock locus with a rapidly diverging sequence but a conserved structure. J Biol Chem. 1986;261:16889–16894. [PubMed] [Google Scholar]
- Gemkow M, Buchenau P, Arndt-Jovin D. FISH in whole mount Drosophilaembryos. RNA: activation of a transcriptional locus; DNA: gene architecture and expression. Bioimaging. 1996;4:107–120. [Google Scholar]
- Haynes SR, Raychaudhuri G, Beyer AL. The Drosophila Hrb98DElocus encodes four protein isoforms homologous to the A1 protein of mammalian heterogeneous nuclear ribonucleoprotein complexes. Mol Cell Biol. 1990;10:316–323. doi: 10.1128/mcb.10.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes SR, Johnson D, Raychaudhuri G, Beyer AL. The Drosophila Hrb87fgene encodes a new member of the A and B hnRNP protein group. Nucleic Acids Res. 1991;19:25–32. doi: 10.1093/nar/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Zeng C, Brinkley BR. Nuclear matrix proteins as structural and functional components of the mitotic apparatus. Int Rev Cytol. 1995;162B:1–74. doi: 10.1016/s0074-7696(08)62614-5. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D, Gautier T. The chromosome periphery during mitosis. Bioessays. 1994;16:179–185. doi: 10.1002/bies.950160308. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF, Parmelee SJ, Rykowski MC, Agard DA, Sedat JW. The onset of homologous chromosome pairing during Drosophila melanogasterembryogenesis. J Cell Biol. 1993;120:591–600. doi: 10.1083/jcb.120.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan NC, Traverse KL, Sullivan DE, Pardue ML. The nucleus-limited Hsr-omega-n transcript is a polyadenylated RNA with a regulated intranuclear turnover. J Cell Biol. 1994;125:21–30. doi: 10.1083/jcb.125.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan NC, Slot F, Traverse KL, Garbe JC, Bendena WG, Pardue ML. Stability of tandem repeats in the Drosophila melanogasterHsromega nuclear RNA. Genetics. 1995;139:1611–1621. doi: 10.1093/genetics/139.4.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovemann B, Walldorf U, Ryseck R-P. Heat-shock locus 93D of Drosophila melanogaster:an RNA with limited coding capacity accumulates precursor transcripts after heat shock. Mol Gen Genet. 1986;204:334–340. [Google Scholar]
- Hovemann BT, Dessen E, Mechler H, Mack E. DrosophilasnRNP associated protein P11 which specifically binds to heat shock puff 93D reveals strong homology with hnRNP core protein A1. Nucleic Acids Res. 1991;19:4909–4914. doi: 10.1093/nar/19.18.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison N, Weintraub H. Localization of DNAse I-sensitive sequences to specific regions of interphase nuclei. Cell. 1985;43:471–482. doi: 10.1016/0092-8674(85)90177-1. [DOI] [PubMed] [Google Scholar]
- Janevski J, Park P, de Boni U. Organization of centromeric domains in hepatocyte nuclei: rearrangement associated with de novo activation of the vitellogenin gene family in Xenopus laevis. . Exp Cell Res. 1995;217:227–239. doi: 10.1006/excr.1995.1082. [DOI] [PubMed] [Google Scholar]
- Kabisch R, Bautz E. Differential distribution of RNA polymerase B and nonhistone chromosomal proteins in polytene chromosomes of Drosophila melanogaster. . EMBO (Eur Mol Biol Organ) J. 1983;2:395–402. doi: 10.1002/j.1460-2075.1983.tb01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallajoki M, Osborn M. Gel electrophoretic analysis of nuclear matrix fractions isolated from different human cell lines. Electrophoresis. 1994;15:520–528. doi: 10.1002/elps.1150150170. [DOI] [PubMed] [Google Scholar]
- Lakhotia SC, Sharma A. RNA metabolism in situ at the 93D heat shock locus in polytene nuclei of Drosophila melanogasterafter various treatments. Chromosome Res. 1995;3:151–161. doi: 10.1007/BF00710708. [DOI] [PubMed] [Google Scholar]
- Leffers H, Dejgaard K, Celis JE. Characterisation of two major cellular poly (rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur J Biochem. 1995;230:447–453. [PubMed] [Google Scholar]
- Lewis CD, Lebkowski JS, Daly AK, Laemmli UK. Interphase nuclear matrix and metaphase scaffolding structures. J Cell Sci Suppl. 1984;1:103–122. doi: 10.1242/jcs.1984.supplement_1.8. [DOI] [PubMed] [Google Scholar]
- Manuelidis L, Borden J. Reproducible compartmentalization of individual chromosome domains in human CNS cells revealed by in situhybridization and three-dimensional reconstruction. Chromosoma (Berlin) 1988;96:397–410. doi: 10.1007/BF00303033. [DOI] [PubMed] [Google Scholar]
- Mattern K, Humbel B, Muijsers A, de Jong L, van Driel R. hnRNP proteins and B23 are the major proteins of the internal nuclear matrix of HeLa S3 cells. J Cell Biochem. 1996;62:275–289. doi: 10.1002/(sici)1097-4644(199608)62:2<275::aid-jcb15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Matunis EL, Matunis MJ, Dreyfuss G. Characterization of the major hnRNP proteins from Drosophila melanogaster. . J Cell Biol. 1992a;116:257–270. doi: 10.1083/jcb.116.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Michael WM, Dreyfuss G. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol Cell Biol. 1992b;12:164–171. doi: 10.1128/mcb.12.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis EL, Matunis MJ, Dreyfuss G. Association of individual hnRNP proteins and snRNPs with nascent transcripts. J Cell Biol. 1993;121:219–228. doi: 10.1083/jcb.121.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Michelotti E, Michelotti G, Aronsohn A, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Sedat JW. Localization of antigenic determinants in whole Drosophilaembryos. Dev Biol. 1983;99:261–264. doi: 10.1016/0012-1606(83)90275-0. [DOI] [PubMed] [Google Scholar]
- Morcillo G, Diez JL, Carbajal ME, Tanguay RM. HSP90 associates with specific heat shock puffs (hsr omega) in polytene chromosomes of Drosophila and Chironomus. . Chromosoma (Berlin) 1993;102:648–659. doi: 10.1007/BF00352313. [DOI] [PubMed] [Google Scholar]
- Osborn M, Weber K. Immunofluorescence and immunocytochemical procedures with affinity purified antibodies: tubulin-containing structures. Methods Cell Biol. 1982;24:97–132. doi: 10.1016/s0091-679x(08)60650-0. [DOI] [PubMed] [Google Scholar]
- Park PC, de Boni U. Spatial rearrangement and enhanced clustering of kinetochores in interphase nuclei of dorsal root ganglion neurons in vitro: association with nucleolar fusion. Exp Cell Res. 1992;203:222–229. doi: 10.1016/0014-4827(92)90058-g. [DOI] [PubMed] [Google Scholar]
- Rabl C. Über Zellteilung. Morphol Jahrbuch. 1885;10:214–330. [Google Scholar]
- Risau W, Symmons P, Saumweber H, Frasch M. Nonpackaging and packaging proteins of hnRNA in Drosophila melanogaster. . Cell. 1983;33:529–541. doi: 10.1016/0092-8674(83)90434-8. [DOI] [PubMed] [Google Scholar]
- Sahlas DJ, Milankov K, Park PC, de Boni U. Distribution of snRNPs, splicing factor SC-35 and actin in interphase nuclei: immunocytochemical evidence for differential distribution during changes in functional states. J Cell Sci. 1993;105:347–357. doi: 10.1242/jcs.105.2.347. [DOI] [PubMed] [Google Scholar]
- Saumweber H, Symmons P, Kabisch R, Will H, Bonhoeffer F. Monoclonal antibodies against chromosomal proteins of Drosophila melanogaster. . Chromosoma (Berlin) 1980;80:253–275. doi: 10.1007/BF00292684. [DOI] [PubMed] [Google Scholar]
- Schmidt, E.R. 1992. A simplified and efficient protocol for nonradioactive in situ hybridization to polytene chromosomes with a DIG-labeled DNA probe. In Nonradioactive in situ Hybridization. Application Manual. S. Grünewald-Junho, J. Keesey, M. Leous, M. Miltenburg, and C. Schroeder, editors. Boehringer GmbH Biochemica, Mannheim, Germany. Second Edition. 97–99.
- Segalat L, Lepesant JA. Spatial distribution of the Sm antigen in Drosophilaearly embryos. Biol Cell. 1992;75:181–185. doi: 10.1016/0248-4900(92)90139-r. [DOI] [PubMed] [Google Scholar]
- Sharma A, Lakhotia SC. In situ quantification of hsp70 and alpha– beta transcripts at 87A and 87C loci in relation to hsr-omega gene activity in polytene cells of Drosophila melanogaster. . Chromosome Res. 1995;3:386–393. doi: 10.1007/BF00710021. [DOI] [PubMed] [Google Scholar]
- Spector D, O'Keefe R, Jimenez-Garcia L. Dynamics of transcription and pre-mRNA splicing within the mammalian cell nucleus. Cold Spring Harbor Symp Quant Biol. 1994;58:799–805. doi: 10.1101/sqb.1993.058.01.087. [DOI] [PubMed] [Google Scholar]
- Strouboulis J, Wolffe AP. Functional compartmentalization of the nucleus. J Cell Sci. 1996;109:1991–2000. doi: 10.1242/jcs.109.8.1991. [DOI] [PubMed] [Google Scholar]
- Stuurman N, de Jong L, van Driel R. Nuclear frameworks: concepts and operational definitions. Cell Biol Int Rep. 1992;16:837–852. doi: 10.1016/s0309-1651(05)80026-8. [DOI] [PubMed] [Google Scholar]
- Swedlow JR, Agard DA, Sedat JW. Chromosome structure inside the nucleus. Curr Opin Cell Biol. 1993;5:412–416. doi: 10.1016/0955-0674(93)90005-b. [DOI] [PubMed] [Google Scholar]
- van Driel R, Wansink DG, van Steensel B, Grande MA, Schul W, de Jong L. Nuclear domains and the nuclear matrix. Int Rev Cytol. 1995;162A:151–189. doi: 10.1016/s0074-7696(08)61231-0. [DOI] [PubMed] [Google Scholar]
- Vazquez J, Pauli D, Tissieres A. Transcriptional regulation in Drosophiladuring heat shock: a nuclear run-on analysis. Chromosoma (Berlin) 1993;102:233–248. doi: 10.1007/BF00352397. [DOI] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink DG, Manders EE, van der Kraan I, Aten JA, van Driel R, de Jong L. RNA polymerase II transcription is concentrated outside replication domains throughout S-phase. J Cell Sci. 1994;107:1449–1456. doi: 10.1242/jcs.107.6.1449. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Dobner PR, Lawrence JB. Higher level organization of individual gene transcription and RNA splicing. Science (Wash DC) 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Zirbel RM, Mathieu UR, Kurz A, Cremer T, Lichter P. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res. 1993;1:93–106. doi: 10.1007/BF00710032. [DOI] [PubMed] [Google Scholar]