Abstract
The Saccharomyces cerevisiae kinesin-related motor Kar3p, though known to be required for karyogamy, plays a poorly defined, nonessential role during vegetative growth. We have found evidence suggesting that Kar3p functions to limit the number and length of cytoplasmic microtubules in a cell cycle–specific manner. Deletion of KAR3 leads to a dramatic increase in cytoplasmic microtubules, a phenotype which is most pronounced from START through the onset of anaphase but less so during late anaphase in synchronized cultures. We have immunolocalized HA-tagged Kar3p to the spindle pole body region, and fittingly, Kar3p was not detected by late anaphase. A microtubule depolymerizing activity may be the major vegetative role for Kar3p. Addition of the microtubule polymerization inhibitors nocodazol or benomyl to the medium or deletion of the nonessential α-tubulin TUB3 gene can mostly correct the abnormal microtubule arrays and other growth defects of kar3 mutants, suggesting that these phenotypes result from excessive microtubule polymerization. Microtubule depolymerization may also be the mechanism by which Kar3p acts in opposition to the anaphase B motors Cin8p and Kip1p. A preanaphase spindle collapse phenotype of cin8 kip1 mutants, previously shown to involve Kar3p, is markedly delayed when microtubule depolymerization is inhibited by the tub2-150 mutation. These results suggest that the Kar3p motor may act to regulate the length and number of microtubules in the preanaphase spindle.
The mechanism by which molecular motors generate movement within the cell is an area of extensive current research. The ATPase “motor” domain of the microtubule motor kinesin (which is the conserved sequence element of the kinesin-related group) is sufficient to make a lateral association with a microtubule and translocate along the microtubule towards the polymer end. This observation, plus the wide divergence of sequence outside of the motor domain, has led to a well accepted model in which the nonmotor domain functions to link intracellular cargo to the moving motor domain (Vale and Goldstein, 1990). This model is strongly supported by ultrastructural studies and is almost certainly correct for at least some types of kinesin-related motors. Another mechanism by which microtubule-based movement could be generated or regulated by motors is by controlling the level of tubulin polymerization and thereby influencing movement of cargo associated with the ends of the microtubules (for review see Desai and Mitchison, 1995). This type of intracellular transport is most likely to be important during mitosis, during anaphase A when the kinetochore microtubules depolymerize, and for anaphase B spindle elongation (Inoue, 1981) when microtubule polymerization is required in many organisms, but may also play a role in other types in intracellular motility as well.
Microtubules are known to be highly dynamic, with tubulin exchange occurring at both ends of the polymer (Salmon et al., 1984; De Brabander, 1986). Microtubule numbers and lengths change during the transition between the interphase and mitotic portions of the cell cycle (Belmont et al., 1990). The switch to the mitotic microtubule array requires cyclin-dependent kinase (Gotoh et al., 1991; Buendia et al., 1992), but the molecular mechanisms by which the cell changes microtubule dynamics in response to this signaling pathway is only beginning to be understood. Some recent observations have suggested a role for microtubule-based motors in influencing the rate of microtubule turnover. Endow et al. (1994) have shown that purified GST–Kar3p fusion protein preferentially destabilizes the minus ends of taxol-stabilized microtubules assembled in vitro. A chimeric kinesin/ncd motor coating latex microspheres was able to stimulate turnover at the plus ends of microtubules grown from tethered Tetrahymena pellicles (Lombillo et al., 1995b ). Antibodies to the kinesin-related motor CENP-E were able to block microtubule disassembly-dependent chromosome movement in vitro (Lombillo et al., 1995a ). A kinesin-related motor was identified in Xenopus that can stimulate tubulin turnover at the plus ends of microtubules assembled in oocyte extracts (Walczak et al., 1996). Immunodepletion of XKCM1 decreased the frequency of catastrophic depolymerization, resulting in a dramatic increase in numbers and lengths of astral microtubules. Importantly, some of these results indicate that motors acting by themselves can influence the rate of microtubule turnover. However, it has not yet been shown whether molecular motors can also influence the rate of microtubule polymerization in vivo, where microtubule ends are often stabilized by association with kinetochores or spindle poles and where microtubule dynamics are different from those of isolated microtubules (Margolis and Wilson, 1978; Sawin and Mitchison, 1991).
In Saccharomyces cerevisiae, the kinesin-related Kar3p is a known minus end–directed motor (Endow et al., 1994; Middleton and Carbon, 1994) and is required for karyogamy (Meluh and Rose, 1990), the union of haploid nuclei during diploid formation. While Kar3p is also likely to play an important role in mitosis, since kar3 mutants have many mitotically arrested or delayed cells (Meluh and Rose, 1990), its function during mitotic division is currently unclear. It has been suggested that Kar3p acts to draw together the preanaphase spindle poles, since loss of Kar3p can partially suppress the spindle collapse observed with loss of the spindle assembly motors Cin8p and Kip1p (Saunders and Hoyt, 1992). Alternatively, the lack of spindle elongation in kar3 mutants has been interpreted as a failure in anaphase B spindle pole separation (Meluh and Rose, 1990; Roof et al., 1992).
We demonstrate here that the Kar3p motor is required for normal microtubule arrays in preanaphase cells. Absence of this motor led to an increase in the number and length of the cytoplasmic microtubules, especially during cell cycle arrest. Arrested controls showed no such change. In synchronized cultures, KAR3 was found to be required for normal microtubule arrays from START until anaphase. During mid to late anaphase, spindles in kar3 mutants appeared normal.
We have used the HA epitope tag to immunolocalize Kar3p to the spindle poles. Tagged Kar3p is apparently lost from the poles at the onset of anaphase, consistent with the observation that it is no longer required for normal microtubule arrays at this time. The kar3-Δ phenotypes of abnormal spindle morphology, partial mitotic arrest, and an observed temperature sensitivity are mostly or completely suppressed by addition of the microtubule destabilizing drugs benomyl or nocodazole or deletion of a nonessential α-tubulin gene, suggesting that excessive microtubule polymerization may be the major vegetative growth defect in kar3 mutants. The spindle collapse phenotype from loss of the spindle assembly motors Cin8p and Kip1p, previously shown to involve Kar3p, can be partially inhibited by a microtubule stabilizing β-tubulin mutation, further suggesting a linkage between Kar3p function and microtubule depolymerization. These results suggest that microtubule turnover before anaphase acts antagonistically to the Cin8p and Kip1p motors and that this microtubule turnover is influenced by Kar3p acting at the spindle poles.
Materials and Methods
Yeast Strains and Media
Yeast strains used in these experiments are derivatives of S288C and are listed in Table I. The kip1-Δ (kip1::HIS3), cin8-Δ (cin8::URA3), cin8-3 (Hoyt et al., 1992), kar3-Δ (kar3-102::LEU2; Meluh and Rose, 1990), tub3-Δ (tub3::TRP1; Schatz et al., 1986), and tub2-150 (Machin et al., 1995) alleles have been previously described. Rich (YPD) and synthetic media were as described (Sherman et al., 1983) with glucose, raffinose, or galactose added to 2%.
Table I.
Genotypes and Plasmids Used in This Study
| Yeast strain | Relevant genotype | n | Average spindle length in microns | Number of trials | Standard error | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| WSY32 | wild-type | 228 | 1.49 | 2 | 0.025 | |||||
| WSY33 | wild-type | 284 | 1.46 | 2 | 0.020 | |||||
| MAY2082 | cin8-3 kip1::HIS3 | 104 | 0.99 | 2 | 0.025 | |||||
| WSY311 | cin8-3 kip1::HIS3 | 251 | 1.17 | 2 | 0.014 | |||||
| WSY319 | tub2-150 | 200 | 1.64 | 2 | 0.025 | |||||
| WSY321 | tub2-150 | 200 | 1.61 | 2 | 0.023 | |||||
| MAY2531 | cin8-3 kip1::HIS3 tub2-150 | 269 | 1.43 | 3 | 0.035 | |||||
| MAY2951 | cin8-3 kip1::HIS3 tub3::TRP1 | 310 | 0.92 | 3 | 0.017 | |||||
| WSY324 | tub3::TRP1 | 200 | 1.01 | 2 | 0.014 | |||||
| WSY325 | tub3::TRP1 | 200 | 1.00 | 2 | 0.042 | |||||
| MAY2056 | cin8::LEU2 | not shown | ||||||||
| MAY2062 | cin8::URA3 | not shown | ||||||||
| WS540 | kar3-102::LEU2 | not shown | ||||||||
| WS288 | kar3-102::LEU2 | not shown | ||||||||
| MAY2371 | kar3-102::LEU2 cin8-3 kip1::HIS3 | not shown | ||||||||
| WSY564 | kar3-102::LEU2 tub3::TRP1 | not shown | ||||||||
| Plasmids Name | Relevant Plasmid LOCI | |||||||||
| pMR798 | KAR3 URA3 (CEN) | |||||||||
| pMR794 | KAR3 URA3 (2μm) | |||||||||
| pGD15 | kar3::HA (CEN) | |||||||||
| pYEp24 | URA3 (2 μm) | |||||||||
| PYCp50 | URA 3 (CEN) |
The genotypes of the reported strains (all haploid) and plasmid genes are indicated. The values for the spindle length from indicated strains are shown. n, the total number of spindles measured for that genotype.
HA Epitope Tagging of KAR3
The 12CA5 epitope sequence was inserted near the 3′ end of the KAR3 coding sequence as follows: a 1.4-kb HindIII-HindIII fragment from pMR798 which contains the full-length KAR3 coding sequence as well as 5′ and 3′ flanking sequences was cloned into the pGEM3Z vector (Promega Biotech, Madison, WI) cut with EcoRI site and the overhangs filled with Klenow polymerase (pGD13). An HA cassette with three HA coding sequences in tandem was PCR amplified from pMR2307 (a gift from Karen Arndt, University of Pittsburgh, Pittsburgh, PA) with an EcoRI site added at each end and cloned into the unique EcoRI site in the KAR3 HindIII fragment. The EcoRI site is located 23 nucleotides before the KAR3 stop codon. To maintain the correct reading frame of the fusion protein, the nucleotides T and GT were included at the 5′ and 3′ ends, respectively, of the HA cassette. The resulting plasmid, pGD14, was sequenced to confirm the expected sequence. The 1.5-kb HindIII-HindIII fragment containing the HA tag was cloned back into pMR798 lacking the 1.4 HindIII-HindIII fragment to yield pGD15. The fusion protein contained a duplication of the amino acids V-N-S (one letter code) from the KAR3 sequence. The last seven amino acids of KAR3 were repeated in frame after the HA epitope. The final sequence of the fusion protein is given below with the partial KAR3 sequence underlined.
| aaa gtg aat tct tac cca tac gat gtt cct gac tat gcg | ||||||||||||
| K | V | N | S | Y | P | Y | D | V | P | D | Y | A |
ggc tat ccc tat gac gtc ccg gac tat
| G | Y | P | Y | D | V | P | D | Y |
| gca gga tcc tat cca tat gac gtt cca gat tac gct gtg | ||||||||||||
| A | G | S | Y | P | Y | D | V | P | D | Y | A | V |
| aat tct acc aga ttg gtt agt aga aaa |
| N | S | T | R | L | V | S | R | K |
The sequence data for KAR3 are available from GenBank/EMBL/DDBJ under accession number M31719.
For immunoblot analysis, yeast cells were lysed with glass beads as described (Harlow and Lane, 1988). Approximately 0.2 OD600 of cells were loaded in each well of a 7.5% SDS-PAGE gel and blotted to a PVDF (polyvinylidene difluoride) membrane (Micron Separations, Inc., Westborough, MA). Blocking, antibody incubation, and washing were essentially as described by Harlow and Lane (1988), with anti-HA diluted 1:1,000 and HRP-conjugated goat anti–mouse IgG (Boehringer Mannheim Corp., Indianapolis, IN) at 25,000-fold dilution. Immunoblots were developed using the Renaissance chemiluminescence system from DuPont NEN (Boston, MA), and the signals were detected with Reflection-nef autoradiography film from DuPont NEN.
Growth curves to test kar3::HA–tagged complementation was performed by growing cells overnight in selective −ura medium, diluting to equal densities of 0.2 OD600, and measuring the change in optical density with time.
Microscopic Analysis of Cells, Imaging, and Spindle Measurements
Anti-tubulin immunofluorescence was performed on formaldehyde-fixed cells using monoclonal antibodies YOL 1/34 (Serotec Ltd., Oxford, UK) and rhodamine-conjugated secondary antibodies (Jackson ImmunoResearch Labs, Inc., West Grove, PA) as described (Pringle et al., 1991; Hoyt et al., 1992). For immunolocalization of Kar3p, kar3-Δ cells were transformed with a CEN plasmid containing HA-tagged kar3 (pGD15). The HA epitope tag is sensitive to formaldehyde fixation. We found that brief fixation times of 7.5 min in 3.7% formaldehyde, with a quick wash in 0.1% BSA/PBS followed by a short 10-min zymolyase treatment and methanol/ acetone fixation, allowed preservation of most microtubules in the cell without losing the kar3::HA staining. Antibodies were added in the following order: mouse anti-HA (Boehringer Mannheim Corp.) at 1:1,000, sheep anti–mouse conjugated to CY3 (Sigma Chemical Co., St. Louis, MO) at 1:500, rat anti-tubulin (Serotec Ltd.) at 1:100, and goat anti–rat conjugated to FITC (Boehringer Mannheim Corp.) at 1:100. When the same procedure was followed without the anti-HA primary antibody or with cells lacking pGD15, essentially all CY3 staining was lost. Careful washing after the anti–mouse CY3 was required to avoid crossreaction with the anti-tubulin antibody, which followed. Cells were also stained with the DNA-specific fluorescent dye DAPI (4,6-diamidino-2-phenylindole; Sigma Chemical Co.) at 1 μg/ml to identify the position of the nucleus. The DAPI staining was typically unremarkable. The slides were examined with a B60 epifluorescence microscope (Olympus Corp., Lake Success, NY) using a 100× objective and digital images captured with an Argus 20 CCD camera and image processor. To increase the percentage of cells in the correct focal plane, all photo images of anti-tubulin and antiHA staining are composites made by combining relevant portions of selected captured images taken from a single sample using cut and paste features of the Adobe Photoshop software (Adobe Systems Inc., Mountain View, CA). Microtubule arrays were chosen based on clarity and uniform plane of focus throughout the spindle, and care was taken to pick a representative sample. Images were processed within the Photoshop program to make a uniform background.
Spindle length measurements were made from captured anti-tubulin fluorescent images using the Argus 20 image processor cursor-based measuring function. Distances were measured from the approximate outside edge of the spindle poles. In many but not all cases, the DAPI staining of chromatin was examined to confirm that the cells were not in anaphase. Calibration of the measurement software program was performed by use of an engraved slide (Olympus Optical Co., Tokyo, Japan).
Cell Cycle Arrests and Inhibitors
Hydroxyurea arrest was performed by treating cells with 0.1 M hydroxyurea (Aldrich Chemical Co., Inc., Milwaukee, WI) in YPD, pH 5.8, or medium selective for the plasmid, at 26°C for 4–5 h. Typically 65–80% of the cells arrested with large buds and a single nucleus, as determined by fixing cells with 70% ethanol and staining with DAPI as described (Hoyt et al., 1992). Large budded cells were defined as those with a bud greater than approximately half the diameter of the mother. α-Factor arrest was achieved by treating cells with pheromone (Bachem California, Torrance, CA) at 6 μg/ml in selective medium, pH 4.0, at 30°C for 3 h. Typically, >80% of the cells arrested without buds, as determined by diffential interference contrast (DIC) microscopy. Release from pheromone was achieved by resuspending the centrifuged and washed cells in the same medium without inhibitor.
The metabolic inhibitor 2, 4-dinitrophenol (Sigma Chemical Co.) was made as a stock of 0.15 M in ethanol and used at a final concentration of 0.9 mM. Cells were synchronized with hydroxyurea as above and resuspended in fresh YPD medium at 23°C for 5 min. Inhibitor was then added for 2 min at 23°C and the cells transferred to 37°C for 10 and 30 min, still in the presence of inhibitor.
Benomyl (a gift from DuPont NEN) was added to the warm (∼65°C) liquid medium to a final concentration of 10 μg/ml from a 10 μg/ml stock in DMSO. We had difficulty reproducing the effective benomyl concentrations due to insolubility. During later experiments, Nocodazole (Sigma Chemical Co.) was used in the liquid cultures at 3.3 μg/ml from a 3.3 μg/ml stock in DMSO. Where tested, both microtubule inhibitors gave similar results.
Results
Deletion of KAR3 Increased Cytoplasmic Microtubule Number and Length in Preanaphase Spindles
To investigate the consequence of loss of KAR3 on mitotic spindle microtubules we carefully compared spindle structure between cells with KAR3-deleted (kar3-Δ) and wildtype cells. (The kar3-Δ strain was a gift of Meluh, P., and M. Rose, Princeton University, Princeton, NJ). Cultures were grown to log phase in YPD medium, fixed, and stained with anti-tubulin antibodies (see Materials and Methods). Spindles in kar3-Δ cells had more microtubules that were not associated with the nuclear spindle and appeared to be cytoplasmic (Fig. 1; kar3-Δ) than in wild-type cells (not shown; see control below). The abnormal microtubule numbers in the kar3 mutants could result from the reported mitotic cell cycle delay (Meluh and Rose, 1990), which may induce an accumulation of extra microtubules. To allow comparison with a mitotically arrested control, we examined kar3-Δ and wild-type cultures treated with the DNA synthesis inhibitor hydroxyurea (Pringle and Hartwell, 1981). Under these conditions most cells arrest in S phase with an assembled spindle but are unable to begin anaphase. Mitotic arrest of the kar3-Δ cells resulted in a marked exaggeration of the abnormal microtubule numbers seen in the asynchronous culture (Fig. 1; kar3-Δ + hu). Strikingly, large microtubule arrays could now be seen radiating from the spindle poles. The majority of cells under these conditions had separated spindle poles as determined by anti–spindle pole body staining (results not shown). Hydroxyurea arrest of the wild-type cells caused little if any change in the numbers of cytoplasmic microtubules (Fig. 1; WT + hu). Many of the extra microtubules seen in the kar3-Δ mutants were clearly cytoplasmic, because they were either too long to be nuclear or pointed away from the nuclear envelope. Others could be either nuclear or cytoplasmic. We carefully counted the numbers of microtubules that appeared to be cytoplasmic by the above criteria and found a marked increase in kar3-Δ cells compared to the wild-type control (Fig. 2; note that all cells are hydroxyurea arrested unless indicated otherwise). Cytoplasmic microtubule numbers in cin8 kip1 mutants were not counted, but they did not show the large microtubule arrays found in kar3-Δ cells following hydroxyurea arrest (not shown).
Figure 1.
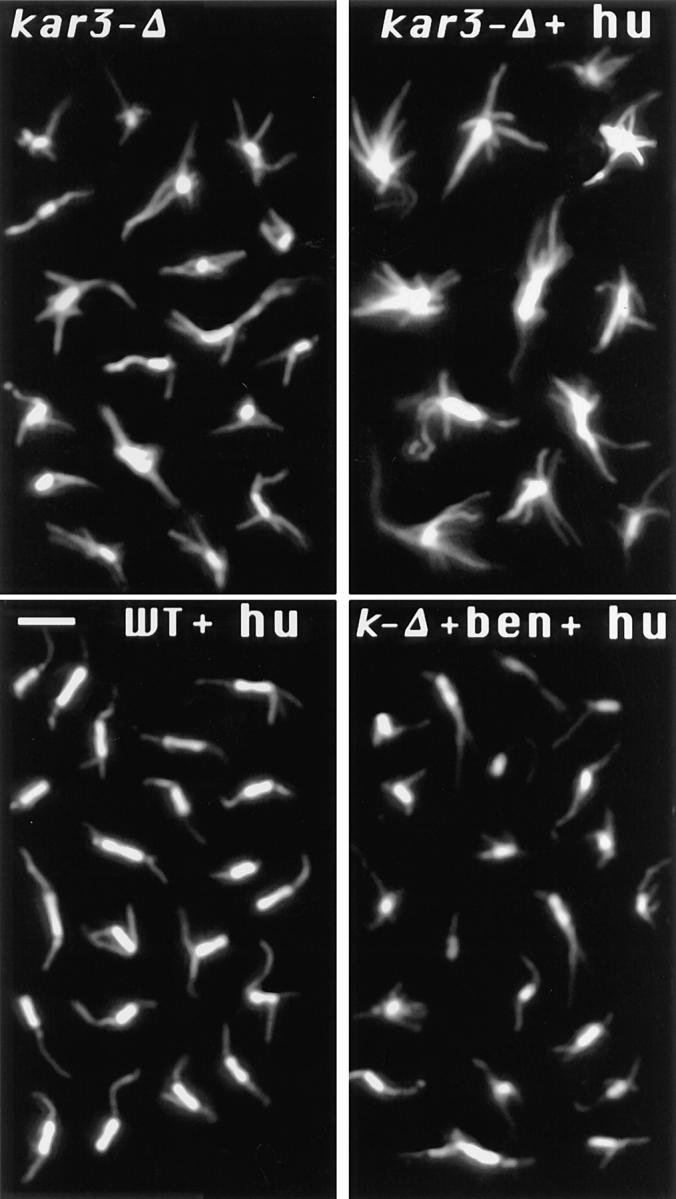
Change in spindle structure with loss of Kar3p. kar3-Δ or wild-type (WT) cells were arrested with hydroxyurea (hu) or grown to log phase without arrest, and samples were fixed and processed for anti-tubulin immunofluorescence. Each panel is a composite image of representative preanaphase spindles from a single sample. As shown, loss of KAR3 led to an increase in cytoplasmic microtubules in unarrested cultures (kar3-Δ). This phenotype was greatly exaggerated after mitotic arrest with hydroxyurea (kar3-Δ + hu). Short spindles in wild-type cells appeared essentially the same with or without hydroxyurea treatment (shown with hydroxyurea treatment). Addition of 10 μg/ml benomyl during the hydroxyurea arrest prevented the abnormal spindle structure in kar3 mutants. Bar, 2 μm.
Figure 2.
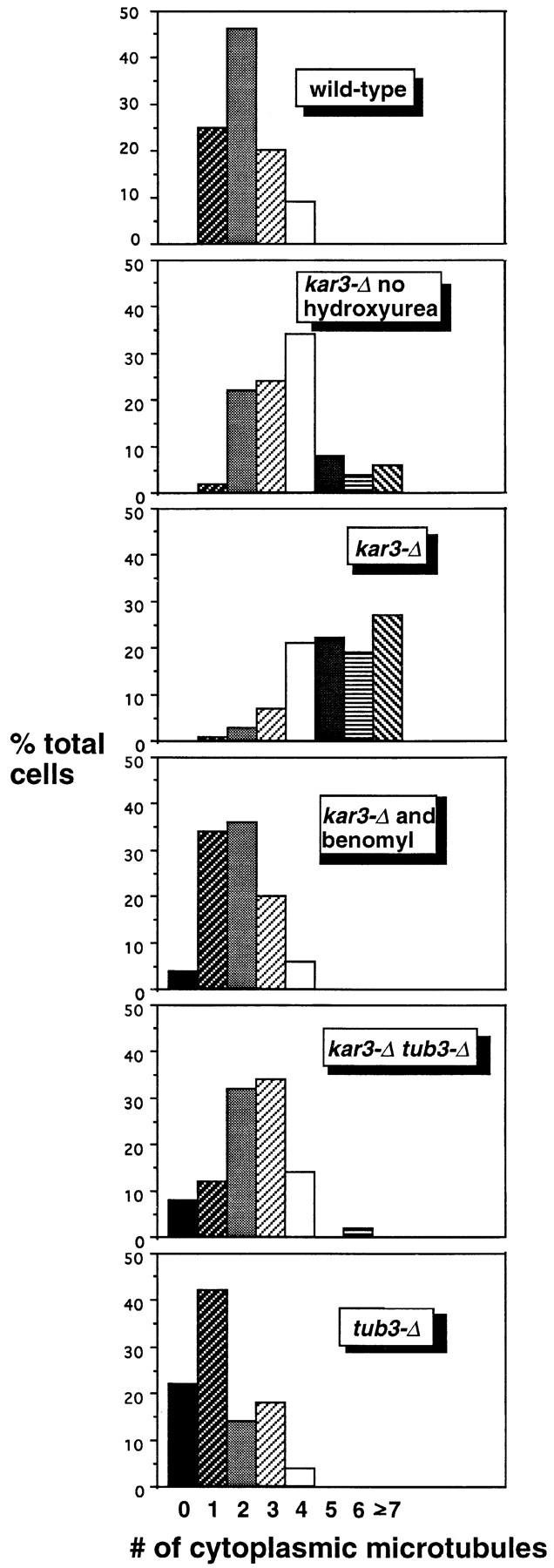
The increased cytoplasmic microtubule number in hydroxyurea-arrested kar3-Δ cells can be corrected by addition of benomyl or deletion of TUB3. All samples were arrested with hydroxyurea (except as indicated) for 4 h at 26°C, fixed, and stained with anti-tubulin antibodies by indirect immunofluorescence (see Materials and Methods). Cells were examined by epifluorescence microscopy, and the approximate number of cytoplasmic microtubules was determined. Cytoplasmic microtubules were defined as those that were too long to be nuclear and/or that clearly pointed away from the nuclear envelope. Loss of Kar3p resulted in an increase in cytoplasmic microtubules, especially in the hydroxyurea-arrested culture. This defect could be mostly corrected by addition of 10 μg/ ml benomyl to the culture medium or by deletion of the TUB3 genomic locus. tub3-Δ single mutants had fewer cytoplasmic microtubules than wild-type cells, while the tub3-Δ kar3-Δ double mutants had a number intermediate between that of tub3-Δ and kar3-Δ single mutants. The number of cytoplasmic microtubules from 100 cells was determined for each sample. Note that these numbers may represent an underestimate of the total, as some short microtubules (∼25% of the total) could not be determined to be nuclear or cytoplasmic. Also, the most abnormal spindles in the kar3-Δ populations were probably not recognized as spindles and not included in the sample.
We also observed an increase in the average length of the microtubules in kar3 mutant cells. In asynchronous culture, microtubules were on average 2.6 μm (n = 100) in length, while in wild-type cells the average length was 1.5 μm (n = 100). Microtubule lengths in arrested cultures were not measured but appeared markedly longer in the kar3 mutant (Fig. 1).
In the hydroxyurea-arrested kar3 culture (but not in the asynchronous culture) the nuclear spindle appeared more robust than in wild-type cells, possibly indicating an increase in nuclear microtubules. However, at least some of this apparent difference could be due to the increase in cytoplasmic microtubules.
To determine if the abnormal microtubule array in kar3 mutants was specific for cells in mitosis, we next compared kar3-Δ and wild-type cells arrested at another point in the cell cycle. Cells exposed to mating pheromone reversibly arrest with a monopolar microtubule array at the START point of the cell cycle, in late G1 (Pringle and Hartwell, 1981). Cultures were treated with the pheromone α-factor for 3 h at 26°C and fixed and stained with anti-tubulin antibodies as above. As observed with hydroxyurea-arrested cells, α-factor–arrested kar3 mutants had microtubules that were more numerous and longer than those found in arrested wild-type cells (Fig. 3 A). These results indicate that loss of KAR3 leads to an increase in cytoplasmic microtubules at both START and S phase of the cell cycle.
Figure 3.
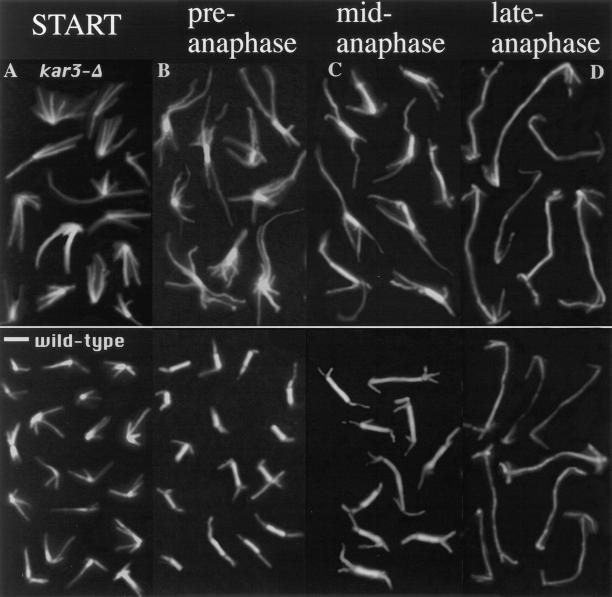
Cytoplasmic microtubule numbers in α-factor–arrested and released kar3-Δ mutants. Wild-type and mutant cells were arrested with α-factor, washed, and released in fresh medium. Samples were fixed and processed for anti-tubulin immunofluorescence for 0 (A) and 90 min (B–D) of release. α-Factor–arrested kar3 mutants had more and longer microtubules than arrested wild-type cells. For the 90 min release samples, selected spindles are shown from the same sample which are short (B), medium (C), or longer in length (D). Short spindles most likely represent cells that have not yet begun or are in the earliest stages of anaphase. kar3 mutant cells at this stage have markedly abnormal microtubule arrays, similar to those seen in the hydroxyurea-arrested cultures (compare to Fig. 1). Medium length spindles are typically from cells that have started but not completed anaphase, and in the kar3 mutants are closer in appearance to those from wild-type cells. Cells with the longest spindles are presumed to be in late anaphase or early telophase. These spindles are almost indistinguishable between wild-type cells and kar3 mutants. Bar, 2 μm.
The observed abnormal spindle microtubule array in asynchronous cultures was clearly exaggerated by the cell cycle arrest. We were interested in whether the abnormal microtubule arrays in the α-factor–arrested cells would form normal spindles when released from the cell cycle arrest. This would allow us to determine whether the arrest was a direct consequence of being blocked in the cell cycle. After 90 min in fresh medium without pheromone, most of the cells escaped the α-factor arrest, as shown by the appearance of assembled spindles. The now cycling cells were not completely synchronous. In some cases the spindles were short, indicating that the cells had not yet entered anaphase. Others had longer spindles, with the length of the spindle being roughly proportional to the time elapsed since the initiation of anaphase. Under these conditions, kar3-Δ mutants with short preanaphase spindles had the same abnormal microtubule arrays as hydroxyurea-arrested cultures (compare Fig. 3 B to Fig. 1) indicating that continuous cell cycle arrest is not required for this phenotype. kar3-Δ mutants in late anaphase, from the same 90 min release timepoint, had spindles that appeared essentially indistinguishable from wild-type cells (Fig. 3 D). Note that all the kar3-Δ cell spindles shown in Fig. 3, B–D were from the same sample and had been released from α-factor arrest for the same time period. Therefore, the differences seen are not due to the total time of release but apparently to the phase of the cell cycle in which the cells reside. It would seem that kar3-Δ mutants were unable to correct the abnormal spindle structure before anaphase but were able to establish a normal microtubule array once anaphase began.
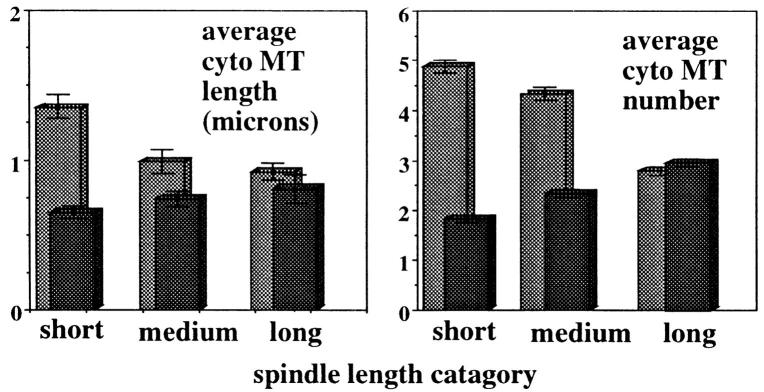
Considering only microtubules that were either too long to be entirely nuclear or that pointed away from the nucleus, the cytoplasmic microtubules in α-factor–arrested and released kar3-Δ mutants were measured and counted (Fig. 4). As observed for hydroxyurea-arrested cells, both the length and number of cytoplasmic microtubules associated with short spindles were higher in α-factor–arrested and released kar3-Δ mutants than in wild-type cells treated in the same manner. As cells entered anaphase, the cytoplasmic microtubules were seen to increase in wild-type cells but to decrease in kar3 mutants. By late anaphase the numbers were very similar.
Figure 4.
Measurements of cytoplasmic microtubule numbers and lengths in α-factor–arrested and released cells. Wild-type and kar3 mutants were arrested and released as in Fig. 3. The numbers and lengths of the cytoplasmic microtubules were counted as in Fig. 2. For this analysis, short spindles were defined as those where no chromatin separation was visible (typically 2.5 μm or less). Medium length spindles were those with spindles between ∼2.5 and 7.0 μm in length and at least some chromatin separation visible. Longer spindles were those greater than ∼7.0 μm in length with well separated chromatin masses. 100 spindles were examined for each category. As shown, the number and length of cytoplasmic microtubules in wild-type cells (black) increased slightly with spindle length. kar3 mutants (gray) with short spindles had many more and longer cytoplasmic microtubules than wild-type cells. Late anaphase spindles had similar numbers of cytoplasmic microtubules in wild-type and kar3 mutants.
Stimulating Microtubule Depolymerization Suppresses kar3-Δ Growth Defects
The increase in cytoplasmic microtubules with loss of KAR3 suggested that Kar3p may function to somehow limit the number and length of the microtubules in the cell. If Kar3p functions primarily to destabilize microtubules then destabilizing microtubules by other means should eliminate the need for Kar3p. To test this hypothesis we used two independent methods of destabilizing microtubules in vivo to determine if these agents could correct for the phenotypes observed from loss of Kar3p. One method of destabilizing microtubules was the use of one of two benzimidazolderived microtubule destabilizing drugs: benomyl, which is often used to reduce microtubule polymerization in yeast (Davidse, 1986), or nocodazole, a widely used microtubule inhibitor (De Brabander, 1986). The second method for destabilizing microtubules was deletion of the redundant nonessential α-tubulin TUB3 gene (Schatz et al., 1986). Loss of the TUB3 gene increases the sensitivity of cells to benomyl and nocodazole, presumably by reducing the levels of α-tubulin.
The kar3-Δ phenotypes tested were the abnormal spindle morphology introduced above, the kar3-Δ–induced mitotic delay described previously (Meluh and Rose, 1990), and an observed temperature sensitivity caused by KAR3 deletion. Benomyl at 10 μg/ml or deletion of TUB3 mostly corrected the observed microtubule defect of kar3-Δ mutants (Figs. 1 and 2; benomyl at around 12–15 μg/ml is lethal). KAR3 deletion was found to cause a slight temperature sensitivity that was completely suppressed by benomyl at 5 μg/ml (Fig. 5). In contrast, cells with a deletion of CIN8, which also causes temperature sensitivity (Hoyt et al., 1992), were not rescued by benomyl (Fig. 5; even at higher concentrations not shown). To test the effect of tub3-Δ on temperature sensitivity, we crossed kar3-Δ and tub3-Δ strains. The kar3-Δ tub3-Δ double mutants were less temperature sensitive than the kar3-Δ parent (Fig. 5), but there was much variability in temperature sensitivity among the progeny. The cause of this variation is unknown, but both the tub3 and kar3 mutations can cause high inviability among meiotic spores (Schatz et al., 1986; and Saunders, W., unpublished observations), a phenotype often caused by aneuploidy, which may also contribute to the observed variation in temperature sensitivity. To confirm the genetic suppression by tub3-Δ, eight kar3-Δ tub3-Δ double mutants were transformed with the TUB3 gene (a gift of D. Botstein, Stanford University, Palo Alto, CA). In each example, addition of the wild-type TUB3 gene significantly increased the temperature sensitivity of the kar3 tub3 double mutants. It is notable that addition of the wild-type allele made the double mutants more temperature sensitive and strongly suggests that loss of TUB3 improved the growth rates of kar3 mutants. The kar3-Δ allele did not change the benomyl sensitivity of cells containing the tub3-Δ allele (results not shown).
Figure 5.

Temperature sensitivity of strains containing the kar3-Δ allele was mostly corrected by addition of benomyl to the medium or deletion of TUB3. Cells of the indicated genotypes were suspended in water and serial dilutions placed on YPD plates with or without 5 μg/ml benomyl, at 30° or 37°C, for 2–3 d. Growth of kar3-Δ cells was observed to be partially inhibited at 37°C. This temperature sensitivity was eliminated by addition of benomyl to the medium or by deletion of TUB3. The benomyl sensitivity caused by the tub3-Δ allele was not rescued by kar3-Δ (not shown). The kar3-Δ, tub3-Δ, and tub3-Δ kar3-Δ double mutants are all from the same tetrad. (Growth of kar3-Δ mutants at 37°C did not cause a noticeable cell cycle arrest phenotype, results not shown.) In contrast, deletion of CIN8, which also causes slight temperature sensitivity, could not be rescued by benomyl.
A third phenotype from loss of KAR3 is a partial mitotic arrest or delay. More than 30% of kar3 cells are large budded with short preanaphase spindles and single nuclei (Meluh and Rose, 1990). It was difficult to determine the exact percentage of cells with spindles in a kar3-Δ strain because of the increased cytoplasmic microtubule numbers (Fig. 1). To allow a more accurate quantitative analysis of the percentage of cells with bipolar spindles, an antibody to the 90-kD spindle pole antigen (Rout and Kilmartin, 1990) was used. Before spindle assembly, spindle pole staining with this antibody appears as a single dot and after assembly as two dots (Saunders and Hoyt, 1992). kar3-Δ, kar3-Δ tub3-Δ, and wild-type cells were grown to log phase in YPD with or without added benomyl, fixed, stained with the anti– spindle pole body antibody, and the number of cells with separated spindle poles determined. Our results confirm the increase in mitotic cells in kar3-Δ cultures observed by Meluh and Rose (1990), shown here as an increase in the percentage of cells with separated spindle poles (Table II; left column). Addition of 3.3 μg/ml nocodazole to the medium, or TUB3 deletion, reduced the number of kar3-Δ cells in mitosis to a percentage close to wild-type cells treated in the same manner. Samples from the same cultures were also fixed with ethanol and stained with DAPI to estimate the number of cells arrested in mitosis. For this analysis we counted the number of large budded mononuclear cells. This phenotype is uncommon in wild-type populations, because the cells usually divide before the bud is larger than half the size of the mother. Mutants unable to complete the preparation for anaphase typically arrest with a large budded mononuclear phenotype (Pringle and Hartwell, 1981). Similar to a previous report (Meluh and Rose, 1990), we observed about one third of the kar3-Δ cells to be large budded with a single nucleus, suggesting a mitotic delay. This arrest phenotype was mostly corrected by addition of nocodazole or deletion of TUB3. This is in sharp contrast to wild-type cells, which actually showed a slight increase in mitotically arrested cells with the addition of nocodazole, most likely due to a checkpoint arrest in response to the microtubule inhibitor (Hoyt et al., 1991; Li and Murray, 1991). The doubling times of kar3-Δ mutant cells also improved with the addition of nocodazole. The cells doubled in 200 min at 30°C in YPD and in 160 min at 30°C in YPD supplemented with 3.3 μg/ml nocodazole, as determined by the change in optical density at 600 nm. In summary, all of the tested vegetative growth defects of kar3-Δ cells, including partial mitotic arrest, abnormal spindle structure, and temperature sensitivity, could be mostly or completely corrected by increasing microtubule depolymerization by either of two independent means. These results strongly suggest a linkage between the requirement for KAR3 and microtubule depolymerization during vegetative growth. In contrast, the karyogamy defect of kar3-Δ cells was not improved by benomyl, nocodazole, or deletion of TUB3, as determined by the ability of kar3-Δ parents to form diploids (results not shown).
Table II.
Suppression of kar3-Δ Mitotic Delay by Treatments that Destabilize Microtubules
| Genotype | Cells in mitosis (anti-spb staining) | Cells arrested in mitosis (DAPI staining) | ||
|---|---|---|---|---|
| Wild-type | 24% ± 5.4 SD | 4.7% ± 1.6 SD | ||
| kar3-Δ | 46% ± 7.1 SD | 34% ± 5.2 SD | ||
| kar3-Δ tub3-Δ | 31% ± 9.6 SD | 7% ± 3 SD | ||
| Wild-type + nocodazole | 25% ± 6.9SD | 11.5% ± 2.9 SD | ||
| kar3-Δ + nocodazole | 29% ± 6.6 SD | 10% ± 4.4 SD | ||
| tub3-Δ | 22% ± 4.2 SD | 2.2% ± 0.8 SD |
As observed previously (Meluh and Rose, 1990), loss of KAR3 produced an increase in the percentage of cells in the preanaphase mitotic phase of the cell cycle, suggesting a delay in initiation or completion of nuclear division. Unsynchronized, log phase cells grown at 30°C were fixed and stained with anti-spindle pole antibodies and DAPI and examined by epifluorescence microscopy. The percentage of cells with separated spindle poles as determined by anti-spb staining and with large buds and a single nucleus as determined by DAPI staining is shown. Cells with separated spindle pole bodies were considered to be in mitosis. Large budded mononuclear cells were considered to be delayed or arrested in mitosis. The increase in mitotically arrested kar3 mutants was mostly or completely corrected if the TUB3 gene was deleted or if 3.3 μg/ml nocodazole was added to the culture. Addition of nocodazole to the wildtype population resulted in a slight increase in mitotically arrested cells, mostly likely indicating a checkpoint arrest response to this microtubule inhibitor. Large buds were defined as greater than ∼50% the size of the mother. 200–400 cells/sample were counted on each of three separate trials; the average is given.
Microtubule Depolymerization Acts Antagonistically to Cin8p and Kip1p
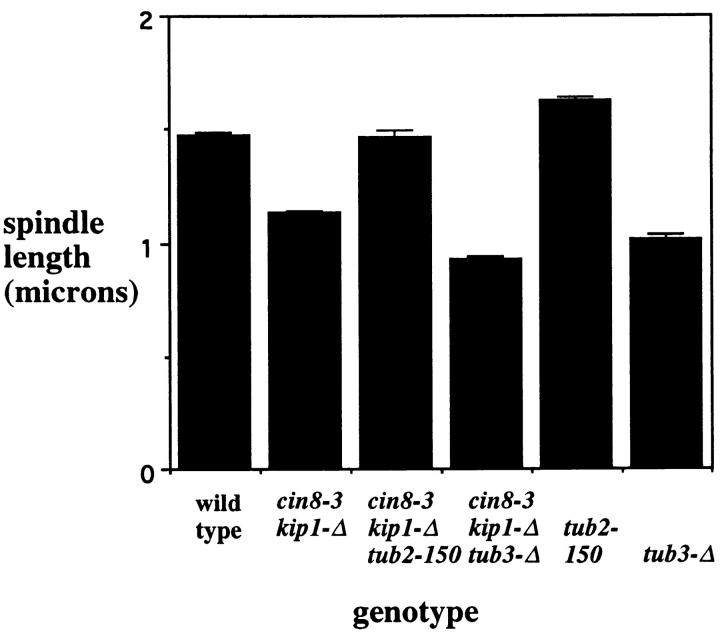
If Kar3p acts antagonistically to Cin8p and Kip1p and Kar3p function is associated with microtubule destabilization, then we should be able to show directly that microtubule depolymerization acts antagonistically to Cin8p and Kip1p. To investigate the effect of changing tubulin polymerization on cin8 kip1 mutants, two related mutant phenotypes were considered. We have observed previously that cin8 kip1 mutants had spindles slightly shorter than wild-type cells (Saunders and Hoyt, 1997). We were able to use the short spindle cin8 kip1 phenotype to investigate the relationship between loss of Cin8p/Kip1p activity and tubulin depolymerization. Two different tubulin mutations were used, the previously described TUB3 deletion and a mutation in the sole β-tubulin gene of S. cerevisiae, tub2-150 (Neff et al., 1983). The tub2-150 mutation acts in an opposite manner as TUB3 deletion, raising the resistance of mutant cells to microtubule depolymerizing agents, most likely by mutationally inhibiting depolymerization. The tub2-150 allele corrected the short spindle defect in cin8 kip1 cells while deletion of TUB3 made the phenotype worse (Fig. 6). These results suggest that spindle length is dependent on rates of microtubule polymerization, and at least this one defect in cin8 kip1 mutants could be corrected by stabilizing microtubules with the tub2-150 mutation.
Figure 6.
The short spindle phenotype from cin8 kip1 mutations is sensitive to tubulin mutations. Cells with the indicated genotypes were arrested with hydroxyurea at 26°C (a permissive temperature for growth of these strains), fixed, stained with antitubulin antibody, and the length of the spindles measured (see Materials and Methods). Mutational inhibition of the Cin8p and Kip1p motors produced a decrease in spindle length. This defect could be corrected by the tub2-150 mutation and was worsened by the tub3-Δ mutation. The numbers given represent the combined averages for two separate strains containing the indicated allele(s). Complete data for the individual strains are given in Table I. Each value represents at least 200 spindles counted during two to four separate experiments.
The observed differences were not due to differences in cell size. Spindle length was proportional to cell size; wildtype cells longer than 7.5 μm had an average spindle length of 1.8 μm, while those cells shorter than 7.5 μm had an average spindle length of 1.4 μm. But the average cell length of cin8-3 kip1-Δ tub3-Δ mutants was 7.9 μm, while wild type was 7.2 μm. Therefore, the shorter spindles in cin8-3 kip1-Δ tub3-Δ mutants were not due to a smaller cell volume. The ratio of cell length to spindle length in wild-type cells was 0.23 and in cin8-3 kip1-Δ tub3-Δ mutants 0.16, again suggesting that the short spindle phenotype was not solely due to changes in cell size.
We also tested the role of microtubule polymerization in a related cin8 kip1 phenotype spindle collapse. An inward collapse of the preanaphase spindles is observed when cells are deprived of Cin8p and Kip1p activity (Saunders and Hoyt, 1992). Cultures were arrested with hydroxyurea and shifted to 37°C, a nonpermissive temperature for the cin8-3 allele, for 10 and 30 min (Table III). The structure of the spindle and the position of the spindle poles were determined by staining with anti-spindle pole or anti-tubulin antibodies (see Materials and Methods). As reported previously, mutational inactivation of CIN8 and KIP1 resulted in an inward collapse of preanaphase spindles producing monopolar staining, and this collapse was partially suppressed by loss of KAR3 (Saunders and Hoyt, 1992). We found that the tub2-150 allele also partially inhibited the spindle collapse resulting from loss of Cin8p and Kip1p motor function (Table III). Thus by inhibiting microtubule depolymerization with the tub2-150 allele, we were able to simulate the loss of KAR3.
Table III.
Delay of Spindle Collapse in cin8 kip1 Mutants by the tub2-150 Allele
| Genotype | Cells at 26°C | Percent remaining after 10 min at 37°C | Percent remaining after 30 min at 37°C | |||
|---|---|---|---|---|---|---|
| With preanaphase spindles | ||||||
| A. Anti-tubulin antibodies | ||||||
| Wild-type | 54 (100%) | 104 | 111 | |||
| cin8-3 kip1-Δ | 42 (100%) | 8 | 2 | |||
| cin8-3 kip1-Δ tub2-150 | 77 (100%) | 90 | 10 | |||
| B. Anti-spindle pole antibodies | ||||||
| With separated spindle poles | ||||||
| Wild-type | 47 (100%) | 102 | 109 | |||
| cin8-3 kip1-Δ | 46 (100%) | 26 | 15 | |||
| cin8-3 kip1-Δ kar3-Δ | 58 (100%) | 70 | 35 | |||
| cin8-3 kip1-Δ tub2-150 | 69 (100%) | 113 | 17 | |||
| cin8-3 kip1-Δ and DNP | 50 (100%) | 101 | 94 | |||
Inhibition of spindle collapse in cin8 kip1 mutants. Cells of the indicated genotypes were arrested with hydroxyurea at 26°C and a portion of the cells shifted to 37°C for 10 and 30 min. Samples were fixed and treated for immunofluorescence, and the percentage of intact spindles determined by counting the number of cells with separated spindle poles using anti-spindle pole body staining or visibly intact spindles by antitubulin staining. (The same samples were treated with each antibody in separate experiments.) The two rightmost columns represent the percentage of spindles from the 26°C culture that remained after 10 and 30 min at 37°C. The spindle collapse of cin8 kip1 mutants was partially inhibited by loss of KAR3 as described previously (Saunders and Hoyt, 1992) and also by the tub2-150 mutation. Both the kar3-Δ and tub2- 150 mutations were most inhibitory at the earlier time point. The ATP synthesis inhibitor DNP at 0.9 μM essentially completely blocked spindle collapse (even to 60 minutes, not shown). The slight increase in wild-type cells with spindles at 37°C is reproducible and may represent a more complete arrest at the later time point. The numbers for the cin8-3 kip1-Δ tub2-150 mutant and the DNP treatment are an average of three separate experiments.
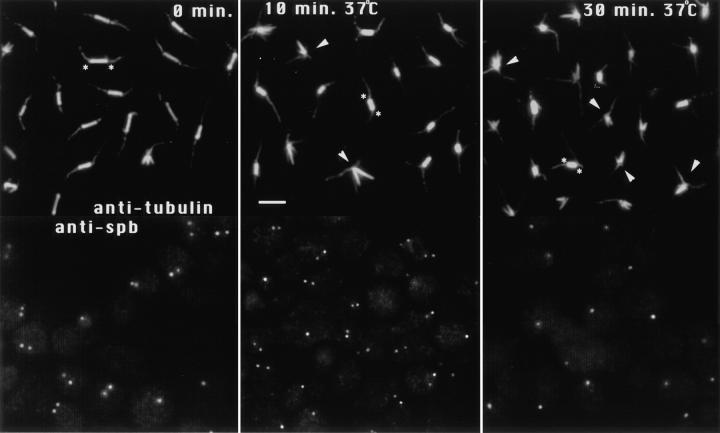
By looking at the later time point of 30 min, we found that both the kar3-Δ and tub2-150 mutations mostly delayed rather than indefinitely blocked the spindle collapse. The tub2-150 allele especially appeared to have the effect of slowing down the spindle collapse in the cin8 kip1 mutants. After 10 min at 37°C, most of the spindles in cin8 kip1 mutants had collapsed, but few if any of the spindles in cin8-3 kip1-Δ tub2-150 mutants had. However, the spindles of the cin8-3 kip1-Δ tub2-150 mutant were decreasing in length (Fig. 7). Before the temperature shift, cin8-3 kip1-Δ tub 2-150 mutants had spindle lengths of 1.8 μm ± 0.4 SD (n = 187), after 10 min at 37°C the average length was 1.3 ± 0.3 SD (n = 201), and after 30 min at 37°C the average length (of the remaining spindles) was 1.1 ± 0.4 SD (n = 80). These results suggest that the spindle collapse requires microtubule depolymerization and that the speed of collapse can be slowed by mutational changes in tubulin depolymerization rates. Note that microtubule depolymerization is presumably not completely blocked by the tub 2-150 allele, as this would certainly cause lethality. The ATPase inhibitors DNP or sodium azide (Table III, sodium azide not shown) essentially completely blocked the spindle collapse, even to 60 min (the latest time point tested, data not shown), showing that the collapsing process apparently requires ATP.
Figure 7.
The tub2-150 allele slowed down the spindle collapse in cin8 kip1 mutants. cin8-3 kip1-Δ tub2-150 mutants were arrested with 0.1 M hydroxyurea for 4 h and transferred to 37°C for 10 and 30 min, still in the presence of hydroxyurea. Samples were fixed and stained with anti-tubulin or anti-spindle pole body antibodies. At 0 min, most cells had intact spindles and duplicated and separated spindle poles. After 10 min at 37°C most of the spindles remained intact but were noticeably shorter than before the temperature shift. After 30 min, most of the spindles had collapsed, and the remaining spindles were shorter still. At this time point only single dots of spindle pole staining were typically observed. The anti-tubulin staining images are composites taken from different fields of the same sample (see Materials and Methods). The anti–spindle pole staining images are not composites and represent single fields. For selected examples, asterisks indicate the position of the spindle poles of intact spindles, and arrowheads point to collapsed spindles. Bar, 2 μm.
cin8-3 kip1-Δ tub 2-150 mutants were not able to divide efficiently at 37°C after release from hydroxyurea (data not shown). Also, the temperature sensitivity of the cin8 kip1 mutants was unchanged by the tub2-150 allele (not shown), suggesting that other requirements for CIN8 and KIP1 could not be eliminated by the tub2-150 allele. It is important to note that unlike the tub2-150 allele, deletion of KAR3 does lower the temperature sensitivity of the cin8 kip1 mutants (Saunders and Hoyt, 1992), which shows that the effects of the tub2-150 and kar3-Δ alleles, while related, are not identical.
Taken together these results suggest that the length of the preanaphase mitotic spindle is dependent on the rates of tubulin polymerization as well as the Cin8p/Kip1p motors. Increasing the stability of the microtubules in the cell can correct the short spindle defect and delay the spindle collapse caused by a loss of Cin8p and Kip1p motor activity.
Immunolocalization of HA Epitope-tagged Kar3p
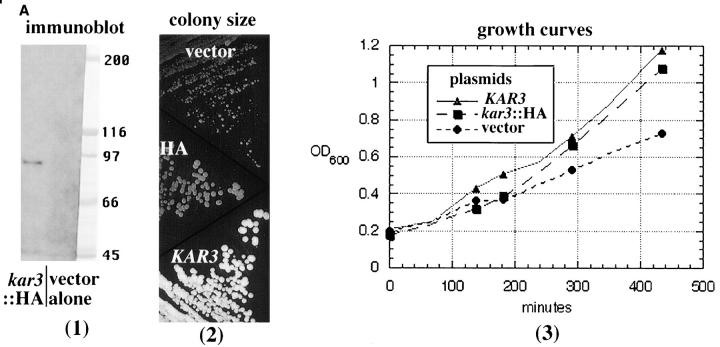
Functional Kar3p in vegetatively growing cells has not previously been immunolocalized. For this purpose we constructed a kar3::HA fusion protein by inserting three HA epitope tags in the KAR3 reading frame 23 nucleotides from the 3′ end of the gene. The insertion resulted in a duplication of the amino acids V-N-S after the epitope tag, and the last seven amino acids of Kar3p were present in frame after this duplication (see Materials and Methods). The immunostaining with anti-HA antibodies appeared specific for Kar3p. A single band of appropriate size was observed by immunoblotting that was not detected with vector alone (Fig. 8 A, 1). The kar3::HA fusion complemented the vegetative kar3-Δ growth defects. The irregular colony size observed in kar3-Δ mutants was lost with addition of HA-tagged KAR3 but not with vector alone (Fig. 8 A, 2). Growth curves of kar3-Δ strains were essentially the same with HA-tagged or wild-type KAR3 and better than vector alone (Fig. 8 A, 3). However, the HA-tagged kar3 was greatly inhibited for karyogamy. kar3-Δ parents were grown overnight to equal density and mixed together at 30°C for 6 h, and the percentage of diploids was determined by growth on selective plates (not shown). Whereas ∼20% of kar3 mutants formed diploids with KAR3 on a plasmid, only ∼2% did with HA-tagged KAR3, although this was much better than the 0.02% seen with vector alone.
Figure 8.
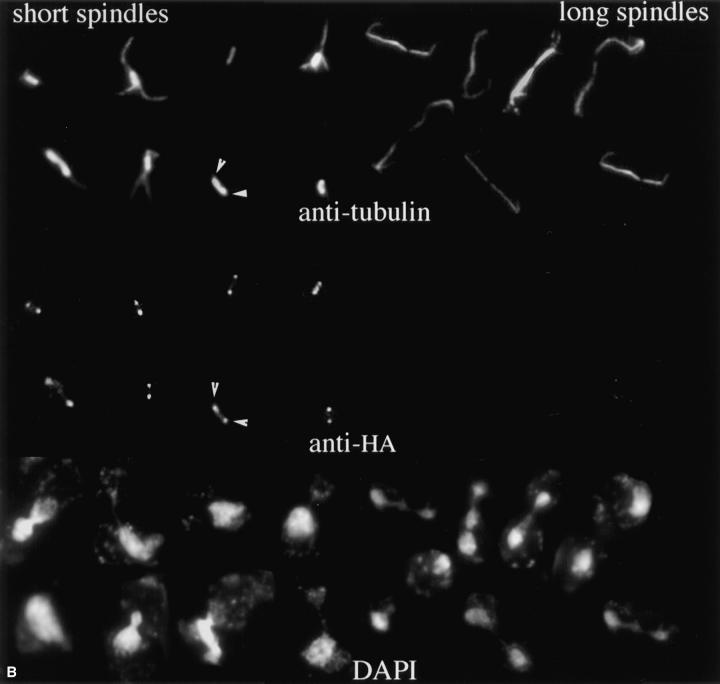
Immunolocalization of HA-tagged Kar3p to the spindle poles. A triple HA epitope tag was inserted near the 3′ end of the KAR3 coding sequence (see Materials and Methods) and transformed into a kar3-Δ strain. (A; 1) Immunoblot with anti-HA antibodies. kar3-Δ cells with the HA-tagged KAR3 (pGD15) or with vector alone (Ycp50) were grown in selective medium. Lysed cells were run on an SDS-PAGE gel, blotted to membrane, and probed with anti-HA antibodies (see Materials and Methods). Ponceau S–stained markers from the same gel are also shown. The predicted size of Kar3p is 84 kD. Similar results were seen in multiple experiments. (2) The colony size of kar3 mutants with vector alone (YCp50) was observed to be smaller and more varied in size than strains with KAR3 on a plasmid (pMR798). When HA-tagged KAR3 (pGD15) was transformed into kar3-Δ mutants the plasmid allowed normal colony size. Cells were grown in selective −ura medium overnight, streaked onto the same YEPD plates, and grown at 30°C for 3 d. Different kar3Δ strains gave similar results. (3) Growth curves of kar3 mutants. kar3-Δ cells were observed to double at similar rates with plasmids containing KAR3 (▴) or HA-tagged kar3 (▪), while those containing vector alone (•) divided more slowly. This test was repeated twice with similar results. (B) Cells grown to log phase were fixed briefly with formaldehyde and subjected to triple staining with antibodies to HA and tubulin and with DAPI. As shown, most of the HA-tagged Kar3p was observed to be associated with the spindle poles. The spindle pole bodies in a sample spindle are marked with arrowheads. Some intrapolar staining could be seen. Staining of intermediate length spindles varied (not shown), but long spindles from anaphase cells were invariably negative. Images shown are a composite of cells from the same sample.
The HA epitope was sensitive to formaldehyde fixation, and a short fixation time of 7.5 min was required for optimal preservation of microtubules and HA antigenicity. With careful processing (see Materials and Methods), most cells had microtubules, and spindles with intact nuclear and cytoplasmic microtubules could be readily found. However, the intensity of immunostaining of the cytoplasmic microtubules with anti-tubulin antibodies was significantly weaker than with fully fixed cells. About half of the cells showed what appeared to be specific anti-HA staining, and this staining was found to be predominantly associated with spindle poles (Fig. 8 B). Some staining could be seen along the length of the nuclear spindle microtubules, but this was weaker than the spindle pole staining. No cytoplasmic microtubule staining was seen. Cells with spindles of different lengths showed significantly different degrees of staining. Short spindles were usually positive with bright pole staining. The staining of the intermediate length spindles was variable, with some spindles showing spindle pole staining and others completely negative staining. Longer anaphase spindles were essentially always negative for anti-HA reactivity. Paired cells that were apparently in late telophase or in early G1 were negative for anti-HA binding (not shown). When cells were arrested with α-factor, we observed staining in nearly all cells with essentially all staining at the spindle poles (results not shown).
Discussion
Microtubule Turnover and Motor Function
Loss of the mitotic motor Kar3p produced a marked increase in the number and length of the cytoplasmic microtubules, suggesting a role for this motor in limiting the growth and extent of cytoplasmic microtubules in the cell. Limiting the number of microtubules may be one of the major functions of this motor during vegetative growth. All of the tested vegetative growth defects associated with loss of KAR3, including temperature sensitivity, slow growth, mitotic arrest/delay, and the increase in cytoplasmic microtubules, were mostly or completely corrected by stimulating microtubule depolymerization with the inhibitors benomyl or nocodazol, or by the deletion of the partially redundant α-tubulin gene TUB3. These results suggest a requirement for Kar3p in depolymerizing or otherwise destabilizing microtubules during vegetative growth.
We were unable to see any improvement in the kar3 karyogamy defect with addition of microtubule inhibitors or deletion of TUB3. This result emphasizes the difference between Kar3p function during vegetative growth and karyogamy. Previous work with a Kar3p-associated protein Cik1p has suggested a change in function during mitosis and karyogamy. Cik1p physically associates with Kar3p and is required for Kar3::βgal localization during karyogamy but not during vegetative growth. Strikingly, loss of Cik1p also stimulated an increase in cytoplasmic microtubules in α-factor–arrested cells (Page et al., 1994). Although Cik1p was not essential for Kar3p localization in the spindle, the similar phenotypes of increased microtubule arrays suggests that Cik1p may still be an important part of the Kar3p mitotic pathway.
The defect in microtubule arrays in kar3 mutants is greatly worsened by either a START or S phase cell cycle arrest. However, continuous arrest is not required, as cells released from arrest at START went on to produce markedly abnormal spindles. In these synchronized cultures, abnormal microtubule arrays are seen until mid to late anaphase. At late anaphase the spindles in kar3 mutants and wild-type controls treated in the same manner are indistinguishable. There appears to be a portion of the cell cycle, between START and the onset of anaphase, when KAR3 is especially important for normal microtubule dynamics. By late anaphase Kar3p is no longer essential for normal microtubule arrays. When cycling kar3 mutant cells pass through anaphase, they are probably able to correct the abnormal microtubule numbers, preventing the extreme phenotype observed in the arrested cells.
It is difficult in this in vivo analysis to determine whether the observed kar3 mutant phenotype is a direct or indirect consequence of loss of KAR3. For example, Kar3p has a known microtubule crosslinking domain outside of the motor region (Meluh and Rose, 1990). It is possible that the appearance of extra cytoplasmic microtubules could be a consequence of loss of cytoplasmic microtubule crosslinking. We believe this is unlikely to completely explain the observed phenotype for the following reasons. The total amount of cytoplasmic anti-tubulin staining, especially in the arrested or synchronized cultures, was much greater than in wild-type cells, implying an increase in cytoplasmic microtubule mass (compare short spindles in Fig. 3 B). Also, it appears that most cytoplasmic microtubules are not crosslinked in wild-type cells (Kilmartin, J., personal communication). Loss of crosslinking is also unlikely to explain the observed increase in microtubule length in the kar3 mutants. Finally, it would appear that most of the Kar3p is located at the spindle pole, a good spot to influence polymerization at the minus ends of the microtubules, but a poor one to influence microtubule crosslinking along the length of the microtubule.
Other indirect effects of loss of Kar3p are also possible and cannot be ruled out. Movement by the motor towards the minus end of the microtubule may partially dislodge the end from a stabilizing association with the spindle pole, indirectly promoting microtubule depolymerization. Alternatively, Kar3p could function to stabilize or organize the spindle poles, and loss of this putative organizing activity could indirectly lead to an increase in microtubule polymerization. However, we believe the most straight forward explanation of these in vivo observations and related results in vitro (Endow et al., 1994) is that Kar3p is directly stimulating microtubule depolymerization in the cell.
Whatever the mechanism of Kar3 function, loss of Kar3p has a different consequence for different types of microtubules. Most of the observed increase was for cytoplasmic microtubules, and little if any change was observed in the nuclear microtubules. This is unexpected as the spindle collapse of cin8 kip1 mutants, thought to be driven by Kar3p, represents a change in the nuclear spindle. One possible explanation for this paradox is that the length and number of the nuclear microtubules may be more constrained than for cytoplasmic microtubules. For example, kinetochore association and/or abutment of the nuclear microtubules with the opposite spindle pole body could limit the length of the spindle microtubules in kar3 mutants. It could also be difficult to visualize a subtle increase in the number of the tightly crosslinked nuclear microtubules compared to cytoplasmic microtubules. In the hydroxyurea arrested or synchronized kar3 cultures there is a hint of increased nuclear microtubule mass, possibly representing an increase in nuclear microtubules similar to that observed for the cytoplasmic microtubules. We think it likely that a similar increase in spindle microtubule number is occurring in the arrested kar3 mutants, but it may not be as readily apparent because of physical constraints within the spindle. It is also possible that Kar3p may act in a fundamentally different way on the nuclear and cytoplasmic microtubules (discussed further below).
Previously, kar3-Δ has been shown to produce an increase in preanaphase cells, suggesting a defect in anaphase spindle elongation (Meluh and Rose, 1990; Roof et al., 1991). The results presented here suggest an alternative explanation for this kar3 mutant phenotype. The mitotic arrest may be due to the recognition of abnormal microtubule polymerization rather than the loss of spindle elongating motor activity. Eukaryotic cells have cell cycle regulatory mechanisms that can detect abnormal microtubule dynamics and in response arrest the cells in G2/M (Hoyt et al., 1991; Li and Murray, 1991). Some of the genes, MAD and BUB, required for this arrest in S. cerevisiae have been identified (Hoyt et al., 1991; Li and Murray, 1991). Mutations in the MAD and BUB genes are known to be lethal when combined with the kar3-Δ mutation (Roof et al., 1991). The mitotic delay in kar3 mutants may be a consequence of a MAD/BUB checkpoint response, and loss of this delay could result in lethality.
If kar3 mutants are delayed from entering anaphase by a MAD/BUB-mediated arrest, could this checkpoint arrest explain the observed differences in cytoplasmic microtubule numbers in synchronized kar3 cells during the cell cycle? Could the differences be a manifestation of a requirement to correct the defect before the cells can enter anaphase? In this model, the abnormal preanaphase spindles would be in cells that had not yet performed this correction, while anaphase spindles would only be seen in cells that had performed the correction. We cannot rule out this interpretation, but the microtubule numbers in synchronized cells suggest it may be unlikely. We do not see a range of spindle phenotypes in the kar3 preanaphase spindles suggesting a correction mechanism in operation at this time. Essentially all the spindles appear as shown in Fig. 3 B; i.e., with markedly higher cytoplasmic microtubule numbers than late anaphase spindles. In contrast, we do see a gradual change in cells that have entered anaphase, from markedly abnormal in early anaphase to indistinguishable from wild type in late anaphase. We believe these observations are most consistent with the acquisition of a correcting mechanism, coincident with the onset of anaphase, rather than a gating mechanism at the metaphase/ anaphase boundary.
If there is not a gating mechanism for microtubule number, what causes the mitotic delay in the kar3 mutants? It is possible that the MAD/BUB checkpoint recognizes other aspects of the kar3 phenotype rather than the excess microtubule number. Recently, the MAD genes have been shown to be required for the mitotic delay in yeast in response to short linear centromeric chromosomes (Wells and Murray, 1996). We note that the midanaphase spindles in synchronized kar3 cells (Fig. 3) had comparable microtubule numbers and lengths to the asynchronous kar3 culture (Fig. 1), suggesting that this level of microtubule number was not by itself sufficient for the observed G2/M arrest in the asynchronous culture. In summary, while we believe it is likely that the kar3 mitotic delay phenotype is a cell cycle checkpoint arrest most likely mediated by the MAD/BUB genes, it is unlikely that this arrest is the direct cause of the differences in cytoplasmic microtubule numbers between preanaphase and anaphase in the synchronized kar3 cells observed here.
HA epitope–tagged Kar3p was found to be located primarily at the spindle poles in cells from α-factor arrest until early anaphase. The HA epitope is sensitive to formaldehyde fixation; therefore we cannot rule out additional Kar3p sites on the nuclear or cytoplasmic microtubules that are lost with fixation. However, in spindles where nuclear and cytoplasmic microtubules were preserved, the most staining was observed at the spindle poles. This is the first localization of complementing Kar3p in vegetatively growing cells, but the results are very similar to immunolocalization of a truncated version of Kar3p lacking the motor domain (Page et al., 1994). With the motor domain intact, we saw what may be lower levels of Kar3p in the region of the spindle between the spindle poles than reported by Page et al. One possibility suggested by these results is that Kar3p binds to microtubules in the absence of motor activity and uses the motor activity to move to the poleward end of the microtubule where it exerts its effect on microtubule numbers. In this model Kar3p would use its motor domain for transporting itself to the required site of action, rather than transporting a distinct cargo.
Importantly, HA-tagged Kar3p was lost from the spindle poles as the cells entered anaphase. Mid-anaphase spindles occasionally revealed some staining, but late anaphase spindles were exclusively negative. This was also the time during the cell cycle when Kar3p was found to be no longer required for normal microtubule arrays (see above). We have observed previously that the spindle collapse in cin8 kip1 mutants, which is stimulated by Kar3p, also ceases at the onset of anaphase (Saunders and Hoyt, 1992). These results suggest that Kar3p is most important before anaphase and less so, or not at all, after the onset of anaphase.
The proposed microtubule depolymerizing activity of Kar3p is likely to be the method by which this motor antagonizes the activity of the anaphase B motors, Cin8p and Kip1p. Both the short spindle phenotype of the cin8 kip1 mutant and the spindle collapse from complete loss of function could be suppressed or delayed by the microtubule stabilizing tub2-150 mutation. Therefore, at least part of the defect caused by loss of these motors may be attributable to microtubule depolymerization. However, it is unknown if the tub2-150 mutation has an effect on motor activity independent of its inhibition of depolymerization.
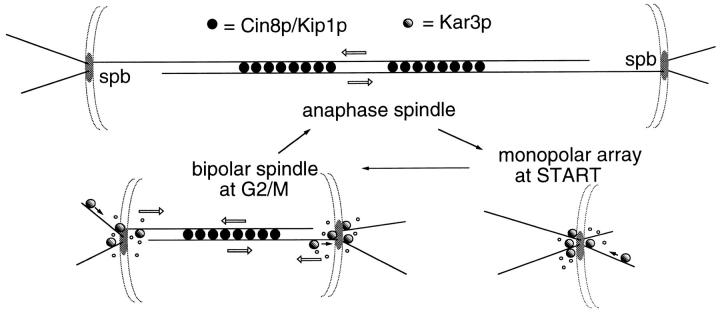
The results reported here support the following model (Fig. 9): Kar3p is proposed to use its minus end–directed motor capacity to move to the spindle pole region of the cells before or at the START point of the cell cycle. At this time Kar3p functions to stimulate microtubule depolymerization at the minus ends associated with the spindle pole. As the bipolar spindle forms, the two poles become physically linked by intrapolar microtubules via kinetochore attachments and polar microtubule crosslinking. Microtubule depolymerization now would have the consequence of pulling the linked spindle poles together. (This model assumes a net polymerization at the kinetochore or plus end of the spindle microtubules and a net depolymerization at the minus end. This type of spindle flux has been observed in spindles assembled in Xenopus oocyte extracts [Sawin and Mitchison, 1991] but has not yet been documented in the smaller yeast spindles.) In the normal spindle the assembly motors Cin8p and Kip1p resist this collapsing action producing a balanced level of motor function in the spindle. As anaphase begins, the microtubule dynamics in the cell change. Now there is a net polymerization of the nuclear microtubules (and to a lesser extent of the cytoplasmic microtubules, as shown here) to produce the long anaphase spindle necessary for proper chromosome segregation. During this time of net microtubule polymerization, the depolymerizing activity of Kar3p is no longer useful, and Kar3p is lost from the spindle poles. The activity of Cin8p and Kip1p is retained, and this shift in the balance of motor activity is proposed to make a major contribution to the timing of the onset of anaphase B. Further experimentation will be required to confirm and expand upon this model. For example, it is currently unknown whether Kar3p acts directly to depolymerize microtubules in vivo and whether it acts on all microtubules as shown, or on a more limited set.
Figure 9.
Model for Kar3p function in microtubule arrays. The Kar3p motor (shown as shaded circles) is proposed to function predominantly at spindle poles to stimulate microtubule turnover. At START of the cell cycle, the activity of Kar3p is required to limit microtubule number and length at the single spindle pole body (spb). During spindle assembly, the now duplicated spindle poles become linked by microtubule crosslinking and chromosome attachment (latter not shown). Depolymerization of the microtubules linking the spindle poles would now have the effect of pulling the poles together. Cin8p and Kip1p (dark circles) apparently function to counter this inwardly directed force, most likely by crosslinking and sliding the nuclear microtubules, using the microtubules to push out on the spindle poles (Hoyt et al., 1992) though other mechanisms are also possible. The bipolar spindle now has two types of forces acting in opposition on the spindle poles, represented by open arrows. Once anaphase begins, the antagonistic relationship between Cin8p and Kar3p changes. Kar3p-driven spindle collapse no longer occurs in cin8 kip1 mutants (Saunders et al., 1992); Kar3p is no longer found at the spindle poles, and there is a net polymerization of the spindle microtubules (and to a lesser extent the cytoplasmic microtubules) resulting in a great increase in spindle length. Thus the outwardly directed force of Cin8p and Kip1p are retained, but the inward pull of Kar3p is lost. It is proposed that this transition is a major contributor to the onset of anaphase B and can occur in the absence of chromosomes (Zhang and Nicklas, 1996). Spindle pole bodies are shown as discs embedded in a nuclear envelope, small open circles represent microtubule depolymerization, and dark arrows the presumed direction of Kar3p movement.
Potential Roles for Kar3p during Mitosis
It is not immediately obvious why the spindle would need an inwardly directed motor acting on the spindle poles as proposed for Kar3p. One possibility is that Kar3p may act as an inhibitor to delay spindle elongation until anaphase has begun. Recently it has been demonstrated that spindles in grasshopper spermatocytes are capable of initiating spindle elongation at the appropriate time in anaphase without chromosomes, suggesting a timing mechanism that is intrinsic to the spindle itself (Zhang and Nicklas, 1996). We have observed that overexpression of KAR3 can cause spindle compression and block spindle elongation in cin8 kip1 (but not wild-type) strains (Saunders and Hoyt, 1997). If the anaphase B motors act to separate the poles by sliding polar microtubules in opposite directions, as demonstrated for isolated Schizosaccharomyces pombe spindles (Masuda et al., 1988), then a protein that acts to depolymerize these same microtubules, as suggested here for Kar3p, would be expected to have such an inhibitory activity.
In a related model, Kar3p-like motors may play a role in the accelerated microtubule treadmilling observed in spindles assembled in Xenopus oocyte extracts. Microtubule treadmilling is the movement of tubulin subunits within the lattice towards the minus ends, due to the unequal rates of tubulin addition and subtraction at the two ends. Treadmilling or flux in the spindle is inhibited by AMP– PNP in assembled Xenopus spindles (Sawin and Mitchison, 1991) and is two orders of magnitude faster than treadmilling of isolated microtubules (Margolis and Wilson, 1978; Sawin and Mitchison, 1991), suggesting an ATP-dependent enhancement. Microtubule treadmilling plays an important role in chromosome segregation, contributing to roughly one third of the anaphase A type movement. Flux in Xenopus spindles in vitro occurs at a rate of 2.5–3 μm/min (Mitchison, 1989), rapid enough to cause the observed spindle collapse in 3–5 min in S. cerevisiae (Saunders and Hoyt, 1992). Inhibition of microtubule depolymerization by the tub2-150 allele or as suggested, loss of Kar3p, may reduce the rate of tubulin flux and therefore the spindle collapse after inactivation of Cin8p and Kip1p. Consistent with this type of observation, we show here that the spindle collapse is mostly delayed rather than completely blocked by the tub2-150 allele or loss of KAR3 but does absolutely require ATP. Loss of a component of the chromosome segregation machinery would explain the high rate of aneuploidy and meiotic inviability commonly observed in kar3-Δ strains. However, microtubule flux has not yet been demonstrated in yeast, and it is not known if this process is conserved in smaller eukaryotes.
Previously it has been suggested that Kar3p produces the minus end–directed motor activity purified from S. cerevisiae extracts by association with centromere DNA (Middleton and Carbon, 1994), suggesting a kinetochore role for this motor. While it may seem that the predominant poleward location of Kar3p from the present manuscript may argue against a kinetochore role, we note that known centromere binding proteins sometimes appear clustered around the pole in S. cerevisiae (Goh and Kilmartin, 1993). However, our observation of increased cytoplasmic microtubules in kar3-Δ strains is not readily explained by a strict kinetochore role for this motor.
The above models assume a role for Kar3p in influencing nuclear microtubules. However, as discussed above, the predominant change observed here in kar3 mutants was in the cytoplasmic microtubules. An alternative model is that the cell uses Kar3p activity to ensure a rapid turnover of the cytoplasmic microtubules. Enhanced cytoplasmic microtubule dynamics may facilitate the association with subcellular structures, for example, the cell cortex, to ensure proper spindle positioning or elongation. The increased cytoplasmic microtubules of kar3 mutants may reflect reduced dynamics and hyperstable microtubules. We have not noticed a major defect in spindle or nuclear alignment in kar3 mutants; however, we note unpublished observations (Satterwhite, L., and M. Rose) that KAR3 becomes essential (or nearly so) in the absence of cytoplasmic dynein. The yeast cytoplasmic dynein gene DHC1/DYN1 is required for normal nuclear and spindle positioning, and this synthetic relationship may suggest a redundant role between these motors in spindle positioning.
Acknowledgments
This work was supported by American Cancer Society grant CB-171 awarded to W. Saunders.
Footnotes
The authors gratefully acknowledge John Kilmartin for the anti–spindle pole antibodies and communicating unpublished results, David Botstein for tubulin alleles and plasmids, and Pamela Meluh, Lisa Satterwhite, and Mark Rose for KAR3 alleles and plasmids.
Please address all correspondence to Bill Saunders, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA 15260; Tel.: (412) 624-4320; Fax: (412) 624-4759; E-mail:wsaund@vms.cis.pitt.edu
References
- Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. Real-time visualization of cell cycle dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–589. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- Buendia B, Draetta G, Karsenti E. Regulation of the microtubule nucleating activity of centrosomes in Xenopusegg extracts: role of cyclin A–associated protein kinase. J Cell Biol. 1992;116:1431–1442. doi: 10.1083/jcb.116.6.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidse LC. Benzimadazole fungicides: mechanism of action and biological impact. Annu Rev Phytopathol. 1986;24:43–65. [Google Scholar]
- De Brabander M. Microtubule dynamics during the cell cycle: the effects of taxol and nocodazole on the microtubule system of PtK2 cells at different stages of the mitotic cycle. Int Rev Cytol. 1986;101:215–274. doi: 10.1016/s0074-7696(08)60250-8. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison T. A new role for motor proteins as couplers to depolymerizing microtubules. J Cell Biol. 1995;128:1–4. doi: 10.1083/jcb.128.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S, Kang S, Satterwhite L, Rose M, Skeen V, Salmon E. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO (Eur Mol Biol Organ) J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. . J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh Y, Nishida E, Matsuda S. In vitro effects on microtubule dynamics of purified XenopusM phase–activated MAP kinase. Nature (Lond) 1991;349:251–254. doi: 10.1038/349251a0. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Hoyt MA, Totis L, Roberts BT. S. cerevisiaegenes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiaekinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S. Cell division and the mitotic spindle. J Cell Biol. 1981;91:131s–147s. doi: 10.1083/jcb.91.3.131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization–dependent motion of chromosomes in vitro. J Cell Biol. 1995a;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature (Lond) 1995b;373:161–165. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- Machin NA, Lee JM, Barnes G. Microtubule stability in budding yeast: characterization and dosage suppression of a benomyl-dependent tubulin mutant. Mol Biol Cell. 1995;6:1241–1259. doi: 10.1091/mbc.6.9.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis RL, Wilson L. Opposite end assembly and disassembly of microtubules at steady state in vitro. Cell. 1978;13:1–8. doi: 10.1016/0092-8674(78)90132-0. [DOI] [PubMed] [Google Scholar]
- Masuda H, McDonald K, Cande WZ. The mechanism of anaphase spindle elongation: uncoupling of tubulin incorporation and microtubule sliding during in vitro spindle elongation. J Cell Biol. 1988;107:623–633. doi: 10.1083/jcb.107.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Middleton K, Carbon J. Kar3-encoded kinesin is a minus-enddirected motor that functions with centromere binding proteins (CBF3) on an in vitro yeast kinetochore. Proc Natl Acad Sci USA. 1994;91:7212–7216. doi: 10.1073/pnas.91.15.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J Cell Biol. 1989;109:637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff NF, Thomas JH, Grisafi P, Botstein D. Isolation of the β-tubulin gene from yeast and demonstration of its essential function in vivo. Cell. 1983;33:211–219. doi: 10.1016/0092-8674(83)90350-1. [DOI] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of the Kar3 kinesin heavy chain-like protein requires the CIK1 interacting protein. J Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JB, Ris H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. . J Cell Sci. 1976;22:219–242. doi: 10.1242/jcs.22.2.219. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., and L.H. Hartwell. 1981. The Saccharomyces cerevisiae Cell Cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory. pp.97–142.
- Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Roof, D.M., P.B. Meluh, and M.D. Rose. 1991. Multiple kinesin-related proteins in yeast mitosis. Cold Spring Harbor Symp. Quant. Biol. 56:693–703. [DOI] [PubMed]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ED, McKeel M, Hays T. Rapid rate of tubulin dissociation from microtubules in the mitotic spindle in vivo measured by blocking polymerization with colchicine. J Cell Biol. 1984;99:1066–1075. doi: 10.1083/jcb.99.3.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Saunders, W.S., and M.A. Hoyt. 1997. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Cell. Biol. In press. [DOI] [PMC free article] [PubMed]
- Sawin KE, Mitchison TJ. Poleward microtubule flux in mitotic spindles assembled in vitro. J Cell Biol. 1991;112:941–954. doi: 10.1083/jcb.112.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz PJ, Solomon F, Botstein D. Genetically essential and nonessential α-tubulin genes specify functionally interchangeable proteins. Mol Cell Biol. 1986;6:3722–3733. doi: 10.1128/mcb.6.11.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1983. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Vale RD, Goldstein LSB. One motor, many tails: an expanding repertoire of force-generating enzymes. Cell. 1990;60:883–885. doi: 10.1016/0092-8674(90)90334-b. [DOI] [PubMed] [Google Scholar]
- Walczak C, Mitchison T, Desai A. XKCM1: a Xenopuskinesinrelated protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Wells WA, Murray AW. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Nicklas RB. “Anaphase” and cytokinesis in the absence of chromosomes. Nature (Lond) 1996;382:466–468. doi: 10.1038/382466a0. [DOI] [PubMed] [Google Scholar]