Abstract
Biochemical and genetic evidence suggest that the SWI/SNF complex is involved in the remodeling of chromatin during gene activation. We have used antibodies specific against three human subunits of this complex to study its subnuclear localization, as well as its potential association with active chromatin and the nuclear skeleton. Immunofluorescence studies revealed a punctate nuclear labeling pattern that was excluded from the nucleoli and from regions of condensed chromatin. Dual labeling failed to reveal significant colocalization of BRG1 or hBRM proteins with RNA polymerase II or with nuclear speckles involved in splicing. Chromatin fractionation experiments showed that both soluble and insoluble active chromatin are enriched in the hSWI/SNF proteins as compared with bulk chromatin. hSWI/SNF proteins were also found to be associated with the nuclear matrix or nuclear scaffold, suggesting that a fraction of the hSWI/SNF complex could be involved in the chromatin organization properties associated with matrix attachment regions.
It is becoming clear that structural aspects of chromatin and the architecture of the nucleus are important factors in the control of gene expression (Felsenfeld, 1992; Paranjape et al., 1994; Stroboulis and Wolffe, 1996). DNA in the nucleus is organized into a hierarchy of structures where the basic building block is the nucleosome. The packaging of DNA in nucleosomes and higher order structures represents an obstacle to the binding of specific proteins (Laybourn and Kadonaga, 1991) and RNA polymerases (Williamson and Felsenfeld, 1978) to control elements and transcription initiation sites. The same is true for transcriptional elongation. A convergence of biochemical and genetic studies has identified several ATP-dependent multiprotein complexes that may help transcriptional activators to overcome chromatin-mediated repression (Kingston et al., 1996). The SWI/SNF complex is one of these complexes. The SWI (for mating type switching) and SNF (for sucrose nonfermenting) genes were first identified in yeast (SWI/SNF gene complex) by mutations interfering with the activation of several inductible promoters (for review see Carlson and Laurent, 1994; Peterson and Tamkun, 1995; Peterson, 1996). Alterations in each of the four histones and in some presumed chromatin structural proteins all result in partial suppression of the impaired transcriptional phenotype of swi or snf mutations (Winston and Carlson, 1992). Genetic and biochemical analysis demonstrated that SWI and SNF proteins function together as one multisubunit complex (Peterson and Hershkowitz, 1992; Cairns et al., 1994; Peterson et al., 1994) able to facilitate the binding of activator proteins to nucleosomal DNA in an ATP-dependent way (Côté et al., 1994). Within SWI/SNF proteins, SNF2 is of special interest. It contains sequence motifs closely related to those found in DNA-dependent ATPases and helicases (Laurent et al., 1993), and, indeed, bacterially expressed SNF2 protein shows DNA-dependent ATPase activity. These data suggest that SNF2 is an essential component of the ATPase-dependent remodeling activity of the complex.
Several groups have demonstrated that a complex homologous to the yeast SWI/SNF complex exists in higher eukaryotes (Kwon et al., 1994; Wang et al., 1996a ,b). One of the first relatives of SWI2/SNF2 to be discovered was brahma (brm), an activator of Drosophila homeotic genes (Tamkun et al., 1992). brm was identified in a screen for suppressors of mutations in polycomb, a repressor of homeotic genes that is thought to act by altering chromatin structure. Humans have at least two genes closely related to SWI2/SNF2: hBRM (human brahma or hSNF2α) and BRG1 (brahma-related gene 1 or hSNF2β) (Muchardt and Yaniv, 1993; Khavari et al., 1993; Chiba et al., 1994). Both hBRM and BRG1 have been shown to enhance transcriptional activation by the glucocorticoid receptor (Muchardt and Yaniv, 1993; Chiba et al., 1994) through a mechanism that may also involve the retinoblastoma (Rb)1 protein (Dunaief et al., 1994; Singh et al., 1995). Human homologues of SNF5 and SWI3 have also been cloned (Kalpana et al., 1994; Muchardt et al., 1995; Wang et al., 1996b ). In addition, human SWI/SNF complexes containing hSNF5, either BRG1 or hBRM proteins, and seven to ten additional BRG1-associated factors have been purified from different cell lines (Wang et al., 1996a ). Like the yeast complex, the human SWI/SNF complexes show, in vitro, nucleosome remodeling activity and facilitate the binding of activators to nucleosomal DNA (Imbalzano et al., 1994; Kwon et al., 1994; Wang et al., 1996a ).
The high degree of similarity between BRG1 and hBRM raises the possibility that they may be functionally redundant. On the other hand, we have observed that BRG1 and hBRM are phosphorylated during mitosis, and, while hBRM is partially degraded, the levels of BRG1 remain constant throughout the cell cycle (Muchardt et al., 1996). These results suggest that the two proteins may play distinct roles during the cell cycle.
Recently, it has been proposed that the yeast SWI/SNF complex copurifies with the holoenzyme fraction of RNA polymerase II (RNAP II), suggesting that interaction with RNAP II may be the mechanism by which SWI/SNF complex is recruited to its genomic targets (Wilson et al., 1996). However, the SWI/SNF complex can also be isolated from yeast without an associated holoenzyme (Cairns et al., 1994; Côté et al., 1994). At present, it is unclear whether the human SWI/SNF proteins are also associated with the RNAP II holoenzyme. In addition, it was also demonstrated that the yeast SWI/SNF complex contains a DNA binding activity that may target it directly to specific DNA structures (Quinn et al., 1996). These examples demonstrate that the mechanism involved in the recruitment of the SWI/SNF complex to its targets in the chromatin, in vivo, remains unclear.
Nuclear fractionation studies have shown that transcriptionally active DNA is tightly associated with the nuclear skeleton or nuclear matrix, while inactive loci are not (Stratling, 1987; Jackson and Cook, 1985; Jackson et al., 1993). The nuclear matrix is operationally defined as the nuclear structure that remains after the salt extraction of nucleasetreated nuclei. It consists of a peripheral lamina–pore complex and an internal filamentous ribonucleoprotein network that has not been well characterized (for review see Berezney et al., 1995). This component of the nuclear architecture provides the internal scaffold of the nucleus. The matrix- or scaffold-associated DNA regions (MARs or SARs) may function as boundary elements for transcriptional domains and confer position-independent expression of associated genes in transgenic mice (Laemmli et al., 1992; Jenuwein et al., 1993). Biochemical studies have also demonstrated that transcriptionally active chromatin exhibit increased sensitivity to nucleases. These observations explain why a fraction of actively transcribed chromatin is easily solubilized after nuclease treatment of isolated nuclei while the rest is tightly bound to the nuclear matrix (Kamakaka and Thomas, 1990; Davie, 1995).
In the present study we have used specific antibodies that recognize BRG1, hBRM, and hSNF5 to study the subnuclear localization of the human SWI/SNF complex. Immunofluorescent staining revealed a punctate distribution of BRG1 and hBRM in interphase HeLa nuclei, the lowest concentration appearing in regions of high DNA density. However, we failed to show strong overlap between hBRM or BRG1 and RNA polymerase II staining. Chromatin fractionation demonstrated that three components of the hSWI/SNF complexes (BRG1, hBRM, and hSNF5) were enriched in highly transcribed chromatin and associated with the nuclear matrix. Association with chromatin or nuclear structures was abolished during mitosis. Finally, we show that hSWI/SNF complexes can be released from the nucleus by incubation with ATP and that this process is inhibited by protein kinase inhibitors.
Materials and Methods
Cell Culture and Cell Cycle Synchronization
HeLa cells were grown at 37°C in 7% CO2 in DME (GIBCO BRL, Gaithersburg, MD) supplemented with 7% FCS. For cell cycle synchronization, exponentially growing cells were treated with 5 mM thymidine for 40 h. Cells were then released from the block, and samples were taken at different times for flow cytometry analysis and for extract preparation. 12 h after the thymidine block release, 0.1 μg/ml nocodazole was added to the medium to block the cells in mitosis.
Isolation of Cell Nuclei and Total Extract
The procedure was adapted from Cereghini et al. (1988). 107 cells were resuspended in 200 μl of HNB (0.5 M sucrose, 15 mM Tris-HCl, pH 7.5, 60 mM KCl, 0.25 mM EDTA, pH 8, 0.125 mM EGTA, pH 8, 0.5 mM spermidine, 0.15 mM spermine, 1 mM DTT, 0.5 mM PMSF, 5 μg/ml pepstatin, 5 μg/ml leupeptin, and 5 μg/ml aprotinin). Then 100 μl of HNB supplemented with 1% NP-40 was added dropwise. After 5 min of incubation at 4°C, nuclei were pelleted by centrifugation at 6,000 rpm for 3 min.
When total extract was required, 107 cells were lysed in 200 μl of urea buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris, pH 8). The protein concentration was determined by Bradford assays (Bradford, 1976).
Nuclear Matrix Isolation
High salt isolation of nuclear matrix was carried out essentially as described (He et al., 1990). After wash in PBS, cells were extracted in cytoskeleton buffer (CSK): 10 mM Pipes, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, supplemented with leupeptin, aprotinin, and pepstatin (1 μg/ml each), 1 mM PMSF, 1 mM DTT, and 0.5% (vol/ vol) Triton X-100. After 3 min at 4°C, the cytoskeletal frameworks were separated from soluble proteins by centrifugation at 5,000 g for 3 min. Chromatin was solubilized by DNA digestion with 1 mg/ml of RNase-free DNAase I in CSK buffer plus proteinases inhibitors for 15 min at 37°C. Then ammonium sulfate was added from a 1 M stock solution in CSK buffer to a final concentration of 0.25 M and, after 5 min at 4°C, samples were pelleted again. The pellet was further extracted with 2 M NaCl in CSK buffer for 5 min at 4°C, and then centrifuged. This treatment removed all the DNA and the histones from the nucleus, as shown by agarose gel electrophoresis and SDS-PAGE, respectively. The remaining pellet was solubilized in urea buffer and was considered the nuclear matrix– containing fraction.
For low salt preparation of nuclear matrix and scaffolds, nuclei were isolated as described by Mirkovitch et al. (1984). Nuclei were then stabilized in isolation buffer (3.75 mM Tris HCl, pH 7.5, 0.05 mM spermine, 0.125 mM spermidine, 0.5 mM EDTA, 5 mM MgCl2, 20 mM KCl) for 20 min at 37°C, and then DNA was digested by incubation under the same conditions for another 15 min in the presence of 1 mg/ml of RNase-free DNAase I. After centrifugation, nuclei were washed with isolation buffer and extracted with the same buffer supplemented with 25 mM 3,5-diiodosalicilic acid, lithium salt (LIS) for 5 min at room temperature. Chromatin-depleted nuclei were recovered by centrifugation and the pellet was solubilized in urea buffer.
Chromatin Fractionation
Isolation of S1, S2, and P fractions from HeLa nuclei chromatin was performed according to the method of Rose and Garrard (1984) with minor modifications. 107 nuclei were isolated as described above and resuspended in 200 μl of nuclear buffer (20 mM Tris-HCl, pH 7.5, 70 mM NaCl, 20 mM KCl, 5 mM MgCl2, and 3 mM CaCl2 supplemented with protease inhibitors). Nuclei suspension was incubated with 3 U of micrococcal nuclease (Sigma Chemical Co., St. Louis, MO) at room temperature during the indicated time. The digestion was terminated by the addition of EDTA and EGTA to 5 mM each, the mixture was then centrifuged at 5,000 g for 3 min, and the supernatant was designated the S1 fraction. The nuclear pellet was lysed in 2 mM EDTA for 15 min at 4°C. This was followed by centrifugation, and the supernatant and the pellet were designated the S2 and P fractions, respectively. For DNA analysis of S1, S2, and P fractions, aliquots were treated with lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM EDTA, 0.5% SDS) for 1 h at 37°C and with phenolCHCl3 extraction. Proteins were analyzed by SDS-PAGE. After 2 min of micrococcal nuclease digestion, S1, S2, and P contained ∼5%, 80%, and 15% of the total DNA, respectively.
Immunoblotting and Antibodies
Immunoblotting was carried out according to standard procedures. Proteins were separated by SDS-PAGE (Laemmli, 1970) and transferred to nitrocellulose. The membrane was then blocked with PBS/0.2% Tween20/10% horse serum and incubated with the various antibodies. Enhanced chemiluminescence reagents (ECL; Amersham, Les Ulis, France) were used for detection. The mAb pol 3/3 recognizes the RNAPII largest subunit at an evolutionary conserved epitope located outside the carboxy-terminal domain (CTD) (Krämer et al., 1980) and was kindly provided by O. Bensaude (Ecole Normal Superieure, Paris, France). The monoclonal CC3 antibody was raised against a nuclear p255 phosphoprotein (Thibodeau et al., 1989) and provided by M. Vincent (Université Laval, Sainte-Foy, Quebec, Canada). Recently, it has been demonstrated that the epitope recognized by this antibody corresponds to a subpopulation of hyperphosphorylated RNAPII largest subunit (Vincent et al., 1997; Dubois et al., 1997). Antibodies against BRG1, hBRM, hSNF5, and JunB have been previously described (Muchardt et al., 1995, 1996; Pfarr et al., 1994). To prepare anti–glucocorticoid receptor antibodies, rabbits were immunized with an Escherichia coli–expressed chimeric protein spanning amino acids 106–318 plus 407–556 from the human glucocorticoid receptor protein.
Indirect Immunofluorescence Analysis
HeLa cells, grown on coverslips, were fixed in 3.5% freshly prepared paraformaldehyde in PBS for 10 min at room temperature, and then permeabilized with 0.05% Triton X-100 in PBS for 30 min at room temperature. Alternatively, cells were permeabilized with ice-cold CSK buffer supplemented with 0.5% Triton X-100 for 3 min, before fixation. For in situ isolation of nuclear matrix, cells were extracted directly on the coverslips with the high salt method described above, and then fixed with 4% paraformaldehyde in PBS. To enhance exposure of hidden epitopes, cells were treated with 6 N guanidine hydrochloride as described previously (Peränen et al., 1993). In all cases, the coverslips were incubated overnight with the appropriate antibodies in PBS/10% horse serum/0.1% Tween-20. Fluorescein- or Texas red–linked anti–rabbit or anti–mouse antibodies from Amersham were used for detection. The cellular DNA was labeled with 0.05% 4′,6-diamidino-2-phenylindole (DAPI) or propidium iodide. The preparations were observed at ×100 magnification with an Axiophot microscope (Zeiss, Oberkochen, Germany) or at ×100 magnification with a confocal microscope using a 488-nm laser and a 535-nm narrow band filter for the FITC signal, and a 568-nm laser and a 590-nm long band filter for Texas red and propidium iodide signal.
ATP-dependent Release of hSWI/SNF from Cell Nuclei
Nuclei from HeLa cells were isolated as described above. 107 nuclei were washed in 1 ml of nuclear buffer, centrifuged, and resuspended in 100 μl of the same buffer. 10-μl aliquots were incubated for 1 h at 30°C in the presence or absence of 3 mM ATP. When indicated, 3 mM of adenosine 5′-(β, γ-methylene) triphosphate (AMP-PCP) or adenosine 5′-O-(3-thiotriphosphate) (ATPγS) was added to the reaction.
For in situ ATP-dependent release of BRG1 and hBRM, HeLa cells grown on coverslips were permeabilized with 0.5% Triton X-100–CSK buffer as previously described and incubated in nuclear buffer (see above) with or without ATP for 1 h at room temperature. Afterward, cells were washed with PBS and fixed with paraformaldehyde.
Results
Subnuclear Immunolocalization of hBRM and BRG1
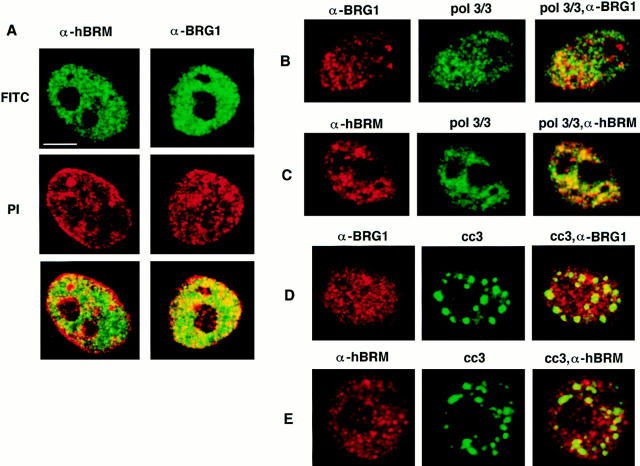
We have previously described the characterization of affinity-purified polyclonal antibodies that specifically recognize BRG1 or hBRM proteins (α-BRG1 and α-hBRM, respectively). In immunofluorescence studies, these antibodies revealed a strictly nuclear signal except in mitotic cells where hBRM and BRG1 signals were found to be excluded from the regions containing the condensed chromosomes (Muchardt et al., 1996). A more detailed analysis of the labeling pattern of HeLa cells, using confocal microscopy, revealed a diffuse nucleoplasmic signal and a variable number of bright dots or foci (Fig. 1 A). Furthermore, hBRM and BRG1 staining was excluded from the nucleoli and from regions of highly condensed heterochromatin, which are stained brightly by propidium iodide. hBRM and, to a lesser extent, BRG1 were also excluded from the periphery of the nucleus. It is known that transcriptionally inactive heterochromatin is adjacent to the inner nuclear membrane (Mathog et al., 1984). These data suggest that hSWI/SNF complex is excluded from heterochromatin. The intranuclear distribution of both proteins was the same in cells that were extracted with Triton X-100 before fixation or in nonextracted cells.
Figure 1.
Subnuclear distribution of hBRM and BRG1 proteins in HeLa cells. HeLa cells grown on coverslips were fixed with 3% paraformaldehyde. (A) After fixation, cells were labeled with α-hBRM (top left) or α-BRG1 (top right) and detected with a fluoresceinconjugated secondary antibody, using confocal microscopy. Cellular DNA was stained with propidium iodide (PI) (middle). Superimposition of antibody staining and PI (bottom). (B and C) Fixed cells were double labeled with pol 3/3 mAbs against the RNAP II-LS (visualized with fluorescein; middle) and α-BRG1 (B) or α-hBRM (C) (visualized with Texas red; left). Confocal images from each fluorochrome were recorded and superimposed (green staining for RNAP II-LS protein and red staining for hBRM or BRG1; right). Regions of colocalization (yellow). (D and E) Fixed cells were double labeled with cc3 mAbs against CTD-hyperphosphorylated RNAP II (visualized with fluorescein; middle) and α-BRG1 (D) or α-hBRM (E) (visualized with Texas red; left). Confocal images from each fluorochrome were recorded and superimposed (green staining for CTD-hyperphosphorylated RNAP II protein and red staining for hBRM or BRG1; right). Regions of colocalization (yellow). Intensities were adjusted for maximum contrast. Bar, 5 μm.
It has been reported that the yeast SWI/SNF proteins are integral components of the RNAP II holoenzyme, being part of the SRB (suppressor of RNA polymerase B) complex. This complex is tightly associated with the RNAP II large subunit CTD (Wilson et al., 1996). To test if human SWI/SNF complexes are associated with RNAP II in situ, we looked for potential colocalization between hBRM or BRG1 and RNAP II. Two mAbs against the largest subunit of RNAP II (RNAP II-LS) were used: the pol 3/3 antibody recognizes nonphosphorylated and phosphorylated forms of RNAP II-LS, and the cc-3 antibody recognizes a subpopulation of hyperphosphorylated RNAP II-LS (Vincent et al., 1997; Dubois et al., 1997) located in 20–30 discrete nuclear foci (Bisotto et al., 1995). These foci are closely linked to speckle domains (irregularly shaped intranuclear regions that contain splicing factors) and colocalize with the splicing factor SC-35 (Bisotto et al., 1995; Bregman et al., 1995; Mortillaro et al., 1996). Combination of α-hBRM or α-BRG1 labeling and pol 3/3 or cc-3 labeling was performed and analyzed by confocal microscopy (Fig. 1, B–E). Pol 3/3 antibodies revealed a punctate staining pattern throughout the nucleus that was excluded from the nucleoli. Although some colocalization could be observed (Fig. 1, B and C, right panels, yellow), a large fraction of the hBRM or BRG1 dots showed little or no RNAP II staining and vice versa. We analyzed the degree of colocalization quantitatively by plotting the pixel intensity distribution of a dual-channel section (two-dimensional scatter plot diagram) or by plotting the signal intensities of the red and the green signals along a single line drawn through the image of a doubly labeled nucleus (not shown). These plots show that there is no clear correlation between the spatial distribution of hBRM or BRG1 and RNAP II. The cc3 antibody recognized 20–30 brightly stained foci. hBRM- and BRG1-containing dots did not colocalize with these foci either (Fig. 1, D and E). It has also been reported that RNAP II inhibitors like α-amanitin or actinomycin D provoke a redistribution of RNA pol II and an increased concentration of the enzyme in the speckles (Bregman et al., 1995). Treatment of HeLa cells with such inhibitors provoked an enlargement of the speckles (around twofold surface increase), revealed by the cc-3 antibody, but did not produce any reorganization of the signals revealed by the α-BRG1 or α-hBRM antibodies (data not shown). These data suggest that a large fraction of the hSWI/SNF complex is not associated with RNAP II and vice versa.
hSNF/SWI Complex Is Partially Associated with the Nuclear Matrix
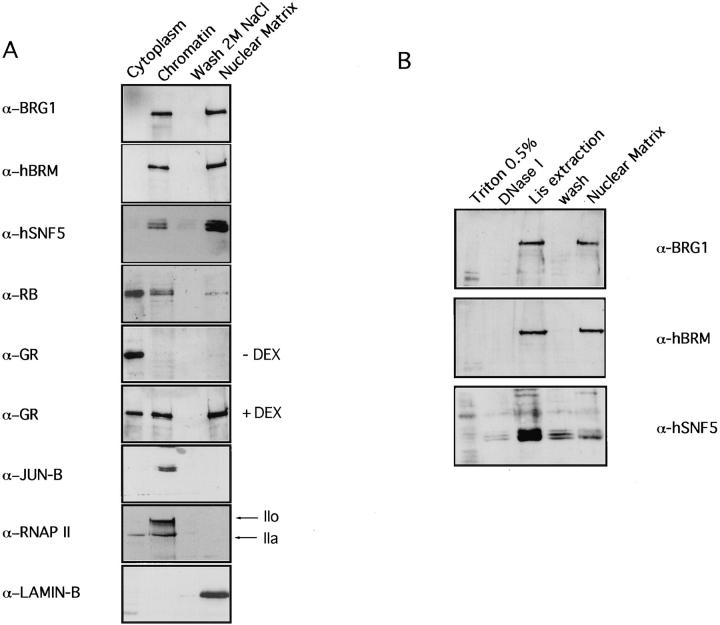
To get further insight on the association of hSWI/SNF components with the nucleus, different procedures were tested for their ability to release the hSWI/SNF complex from HeLa nuclei. More than 60% of the amount of hBRM, BRG1, and hSNF5 proteins was found to be resistant to extraction with detergents and 0.45 M NaCl or 100 mM MgCl2, even after total DNA digestion with DNase I (data not shown). Such treatments are normally used to extract chromatin components including high mobility group proteins, transcription factors, or histones. To investigate the possibility that the hSWI/SNF complex may bind to the nuclear skeleton or nuclear matrix, we isolated the nuclear matrix by the sequential extraction procedure of He et al. (1990). In the first fractionation step, soluble proteins are removed by extraction with Triton X-100. Chromatin proteins are then released by DNase I digestion and extraction with 0.25 M ammonium sulfate. 95% of the histones were extracted in this step (data not shown). After washing with 2 M NaCl, the last fraction is composed of structural nuclear matrix proteins and nuclear matrix–associated proteins. Supernatants from each extraction step and the final nuclear matrix pellet were analyzed by SDSPAGE and immunoblotting. As shown in Fig. 2 A, ∼50% of the BRG1, hBRM, and hSNF5 proteins were released with the chromatin proteins, while the remainder was tightly associated with the nuclear matrix proteins. This was compared with the distribution of other nuclear proteins. It has been shown previously that a fraction of hypophosphorylated Rb protein is associated with the nuclear matrix (Mancini et al., 1994). In our extracts, hyperphosphorylated Rb appeared in the first fraction together with soluble proteins. Most hypophosphorylated Rb was released in the Triton X-100 extraction or in the chromatin fraction, while ∼20% was found in the insoluble nuclear matrix pellet (Fig. 2 A). It has also been reported that glucocorticoid receptor and other nuclear receptors are associated with the nuclear matrix (van Steensel et al., 1995b). Indeed, our nuclear matrix preparations contained ∼50% of the nuclear glucocorticoid receptor (GR) from dexamethasone-treated HeLa cells. However, in nontreated cells, all the GR was found in the cytoplasmic fraction (Fig. 2 A). In contrast, the transcription factor JunB and the large subunit of the RNAP II (revealed by the pol 3/3 antibody) were fully extracted in the chromatin fraction, indicating that nuclear matrix preparations were not contaminated with all transcription factors. Finally, we also used an antibody against lamin B, one of the major components of the nuclear matrix, as a marker to verify that cytoplasmic and chromatin fractions were not contaminated with nuclear matrix proteins. As expected, lamin B was solely found in the nuclear matrix fraction (Fig. 2 A).
Figure 2.
A fraction of the hSWI/SNF complex is associated with nuclear matrix. (A) HeLa nuclear matrix was prepared by the high salt method as described in Materials and Methods. Cells were sequentially extracted with 0.5% Triton X-100 (Cytoplasm), DNase I and 0.25 M (NH4)2SO4 (Chromatin), and 2 M NaCl (Wash 2M NaCl), and the remaining pellet was solubilized in 8 M urea (Nuclear Matrix). An equivalent aliquot of each step of the extraction protocol was subjected to SDSPAGE and immunoblotted with the indicated antibodies. RNAP II-LS was detected with pol 3/3 antibodies. When indicated, HeLa cells were treated with 10−6 M of dexamethasone (DEX). (B) HeLa nuclear matrix was prepared by the LIS method as described in Materials and Methods. Cells were sequentially extracted with 0.5% Triton X-100, DNase I, 25 mM LIS in low ionic strength buffer, and low-ionic strength buffer, and finally the pellet was resuspended in urea buffer (Nuclear Matrix). An equivalent aliquot of each step of the extraction protocol was subjected to SDSPAGE and immunoblotted with the indicated antibodies.
The possibility has been raised that the high salt extraction procedure used for nuclear matrix preparations induces the precipitation of some nuclear proteins (Kirov and Tsanev, 1986). To verify the observations described above with a method that avoids high salt conditions, we used the procedure of Mirkovich et al. (1984). This protocol entails stabilization of nuclei by a brief incubation at elevated temperature, followed by extraction of histones and other chromatin components with detergent-like LIS and digestion of the genomic DNA. The resulting structures have been termed nuclear scaffolds (Mirkovich et al., 1984). When nuclear scaffolds were isolated by this method, ∼50% of hBRM and BRG1 proteins were extracted in the LIS-soluble fraction, together with chromatin proteins, whereas the other 50% appeared in the nuclear scaffold preparation (Fig. 2 B). However, LIS solubilized a large fraction of the hSNF5 protein, suggesting that the LIS treatment may disrupt the hSWI/SNF complex or the association of hSNF5 with other members of the complex (Fig. 2 B). Taken together, these data demonstrate that the hSWI/ SNF complex is distributed among different compartments of the nucleus. The existence of several hSWI/SNF complexes with distinct subunit composition (Wang et al., 1996a ) opens the possibility that nuclear matrix–associated and nonassociated complexes may have a partially different subunit composition.
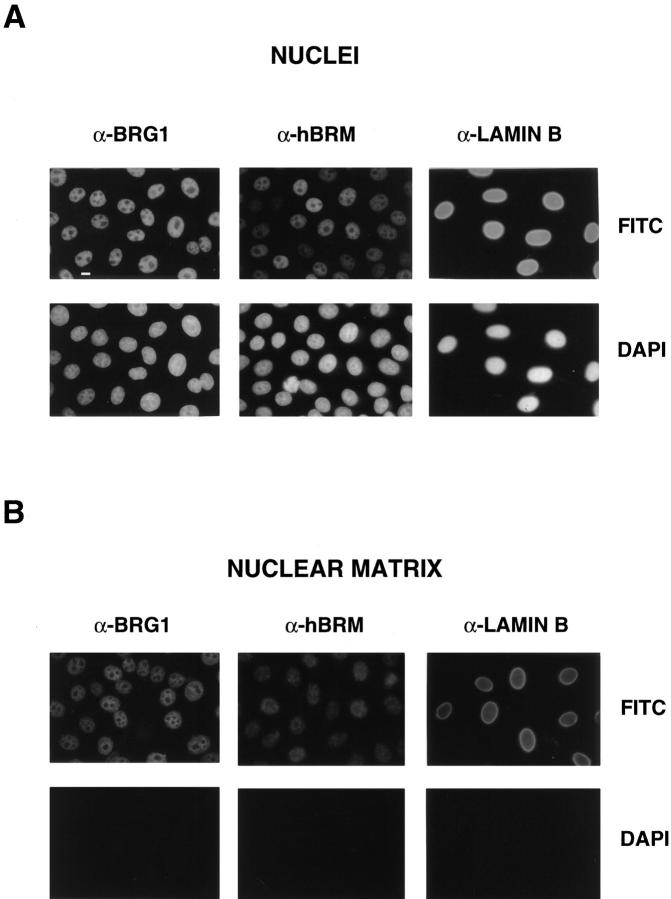
To investigate whether the distribution of hBRM and BRG1 proteins is similar in whole nuclei and in nuclear matrices, we used α-hBRM and α-BRG1 antibodies in indirect immunofluorescence analysis of the nuclear material remaining after in situ sequential DNase I and high salt extraction. As shown in Fig. 3, the labeling pattern was similar for both proteins before and after extraction, but the intensity of the signal decreased in extracted nuclei. As expected, the DAPI signal disappeared completely in the nuclear matrix preparations (Fig. 3 B). α-lamin B antibodies gave a strong signal in the nuclear lamina and a much weaker, diffuse signal in the nucleoplasm. The intensity of the signal was similar in whole nuclei and in extracted nuclei; however, lamina of extracted nuclei appeared slightly deformed or broken, probably as a result of the osmotic shock occurring during the extraction procedure (Fig. 3 B).
Figure 3.
In situ extraction of nuclear matrix. (A) Immunofluorescent staining of nonextracted HeLa cells with α-BRG1, α-hBRM, and α-lamin B antibodies. (B) Immunofluorescent staining pattern of in situ prepared nuclear matrices with α-BRG1, α-hBRM, and α-lamin B antibodies. Microscopy and photography parameters were standardized for all of the images. Bar, 10 μm.
hSWI/SNF Proteins Are Enriched in Active Chromatin
In the previously described nuclear matrix isolation protocol, >95% of chromatin was solubilized in a single fraction. To try to partition between active and inactive chromatin, we have used an established fractionation procedure in which nuclei are subjected to mild digestion with micrococcal nuclease and subsequent extraction with EDTA (Huang and Garrard, 1989; Rose and Garrard, 1984). After nuclease treatment of HeLa cell nuclei for 1, 2, or 4 min and centrifugation, the supernatant fraction S1 contained between 1% and 8% of the total cellular DNA. The pellet was then resuspended in 2 mM EDTA, recentrifuged, and separated into a second supernatant fraction S2 and a pellet P, which contained ∼80% and 15% of the total DNA, respectively. As shown by DNA analysis in agarose gel, the S1 and S2 fractions represent differences in the nuclease accessibility of chromatin (Fig. 4 A). Fraction S1 is mainly composed of mononucleosomal-sized DNA fragments, whereas the S2 fraction contained a typical nucleosomal ladder of DNA fragments. The P fraction contained heterogeneous-sized DNA that remained bound to the nuclear scaffold after the fractionation. This fraction has also been called insoluble chromatin (Xu et al., 1986). It has been shown that S1 and P fractions are enriched in transcriptionally active DNA, whereas fraction S2 is depleted of transcribed sequences (Rose and Garrard, 1984). Aliquots of S1, S2, and P fractions were subjected to SDSPAGE followed by electrotransfer to nitrocellulose membranes and by immunoblot analysis. As shown in Fig. 4 B, BRG1, hBRM, and hSNF5 proteins were enriched in fractions S1 and P with respect to fraction S2. This enrichment appears to be higher for hBRM and hSNF5 than for BRG1 (around two- to threefold for BRG1, 10–15-fold for hBRM, and 20–30-fold for SNF5). Since fraction S2 contained the major part of the DNA, abundance of the hSWI/SNF proteins including BRG1, relative to DNA, was very low in this fraction. These data suggest that the hSWI/SNF complex is mostly concentrated in regions highly sensitive to micrococcal nuclease and in the fraction of the transcriptionally active chromatin that remains bound to the nuclear pellet after digestion. On the contrary, RNAP II was only found in fraction P, consistent with reports that demonstrate that transcriptional activity is associated with an insoluble phase of the nucleus (Jackson and Cook, 1985). Although at first view contradictory, this result is compatible with the fact that RNAP II was found in the soluble fraction in the experiments described in Fig. 2. RNAP II is extractable from the nucleus with the high salt concentration used in the nuclear matrix isolation protocol, but not with the low ionic strength buffers used in the chromatin fractionation protocol.
Figure 4.
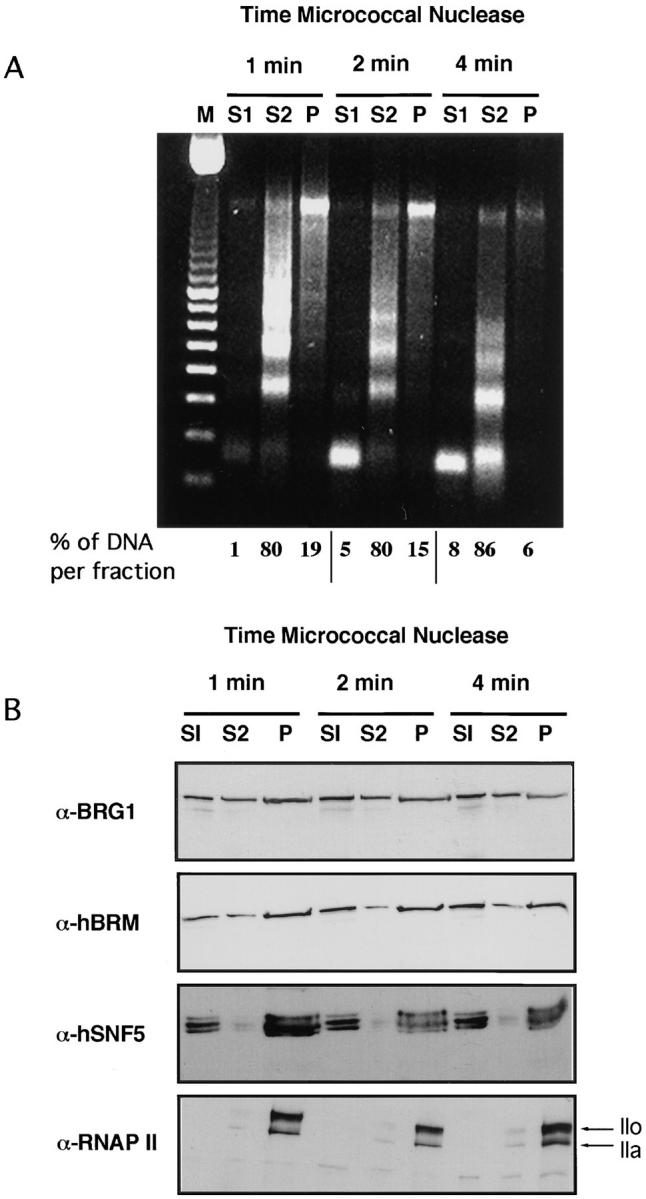
Active chromatin is enriched in hSWI/SNF components. 107 nuclei were subjected to partial digestion by micrococcal nuclease for the indicated times and separated into supernatant (S1) and pellet. The pellet was extracted with EDTA and centrifuged to yield supernatant (S2) and pellet (P) chromatin fractions. (A) DNA from each fraction was electrophoresed in 1% agarose gels and stained with ethidium bromide. M, 100-bp standards. (B) An equivalent volume of each fraction was subjected to SDS-PAGE and immunoblotted with α-BRG1, α-hBRM, α-SNF5, and pol 3/3 (anti-RNAP II-LS) antibodies.
Association of BRG1 and hBRM with the Nuclei during the Cell Cycle
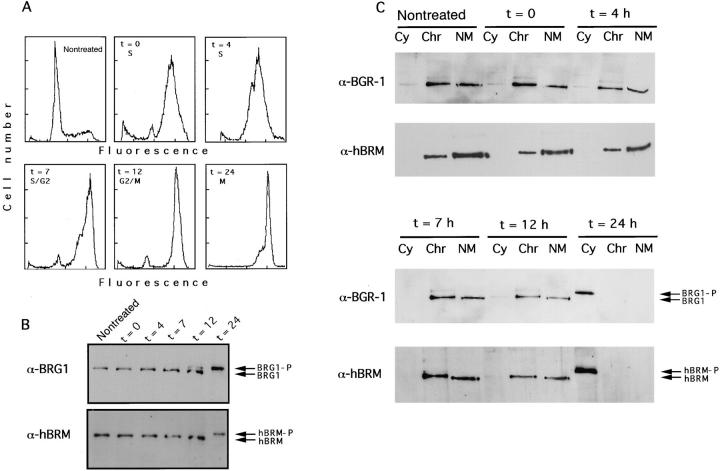
We have recently demonstrated that hBRM and BRG1 are phosphorylated in mitotic cells. The phosphorylated forms of the proteins are localized in the entire cell volume with exclusion from the condensed chromosomes (Muchardt et al., 1996). To determine more accurately the moment at which phosphorylation occurs and to pinpoint whether a correlation exists between the nature of the nuclear association of the hSWI/SNF complex and the position in the cell cycle, we have fractionated nuclei of HeLa cells synchronized at various stages of the cycle. Exponentially growing HeLa cells were blocked with thymidine (S-phase arrest) for 40 h, and then released from the block. Flow cytometry analysis revealed that after 12 h ∼95% of the cells contained a 4 N chromosomal content (Fig. 5 A). However, only ∼15% of the cells had entered mitosis, as determined by phase-contrast microscopy. At that time, nocodazole was added to the medium to block the cells in mitosis. Samples were taken at different times and total extracts were prepared. Alternatively, cells were subjected to the DNase I–high salt fractionation protocol described above. As shown in Fig. 5 B, both BRG1 and hBRM were phosphorylated at the time when cells entered mitosis but not in S and G2 phases. Both BRG1 and hBRM remained associated with the nuclear matrix during G1, S, and G2 phases (Fig. 5 C). However, all of the signal appeared in the cytoplasmic fraction in mitotic cells, consistent with the breakdown of the nuclear structure at mitosis and the exclusion of hBRM and BRG1 from the condensed chromatin as previously published (Muchardt et al., 1996).
Figure 5.
Association of BRG1 and hBRM with the nuclear matrix in synchronized HeLa cells. (A) HeLa Cells were synchronized by treatment with thymidine for 40 h. After release of the block (t = 0), samples were taken at the indicated time for flow cytometry analysis. 12 h later nocodazole was added at 0.1 μg/ml final concentration. (B) Synchronized cells were taken at the indicated time and total extracts were prepared. Equal quantities of protein were subjected to SDS-PAGE in 5% acrylamide gels and immunoblotted with α-BRG1 and α-hBRM antibodies. Phosphorylated and unphosphorylated BRG1 and hBRM are indicated. (C) Synchronized cells were taken at the indicated time and fractionated as described in Fig. 2. Cytoplasmic (Cy), chromatin (Chr), and nuclear matrix (NM) fractions were electrophoresed in 5% acrylamide gels and immunoblotted with α-BRG1 and α-hBRM antibodies.
ATP-dependent Release of BRG1 and hBRM from the Nucleus
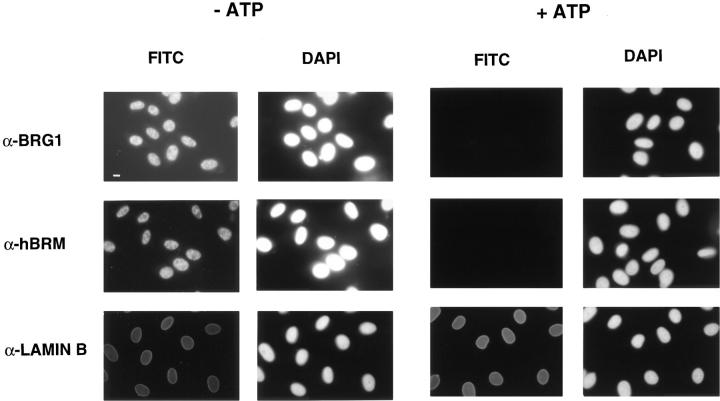
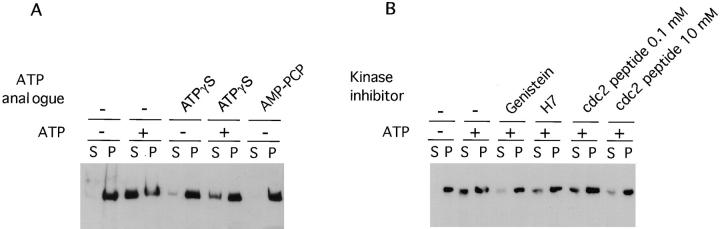
We further investigated the mechanisms that influence the binding of SWI/SNF complex to its targets in chromatin and in the nuclear matrix (Muchardt et al., 1996). BRG1 and hBRM are phosphorylated very late in G2 phase or at the beginning of mitosis, coinciding with their exclusion from the condensed chromatin. Therefore, we tested if incubation of isolated HeLa nuclei with mitotic cell extracts in the presence of ATP would release BRG1 and hBRM from the nuclei. As a control, nuclei were also incubated with ATP in the absence of mitotic cell extract or without ATP. Surprisingly, release of BRG1 and hBRM from the nucleus depended only on the presence of ATP but not on the presence of mitotic extract (data not shown). We therefore investigated the ATP-dependent release of hBRM and BRG1 from the nucleus using immunofluorescence. HeLa cells grown on coverslips were permeabilized in isotonic buffer supplemented with 0.5% Triton X-100, washed, and then incubated for 1 h in the presence or absence of 3 mM ATP before fixation with paraformaldehyde. Immunofluorescent staining of these cells, using α-BRG1, α-hBRM, or α-lamin B antibodies, revealed that BRG1 and hBRM, but not lamin B, were released from the nuclei of the ATP-treated cells (Fig. 6). The release of hBRM after incubation of nuclei with ATP could also be followed by immunoblotting (Fig. 7), demonstrating that hBRM was not degraded. Incubation of the nuclei with nonhydrolyzable ATP analogues like AMP-PCP or ATPγS had essentially no effect on the release of hBRM from the nucleus (Fig. 7 A). In addition, the presence of ATPγS together with ATP decreased the quantity of released hBRM (Fig. 7 A).
Figure 6.
In situ, ATP-dependent release of BRG1 and hBRM from the nuclei. HeLa cells grown on coverslips were permeabilized with CSK 0.5% Triton X-100 buffer and incubated in nuclear buffer with or without 3 mM ATP for 1 h at room temperature. Thereafter, cells were washed with PBS, fixed with 3% paraformaldehyde, and incubated with the indicated antibodies. Bar, 10 μm.
Figure 7.
ATP-dependent release of hBRM from the nuclei requires a hydrolyzable form of ATP. (A) HeLa nuclei were incubated with 3 mM ATP or the same concentration of AMP-PCP or ATPγS. After a 1-h incubation at 30°C, nuclei were pelleted, and proteins from the supernatant (S) or the pellet (P) were electrophoresed and immunoblotted with α-hBRM antibody. (B) HeLa nuclei were incubated as in A with the indicated kinase inhibitor at 0.1 mM final concentration, except in the case of cdc2 peptide that was used at 0.1 and 10 mM.
To verify whether this process involved a protein kinase activity, we have incubated HeLa nuclei with ATP in the presence of several kinase inhibitors: H7, an inhibitor of serine and threonine kinases (Hidaka et al., 1984); genistein, a tyrosine kinase inhibitor (Akiyama and Ogawara, 1991); or a cdc2 peptide that competitively inhibits phosphorylation by cyclin-dependent kinases (Kemp and Pearson, 1991). As shown in Fig. 7 B, genistein and a high concentration of the cdc2 peptide strongly inhibited the release of hBRM from the nuclei (the same results were obtained for BRG1), suggesting that phosphorylation by several distinct protein kinases or by a cascade of activating kinases is involved in the release of BRG1 and hBRM from the nuclear structure. However, ATP-released hBRM protein was not phosphorylated in the same way as in mitotic cells, as deduced by the absence of a change in its electrophoretic mobility (Fig. 7).
Discussion
To investigate the step(s) in gene activation in which the hSWI/SNF complex operates, we have compared the spatial distribution and the characteristics of the association of three hSWI/SNF proteins with other nuclear components. BRG1 and hBRM were not homogeneously distributed in the nucleus of HeLa cells. Indirect immunofluorescent labeling revealed a punctate staining pattern that was excluded from nucleoli and from regions of very condensed chromatin. Little is known about the distribution of transcription factors in the interphase nucleus. The few studies that have been published so far show that transcription factors occur in clusters rather than being diffusely distributed throughout the nucleoplasm. Such intranuclear distribution has been described for Rb (Mancini et al., 1994), p53 (Jackson et al., 1994), GR (van Steensel et al., 1995b), or RNA polymerase II (van Driel et al., 1995). In the case of GR, this punctate distribution has been interpreted as evidence that this factor is present in the nucleus in many hundreds of clusters of 10–100 receptor molecules each (van Steensel et al., 1995a). These clusters are distributed throughout the nucleoplasm and excluded from the nucleoli. Earlier studies have shown that both hBRM and BRG1 proteins enhance transcriptional activation by GR. It is attractive to speculate that those genes that require the interplay of both GR and hSWI/ SNF complex are associated with domains that contain both factors.
Recently, it has been reported that the yeast SWI/SNF complex copurifies with an active fraction of yeast RNAP II (Wilson et al., 1996). However, this complex can be also purified without any associated RNAP II holoenzyme (Cairns et al., 1994; Côté et al., 1994). In addition, RNAP II–free SWI/SNF complex functions in conjunction with nucleosome-bound transcription factors to generate regions of nucleosome disruption (Owen-Hughes et al., 1996). Our double-immunolabeling experiments using α-BRG1 and α-hBRM antibodies and two antibodies that recognize two subpopulations of RNAP II did not reveal strong colocalization between the hSWI/SNF complex and RNAP II (Fig. 1). We cannot rule out the possibility that a fraction of the hSWI/SNF complex interacts with a fraction of RNAP II holoenzyme. However, our experiments indicate that a large fraction of RNAP II is not associated with the majority of the hSWI/SNF complex. This result suggests that the hSWI/SNF complex can be targeted to particular regions by interactions with specific transcription factors or DNA sequences without requiring interaction with RNAP II.
To further monitor the partition of the SWI/SNF complex in the nucleus, we applied several subnuclear fractionation procedures directed towards the isolation of the nuclear matrix or nuclear scaffold. DNA binds to nuclear matrix through regions called MAR or SAR (for review see Laemmli et al., 1992). These regions are distributed throughout the genome and organize the chromatin in loops of variable size. Numerous experiments extend this notion, suggesting a functional role of SARs in gene expression. SARs are frequently observed in close association with enhancer elements. In flanking positions, SARs confer elevated and position-independent expression of genes in homologous as well as heterologous systems (Laemmli et al., 1992). How SARs activate transcription is not clear; however, it has been suggested that it may relate to changes in chromatin structure, affected by factors that bind to them. Studies by Forrester et al. (1994) indicate that the IgH locus becomes remodeled and DNase I hypersensitive at the pre–B cell stage, and that the Eμ SARs are necessary for this event (Jenuwein et al., 1997). In a number of cases, SARs have been found at the boundaries of chromatin domains as defined by the level of general DNase I sensitivity (Loc and Strätling, 1988; Levy-Wilson and Fortier, 1990). Taken together, all these data suggest that SARs and/or their associated proteins are involved in chromatin remodeling activity. Our experiments clearly demonstrate that ∼50% of BRG1 and hBRM and a slightly lower percentage of hSNF5 proteins are strongly associated with the nuclear matrix, suggesting that a fraction of the hSWI/SNF complex is attached to this nuclear structure. There are several well-characterized SAR binding proteins. One of these proteins called Bright binds to SARs placed proximal to the heavy chain enhancer Eμ of the IgH gene (Herrscher et al., 1995). Bright binds to the minor grove of DNA through a DNA binding domain that shows a significant sequence identity with the yeast SWI1 protein, a component of the yeast SWI/SNF complex (Herrscher et al., 1995). An homologous domain has also been identified in other SAR binding proteins. Furthermore, a DNA minor grove binding activity has been recently reported for the yeast SWI/SNF complex (Quinn et al., 1996). Cross-linking experiments suggest that SWI1 is the protein responsible for this binding activity. It is unknown if a homologue of SWI1 exists in the human SWI/ SNF complex. However, our results demonstrate that hSWI/ SNF components are present in scaffold preparations, suggesting that the hSWI/SNF complex could contain a SAR binding protein. An attractive hypothesis is that a fraction of the hSWI/SNF complex may contain a protein homologous to SWI1 or Bright that can bind SARs. This SARbound hSWI/SNF complex could be responsible for the chromatin remodeling activity associated with SAR elements. Recently, it has been demonstrated that multiple forms of hSWI/SNF complexes exist, with different subunit composition, in the same cell, as well as different complexes in different cell types (Wang et al., 1996a ). Our data suggest the existence of two subpopulations of the hSWI/ SNF complex, one associated with the nuclear matrix and another not associated with this nuclear structure.
We further investigated the distribution of the hSWI/ SNF proteins in active and inactive fractions of chromatin. hBRM, hSNF5 and, to a lesser extent, BRG1 were enriched both in chromatin highly accessible to micrococcal nuclease and in “insoluble chromatin,” or chromatin that remains bound to the nuclear skeleton. These two fractions (termed S1 and P) were shown to be enriched in transcriptionally active genes (Rose and Garrard, 1984). These data agree with our immunolocalization experiments, showing that BRG1 and hBRM are excluded from heterochromatin. In the fractionation procedure, RNAP II was found almost exclusively in the P fraction, suggesting that DNA sequences found in this fraction are being actively transcribed. We should outline that, even though fraction S1 was shown to contain DNA from actively transcribed genes, it lacks RNAP II. Easily solubilized mononucleosomes may originate from regions with open chromatin that are not being transcribed, like enhancers, or from regions in transcribed templates that precede or follow the actively transcribing RNAP II. Our results suggest that the hSWI/SNF complex is associated in vivo with regions of open chromatin where RNAP II is not present, as well as with DNA segments directly bound by RNAP II. Taken together, our subnuclear fractionation studies clearly demonstrate that components of the hSWI/SNF complexes may be associated with several distinct steps in gene activation. We can speculate that it may participate in SAR or MAR function, global chromatin remodeling during gene activation, and perhaps the transcription initiation process itself. Further studies will be important to prove our hypothesis and to clarify the exact biochemical role of the hSWI/SNF complex in transcription control.
We have also investigated the conditions that determine the binding of hSWI/SNF complex to its targets in the nucleus. The release of SWI/SNF proteins from the tight nuclear interaction coincides temporally with the start of mitosis (Fig. 5). This corresponds in time with the onset of BRG1 and hBRM hyperphosphorylation, suggesting that either phosphorylation itself or an event linked to this modification is the cause of the dissociation event. We found that treatment of permeabilized nuclei with hydrolyzable ATP leads to the release of BRG1 and hBRM from the nuclei. ATP-dependent release was inhibited by a protein tyrosine kinase inhibitor and a cdc2 substrate peptide, suggesting that phosphorylation of some nuclear components is involved in this process. However, released BRG1 and hBRM were not hyperphosphorylated after ATP treatment in the same way than they are during mitosis. Changes in the activity and morphology of the nucleus throughout the cell cycle require the nuclear structure to be dynamic. Phosphorylation appears to be a major control mechanism of the nuclear skeleton. For example, nuclear disassembly is driven by the hyperphosphorylation of lamin filaments (Fisher, 1987). There is also evidence of both casein kinase and tyrosine kinase activity associated with the nuclear matrix (Ohmuda et al., 1986; Tawfic and Ahmed, 1994). Since both tyrosine kinases and serine- and threonine-phosphorylating kinases seem to be implicated, it is possible that ATP-dependent release of hSWI/SNF complex from its nuclear targets is a consequence of a cascade of kinase activities. It is also possible that this cascade is involved in the normal cell cycle regulation of the nuclear structure and may not be specific for the SWI/SNF complex.
Finally, in this work we have analyzed the distribution of components of the hSWI/SNF complex in different fractions of the nucleus and the chromatin. Numerous observations indicate a very high level of structural organization and compartmentalization for the activities occurring within the nucleus. We think that these structural aspects will be important factors for the understanding of the mechanism by which the SWI/SNF complex operates.
Acknowledgments
We are grateful to O. Bensaude for pol 3/3 antibodies and valuable discussion of the manuscript. We thank M. Vincent, J.C. Courvalin, and D. Lallemand for cc-3, anti–lamin B, and anti-JunB antibodies, respectively. We are grateful to R. Hellio for performing confocal microscopy, and to J. Weitzman for critical reading of the manuscript.
Abbreviations used in this paper
- CTD
carboxy-terminal domain
- DAPI
4′,6-diamidino-2-phenylindole
- GR
glucocorticoid receptor
- LIS
3,5-diiodosalicilic acid, lithium salt
- MAR
matrix-associated region
- SAR
scaffold-associated region
- Rb
retinoblastoma
- RNAP II
RNA polymerase II
Footnotes
J.C. Reyes was the recipient of European Molecular Biology Organization and Human Frontier Science Program Organization postdoctoral fellowships. The work was supported by Association pour la Recherche sur le Cancer, Ligue Nationale Française Contre le Cancer, and the Actions Concertées des Sciences du Vivant program of the French Ministry of Science.
Please address all correspondence to Moshe Yaniv, Unité des Virus Oncogènes, Département des Biotechnologies, 25, rue Dr. Roux, 75724 Paris Cedex 15, France. Tel.: (33) 1-45-68-85-13. Fax: (33) 1-40-61-30-33.
References
- Akiyama T, Ogawara H. Use and specificity of genistein as inhibitor of protein-tyrosine kinases. Methods Enzymol. 1991;201:362–370. doi: 10.1016/0076-6879(91)01032-w. [DOI] [PubMed] [Google Scholar]
- Berezney R, Mortillaro MJ, Ma H, Wei X, Samarabandu J. The nuclear matrix: a structural milieu for genomic function. Int Rev Cytol. 1995;162:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- Bisotto S, Lauriault P, Duval M, Vincent M. Colocalization of a high molecular mass phosphoprotein of the nuclear matrix (p255) with spliceosomes. J Cell Sci. 1995;108:1873–1882. doi: 10.1242/jcs.108.5.1873. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcriptiondependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Laurent BC. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Cereghini S, Blumenfeld M, Yaniv M. A liver-specific factor essential for albumin transcription differs between differentiated and dedifferentiated rat hepatoma cells. Genes & Dev. 1988;2:957–974. doi: 10.1101/gad.2.8.957. [DOI] [PubMed] [Google Scholar]
- Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahmaare transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science (Wash DC) 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Davie JR. The nuclear matrix and the regulation of chromatin organization and function. Int Rev Cytol. 1995;162:191–250. doi: 10.1016/s0074-7696(08)61232-2. [DOI] [PubMed] [Google Scholar]
- Dubois MF, Vincent M, Adamczewski J, Egly JM, Bensaude O. Heat-shock inactivation of the THIIH-associated kinase and change in the phosphorylation sites on RNA polymerase II largest subunit. Nucleic Acids Res. 1997;25:694–700. doi: 10.1093/nar/25.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Ålin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature (Lond) 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Fisher P. Disassembly and reassembly of nuclei in cell-free systems. Cell. 1987;48:175–186. doi: 10.1016/0092-8674(87)90417-x. [DOI] [PubMed] [Google Scholar]
- Forrester WC, van Genderen C, Jenuwein T, Grosschedl R. Dependence of enhancer-mediated transcription of the immunoglobulin μ gene on nuclear matrix attachment regions. Science (Wash DC) 1994;265:1221–1225. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- He D, Nickerson AN, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheuermann R, Tucker PW. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes & Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- Hidaka H, Inagaki M, Kawamoto S, Sasaki Y. Isoquinolinesulfonamides, novel and potent inhibitors of cyclic nucleotide dependent protein kinase and protein kinase C. Biochemistry. 1984;23:5036–5041. doi: 10.1021/bi00316a032. [DOI] [PubMed] [Google Scholar]
- Huang SY, Garrard WT. Electrophoretic analyses of nucleosomes and other protein-DNA comlexes. Methods Enzymol. 1989;170:116–142. doi: 10.1016/0076-6879(89)70044-6. [DOI] [PubMed] [Google Scholar]
- Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature (Lond) 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Cook PR. Transcription occurs at a nucleoskeleton. EMBO (Eur Mol Biol Organ) J. 1985;4:919–925. doi: 10.1002/j.1460-2075.1985.tb03719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO (Eur Mol Biol Organ) J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Sites in human nuclei where damage induced by ultraviolet light is repaired: localization relative to transcription sites and concentrations of proliferating cell nuclear antigen and the tumour suppressor protein, p53. J Cell Sci. 1994;107:1753–1760. doi: 10.1242/jcs.107.7.1753. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Forrester WC, Qiu RG, Grosschedl R. The immunoglobulin μ enhancer core establishes local factor access in nuclear chromatin independent of transcriptional stimulation. Genes & Dev. 1993;7:2016–2032. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Forrester WC, Fernandez-Herrero LA, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature (Lond) 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science (Wash DC) 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- Kamakaka RT, Thomas JO. Chromatin structure of transcriptionally competent and repressed genes. EMBO (Eur Mol Biol Organ) J. 1990;9:3997–4006. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp BE, Pearson RB. Design and use of peptide substrates for protein kinases. Methods Enzymol. 1991;200:121–134. doi: 10.1016/0076-6879(91)00134-i. [DOI] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature (Lond) 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Bunker CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes & Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- Kirov N, Tsanev R. Activated murine alpha-globin gene is not preferentially associated with the nuclear matrix. Int J Biochem. 1986;18:155–159. doi: 10.1016/0020-711x(86)90148-5. [DOI] [PubMed] [Google Scholar]
- Krämer A, Haars R, Kabisch R, Will H, Bautz FA, Bautz EK. Monoclonal antibody directed against RNA polymerase II of Drosophila melanogaster. . Mol Gen Genet. 1980;180:193–199. doi: 10.1007/BF00267369. [DOI] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature (Lond) 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK, Kas E, Poljak L, Adachi Y. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr Opin Genet Dev. 1992;2:275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes & Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- Laybourn PJ, Kadonaga JT. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science (Wash DC) 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Levy-Wilson B, Fortier C. The limits of the DNase I-sensitive domain of the human apolipoprotein B gene coincide with the locations of chromosomal anchorage loops and define the 5′ and 3′ boundaries of the gene. J Biol Chem. 1989;264:21196–21204. [PubMed] [Google Scholar]
- Loc PV, Strätling WH. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO (Eur Mol Biol Organ) J. 1988;7:655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini MA, Shan B, Nickerson JA, Penman S, Lee WH. The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc Natl Acad Sci USA. 1994;91:418–422. doi: 10.1073/pnas.91.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathog D, Hochstrasser M, Gruenbaum Y, Saumweber H, Sedat J. Characteristic folding pattern of polytene chromosomes in Drosophilasalivary gland nuclei. Nature (Lond) 1984;308:414–421. doi: 10.1038/308414a0. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J, Mirault M-E, Laemmli UK. Organization of the higher order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brmgenes potentiates transcriptional activation by the glucocorticoid receptor. EMBO (Eur Mol Biol Organ) J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein with homology to S. cerevisiaeSNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Reyes JC, Bourachot B, Legouy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO (Eur Mol Biol Organ) J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- Owen-Hughes T, Utley RT, Cote J, Peterson CL, Workman JL. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science (Wash DC) 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- Paranjape SM, Kamakaka RT, Kadonaga JT. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- Peränen J, Rikkonen M, Kääriäinen L. A method for exposing hidden antigenic sites in paraformaldehyde-fixed cultured cells, applied to initially unreactive antibodies. J Histochem Cytochem. 1993;41:447–454. doi: 10.1177/41.3.8429208. [DOI] [PubMed] [Google Scholar]
- Peterson CL. Multiple switches to turn on chromatin. Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Tamkun JW. The SWI-SNF complex: a chromatin remodeling machine? . Trends Genet. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Dingwall A, Scott MP. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement . Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr CM, Mechta F, Spyrou G, Lallemand D, Carillo S, Yaniv M. Mouse JunD negatively regulates fibroblast growth and antagonizes transformation by ras. Cell. 1994;76:747–760. doi: 10.1016/0092-8674(94)90513-4. [DOI] [PubMed] [Google Scholar]
- Quinn J, Fyrberg AM, Ganster RW, Schmidt MC, Peterson CL. DNA-binding properties of the yeast SWI/SNF complex. Nature (Lond) 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- Rose SM, Garrard WT. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J Biol Chem. 1984;259:8534–8544. [PubMed] [Google Scholar]
- Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature (Lond) 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- Stratling WH. Gene-specific differences in the supranucleosomal organization of rat liver chromatin. Biochemistry. 1987;26:7893–7899. doi: 10.1021/bi00398a053. [DOI] [PubMed] [Google Scholar]
- Stroboulis J, Wolffe A. Functional compartmentalization of the nucleus. J Cell Sci. 1996;109:1991–2000. doi: 10.1242/jcs.109.8.1991. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: a regulator of Drosophilahomeotic genes structurally related to the yeast transcriptional activator SNF2/ SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Tawfic S, Ahmed K. Association of casein kinase 2 with nuclear matrix. Possible role in nuclear matrix protein phosphorylation. J Biol Chem. 1994;269:7489–7493. [PubMed] [Google Scholar]
- Thibodeau A, Duchaine J, Simard JL, Vincent M. Localization of molecules with restricted patterns of expression in morphogenesis: an immunohistochemical approach. Histochem J. 1989;21:348–356. doi: 10.1007/BF01798498. [DOI] [PubMed] [Google Scholar]
- van Driel R, Wansink DG, van Steensel B, Grande MA, Schul W, de Jong L. Nuclear domains and the nuclear matrix. Int Rev Cytol. 1995;162A:151–189. doi: 10.1016/s0074-7696(08)61231-0. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Brink M, van der Meulen K, van Binnendijk EP, Wansink DG, de Jong L, de Kloet ER, van Driel R. Localization of the glucocorticoid receptor in discrete clusters in the cell nucleus. J Cell Sci. 1995a;108:3003–3011. doi: 10.1242/jcs.108.9.3003. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Jenster G, Damm K, Brinkmann AO, van Driel R. Domains of the human androgen receptor and glucocorticoid receptor involved in binding to the nuclear matrix. J Cell Biochem. 1995b;57:465–478. doi: 10.1002/jcb.240570312. [DOI] [PubMed] [Google Scholar]
- Vincent M, Lauriault P, Duboise MF, Lacroie S, Bensaude O, Chaboh B. The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with the spliceosome. Nucleic Acids Res. 1997;24:4649–4652. doi: 10.1093/nar/24.23.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Côté J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. Identification of multiple forms of SWI/SNF complexes in mammalian cells: implication for its diverse functions in developmental and tissue-specific gene expression. EMBO (Eur Mol Biol Organ) J. 1996a;15:5370–5382. [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complex. Genes & Dev. 1996b;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Williamson P, Felsenfeld G. Transcription of histone-covered T7 DNA by Escherichia coliRNA polymerase. Biochemistry. 1978;17:5695–5705. doi: 10.1021/bi00619a015. [DOI] [PubMed] [Google Scholar]
- Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- Xu M, Barnard MB, Rose SM, Cockerill PN, Huang SY, Garrard WT. Transcription termination and chromatin structure of the active immunoglobulin kappa gene locus. J Biol Chem. 1986;261:3838–3845. [PubMed] [Google Scholar]