The observation that almost all malignant cancers have telomerase activity has been explained by the assumption that telomerase is crucial for the progression of malignancy. Surprisingly, cells from mice without a functional telomerase RNA gene and no telomerase activity are indistinguishable from normal cells in different transformation and immortalization assays. However, detailed analysis of chromosomes in telomerase null cells has revealed multiple defects that point to the role of telomerase in normal biology and raise well-defined questions for future research.
Telomere Biology
Telomeres are specialized structures at the end of chromosomes composed of DNA and proteins that are essential for maintaining the stability of eukaryotic genomes (for review see references 4, 34). The major functions of telomeres are (a) to protect chromosome ends from recombination, fusion, and degradation (cap function), (b) to position and move chromosomes during various stages of the meiotic and mitotic cell cycle (anchor function), and (c) to counter telomere erosion resulting from incomplete replication of chromosome ends (end-replication “buffer” function). The latter is essential because conventional DNA polymerases are unidirectional and cannot copy all bases at the 3′ end of a linear duplex (33), resulting in the slow loss of genetic material from the ends of chromosomes with each replication round. In all vertebrates, including humans, the most terminal DNA consists of extended (up to 100 kb) arrays of TTAGGG repeats (26, 28), and the end-replication “buffer” function is accomplished in two fundamentally different ways. In cells of the germ line and most immortal cell lines, repeats are added to the 3′ end by telomerase, a multimeric enzyme with reverse transcriptase activity and an RNA template encoding for terminal repeats (for review see reference 15). In most if not all normal somatic cells, telomerase is either not expressed or not capable of extending chromosome ends, and as a result, telomere repeats are lost with each replication round (10, 13, 16, 19). A minimum telomere length appears to be required to maintain the structural integrity of the chromosomes. Shortening beyond this point has been implicated in replicative senescence of cells (for review see reference 17), and (re)activation of functional telomerase has been proposed as an important step in the development of tumors (11, 20; for review see reference 31). In this scenario, the loss of telomere repeats with each replication round represents a mechanism to suppress the uncontrolled proliferation of premalignant cells in long-lived species.
Murine Models
The putative tumor-suppressor function and cell senescence function of telomeres have long been challenged by observations in the mouse (22; for review see reference 21). On average, telomeres are 5–10 times longer in murine cells than in human cells, and yet murine cells show much more rapid senescence in culture. Nevertheless, like in human cells, measurable telomerase activity is upregulated in mouse tumors (3, 6, 7, 9), suggesting a role for telomerase in murine tumor formation after all. These and other conflicting data have added to a large degree of confusion regarding the role of telomeres and telomerase in cellular senescence and tumor formation in mouse and man. In view of this situation, data from mice without telomerase were eagerly awaited by investigators both inside and outside the telomere field. Such data have now been obtained by Blasco et al. (5), who developed mice in which the telomerase RNA template gene has been removed from the germ line using standard gene knock-out (KO)1 techniques. The results obtained with (the cells from) the telomerase KO mice contain valuable lessons for anyone interested in telomere biology.
The Hidden Phenotype of Telomerase KO Mice
As expected, no telomerase could be detected in cells derived from homozygous KO animals, supporting the notion of a single telomerase RNA gene in the murine genome. The fact that the KO mice were born alive and apparently normal was the first surprise, since it indicates that telomerase is not essential for maintaining telomeres in somatic (stem) cells of renewing tissues such as skin, gut, and blood during development and normal steady-state tissue homeostasis. It has been suggested that cellular defects may be demonstrated upon challenges to such tissues or with aging of the mice (24), but details of such defects are currently not known. If the cells of self-renewing tissues are affected, it will be important to distinguish between the absence of telomerase and a decreased proliferative potential (resulting from overall shorter telomeres; see below) as primary defects. The observation that the telomerase null mice were fertile (resulting in multiple subsequent generations of KO animals) was the second surprise, initially suggesting that telomerase was not necessary to maintain telomeres in the germ line. Furthermore, cells from telomerase-deficient mice were as efficiently transformed into immortal and in vivo tumor-forming cells as cells from telomerase-positive animals, demonstrating conclusively that, in the mouse, telomerase is not an essential requirement for the establishment of cell lines, oncogenic transformation, or tumor formation. The surprises did not end there. All of the above could still be explained by assuming that telomere shortening was perhaps occurring at a very slow rate in cells known to have very long initial telomeres (22), perhaps together with alternative pathways for telomere maintenance in immortal tumor cells (see below). The studies by Blasco et al. (5) have revealed that the situation is more complex. Continued inbreeding has produced up to six generations of KO animals. Although conventional telomere length analysis by Southern essentially failed to detect telomere shortening, clear differences in telomere length between cells from subsequent generations of KO animals were observed using quantitative fluorescence in situ hybridization (Q-FISH) (25, 36). The Q-FISH observations on the ends of individual chromosomes in telomerase KO cells has provided some answers to several of the long-standing questions about telomeres in the mouse and provide a focus for future studies. Such observations are summarized below.
Germ Cells Need Telomerase to Maintain Telomere Length
The first lesson from Q-FISH is that telomerase is essential to maintain telomere length in the germ line. Without telomerase, telomere repeats are lost at a variable rate of 2–7 kb per generation of mice. Assuming a loss of 75–150 or 100 bp/cell division (1, 16, 29), this translates into 20–70 cell divisions from germ cell to germ cell in subsequent generations of animals. It has been estimated that sperm cells undergo an average of 62 cell divisions from the zygote, whereas from zygote to oocyte takes on average only 25 divisions (14). The agreement between experimental data and theoretical predictions is striking and strongly supports the original suggestion that telomere shortening in somatic cells results from the absence of telomerase in such cells (10).
Uncapped Chromosomes?
The next surprise is related to telomeres apparently lacking TTAGGG repeats altogether. Chromosomes without detectable TTAGGG on at least one end were observed at increased frequencies from generation 2 and higher (several per metaphase spread in later generations). That such ends are unstable is demonstrated by the increasing frequency of aneuploid cells and end-to-end associations of chromosomes with each subsequent generation of the KO mice. This observation provides direct and formal proof that chromosome ends without TTAGGG are unstable and predisposed to chromosomal abnormalities, as was predicted (18; for review see reference 12). More surprising is that such uncapped ends are so readily observed. Using plasmids with TTAGGG inserts of variable size, the sensitivity of Q-FISH for the detection of TTAGGG repeats has been determined to be in the order of a few hundred base pairs or less (36). If sensitivity is similar for chromosomes in metaphase spreads, ends without detectable Q-FISH signals may contain less than a few hundred base pairs of TTAGGG. What is the history and what are the implications of such apparently uncapped chromosome ends? Did or do such ends signal a prolonged but transient cell cycle arrest as in yeast (see below), or are they at least temporarily ignored, perhaps similarly to uncapped ends of Drosophila chromosomes (27)? If loss of TTAGGG by itself is not sufficient to make chromosomes become fusogenic and/or recombinogenic, what other factors are involved in this transition?
Uncapped Chromosomes: Are Adaptive Responses Involved?
It has been shown that a single break at the end of a yeast chromosome will trigger a RAD9-mediated cell cycle arrest (30). Interestingly, in that study many of the yeast cells recovered from the initial arrest without repairing the damaged chromosome and resumed cell divisions. Although uncapped chromosomes in yeast were destined for eventual loss, they were apparently no longer recognized by the RAD9 pathway, or this pathway was shut off. A similar adaptation of signal transduction systems may apply to cells of telomerase KO mice. It is possible that cells without TTAGGG on one or more chromosomes, perhaps also after a transient cell cycle arrest, adapted to signaling by uncapped chromosomes, and this possibility warrants further investigation. In any event, it seems that murine cells with one or more uncapped chromosomes can divide for many times with or without an initial arrest. Indeed, normal development and even fertility does not seem to be immediately affected by the presence of such uncapped chromosomes.
Telomere Elongation by Recombination?
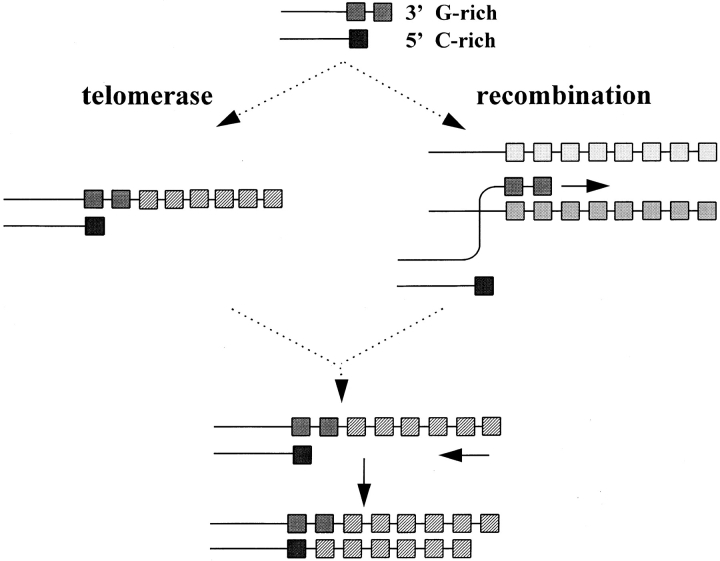
Conceivably, chromosomes without TTAGGG repeats could also be “healed” by recombinational repair. Based on studies of yeast (for review see reference 35), many alternative scenarios for recombinational telomere repair are possible. Uncapped chromosomes in yeast can acquire a new telomere by homologous recombination, both reciprocal and nonreciprocal, via RAD52-mediated gene conversion (32). Nonreciprocal recombination or gene conversion between telomeres on homologous or nonhomologous chromosomes (as depicted in Fig. 1) could occur in mammalian cells as well. Indeed, the overall long telomeres in the mouse could favor the use of such a nonreciprocal recombination: a critically short telomere in a murine cell could perhaps easily recombine with remaining long telomeres on other chromosomes. As the overall telomere length decreases in subsequent generations of the telomerase KO animals, the efficiency of this pathways could decrease, possibly with the increased frequencies of chromosomal abnormalities seen at later generations as a result.
Figure 1.
Pathways to maintain telomere length. Telomerase, a reverse transcriptase, can extend the 3′ end of telomeres using an RNA component complimentary to the G-rich repeats of the 3′ strand. Are recombination pathways important to maintain telomere length in telomerase KO mice? Shown is nonreciprocal recombination proposed for telomere elongation in yeast (32). Continued shortening of telomeres and abundant chromosome ends without detectable telomere repeats in cells from telomerase KO mice suggest that such recombination pathways are either unable to prevent overall telomere loss or are not active at all. Each box represents a repeat unit. Hatched boxes represent newly synthesized repeats. The figure was adapted from reference 32.
Chromosomal Abnormalities in Telomerase KO Cells
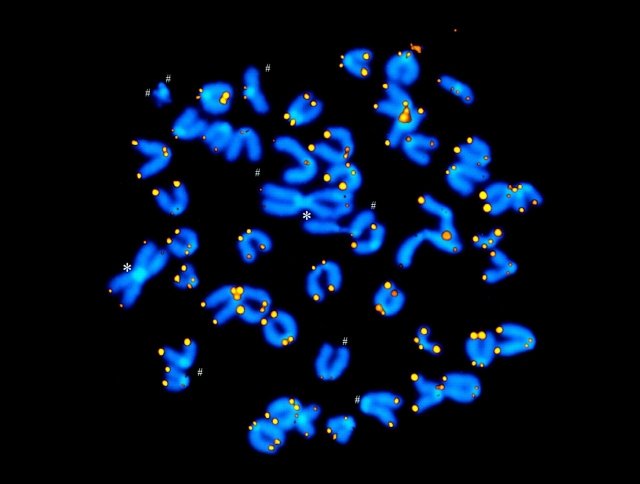
The presence of the Robertsonian fusions observed in later generations of the telomerase KO mice deserves special mention (Fig. 2). Given that telomeres on the short P arm of acrocentric murine chromosomes are, in general, significantly shorter than q arm telomeres (36), a gradual loss of telomere repeats would be expected to predispose to this type of chromosomal abnormality. Are the Robertsonian chromosomes in the telomerase KO mouse stable? If the primary fusion event was indeed between two different uncapped chromosomes, the prediction would be that they are not, as such chromosomes are expected to have two functional centromeres (23). Further studies of these Robertsonian chromosomes should reveal whether they represent unique cytogenetic abnormalities in telomerase KO mice.
Figure 2.
Chromosomal abnormalities in embryonic fibroblasts derived from the sixth generation of telomerase null mice. Results were obtained by Q-FISH using peptide nucleic acid (PNA) probes as described (25, 36). Yellow and orange represent telomere signals obtained with Cy3-labeled (CCCTAA)3 PNA, and blue represents DAPI-stained chromosomal DNA. Pseudocolors were assigned using Adobe Photoshop software (San Jose, CA). Asterisks indicate metacentric Robertsonian fusion products between acrocentric chromosomes. Number signs indicate chromosome arms without detectable TTAGGG repeats.
Implications for Models of Tumor Growth and Telomerase Inhibition Therapy
The observation that telomerase appears completely dispensable for tumorigenesis in the mouse should be discussed in relation to models of tumor cell proliferation and the possible use of telomerase inhibitors in cancer therapy (17). Approximately a quarter of immortalized human cell lines lack detectable telomerase activity (for review see reference 8), indicating that alternatives to telomerase for the maintenance of telomeres also exist in human cells. Furthermore, evidence is accumulating that in different somatic cell types, including tumor cells, telomerase may be best correlated with proliferation rate as telomeres continue to shorten in many telomerase-positive cells (reviewed in 2). Apparently, measurable enzyme activity is frequently not associated with elongation or static maintenance of telomere length, perhaps because telomeres are inaccessible to telomerase in most somatic and tumor cells. The observations with the telomerase KO mice clearly show that telomere shortening, lack of telomerase, and even uncapped chromosomes are not incompatible with continued and extensive proliferation. However, caution in the extrapolation of the murine data to the human situation is warranted, in view of the well-known observation that human cells are less efficiently immortalized than murine cells. Could less efficient adaptive responses and/or recombination (as shown in Fig. 1) explain the differences in immortalization rates between the species? The observation that cells from M. spretus (with telomeres of comparable length to human cells) will spontaneously, be it less efficiently, immortalize in culture (29) indicates that overall telomere length cannot be a major factor predisposing cells towards spontaneous immortalization. Differences in the signaling and/or processing of critically short telomeres between murine and human cells could conceivably make continued proliferation in human cells more dependent on telomerase. In general, many questions about the role of telomerase and recombination pathways in the proliferation, senescence, and immortalization of normal and malignant cells from different murine and human tissues remain. For this reason caution in extrapolating findings with murine cells too directly to human cells remains warranted. However, with the current rate of progress, answers to many of these questions should be available in the near future.
Footnotes
1. Abbreviations used in this paper: KO, knock-out; Q-FISH, quantitative fluorescence in situ hybridization.
I thank anonymous reviewers for their helpful comments and Dr. Prakash Hande (Terry Fox Laboratory, Vancouver, Canada) for the image files used for preparation of Fig. 2.
Work in my lab is supported by grants RO1A129524 and GM56162 from the National Institutes of Health and by a grant from the National Cancer Institute of Canada with funds from the Terry Fox Run.
Address all correspondence to Peter M. Lansdorp, Terry Fox Laboratory, British Columbia Cancer Research Centre, Vancouver, BC, Canada, V5Z 1L3. Tel.: (604) 877-6070, Ext. 3026. Fax: (604) 877-0712. E-mail: peter@terryfox.ubc.ca
References
- 1.Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Autexier C, Greider CW. Telomerase and cancer: revisiting the telomere hypothesis. Trends Biochem Sci. 1996;21:387–391. [PubMed] [Google Scholar]
- 3.Bednarek A, Budunova I, Slaga T, Aldez CM. Increased telomerase activity in mouse skin premalignant progression. Cancer Res. 1995;55:4566–4569. [PubMed] [Google Scholar]
- 4.Blackburn EH. Telomeres: no end in sight. Cell. 1994;77:621–623. doi: 10.1016/0092-8674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 5.Blasco MA, Lee H-W, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 6.Blasco MA, Rizen M, Greider CW, Hanahan D. Differential regulation of telomerase activity and telomerase RNA during multi-stage tumorigenesis. Nat Genet. 1996;12:200–204. doi: 10.1038/ng0296-200. [DOI] [PubMed] [Google Scholar]
- 7.Broccoli D, Godley LA, Donehower LA, Varmus HE, de Lange T. Telomerase activation in mouse mammary tumors: lack of detectable telomere shortening and evidence for regulation of telomerase RNA with cell proliferation. Mol Cell Biol. 1996;16:3765–3772. doi: 10.1128/mcb.16.7.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan TM, Reddel RR. Telomere dynamics and telomerase activity in in vitroimmortalised human cells. Eur J Cancer. 1997;33:767–773. doi: 10.1016/S0959-8049(97)00065-8. [DOI] [PubMed] [Google Scholar]
- 9.Chadeneau C, Siegel P, Harley CB, Muller WJ, Bacchetti S. Telomerase activity in normal and malignant murine tissues. Oncogene. 1995;11:893–898. [PubMed] [Google Scholar]
- 10.Cooke HJ, Smith BA. Variability at the telomeres of the human X/Y pseudoautosomal region. Cold Spring Harbor Symp Quant Biol. 1986;51:213–219. doi: 10.1101/sqb.1986.051.01.026. [DOI] [PubMed] [Google Scholar]
- 11.Counter CM, Hirte HW, Bacchetti S, Harley CB. Telomerase activity in human ovarian carcinoma. Proc Natl Acad Sci USA. 1994;91:2900–2904. doi: 10.1073/pnas.91.8.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lange, T. 1995. Telomere dynamics and genome instability in human cancer. In Telomeres. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 265–293.
- 13.de Lange T, Shiue L, Myers R, Cox DR, Naylor SL, Killery AM, Varmus HE. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drost JB, Lee WR. Biological basis of germline mutation: comparison of spontaneous germline mutation rates in Drosophila, mouse and human. Environ Mol Mutagen. 1995;25:48–64. doi: 10.1002/em.2850250609. [DOI] [PubMed] [Google Scholar]
- 15.Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 16.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature (Lond) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 17.Harley CB, Kim NW, Prowse KR, Weinrich SL, Hirsch KS, West MD, Bacchetti S, Hirte HW, Counter CM, Greider CW, et al. Telomerase, cell immortality, and cancer. Cold Spring Harbor Symp Quant Biol. 1994;59:307–315. doi: 10.1101/sqb.1994.059.01.035. [DOI] [PubMed] [Google Scholar]
- 18.Hastie ND, Allshire RC. Human telomeres: fusion and interstitial sites. Trends Genet. 1989;5:326–330. doi: 10.1016/0168-9525(89)90137-6. [DOI] [PubMed] [Google Scholar]
- 19.Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere reduction in human colorectal carcinoma and with ageing. Nature (Lond) 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 20.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science (Wash DC) 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 21.Kipling D. Telomere structure and telomerase expression during mouse development and tumorigenesis. Eur J Cancer. 1997;33:792–800. doi: 10.1016/S0959-8049(97)00060-9. [DOI] [PubMed] [Google Scholar]
- 22.Kipling D, Cooke HJ. Hypervariable ultra-long telomeres in mice. Nature (Lond) 1990;347:347–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 23.Kipling D, Wilson HE, Mitchell AR, Taylor BA, Cooke HJ. Mouse centromere mapping using oligonucleotide probes that detect variants of the minor satellite. Chromosoma (Berl) 1994;103:46–55. doi: 10.1007/BF00364725. [DOI] [PubMed] [Google Scholar]
- 24.Kolberg R. No neat endings yet in the tale of telomerase KO mice. J NIH Res. 1997;9:24–26. [Google Scholar]
- 25.Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little M-T, Dirks RW, Raap AK, Tanke HJ. Heterogeneity in telomere length of human chromosomes. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 26.Lejnine S, Makarov VL, Langmore JP. Conserved nucleoprotein structure at the ends of vertebrate and invertebrate chromosomes. Proc Natl Acad Sci USA. 1995;92:2393–2397. doi: 10.1073/pnas.92.6.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason JM, Biessmann H. The unusual telomeres of Drosophila. . Trends Genet. 1995;11:58–62. doi: 10.1016/s0168-9525(00)88998-2. [DOI] [PubMed] [Google Scholar]
- 28.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu J-R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prowse KR, Greider CW. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc Natl Acad Sci USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 31.Shay JW, Wright WE. Telomerase activity in human cancer. Curr Opin Oncol. 1996;8:66–71. doi: 10.1097/00001622-199601000-00012. [DOI] [PubMed] [Google Scholar]
- 32.Wang S-S, Zakian VA. Telomere-telomere recombination provides an express pathway for telomere acquisition. Nature (Lond) 1990;345:456–458. doi: 10.1038/345456a0. [DOI] [PubMed] [Google Scholar]
- 33.Watson JD. Origin of concatameric T4 DNA. Nature New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 34.Zakian VA. Telomeres: beginning to understand the end. Science (Wash DC) 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 35.Zakian VA. Telomere functions: lessons from yeast. Trends Cell Biol. 1996;6:29–33. doi: 10.1016/0962-8924(96)81035-x. [DOI] [PubMed] [Google Scholar]
- 36.Zijlmans JM, Martens UM, Poon SS, Raap AK, Tanke HJ, Ward RK, Lansdorp PM. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]