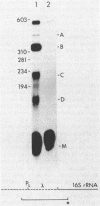
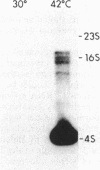
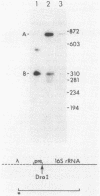
Abstract
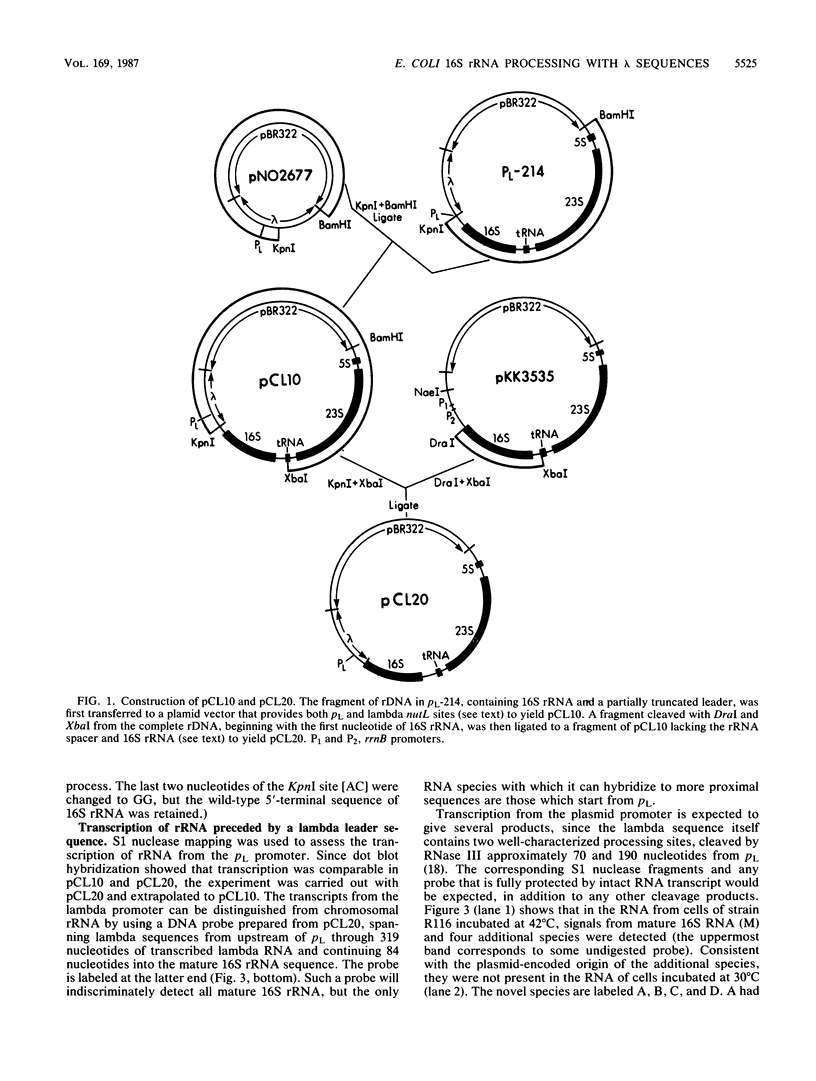
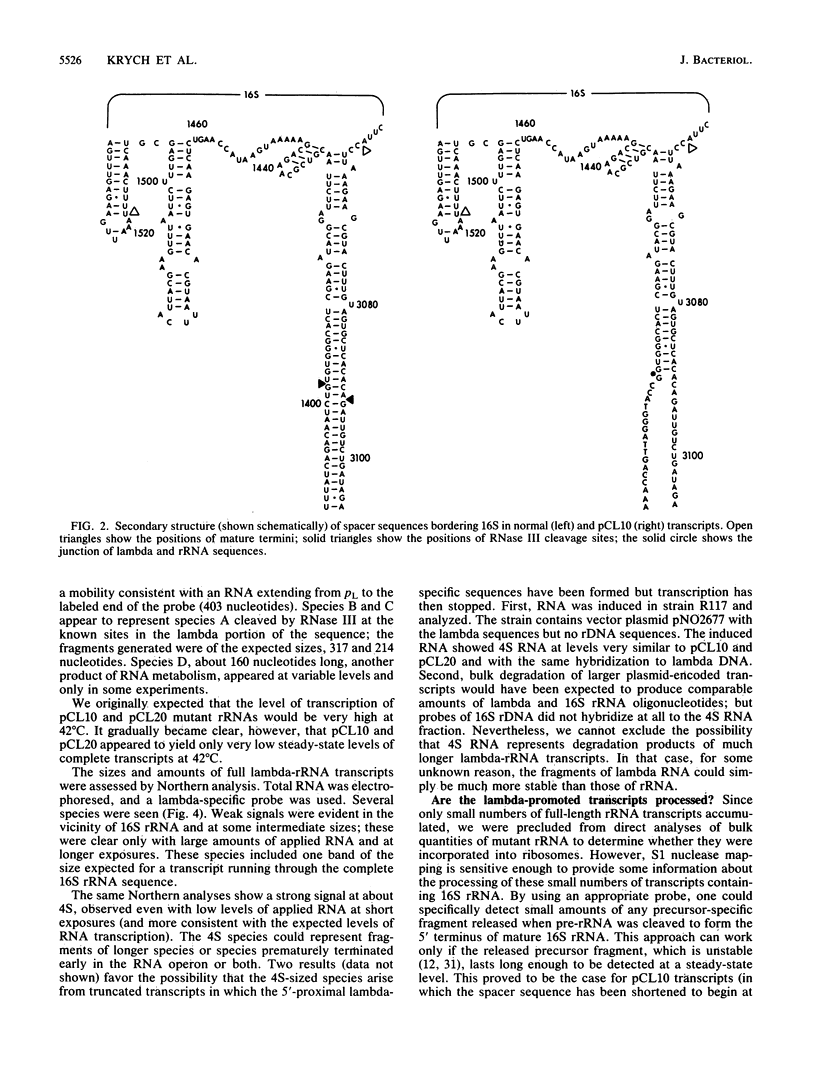
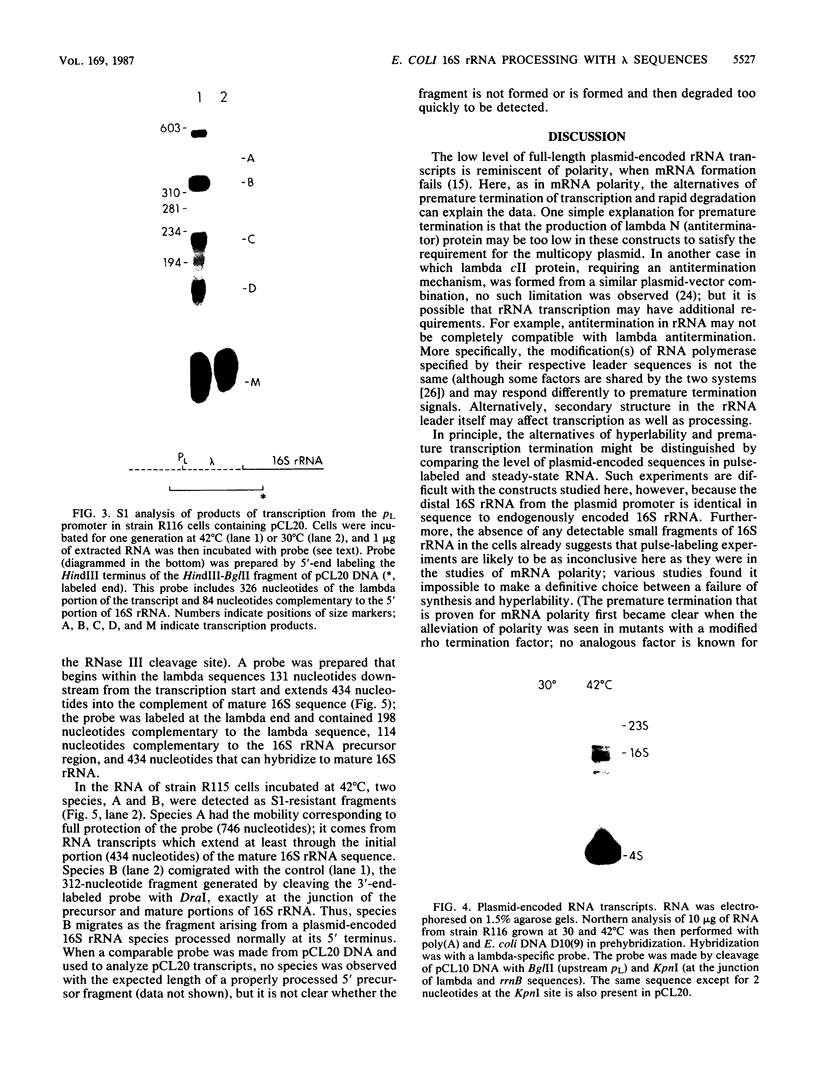
To test whether any specific 5' precursor sequences are required for the processing of pre-16S rRNA, constructs were studied in which large parts of the 5' leader sequence were replaced by the coliphage lambda pL promoter and adjacent sequences. Unexpectedly, few full-length transcripts of the rRNA were detected after the pL promoter was induced, implying that either transcription was poor or most of the rRNA chains with lambda leader sequences were unstable. Nevertheless, sufficient transcription occurred to permit the detection of processing by S1 nuclease analysis. RNA transcripts in which 2/3 of the normal rRNA leader was deleted (from the promoter up to the normal RNase III cleavage site) were processed to form the normal 5' terminus. Thus, most of the double-stranded stem that forms from sequences bracketing wild-type 16S pre-rRNA is apparently not required for proper processing; the expression of such modified transcripts, however, must be increased before the efficiency of processing of the 16S rRNA formed can be assessed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aksoy S., Squires C. L., Squires C. Evidence for antitermination in Escherichia coli RRNA transcription. J Bacteriol. 1984 Jul;159(1):260–264. doi: 10.1128/jb.159.1.260-264.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apirion D., Dallmann G., Miczak A., Szeberenyi J., Tomcsanyi T. Processing and decay of RNA in Escherichia coli: the chicken and egg problem. Biochem Soc Trans. 1986 Oct;14(5):807–810. doi: 10.1042/bst0140807. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Brosius J., Ullrich A., Raker M. A., Gray A., Dull T. J., Gutell R. R., Noller H. F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981 Jul;6(1):112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. R., Georgopoulos C., Tilly K., Herskowitz I., Banuett F. Interactions of bacteriophage and host macromolecules in the growth of bacteriophage lambda. Microbiol Rev. 1984 Dec;48(4):299–325. doi: 10.1128/mr.48.4.299-325.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R. F. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966 Mar;16(1):67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., Takebe Y., Sharrock R. A., Nomura M. Feedback regulation of rRNA and tRNA synthesis and accumulation of free ribosomes after conditional expression of rRNA genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1069–1073. doi: 10.1073/pnas.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., de Boer H. A., Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986 Jan 17;44(1):197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- Kano Y., Silengo L., Imamoto F. Stability of "spacer" sequences of pre-ribosomal RNA in Escherichia coli. Mol Gen Genet. 1976 Aug 2;146(3):275–283. doi: 10.1007/BF00701251. [DOI] [PubMed] [Google Scholar]
- King T. C., Schlessinger D. S1 nuclease mapping analysis of ribosomal RNA processing in wild type and processing deficient Escherichia coli. J Biol Chem. 1983 Oct 10;258(19):12034–12042. [PubMed] [Google Scholar]
- King T. C., Sirdeshmukh R., Schlessinger D. RNase III cleavage is obligate for maturation but not for function of Escherichia coli pre-23S rRNA. Proc Natl Acad Sci U S A. 1984 Jan;81(1):185–188. doi: 10.1073/pnas.81.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T. C., Sirdeskmukh R., Schlessinger D. Nucleolytic processing of ribonucleic acid transcripts in procaryotes. Microbiol Rev. 1986 Dec;50(4):428–451. doi: 10.1128/mr.50.4.428-451.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein B. K., Staden A., Schlessinger D. Alternative conformations in Escherichia coli 16S ribosomal RNA. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3539–3542. doi: 10.1073/pnas.82.11.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. C., Squires C. L., Squires C. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell. 1984 Oct;38(3):851–860. doi: 10.1016/0092-8674(84)90280-0. [DOI] [PubMed] [Google Scholar]
- Lozeron H. A., Dahlberg J. E., Szybalski W. Processing of the major leftward mRNA of coliphage lambda. Virology. 1976 May;71(1):262–277. doi: 10.1016/0042-6822(76)90111-2. [DOI] [PubMed] [Google Scholar]
- Mangiarotti G., Turco E., Perlo C., Altruda F. Role of precursor 16S RNA in assembly of E. coli 30S ribosomes. Nature. 1975 Feb 13;253(5492):569–571. doi: 10.1038/253569a0. [DOI] [PubMed] [Google Scholar]
- Meyhack B., Pace B., Uhlenbeck O. C., Pace N. R. Use of T4 RNA ligase to construct model substrates for a ribosomal RNA maturation endonuclease. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3045–3049. doi: 10.1073/pnas.75.7.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E. A. Antitermination mechanisms in rRNA operons of Escherichia coli. J Bacteriol. 1986 Oct;168(1):1–5. doi: 10.1128/jb.168.1.1-5.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F., Woese C. R. Secondary structure of 16S ribosomal RNA. Science. 1981 Apr 24;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Ho Y. S., Shatzman A. The use of pKc30 and its derivatives for controlled expression of genes. Methods Enzymol. 1983;101:123–138. doi: 10.1016/0076-6879(83)01009-5. [DOI] [PubMed] [Google Scholar]
- Sharrock R. A., Gourse R. L., Nomura M. Defective antitermination of rRNA transcription and derepression of rRNA and tRNA synthesis in the nusB5 mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5275–5279. doi: 10.1073/pnas.82.16.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock R. A., Gourse R. L., Nomura M. Inhibitory effect of high-level transcription of the bacteriophage lambda nutL region on transcription of rRNA in Escherichia coli. J Bacteriol. 1985 Aug;163(2):704–708. doi: 10.1128/jb.163.2.704-708.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirdeshmukh R., Schlessinger D. Ordered processing of Escherichia coli 23S rRNA in vitro. Nucleic Acids Res. 1985 Jul 25;13(14):5041–5054. doi: 10.1093/nar/13.14.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M. J., Gourse R. L., Jemiolo D. K., Dahlberg A. E. A mutation in an Escherichia coli ribosomal RNA operon that blocks the production of precursor 23 S ribosomal RNA by RNase III in vivo and in vitro. J Mol Biol. 1985 Mar 20;182(2):205–216. doi: 10.1016/0022-2836(85)90339-0. [DOI] [PubMed] [Google Scholar]
- Stark M. J., Gregory R. J., Gourse R. L., Thurlow D. L., Zwieb C., Zimmermann R. A., Dahlberg A. E. Effects of site-directed mutations in the central domain of 16 S ribosomal RNA upon ribosomal protein binding, RNA processing and 30 S subunit assembly. J Mol Biol. 1984 Sep 15;178(2):303–322. doi: 10.1016/0022-2836(84)90146-3. [DOI] [PubMed] [Google Scholar]
- Wilder D. A., Lozeron H. A. Differential modes of processing and decay for the major N-dependent RNA transcript of coliphage lambda. Virology. 1979 Dec;99(2):241–256. doi: 10.1016/0042-6822(79)90004-7. [DOI] [PubMed] [Google Scholar]