Abstract
The structure of mitotic chromosomes in cultured newt lung cells was investigated by a quantitative study of their deformability, using micropipettes. Metaphase chromosomes are highly extensible objects that return to their native shape after being stretched up to 10 times their normal length. Larger deformations of 10 to 100 times irreversibly and progressively transform the chromosomes into a “thin filament,” parts of which display a helical organization. Chromosomes break for elongations of the order of 100 times, at which time the applied force is around 100 nanonewtons. We have also observed that as mitosis proceeds from nuclear envelope breakdown to metaphase, the native chromosomes progressively become more flexible. (The elastic Young modulus drops from 5,000 ± 1,000 to 1,000 ± 200 Pa.) These observations and measurements are in agreement with a helix-hierarchy model of chromosome structure. Knowing the Young modulus allows us to estimate that the force exerted by the spindle on a newt chromosome at anaphase is roughly one nanonewton.
Mitosis involves gross physical reorganization of chromosomes; the duplicated chromatids are condensed, resolved, and finally segregated. These processes can be expected to change the material properties of chromosomes, notably their elasticity. Elasticity indicates the nature and strength of the interactions holding materials together, and thus can be used to probe chromosome structure. Given the poor state of understanding of chromosome structure, it is therefore remarkable how little this subject has been studied. In a pioneering work, Nicklas (1983) measured that the force applied to grasshopper chromosomes during anaphase was 700 piconewtons, from which he inferred the chromosome stiffness. More recently, Claussen et al. (1994) stretched human metaphase chromosomes spread on a cover glass. They found that after stretching of up to 10 times, the chromosomes returned to their original shape. However, these studies did not address the question of chromosome architecture.
An often-discussed model is one in which the “thick” metaphase chromosome is composed of a “thin filament” of diameter 200–300 nm (Sedat and Manuelidis, 1978; Manuelidis, 1990). In fact, Bak et al. (1977) reported that as isolated human metaphase chromosomes disintegrate, they can change into a thin filament of diameter 400 nm, five times the original chromosome length. They suggested that metaphase chromosomes were formed by helical wrapping of this thin fiber. On the basis of electron microscopy, they further proposed that the thin fiber had a helical structure.
The proposal for a helical structure of metaphase chromosomes is old. Observations of “spiral chromatonema” during meiotic metaphase I date to at least 1926. Ohnuki (1968) established that hypotonic treatment stabilized spiral structure in human mitotic metaphase chromosomes. Boy de la Tour and Laemmli (1988) observed that fluorescent anti–topoisomerase II was helically organized when bound to histone H1–depleted chromosomes. Recent work by Hirano and Mitchison (1994) revealed that a protein heterodimer required for chromosome condensation in vitro (XCAP-C/E) was localized along a helical track along the metaphase-like chromatids. These and other studies (Belmont et al., 1987, 1989; Saitoh and Laemmli, 1993) suggest a chromosome with an internal structure made of a coiled or folded fiber. However, the spiral structures observed may be the result of chemical treatments of the chromosomes (Cook, 1995).
In this paper, we report a simple mechanical study of mitotic chromosomes in living cultured newt lung cells using micropipette aspiration and manipulation. First, we find that chromosomes display remarkable elasticity, returning to their initial shape after being extended by up to 10 times. For larger deformations, the chromosome no longer returns to its initial length. Instead, the thick native chromosome is progressively converted into a thin filament 15 times the length of the original chromosome. This thin filament is itself elastic; it can be stretched six times before breaking. After the filament is released, it takes on an irregular but unmistakably helical form. Furthermore, by measuring force versus deformation, we have determined the Young elastic modulus of the metaphase chromosome, the force at which the metaphase chromosome begins to be converted to thin fiber, and the force required to break the thin fiber. These measurements reveal the strength of interactions that stabilize the different levels of structure. Finally, we have observed that the Young modulus drops by about fivefold during the interval from nuclear envelope breakdown to metaphase.
Our results lead to a simple unifying picture of chromosome elasticity and structure: Our conclusion is that metaphase chromosomes are composed of an underlying thin filament. The large range over which the metaphase chromosome is elastic, the scale of its Young modulus, and the helical structure of the filament all argue in favor of its helical folding. By the same line of reasoning, the fact that the thin filament is elastic over a large range of extensions suggests that it also has a folded or helical structure. We also show how the force exerted by the mitotic spindle on a chromosome and the resistance of the cytoplasm to chromosome movement can be deduced from the Young modulus measurement and chromosome shape at anaphase.
MATERIALS AND METHODS
Tissue Culture and Solutions
Newt lung cultures were prepared in a Rose chamber following standard procedures (Rieder and Hard, 1990). Newts (Notophthalamus viridescens, Connecticut Valley L500) were killed by immersion for 20 min in 1 mg/ml Tricaine (A-5040; Sigma Chemical Co., St. Louis, MO) in distilled water. The lungs were immediately dissected and cut into 1-mm2 pieces under sterile conditions and then soaked for 24 h in culture buffer (50% L-15, L-5520 [Sigma Chemical Co.]; 42% distilled water; 8% FBS, F-2442 [Sigma Chemcial Co.]; 40 U/ml Pen-strep, 15075-013 [GIBCO BRL, Gaithersburg, MD]; 1 μg/ml Fungizone, 0437-60 [Bristol-Myers Squibb, New York]). The lung fragments were then lightly squashed between the lower glass of the Rose chamber and a dialysis filter (SpectraPor 12–14 kD, 08-667E [Fisher Scientific, Pittsburgh, PA]); the Rose chamber was filled with culture buffer. A monolayer of epithelial cells formed after 3 or 4 d. The dialysis filter was removed after 7 d, leaving the monolayer in the bottom of a shallow dish suitable for micromanipulation. Mitotic activity was usually most intense over the next 48 h. Almost all experiments were done before the cultures were 12 d old.
Microscopy and Micromanipulation
All microscopy was done with an inverted microscope (Carl Zeiss, Inc., Thornwood, NY) using a 60×, 1.4 NA differential interference contrast objective and an XY motorized stage. Glass micropipettes were prepared using a puller (model 87; Sutter Instrument Co., Novato, CA) and were then cut using a forge to obtain sharp edges. A hydraulic micromanipulator was used to move the pipettes. All mechanical experiments were carried out using a combination of stage and manipulator movement. When strong adhesion of the chromosome to the micropipette was required, the micropipette was filled with culture buffer. For elasticity measurements using aspiration, 10 g/liter BSA (A3156; Sigma Chemical Co.) was added to the filling solution to prevent sticking of the chromosome to the glass micropipette.
Stretching Single Chromosomes Using a Micropipette
The micropipette diameter (2 μm) must be chosen to be slightly less than the total chromosome diameter (3 μm) to ensure strong chromosome– pipette contact. After insertion of the pipette into a mitotic cell while positive pressure difference (relative to atmospheric pressure) of 100 Pa was maintained, the first 2–4 μm of a chromosome was aspirated using 100 Pa of suction. After 30 s, the chromosome was stuck to the glass, and further aspiration was not necessary. The pipette could then be moved from the cell to stretch the chromosome, while it was anchored at its other end to the mass of other chromosomes.
The bending of the pipette, when translated perpendicular to its axis, measured the force it exerted on the chromosome. However, since one uses a stiff pipette to penetrate the cell, this measurement is not very sensitive. The elasticity of the pipette is 10−9 ± 30% Newton/μm; this was determined by a two-step calibration procedure. First, the elasticity of a thin wire was measured using a balance. This wire was then used, under the microscope, to bend the micropipette.
Measurements of Young Modulus
Chromosome elasticity for small deformations was measured using aspiration into the micropipette. Contrary to the stretching experiment described above, the pipette was this time fixed and maintained inside the cell, and pressure difference inside was used to extend the chromosome. As before, a pipette is introduced inside the mitotic cell, and weak suction is used to grab a chromosome. At some point as the chromosome slides in, a seal is made at the end of the pipette. From this point on, the only part of the chromosome that can be deformed is the portion inside the micropipette. The length of that portion is then measured as a function of pressure. Note that to avoid adhesion of the chromosome inside the pipette, the medium filling the pipette contained BSA.
The length measurement was made by digitizing images on a computer and has a precision of 5%. The uncertainty on the pressure measurement is also in the range of 5%, mainly because of drift problems. Finally, possible sticking of chromosomes to the pipette glass added another uncertainty. After the release of pressure, if the measured length was different from the original one by more than 10%, the measurement was discarded.
Elasticity Definitions and Relations
Strain.
The strain used in this paper is longitudinal deformation and is defined as ε = ΔL/L, where L is the original length of the chromosome and ΔL is the added length due to the application of tension. (Note that the precise definition of the strain includes a term proportional to (ΔL)2, which may be ignored for measurements of the Young modulus as long as data for ΔL/L < 1 are used; see Landau and Lifshitz [1986].)
Young Modulus.
The Young modulus Y is the proportionality factor between the force per unit area (or pressure) exerted and the resulting dimensionless deformation: P = Yε. Typical hard materials (e.g., metals and glass) have a Young modulus of the order of 1010 Pa. (1 Pascal = 1 Newton/m2 is the unit of pressure). For synthetic polymer gels, Y is in the range of 104–105 Pa (Horkay et al., 1989).
Poisson Ratio.
A longitudinal deformation of a solid is always accompanied by a change (almost always a decrease) in its thickness. The Poisson ratio σ relates relative change in the thickness, εthick, to relative change in length: εthick = −σε. A typical value is σ = 0.3.
Bending Modulus.
The bending of a thin rod is characterized by one elastic coefficient, called the bending modulus B (Love, 1944). The elastic energy E per length of rod is proportional to the square of its curvature κ: E/L = B κ2/2. Curvature is just the inverse of the radius of the circle that describes the bend. If the curvature varies with arc length s along the rod, the total energy is just given by the integral E = (B/2)∫κ2 ds. Thus, the minimum-energy shape is straight (κ = 0). The bending modulus in terms of the Young modulus of the rod and the rod radius R is B = (π/4)YR 4 (Love, 1944). B has dimensions of energy times length.
Polymer Definitions and Relations
Persistence Length.
A thin rod can be bent by thermal fluctuations. The persistence length of a thin rod is defined as L p = B/k B T, where k B = 1.4 × 10−23 J/K is Boltzmann's constant, and T is absolute temperature. To see what this means, consider that a typical thermal fluctuation has energy k B T; thus, given the bending energy above, the length of rod L that can be bent through an angle of one radian by thermal fluctuation satisfies k B T ≈ B/L. So the persistence length is the length of rod that is typically deflected by about a radian by thermal fluctuations. Roughly speaking, L p is the smallest length scale over which bending fluctuations can be observed; a rod is essentially straight on scales smaller than its L p. For double-stranded DNA, L p ≈ 0.05 μm; for polymerized actin, L p ≈ 0.5 μm; for microtubules L p ≈ 5 mm.
End-to-End Size of Thermally Bent Rod.
Consider a rod of length L ≫ L
p. Thermal fluctuations produce random bends along the rod. Thus, the rod becomes a “random walk” of L/L
p steps, each of length L
p. The average distance between ends of a random walk grows as the square root of the number of steps in the walk. Thus the mean distance between the ends of the rod will be L
e ≈ L
p
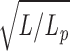 . (The exact result for the average of Le
2 is 2LL
p; note that the average end-to-end displacement in any direction is zero since the polymer may reorient in any direction.) Applied to a polymer (e.g., a DNA longer than a few kilobuses), L
e corresponds to the size of a typical “random coil” conformation (de Gennes, 1988).
. (The exact result for the average of Le
2 is 2LL
p; note that the average end-to-end displacement in any direction is zero since the polymer may reorient in any direction.) Applied to a polymer (e.g., a DNA longer than a few kilobuses), L
e corresponds to the size of a typical “random coil” conformation (de Gennes, 1988).
Elastic Response of a Single Polymer.
Consider a single polymer subjected to a force F between its ends. Knowing that the spontaneous fluctuation of the end-to-end distance is L e indicates that if we force the ends of the chain to be a distance X apart, there will be a free energy cost of ∼k B T(X/L e)2 (assuming that X is small compared to L). This free energy cost is due to there being fewer configurations available to the chain (its entropy is reduced) if its ends are pulled apart. The derivative of the free energy with X is just the force required to obtain extension X, or F = k B TX/L e 2. A single flexible polymer thus displays linear entropic elasticity that is strictly of thermal origin. Since the unperturbed overall size of the polymer is L e, we define its strain as ε = X/L e; giving us F = (k B T/L e)ε.
Polymer Gel and Young's Modulus.
A polymer gel is a network of cross-linked polymers, swollen in a solvent that has affinity for the chains. (Think of a three-dimensional jungle gym with flexible links.) From the point of view of this paper, it is immaterial whether the gel is composed of many polymers cross-linked together (e.g., an agarose gel as prepared for electrophoresis), or a single tremendously long fiber cross-linked to itself many times (a conceivable model for a condensed chromosome). The relevant polymer length is the amount of chain between successive crosslinks. If this is more than a persistence length, each chain segment in the gel will display entropic elasticity, and therefore the network as a whole will be an elastic medium.
Suppose the average length of chain between cross-links is L. A simple estimate for the elasticity of a gel is obtained by summing up the elasticities of all the inter–cross-link segments, assuming each to be a random walk when no stress is placed on the network and further assuming each to be equally deformed when stress acts. Note that these assumptions immediately limit the maximum strain to ε = L/L
e = 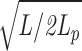 (the ratio of total length of a typical segment to its random walk size).
(the ratio of total length of a typical segment to its random walk size).
Given the distance between cross-links ξ, we can estimate the force per area, or pressure, corresponding to a given strain. Note that ξ may be less than L e; given one cross-link, the nearest cross-link may not be along the same chain. Consider a volume L e on a side. The number of chains in that volume is (Le/ξ)3, and therefore the number of chains per area is L e/ξ3. To have an overall strain ε, each chain must have a strain ε, so the force per area in the gel is p ≈ (k B T/ξ3)ε. Following the definitions above, the gel Young's modulus is Y = k B T/ξ3. Notice that this is precisely one k B T per cross-link, which gives the intuitively reasonable result that there is a free energy cost of k B T per constraint imposed on the network. (In general, “freezing” one microscopic thermally fluctuating degree of freedom costs k B T in free energy.)
If the polymers in the gel are thin filaments of cross-sectional radius R as above, then the fraction of space (volume fraction) φ taken up by the polymer is φ ≈ πR 2 L/ξ3. Thus, the gel Young modulus may be expressed in terms of the length of inter–cross-link fibers and the volume fraction as Y ≈ k B T φ/(πLR 2) (de Gennes, 1988). The latter formula with φ = 1 corresponds to the elasticity of a network after all solvent is removed and is a rough description of a piece of rubber.
Effect of Entanglements on a Gel.
Two chain segments in a gel may be entangled together, meaning that they may not be individually free to explore all random walk conformations thanks to unremovable links or knots. This is a poorly understood area of polymer statistical mechanics, but entanglements may be very roughly taken into account by considering each entanglement to contribute an additional cross-link to the network. If we suppose that there are n entanglements per chain segment in our gel, a very rough estimate of the gel modulus is Y ≈ k B Tφ(1 + n)/(πLR 2). This line of argument indicates that increased entanglement boosts the modulus of a gel (stiffens it); resolution of entanglements in a gel will reduce its modulus.
RESULTS
The large size of newt cells (∼100 μm) and of their mitotic chromosomes (∼20 μm long) make them ideal for micromanipulation. In the following, the term “chromosome” always refers to a pair of sister chromatids; all stretching experiments were performed on the two sisters together.
Two types of experiments are described below. The first was used to study large deformations and forces by stretching the chromosomes between the cell and a micropipette. The micropipette, when moved perpendicular to the stretching direction, was observed to bend. Calibration of this bending allowed simultaneous measurement of force and strain. The second was used to measure small deformation and forces by stretching the chromosome under suction inside the micropipette, the tip of which was corked by the chromosome. The elastic (Young) modulus was thus deduced.
Large Deformation of Chromosomes and Thin Fiber Formation
Large deformations were studied by exploiting the very strong adhesion of the chromosome to the pipette at one end and to the mass of other chromosomes at the other end. The chromosome is suspended between the pipette and the cell, in the culture buffer. The pipette is then moved to deform the chromosome by a given amount and brought back (Fig. 1).
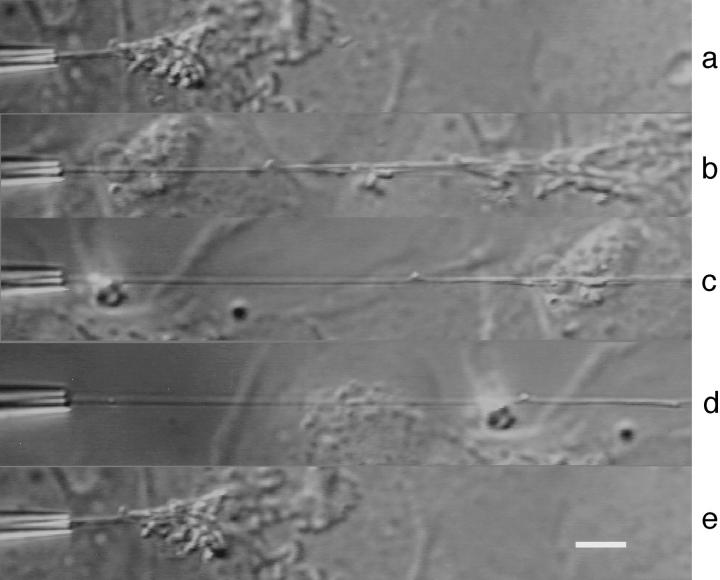
Figure 1.
The tip of a chromosome is grabbed inside the micropipette, and the chromosome is suspended between the ensemble of other chromosomes and the pipette (a). The portion between kinetochore and pipette is then progressively stretched by a factor of 10 (b–d). The micropipette is then brought back to the original position (e). No plasticity is observed, and the chromosome recovers its original length. The duration of the deformation–release cycle was 30 s. Bar, 10 μm.
The resulting shape of the chromosome depends on this deformation. We define the length of the chromosome after each cycle as the minimal length over which the chromosome is straight. (If the pipette is pushed closer to the cell, one observes bends or helicies along the chromosome.) We first observe that the final shape of the chromosome depends only on the imposed deformation. Exposure to the buffer for a long time (1 h) or restretching the chromosome by the same amount does not change the chromosome morphology.
The different chromosome morphologies obtained after different deformations can be categorized as follows:
Elastic Regime.
For ε < 10, the chromosome is highly elastic, relaxing to its initial “native” length and appearance (Fig. 2 a). Such a large range of elasticity is rare (most materials are elastic only for ε < 0.01) but is characteristic of polymer gels (Horkay et al., 1989) or of extensible elastic objects, such as a helical spring (Love, 1944). The elastic Poisson coefficient can be evaluated directly from the images (the diameter of the chromosome decreases as ε increases) giving 0.20 ± 0.05. This regime of perfect elasticity indicates that there is a well-defined elastic modulus for a chromosome.
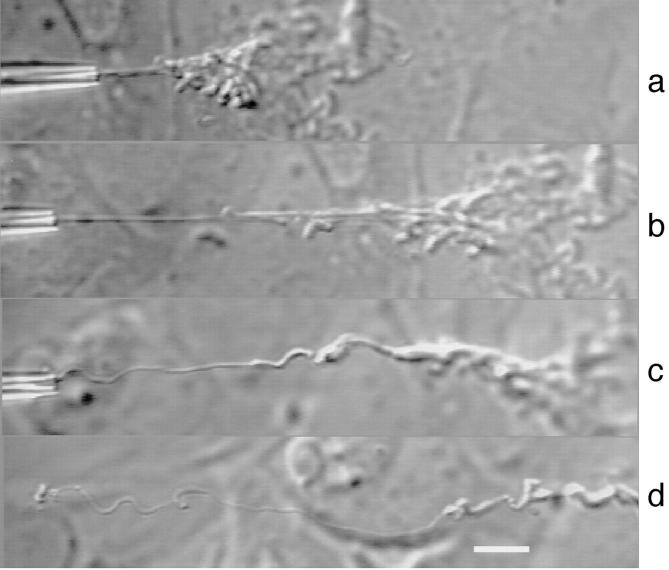
Figure 2.
The state of the chromosome after different cycles of deformation– release, when the pipette is brought back near the cell. (a) ε = 10, similar to Fig. 1 e; (b) ε = 15; (c) ε = 20; and (d) ε = 55. During the last cycle (bottom), the chromosome broke and was released from the pipette. The shape observed is stable over at least 1 h. Bar, 10 μm.
Plastic Regime.
When ε is larger than 10, the relaxed chromosome is longer than its original length; it no longer returns to its native state (Fig. 2, b–d). For example, a chromosome with native length of 20 μm, after stretching to 300 μm (ε = 14), relaxes to a length of about 30 μm.
This increase in length is inhomogeneous: Part of the chromosome remains thick while part of it is thinned. The border between these two regions is usually at the kinetochore. In contrast to the elastic regime, the length and diameter after plastic deformation vary with ε. The thinned chromosome diameter decreases from 3 to ∼0.8 μm as ε is increased from 10 to 25–35 (depending on the rate of deformation).
Fig. 3 shows an interesting aspect of the plastic behavior for ε ≈ 20. When the micropipette tip is brought back near the cell so as to slightly compress the chromosome, transient undulations develop along parts of the chromosome. Those undulating regions relax into a region of thicker straight chromosome after 1 to 2 s. If the pipette is pushed closer, stable helices appear (Fig. 4). Some but not all of the plastic deformation can be reabsorbed by the chromosome.
Figure 3.
After a chromosome elongation of 20 times, the pipette was brought back near the cell. An undulating shape appeared near the tip (a), which relaxed (b and c) to a straight line (d) after 1.5 s (photographs taken every 0.5 s). Bar, 10 μm.
Figure 4.
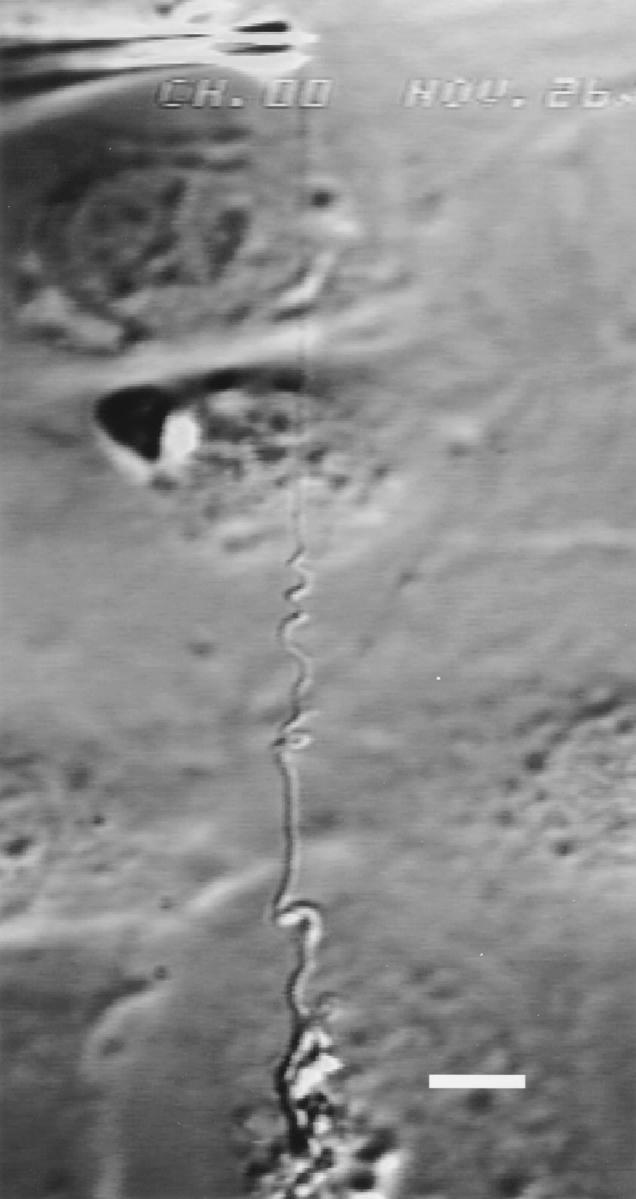
Stable helix formed after a plastic deformation. Bar, 10 μm.
Thin Filament.
For ε larger than ∼ 30, the relaxed chromosome converts to a thin filament that is 15 times the length of the original thick chromosome. Further elongation proceeds by elastic deformation of this thin filament.
The conversion of the chromosome to thin filament is not only irreversible, but it also depends on the rate of deformation. Most of our cycles of deformation–release are done at speeds of less than 20 μm/s. At higher speed, the transition is abrupt, with part of the chromosome suddenly relaxing while the remainder extends to thin filament.
Finally, for ε ≈ 100, the filament breaks. (The precise value of ε at breaking depends on rate of elongation, ranging from 60 to 100; Fig. 5.) The deflection of the micropipette at the breaking point indicates a force between 90 to 150 nanonewtons. It is not easy to deduce the Young modulus of the thin filament from the force measurement. The thickness d of the thin filament when highly stretched is at the resolution limit of the microscope, and its measurement is critical for the Young modulus [Y thin ≈ F/(εd 2)]. Our best estimate is Y thin = 1–5 × 105 Pa.
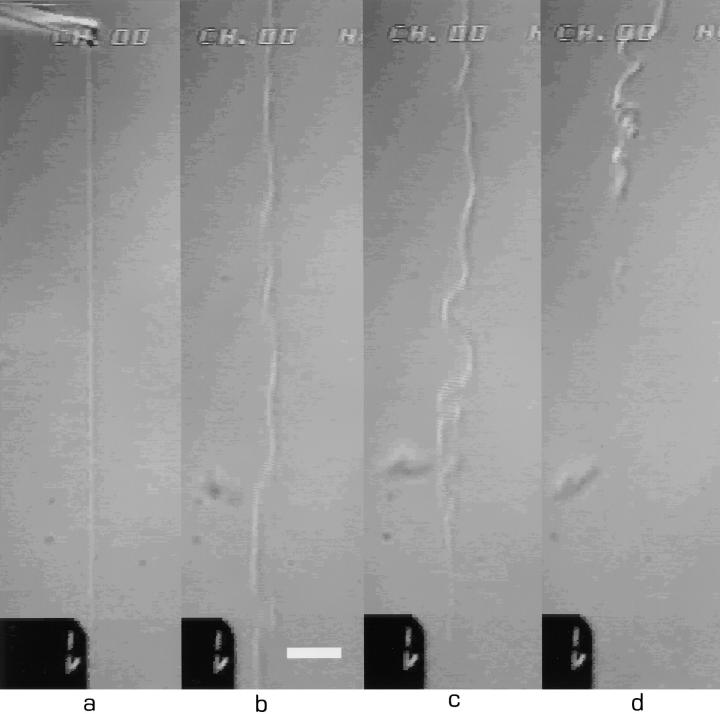
Figure 5.
Breakage of the thin filament after 75 times extension. Note the irregular undulating shape that appeared. (a) t = 0.0 s; (b) t = 0.2 s; (c) t = 0.5 s; and (d) t = 0.8 s. Because of the fast breakage and relaxation dynamics, it is hard to keep the filament in focus. Bar, 10 μm.
When the tension on the thin filament is released, following either partial or total conversion of the chromosome to this form, portions of it take on a helical shape. Each helical turn involves 5 to 10 μm of relaxed thin filament. Both left- and right-handed curls were observed. Fig. 6 a shows a thin filament after its breakage. If we then perturb it with a flow, the helical filament behaves like a spiral spring (Fig. 6, b–d).
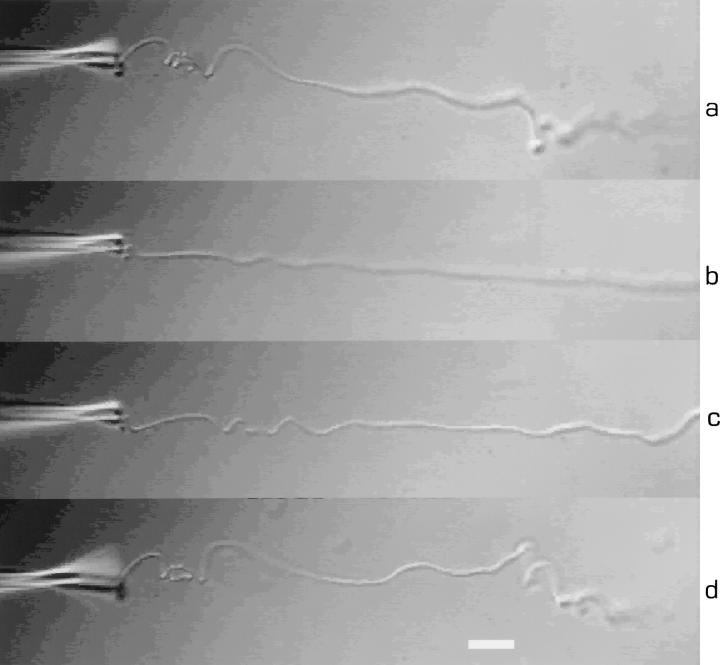
Figure 6.
A broken thin filament attached to the pipette and deformed by flow. From top to bottom: (a) the original shape; (b) the filament stretched by the flow (the flow was produced by movement of the XY stage relative to the pipette); (c) 1.4 s after flow stopped; and (d) 2.8 s after flow stopped. Bar, 10 μm.
Young Modulus Measurements
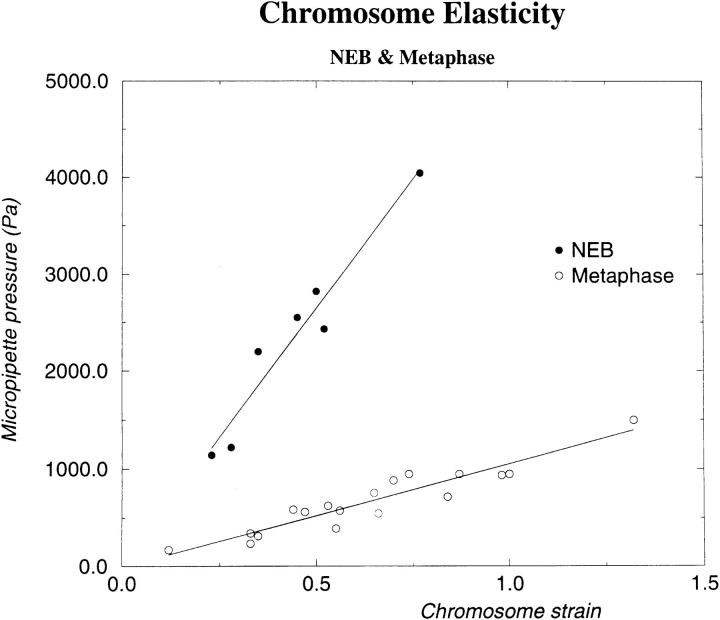
We now return to the elasticity regime where the deformations are relatively small (ε < 2). Using aspiration of chromosomes, we measured the deformation as a function of pressure inside the pipette (Fig. 7). Results for 10 metaphase chromosomes are shown in Fig. 8; at metaphase Y meta = 1,000 ± 200 Pa. As mentioned above, the Poisson ratio of metaphase chromosomes was measured to be 0.20 ± 0.05. Thus, the metaphase chromosome has a Young modulus about two orders of magnitude less than that of the thin filament.
Figure 7.
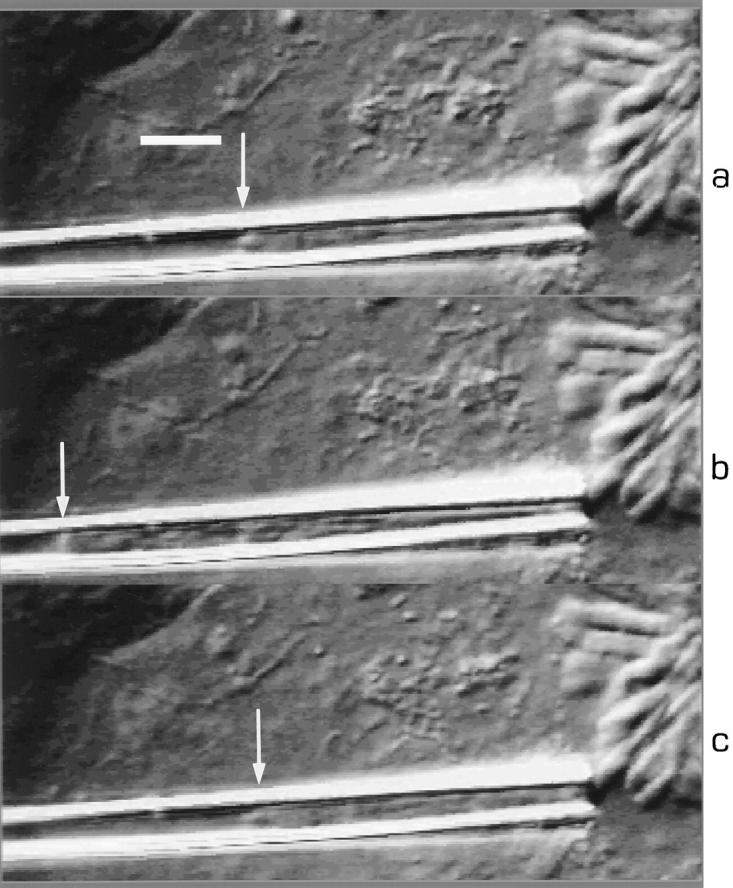
Young modulus measurements: deformation of the chromosome inside the pipette under aspiration. Deformation is measured as a function of the pressure difference (ΔP) between inside the pipette and the culture medium. From top to bottom: (a) ΔP = 0; (b) ΔP = 44 Pa. Arrows indicate the tip of the chromosome. Bar, 5 μm.
Figure 8.
Young modulus measurement. Pressure vs. deformation for chromosome just after NEB (closed circles) and at metaphase (open circles). The slope of the curves is the Young modulus. Y NEB = 5,000 ± 1,000 Pa; Y meta = 1,000 ± 200 Pa.
The same type of experiment was also carried out for cells at prometaphase, immediately following (<10 min) nuclear envelope breakdown (NEB).1 Data from three NEB chromosomes are shown in Fig. 8; Y NEB = 5,000 ± 1,000 Pa. Between NEB and metaphase, the chromosome Young modulus ranges from 5,000 to 1,000 Pa, but it is difficult to follow this variation since the duration of mitosis varies from cell to cell.
DISCUSSION
Let us summarize our observations. Metaphase chromosomes have a Young modulus of 1,000 Pa and are elastically deformable by up to 10 times. Under higher stress, they are transformed into a thin fiber that is 15 times the native chromosome in length. The thin filament is itself elastically deformable by at least a factor of six, and its Young modulus is in the range of 1–5 × 105 Pa.
In the following, we present a model for the structure of thick chromosome and thin filament consistent with our observations. We also discuss the variation of the Young modulus during the cell cycle. We finally show that the shape of the anaphase chromosome allows us to deduce the resistance of the cytoplasm to the motion once the Young modulus is known.
Chromosome Structure
Remarkably, the metaphase chromosome returns to its original size after being lengthened by a factor of 10. There are two classes of materials that exhibit elasticity over such a large range of extension.
Chromatin Gel.
The first possibility for the structure of the metaphase chromosome is that of a polymer gel, i.e., where the length is stored in random thermal fluctuations of a cross-linked network of flexible chains. Since each chromatid is composed of a single contiguous chromatin fiber, chromatin is known to be flexible (recent experiments indicate a persistence length of roughly L p ≈ 50 nm [Castro, 1994]), and it is known that the chromatin is periodically constrained (i.e., to form the “loops” discussed by many authors [Paulson and Laemmli, 1977, Gasser et al., 1986; Cook et al., 1990; Saitoh and Laemmli, 1993]), it behooves us to consider whether chromosome elasticity can be plausibly attributed to elasticity of a chromatin gel, rather than to the rigidity of some internal “scaffold.” Stretching a gel forces the chains to extend, reducing their entropy, thus requiring work to be done. The maximum extension possible for a polymer gel is roughly the ratio of the length of the chains between cross-links to the random walk size of the chains (see Materials and Methods).
Our measurement of the Young modulus (1,000 Pa) rules out the possibility that the initial elasticity is that of a chromatin gel. To see this, consider that the observed extensibility by 10 times requires that the length of fiber between cross-links be L = 200L p ≈ 10 μm (Materials and Methods). Such a gel will have a Young modulus of roughly φk B T/(πLR 2), where φ is the fraction of volume occupied by the polymer. (The remaining fraction 1 − φ is supposed to be occupied by “solvent,” or more precisely the fraction of cytosol that can freely enter and leave the spaces between chromatin fibers.) Plugging in the required L = 10 μm, setting φ = 1 (this maximizes the modulus estimate) and using k B T = 4 × 10−21 J and R = 15 nm, we find Y gel ≈ 0.6 Pa. The large L required by the observed extensibility necessitates a tiny elastic modulus. The observed elasticity is not plausibly due to that of the constituent chromatin and therefore must be due to the springlike elasticity of a coiled or folded internal fiber.
Folded or Coiled Fiber.
The second possibility is that of a fiber with permanent bends along its length. The permanent bends store a large amount of length that can be liberated by tension. The simplest example of such a structure is a regular helical spring, but one could envision a mixture of left- and right-handed helical turns, or a zigzag structure.
This case is compatible with the idea of a folded or coiled chromosome scaffold (Paulson and Laemmli, 1977; Gasser et al., 1986; Boy de la Tour and Laemmli, 1988; Saitoh and Laemmli, 1993; Hirano and Mitchison, 1994) and is consistent with several of our observations. First, we observe a sharp transition from thick chromosome to thin fiber; the transition is sharpened by rapid extension. Second, after relaxation, the thin fiber is mechanically stiff and has a permanent undulating shape. Third, a geometrical argument can be advanced to support the picture of a thin filament that is bent or wrapped into the metaphase chromosome, as follows. Suppose the thin filament of diameter d is wrapped into N successive solenoidal turns of diameter D that form the thick chromosome. The length of the thick chromosome is then L = Nd, while the length of the filament l = NπD, so l/L = πD/d. This is consistent with our observed elongation l/L = 15 and diameter reduction D/d = 4, when the thick chromosome is converted to thin filament.
Thin Filament Structure
For ε > 10, metaphase chromosomes were permanently lengthened. Supposing that the metaphase chromosome is composed of a folded or coiled filament, this plasticity and the extraction of the thin filament is to be interpreted as the progressive failure of structural elements that define its intrinsic shape. Eventually, the entire chromosome is converted into a thin filament that displays elasticity over a sixfold range of extensions and a Young modulus of Y thin = 1–5 × 105 Pa.
We are thus faced with a second case of a large range of elasticity. Since Y thin ≈ 102 Y meta, we can reuse the argument of the previous section to again argue against the thin filament being a polymer gel. The thin filament itself should therefore be constituted of a “basic fiber” that is wrapped or folded so as to again provide a reservoir of length.
In the experiments described above, the two sister chromatids were always deformed together, while our discussion of their unfolding considers them as independent. In the initial elastic regime, the main question is of the contribution of interchromatid entanglement to the Young modulus. In the next subsection, we will see that this is at most a small part of the measured Young modulus. During the plastic deformation, the permanent and irreversible bends that form may indicate that the original compactions of the two underlying sister thin filaments are correlated. An observation due to Boy de la Tour and Laemmli (1988) of opposite helical handedness of sister chromatids indicates the same conclusion. On the other hand, the helical shape of the thin filament may reflect that the two chromatids have been forced to uncoil during their extension (possibly in conjunction with the constraint that they stay alongside one another) and when released have refolded in some way unrelated to their native folding. This direction of argument again suggests that the helical shape of the thin fiber comes from the necessity that the chromatids be uncoiled to be lengthened. Further experiments are needed to clarify this issue. It would be very interesting to study single chromatids assembled from Xenopus egg extracts, for which there is some evidence for a helical “scaffold” (Hirano and Mitchison, 1994).
Decrease of Young Modulus during Mitosis
We observed that the chromosome Young modulus decreased from 5,000 Pa at NEB to 1,000 Pa at metaphase. This gradual weakening of the chromosome can simply be explained by supposing a structural change in the internal scaffold. Just to raise one possibility, it is known than 70% of the topoisomerase II present at NEB is progressively removed before anaphase (Sweldow et al., 1993). This removal may reduce the chromosome stiffness.
A second possibility is that resolution of entanglements of the sister chromatids during NEB to metaphase might weaken the chromosome elasticity. The maximum change in Y that can be possibly obtained is (L/L p)k B T/(πLR 2) = k b T/ (πL p R 2). This supposes maximal entanglement (one per persistence length of chromatin, certainly a large overestimate) at NEB and total disentanglement at metaphase. This extreme estimate only accounts for an ∼100 Pa change in Y. We conclude that the observed change in Y is due to a structural change of the bent scaffold.
Young Modulus and the Shape of Anaphase Chromosomes
This paper was mainly dedicated to the elasticity of the chromosomes as a probe of their internal structure, but our results also allow us to solve an apparent paradox concerning the forces exerted during anaphase.
Nicklas (1983) measured that the spindle exerts a force of ∼700 piconewtons on a chromosome at anaphase. This level of force was puzzling because previous calculations (Nicklas, 1965; Taylor, 1965) had shown that even assuming a cytoplasm viscosity as large as 1 Poise, the force needed to move a chromosome at 0.3 μm/min would be only about 0.1 piconewton. The spindle is thus applying a force 104 times larger than might be expected. To explain this paradox, Nicklas considered a few mechanisms, including a feedback system to limit chromosome velocity.
The basic physical facts indicate that these large forces are really required to move chromosomes. The shape of the chromosome, its Young modulus, and the force exerted by the cytoplasm are closely related. An elastic cylinder pulled by its center and moving through any medium is submitted to a drag force that tends to bend it. At constant speed, the force exerted by the medium equals the force that pulls the object. The bigger the force, the more severe the bending. In the case of high drag force, a cylindrical object takes a U shape, which is what is observed for chromosomes pulled at their kinetochore by the spindle. The distance X between the arms of the U, the exerted force F 0, the Young modulus Y, and the radius R of the chromosome are related by F 0 = 6πYR 4/X 2, which can be obtained using classical elasticity theory (see Appendix).
Note that this result can be estimated by dimensional analysis: The bending modulus B ≈ YR 4 and the distance X are the only quantities in the problem, and the only way to form a force from them is the ratio B/X 2. To obtain this relation, no assumption about the effective viscosity of the cytoplasm, or even whether or not it has Newtonian behavior, is required; only the fact that there is a drag (friction) force in balance with the driving force is invoked.
For newt chromosomes, X ≈ 2 μm, R ≈ 0.7 μm, and Y = 1,000 Pa, so the force exerted by the spindle, and therefore the total resistance presented by the surrounding medium, is ∼1,000 piconewtons. This means that the resistance of the cytoplasm to the movement of the chromosome is orders of magnitude higher than what would be encountered in a 1-Poise viscous fluid, as supposed by Nicklas and Taylor. This is not surprising if one considers that a large object like a chromosome has to move by deformation of a network of cytoplasmic filaments. The drag force on a large object could consequently be much larger than the drag inferred from motion of small molecules.
It should be remarked that the bending modulus of the chromosome has not been directly measured, but rather deduced from the Young modulus. The relation B = πYR 4/4 holds for uniform solids because bending causes the same kind of local deformations as stretching. (Note that on the outer edge of a bend, an object is stretched, while on the inner edge, it is compressed.) However, one can imagine a chromosome with a very flexible yet inextensible filament running down its center, or a chromosome that is predominantly liquid. Our observations of elastic and plastic behavior make these possibilities seem unlikely, but of course they must be checked experimentally. We are planning a direct measurement of the bending modulus to definitively settle this point.
Acknowledgments
This work was supported by National Science Foundation grant PHY9408995 and the Mather Foundation. J. Marko also acknowledges support of the Meyer Foundation at Rockefeller University (New York).
APPENDIX
Computation of the Resistive Force during Anaphase
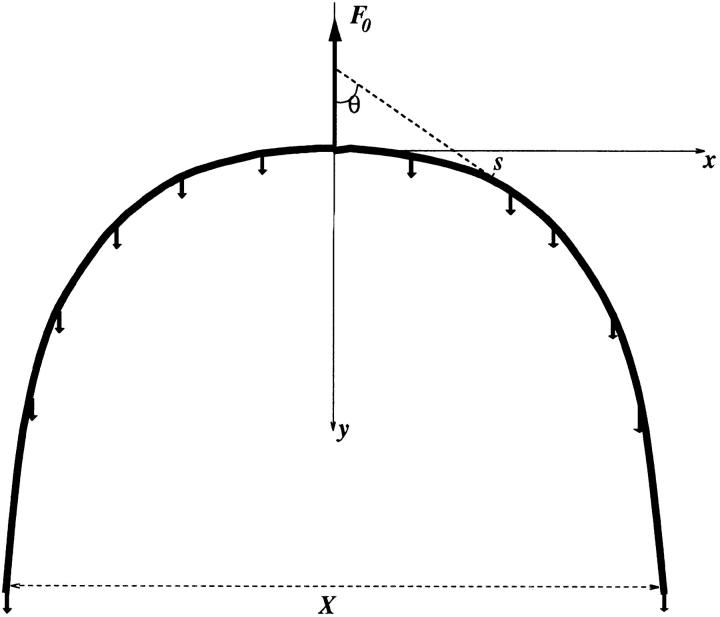
The relation F 0 = 6πYR 4/X 2 introduced in the last section relates drag forces to the U shape of the chromosome during anaphase movement. In this appendix, three routes to this equation are described, in order of increasing detail. We consider an elastic rod of length 2L , pulled at its central point (centromere) by a (spindle) force F 0. The rod experiences resistive (drag) forces K distributed along its length (K is a force per length of rod), which when summed balance F 0 (Fig. A1). Under the action of these forces, the rod bends to form a U shape.
Figure A1.
Bending of an elastic rod due to drag forces distributed along its length.
Our first estimate will be the simplest and least satisfying. We consider the situation where the force is large enough that the rod is tightly bent near its center, with its nearly parallel arms lagging behind. The drag forces will be spread nearly evenly along the arms, so we can estimate that K ≈ F 0/L in order for total drag force to balance the spindle force. Arguing that the deformation and therefore the elastic energy is concentrated at the hairpin bend and the radius of the bend is approximately X, we estimate the elastic energy to be E ≈ B/X. The length of rod in the bend is ∼X and the curvature is ∼1/X (see Materials and Methods). Since the bend is made by forcing the ends of the bent region together by a distance ∼X, the (drag) force that must be applied to make such a bend should be just E/X, or B/X 2. Thus, we arrive at a relation between the total drag force ∼KL, which is in balance with the spindle force F 0, and X: F 0 ≈ KL ≈ YR 4/X 2.
The first estimate is essentially dimensional analysis and does not address the complications that X is not precisely given by the radius of curvature of the hairpin bend and that both drag forces and the deformation are not uniformily spread along the entire length of chromosome. The complete equations of elastic equilibrium for the rod are (Landau and Lifshitz, 1986)
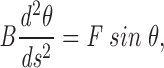 |
A1 |
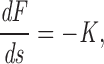 |
A2 |
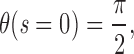 |
A3 |
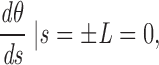 |
A4 |
where s is the contour length along the rod (s = 0 corresponds to the middle of the rod, or centromere), θ is the angle between the y-axis and the tangent to the rod at point s, F(s) is the net external force acting on the rod from point s to its free end L, B is the bending modulus (for a cylinder of radius R, B = πYR4/4; see Materials and Methods), K is the density of external forces acting on the rod at point s, and
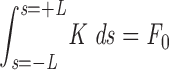
(see Fig. A1). Note that as the applied forces are symmetric, one needs only to solve these equations for one half of the rod (i.e., for 0 < s < L), with the equivalent boundary condition that the rod be “clamped” at s = 0.
Before going to the complicated case in which K is spread along the entire rod, we solve these equations for a simpler case which the drag force is concentrated at the lagging ends. Suppose that instead of resistive forces distributed along the rod, we have drag forces of F 0/2 (note that the total drag must balance the spindle force F 0) applied to the ends (s = ±L). This simple case is the classic problem of a rod clamped to a table and bent by a weight hung from its free end (Landau and Lifshitz, 1965; Sec. 19, problems 1 and 2), which has exact solution
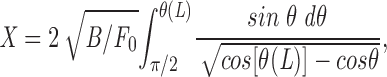 |
A5 |
where θ(L) is the angle between the tangent to the rod at the extremity (S = L). Replacing B and computing the integral,
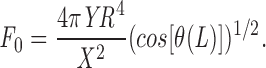 |
A6 |
For strong pulling, the rod ends will be forced to be along the y-axis, giving cos [θ(L)] ≈1. (The precise value of cos [θ(L)] can of course be computed from the exact solution.) So, the magnitude of the resistive force can thus be deduced from the bending of the rod. As before, we find that F 0 ≈ YR 4/X 2.
Finally, consider the case of a chromosome pulled during anaphase toward the pole. As the migration speed is constant, the pulling force and the drag forces balance, and the chromosome bends. The amplitude of these forces can be inferred from the bending since we know the elasticity constant from other measurements. The problem is to choose a reasonable model for the drag forces. As the chromosome has to pass through a network of filaments, the frictional forces applied to a small segment of it plausibly depend on its projected length in the x direction, or
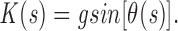 |
A7 |
This accounts for the effect that a segment of chromosome will experience a bigger force if it is parallel to the x-axis (with its tangent perpendicular to velocity) than if it is parallel to the y-axis (tangent parallel to velocity). The coefficient g depends on the deformability of the filament network, on the speed of the chromosome, and other shape-independent parameters.
Using this expression for the drag force density, integrating the differential equations A1 and A2 and using boundary conditions in Eqs. A3 and A4, we find
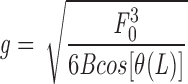 |
A8 |
and
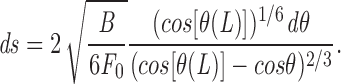 |
A9 |
The distance X between the arms is
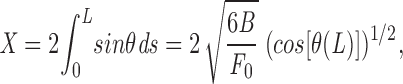 |
A10 |
which again reduces to F 0 ≈ YR4/X 2 for strong pulling (cos [θ(L)] ≈ 1).
The value of the drag force is not very sensitive to the choice of the form of the force density function K(s); the extreme choice of all drag force concentrated at the telomeres gave the same magnitude of spindle force and bending. It should be noted that Nicklas (1965) has observed that the velocity of chromosomes during anaphase is independent of their length. This is in good agreement with our model: The value of the spindle force required to move the chromosomes is independent of the chromosome's length L, as long as L ≫ 2(B/F 0)1/2 ≈ 1 μm for newt chromosomes.
In summary, the force which resists chromosome movement during anaphase is also responsible for chromosome bending and can be deduced by measuring their deformation.
Footnotes
1. Abbreviation used in this paper: NEB, nuclear envelope breakdown.
Address all correspondence to Bahram Houchmandzadeh, CNRS, Laboratorie de Spectrométrie Physique, BP 57, 38402 Saint-Martin-d'Hères Cedex, France. Tel.: (33) 476 51 44 27. Fax: (33) 476 51 45 44. E-mail: bahram@coucou.ujf-grenoble.fr
We warmly thank M. Elbaum who developed the micromanipulation set-up, and S. Childress, M. Goulian, T. Hirano, H. Macgregor, W. Marshall, P. Moens, Y. Rabin, E. Siggia, and J. Swedlow for many helpful discussions.
REFERENCES
- Bak AL, Zeuthen J, Crick FHC. Higher order structure of human mitotic chromosomes. Proc Natl Acad Sci USA. 1977;74:1595–1599. doi: 10.1073/pnas.74.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS, Sedat JW, Agard DA. A three-dimensional approach to mitotic chromosome structure: evidence for a complex hierarchical organization. J Cell Biol. 1987;105:77–92. doi: 10.1083/jcb.105.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS, Braunfeld MB, Sedat JW, Agard DA. Large scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma (Berl) 1989;98:129–143. doi: 10.1007/BF00291049. [DOI] [PubMed] [Google Scholar]
- Boy de la Tour E, Laemmli UK. The metaphase scaffold is helically folded. Cell. 1988;55:937–944. doi: 10.1016/0092-8674(88)90239-5. [DOI] [PubMed] [Google Scholar]
- Castro, C. 1994. Measurement of the elasticity of single chromatin fibers: the effect of histone H1. Ph.D. thesis. University of Oregon, Eugene, OR. 142 pp.
- Claussen U, Mazur A, Rubstov N. Chromosomes are highly elastic and can be stretched. Cytogenet Cell Gen. 1994;66:120–125. doi: 10.1159/000133681. [DOI] [PubMed] [Google Scholar]
- Cook PR, Jackson DA, Dickinson P. The size of chromatin loop in HeLa cells. EMBO (Eur Mol Biol Organ) J. 1990;9:567–571. doi: 10.1002/j.1460-2075.1990.tb08144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR. A chromomeric model for nuclear and chromosome structure. J Cell Sci. 1995;108:2927–2935. doi: 10.1242/jcs.108.9.2927. [DOI] [PubMed] [Google Scholar]
- de Gennes, P.G. 1988. Scaling concepts in polymer physics. Ph. D. thesis. Cornell University, Ithaca, NY.
- Gasser SM, Laroche T, Falquet J, Boy de la Tour E, Laemmli UK. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986;188:613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Horkay F, Hecht AM, Geissler E. The effect of crosslinking on the equation of state of a polymer solution. J Chem Phys. 1989;91:2706–2711. [Google Scholar]
- Landau, L.D., and E.M. Lifshitz. 1986. Theory of Elasticity. Pergamon Press, Tarrytown, NY. 264 pp.
- Love, A.E.H. 1944. A treatise on the mathematical theory of elasticity. Dover, New York. 644 pp.
- Manuelidis L. A view of interphase chromosomes. Science (Wash DC) 1990;250:1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. Chromosome velocity during mitosis as a function of chromosome size and position. J Cell Biol. 1965;25:119–135. doi: 10.1083/jcb.25.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97:542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuki Y. Structure of chromosomes. I. Morphological studies of the spiral structure of human somatic chromosomes. Chromosoma (Berl) 1968;25:402–428. doi: 10.1007/BF02327721. [DOI] [PubMed] [Google Scholar]
- Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Hard R. Newt lung epithelial cells: cultivation, use, and advantages for biomedical research. Int Rev Cytol. 1990;122:153–220. doi: 10.1016/s0074-7696(08)61208-5. [DOI] [PubMed] [Google Scholar]
- Saitoh Y, Laemmli UK. From the chromosomal loops and the scaffold to the classic bands of metaphase chromosomes. Cold Spring Harbor Symp Quant Biol. 1993;58:755. doi: 10.1101/sqb.1993.058.01.083. [DOI] [PubMed] [Google Scholar]
- Sedat J, Manuelidis LA. A direct approach to the structure of mitotic chromosomes. Cold Spring Harbor Symp Quant Biol. 1978;42:331–350. doi: 10.1101/sqb.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- Swedlow JR, Sedat JW, Agard DA. Multiple chromosomal populations of topoisomerase II detected in vivo. Cell. 1993;73:97–108. doi: 10.1016/0092-8674(93)90163-k. [DOI] [PubMed] [Google Scholar]
- Taylor EW. Brownian and saltatory movements of cytoplasmic granules and the movement of anaphase chromosomes. Proc Int Congr Rheology. 1965;4:175–191. [Google Scholar]