Abstract
The chloroplast genome of all higher plants encodes, in its large single-copy region, a conserved open reading frame of unknown function (ycf3), which is split by two group II introns and undergoes RNA editing in monocotyledonous plants. To elucidate the function of ycf3 we have deleted the reading frame from the tobacco plastid genome by biolistic transformation. We show here that homoplasmic Δycf3 plants display a photosynthetically incompetent phenotype. Molecular analyses indicate that this phenotype is not due to a defect in any of the general functions of the plastid genetic apparatus. Instead, the mutant plants specifically lack detectable amounts of all photosystem I (PSI) subunits analyzed. In contrast, at least under low light conditions, photosystem II subunits are still present and assemble into a physiologically active complex. Faithful transcription of photosystem I genes as well as correct mRNA processing and efficient transcript loading with ribosomes in the Δycf3 plants suggest a posttranslational cause of the PSI-defective phenotype. We therefore propose that ycf3 encodes an essential protein for the assembly and/or stability of functional PSI units. This study provides a first example for the suitability of reverse genetics approaches to complete our picture of the coding capacity of higher plant chloroplast genomes.
The complete sequence analysis of two chloroplast genomes ten years ago (20, 29) marks a milestone in plastid genetics and has had a profound influence on our understanding of the structure and function of plant organellar genomes. Detailed computer analyses of the sequence data (41) allowed the identification of numerous regions potentially encoding novel proteins. In the following years, most of these open reading frames could be assigned to functional gene products involved in either genetic system functions or in photosynthesis. However, there are about 10 conserved reading frames left, the functions of which are still elusive. One of them is a reading frame of 168 (tobacco) or 170 (maize) codons located in the large single-copy region of higher plant chloroplast genomes and interrupted by two group II introns. Referring to this remarkable feature, it was initially designated IRF168 (intron-containing reading frame of 168 codons) (28), but later renamed ycf3 (hypothetical chloroplast reading frame No. 3).
Several lines of evidence suggest that ycf3 encodes a functional gene product. First, the reading frame is conserved in all land plant chloroplast genomes (15) and displays a high degree of DNA homology as well as putative protein sequence homology (23). ycf3 homologues are also present in the plastid genomes of several algae (13, 22, 35) and in cyanobacteria (39). Second, ycf3 is actively transcribed, most probably as part of a polycistronic transcription unit, the synthesis of which initiates upstream of rps4 (see Fig. 1 A) (16, 23). Third, the ycf3 primary transcript undergoes a series of mRNA maturation events: cleavage into its monocistronic form, excision of two group II introns, and RNA editing at two sites in Zea mays (23). Both editing events restore conserved amino acid residues and were shown to occur very early after transcription and independent of the other RNA processing steps (23).
Figure 1.
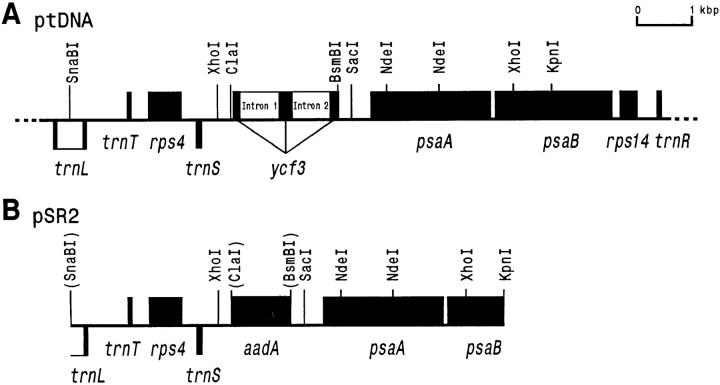
Experimental strategy for targeted replacement of the ycf3 reading frame. (A) Map of the plastid DNA region containing ycf3. Genes above the line are transcribed from left to right; genes below the line are transcribed in the opposite direction. Restriction sites relevant for vector construction, RFLP analysis, or generation of hybridization probes are marked. Introns are shown as open boxes. (B) Map of the plastid DNA fragment in the final transformation vector pSR2. A chimeric spectinomycin resistance gene (aadA) replaces ycf3. Restriction sites eliminated by ligation with different half-sites are shown in parentheses. Note that the aadA gene is transcribed in the same direction as ycf3 in the cognate sequence of the plastid genome.
Though circumstantial, all of this evidence supports the assumption that the mature ycf3 mRNA is translated into a functional polypeptide. However, the lack of homology to any known gene does not allow predictions as to the function of this putative gene product. To address this problem directly, we have taken a reverse genetics approach to reveal the phenotype of plants deficient for ycf3. This approach was made feasible by the development of a technology for genetic transformation of higher plant plastids (36, 37). Over the past few years, a number of studies have demonstrated the great value of chloroplast transformation for investigating virtually all aspects of plastid gene expression in vivo. For example, this technology has been successfully employed to study transcriptional and posttranscriptional regulation (1, 31, 33), RNA editing (5–7), splicing (4) and DNA replication (32). In this study, we have attempted to make use of plastid transformation to uncover the function of the conserved chloroplast open reading frame ycf3.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Sterile tobacco plants (Nicotiana tabacum) were grown on agar-solidified MS medium (19) containing 30 g/liter sucrose. Homoplasmic transplastomic lines were rooted and propagated on the same medium. For protein isolation and physiological measurements, transformed plants were kept under low light conditions (0.4–0.5 W/m2) to minimize photooxidative damage in the mutant chloroplasts.
List of Oligonucleotides
The following synthetic oligonucleotides were employed in this study:
P10 5′-AACCTCCTATAGACTAGGC-3′
P11 5′-AGCGAAATGTAGTGCTTACG-3′
P31 5′-ATGTCACATTCAGTAAAGAT-3′
P32 5′-TCAATAAGCTAGACCCATAC-3′
P33 5′-CCCTTCTATGACAAATTTGA-3′
P34 5′-CCAGCGGATCTAAACAATCT-3′
P35 5′-GGTTTTTCAATGCGAGATCTA-3′
P36 5′-CATGACAATAACTAGAATGAA-3′
Construction of a Δycf3 Plastid Transformation Vector
The region of the tobacco chloroplast genome containing the ycf3 reading frame was excised from a SalI ptDNA clone (provided by P. Maliga, Piscataway, NJ) as a KpnI/SnaBI fragment corresponding to nucleotide positions 40,465–49,586 (29). The fragment was ligated into a Bluescript KS vector (Stratagene, La Jolla, CA) cut with KpnI and Ecl136II, generating plasmid pSR1. The ycf3 reading frame was subsequently deleted by digestion with ClaI and BsmBI. ClaI cuts 116 nucleotides upstream of the ycf3 start codon within the 5′-untranslated region (nucleotide position 46,424). The BsmBI site is located close to the end of the ycf3 coding region, 17 nucleotides upstream of the termination codon. After a fill-in reaction of the recessed ends with Klenow DNA polymerase, a chimeric aadA gene conferring resistance to aminoglycoside antibiotics (36) was inserted to replace ycf3 and to facilitate selection of chloroplast transformants. A plasmid clone carrying the aadA gene in the same orientation as previously ycf3 yielded the final transformation vector pSR2 (see Fig. 1 B).
Plastid Transformation and Selection of Homoplasmic-transformed Tobacco Lines
Young leaves from sterile tobacco plants were bombarded with plasmid pSR2–coated tungsten particles using the DuPont biolistic gun (PDS1000He; BioRad, Hercules, CA) (12, 36). Primary spectinomycin-resistant lines were selected on RMOP regeneration medium containing 500 mg/liter spectinomycin dihydrochloride (37). Plastid transformants were identified by PCR amplification according to standard protocols using the primer pair P10 (complementary to the psbA 3′-untranslated region of the chimeric aadA gene) and P11 (derived from the 3′ portion of the aadA coding region). Three independent transplastomic lines were subjected to four additional rounds of regeneration on RMOP/spectinomycin to obtain homoplasmic tissue. Homoplasmy was verified by DNA gel blot analysis (see Fig. 2).
Figure 2.
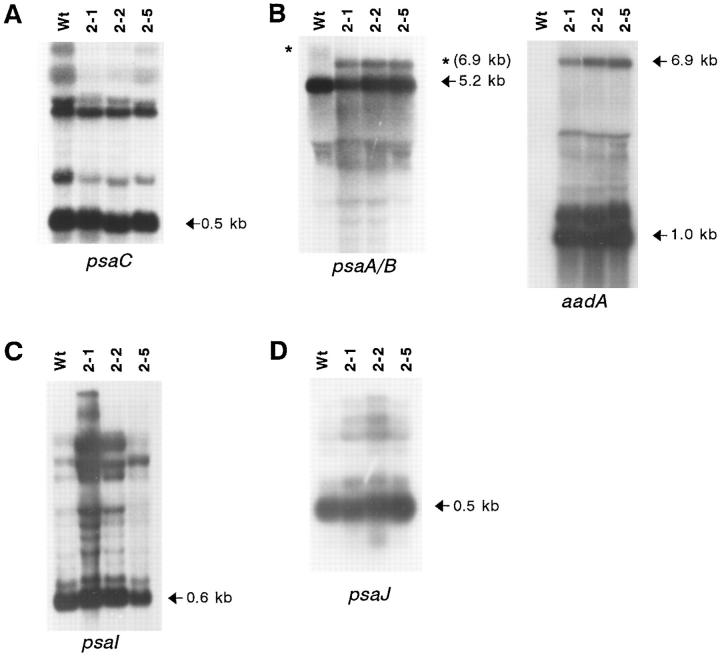
RFLP analysis to verify chloroplast transformation and homoplasmy of the Δycf3 plants. Total cellular DNA from wild-type plants and from three independently transformed lines (Nt-pSR2-1, Nt-pSR2-2, and Nt-pSR2-5, subsequently referred to as 2-1, 2-2, and 2-5) was digested with XhoI and hybridized to the radiolabeled SacI/XhoI fragment covering the region downstream of the ycf3 reading frame (i. e., the psaA gene and the 5′ portion of psaB; Fig. 1). The probe detects a 5.6-kb fragment in wild-type plants (corresponding to nucleotide positions 40,883 to 46,524; 29; Fig. 1) and a 4.9-kb fragment in the transplastomic lines. Absence of the 5.6-kb signal in the lanes representing the Δycf3 plants indicates a uniformly transformed population of plastid DNA molecules.
Isolation of Nucleic Acids and Hybridization Procedures
Total plant DNA was isolated according to a rapid miniprep procedure (8). Total cellular RNA was extracted using the TRIzol reagent (GIBCO BRL, Paisley, Scotland). Restriction enzyme–digested DNA samples were separated on 0.8% agarose gels and blotted onto Hybond N nylon membranes (Amersham Intl., Little Chalfont, UK) using standard protocols (24). Total cellular RNA or polysome-bound RNA was electrophoresed on formaldehyde-containing 0.8–1.5% agarose gels and transferred onto Hybond N+ membranes. For hybridization, α[32P]dATP–labeled probes were generated by random priming (Boehringer Mannheim, Mannheim, Germany) following the instructions of the manufacturer. A radiolabeled SacI/XhoI restriction fragment (corresponding to nucleotide positions 43,807–40,883 in the tobacco chloroplast genome) (29) was used as probe for the restriction fragment–length polymorphism (RFLP) analysis. Tobacco psaC-, psaI-, and psaJ-specific probes were synthesized by radiolabeling PCR products covering the entire coding regions of the genes (obtained by amplification with primer pair P31/P32 for psaC, P33/P34 for psaI, and P35/P36 for psaJ). A psaA probe was prepared from an internal NdeI fragment (corresponding to nucleotide positions 41,479–42,376). Hybridizations were carried out at 65°C in Rapid Hybridization Buffer (Amersham Intl.). A restriction fragment covering the entire coding region was used as an aadA-specific probe.
Isolation of Polysome Fractions and Polysome-associated RNAs
Polysomes were purified as described in reference 3. Young mutant or wild-type leaves (350 mg) from plants grown in sterile culture were ground in liquid nitrogen and treated with 2 ml of polysome extraction buffer (3). After removal of the insoluble material, polysomes were pelleted in a discontinuous sucrose gradient and subsequently fractionated in an analytical (continuous) sucrose gradient (2). As a control, an EDTA-containing sample (20 mM in the resuspension buffer, 1 mM in the gradient) was prepared that causes release of ribosomes from the mRNA chains, resulting in a uniform population of monosomes. The following fractions were collected (from top to bottom, using SW65 ultracentrifuge tubes): (a) 150, (b) 700, (c) 750, (d) 900, and (e) 900 μl. All fractions were diluted with 0.6 vol water before RNA isolation to reduce their sucrose content. RNA was extracted from individual fractions by adding EDTA (final concentration 20 mM) and phenol/chloroform (1:1 vol/vol). Subsequently, the RNA was precipitated with isopropanol after addition of 10 μg glycogen (Boehringer Mannheim). RNA pellets were resuspended in 20 μl sterile distilled water, and aliquots of 3 μl (fractions 2–5) were loaded on denaturing agarose gels for Northern hybridization analysis.
Protein Isolation Procedures
Thylakoid proteins from wild-type and mutant tissue were isolated according to reference 14. For preparation of soluble proteins, leaf samples were homogenized in 2 vol of extraction buffer (300 mM sucrose, 50 mM Tris/HCl, pH 8.0, 10 mM EDTA, 2 mM EGTA, 10 mM DTT, 1 mM Pefabloc [Boehringer Mannheim] and passed through two layers of Miracloth (Calbiochem-Novabiochem, La Jolla, CA). The filtrate was centrifuged for 10 min at 15,000 g, and the supernatant was subsequently subjected to an additional centrifugation step under identical conditions.
SDS-PAGE and Western Blot Analyses
Isolated thylakoid or soluble proteins were separated on tricine-SDS polyacrylamide gels (26) and transferred to Protran nitrocellulose BA83 membranes (Schleicher & Schuell Inc., Keene, NH) using the Trans-Blot® SD semi-dry transfer cell (BioRad Laboratories, Hercules, CA) with a standard transfer buffer (182 mM glycine, 20 mM Tris, 20% methanol, 0.05% SDS). Immunoblot detection was performed using the enhanced chemiluminescence system (ECL) (Amersham Intl.).
Physiological Measurements
Determination of photosystem II (PSII)1 activity was performed on young, dark-adapted leaves from wild-type and mutant plants grown under low light conditions. PSII-dependent chlorophyll fluorescence was recorded at 650 nm with a pulsed amplitude modulation fluorimeter (Walz, Effeltrich, Germany) (27) under illumination of intact leaf tissue with white actinic light (flux density 10 μE/m2s; pulse frequency 100 kHz). For complete reduction of QA, the primary quinone-type acceptor of PSII, leaves were exposed to pulses of saturating light (700 ms; flux density 4,000 μE/m2s) every 20 s.
RESULTS
Deletion of the ycf3 Reading Frame from the Tobacco Chloroplast Genome
An intron-containing reading frame of unknown function designated ycf3 resides in the large single-copy region of all higher plant chloroplast genomes. The position of this open reading frame in relation to adjacent genes in the tobacco plastid DNA is depicted in Fig. 1 A. The evolutionary conservation of ycf3 as well as the existence of a cyanobacterial homologue (39) suggest an important cellular function of this reading frame. However, the gene product has not been identified to date, and no mutants associated with ycf3 are available. We have therefore attempted to shed some light on the function of ycf3 by creating a null allele and introducing it into the tobacco chloroplast genome to replace the functional copy of ycf3.
Construction of the null allele was accomplished by deleting most of the ycf3 coding region and replacing it with a chimeric selectable marker gene (aadA) (36) in a cloned plastid DNA fragment (Fig. 1, A and B). The transformation vector pSR2 was introduced into tobacco plastids using the biolistic protocol. Two homologous recombination events in the flanking plastid DNA sequences result in replacement of ycf3 by aadA (Fig. 1). Since a single leaf cell in higher plants may contain up to 10,000 identical copies of the chloroplast genome, application of high selective pressure is required to amplify transformed plastid DNA molecules and to eliminate wild-type genomes. This can be achieved by regeneration of the bombarded leaf tissue under selection on spectinomycin-containing medium, since the presence of the aadA transgene confers resistance to aminoglycoside antibiotics (36). From the initial round of selection, we obtained several resistant lines harboring the aadA transgene in their chloroplast genome. The primary transformants containing a mixture of wild-type and transformed chloroplast genomes were subjected to several additional rounds of regeneration on selective medium. This eventually resulted in mutant lines with a uniformly altered plastid DNA population. The absence of residual wild-type genome copies was verified by DNA gel blot analysis (Fig. 2).
Δycf3 Plants Exhibit a Photosynthetically Deficient Phenotype
Complete elimination of the ycf3 reading frame results in plants viable on sucrose-containing medium. This indicates that ycf3 is not an essential gene for plastid maintenance and plant development.
Shoots from homoplasmic Δycf3 lines displayed a pale-green phenotype upon regeneration on spectinomycin-containing medium under standard light conditions (3.5–4 W/m2). When transferred to boxes (for rooting on drug- and phytohormone-free medium), the plants bleached out completely within a few days (Fig. 3 A). The phenotype was much less severe under low light conditions (0.4–0.5 W/m2). The plants were now light green (Fig. 3 B), and nearly indistinguishable from wild-type plants kept under identical conditions. However, the mutant plants grew very slowly, and after maintenance for more than 6 wk the lower leaves began to turn white. Only young leaves (up to 3-wk-old) from plants grown under low light conditions were used for the following molecular analyses.
Figure 3.

Phenotype of homoplasmic Δycf3 plants. (A) A mutant plant kept under standard light conditions (3.5–4 W/m2). Massive photooxidative damage in mutant chloroplasts results in completely white plants. (B) A mutant plant grown under low light conditions (0.4–0.5 W/m2). Bars, 1 cm.
The mutant, pigment-deficient phenotype proved to be stable under nonselective conditions, providing additional proof for the complete absence of wild-type genome copies.
Mutant Plastids Specifically Lack Photosystem I
The phenotype of the homoplasmic transformants suggests that the ycf3 gene product is directly or indirectly involved in photosynthetic electron transfer. To test whether the photosynthetic deficiency of the Δycf3 plants can be attributed to a specific complex in the thylakoid membrane we performed immunoblot analyses using various antibodies raised against proteins of PSII, PSI, the cytochrome bf complex, and the plastid ATP synthase complex (Table I). Whereas PSII proteins as well as cytochrome bf complex and ATPase subunits are readily detected in thylakoid membrane protein preparations from Δycf3 plants, PSI proteins appear to be absent or accumulate to levels falling below the sensitivity of our Western blots (Fig. 4). PSI subunits are also undetectable in the soluble protein fraction excluding the possibility that the proteins are stable in the stroma but cannot be incorporated into the thylakoid membrane.
Table I.
Test for Presence of Plastid-localized Proteins in Δycf3 Plants by Immunoblot Analyses
| Gene | Gene Localization | Gene Product | Detectable in Δycf3 Plants | |||
|---|---|---|---|---|---|---|
| aptB | Plastid | ATP synthase CF1 β subunit | Yes | |||
| psbA | Plastid | D1 protein of PSII | Yes | |||
| psbD | Plastid | D2 protein of PSII | Yes | |||
| PsbO | Nucleus | PsbO, manganese-stabilizing protein of PSII | Yes | |||
| PsbP | Nucleus | PsbP, lumen-localized extrinsic protein of PSII | Yes | |||
| Lchb6 | Nucleus | 20-kD apoprotein of CP24 | Yes | |||
| petA | Plastid | Cytochrome f | Yes | |||
| PetE | Nucleus | Plastocyanin | Yes | |||
| psaC | Plastid | PsaC, subunit VII of PSI, FA/FB-binding protein | No | |||
| PsaD | Nucleus | PsaD, subunit, II of PSI, ferredoxin-binding protein | No | |||
| PsaF | Nucleus | PsaF, subunit III of PSI, plastocyanin-docking protein | No |
Figure 4.
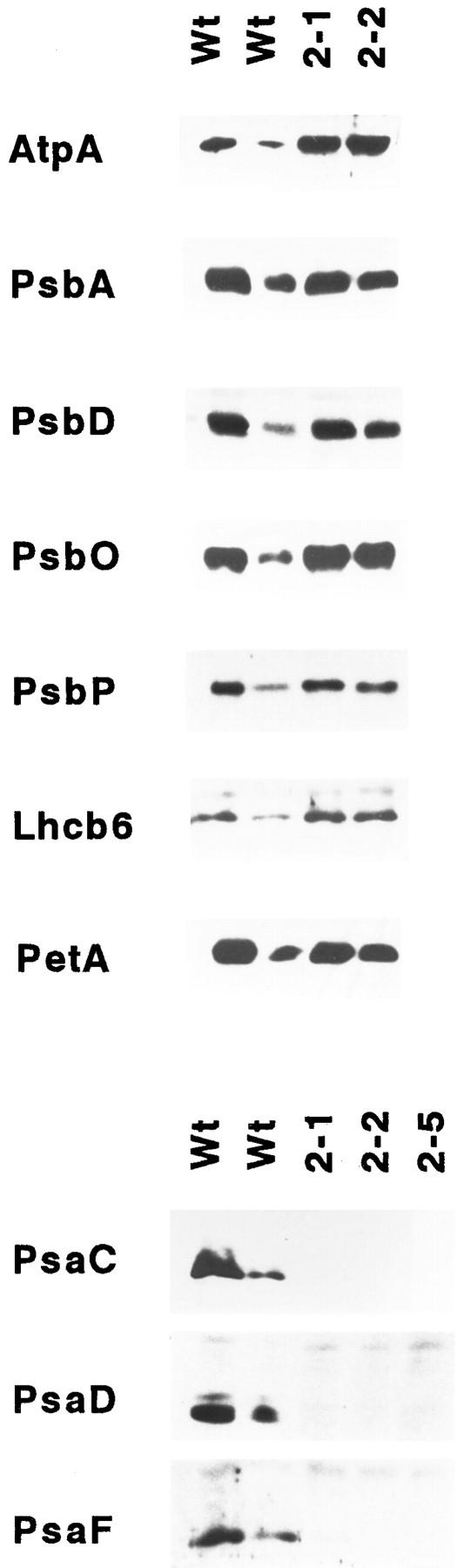
Accumulation of thylakoid proteins in Δycf3 plants. Immunoblots probed with antisera against the ATPase subunit AtpB, the PSII proteins PsbA, PsbD, PsbO, PsbP, and Lhcb6, the cytochrome bf complex subunit PetA, and the PSI proteins PsaC, PsaD, and PsaF are shown for wild-type plants and two or three independently transformed Δycf3 lines. For comparison, a dilution series of the wild-type extract is shown. Chlorophyll concentrations were wild type (higher concentration; first lane)/wild type (lower concentration; second lane)/mutant (1: 0.2:1.) Note that PSI proteins are undetectable in mutant plants whereas all the other protein complexes of the thylakoid membrane appear to be not primarily affected by the absence of the ycf3 gene product.
Immunoblot analysis of soluble proteins revealed that plastocyanin transferring the electrons from cytochrome f to the primary PSI acceptor, P700, is also present at wild-type levels in Δycf3 plants (data not shown). Thus it seems that the lack of ycf3 gene product selectively compromises PSI and does not primarily affect any of the other photosynthetic protein complexes.
Presence of functional PSII units in the mutant plants was further confirmed by measurements of PSII-dependent chlorophyll fluorescence at room temperature (Fig. 5). Even a moderate light flux of as little as 80 μE/m2s (corresponding to ∼6% of normal sunlight) resulted in a completely reduced pool of the primary quinone-type acceptor QA. This finding indicates that the electrons generated by PSII are not efficiently accepted by one of the downstream components of the electron transfer chain.
Figure 5.

Fluorescence measurements as test for PSII activity in Δycf3 plants. Wild-type and mutant plants grown under low light conditions were dark adapted, and leaf samples were illuminated with white actinic light. As a control, a wild-type sample treated with the plastoquinone-reducing herbicide 3-(3,4-dichlorphenyl)- 1,1-dimethylurea (DCMU) was included. PSII activity is clearly detectable in Δycf3 plants. However, comparison of the variable fluorescent yields indicates that the mutant accumulates fewer functional PSII reaction centers than the wild type, which is most likely the result of photooxidative damage as caused by the lack of functional electron acceptors downstream of PSII. The course of the fluorescence curve recorded for the mutant is virtually identical with the one of the wild-type sample treated with DCMU demonstrating that in both cases electrons accumulate in PSII and are not transferred to downstream components of the photosynthetic electron transport chain.
Transcription and RNA Stability of Photosystem I Genes Are not Impaired in Mutant Plastids
Several possible reasons for the lack of PSI protein accumulation in Δycf3 plants can be envisaged: (a) plastid-encoded PSI genes are not transcribed, (b) their mRNAs are not stable, or (c) not translated. Also, the ycf3 gene product could play a posttranscriptional role in either (d) PSI assembly or (e) stability.
To exclude the possibility that the absence of ycf3 protein specifically impairs PSI gene transcription or RNA stability, mRNA accumulation was tested for all plastid-encoded PSI genes: psaA, psaB, psaC, psaI, and psaJ. psaA and psaB (encoding the two P700-chlorophyll a apoproteins of PSI) had to be analyzed also for a second reason: they are located downstream of ycf3, and replacement of ycf3 with the chimeric aadA gene theoretically could exert a negative effect on psaA/B transcription.
psaC is located in the small, single-copy region of higher plant plastid genomes. It is cotranscribed with six genes homologous to NADPH dehydrogenase subunits as part of the plastid ndhH operon (18). Hybridization with a psaC-specific probe detects a complex transcript pattern (Fig. 6 A), most likely resulting from cleavage of the polycistronic precursor transcript into numerous processing intermediates and from splicing of the intron-containing ndhA gene. The major transcript of ∼0.5 kb represents the monocistronic psaC mRNA being one of the final maturation products (18). No differences between wild-type and mutant plants could be detected in mRNA accumulation or transcript pattern (Fig. 6 A) thus excluding a pretranslational defect as the reason for the lack of PsaC protein accumulation in Δycf3 plants.
Figure 6.
Northern blot analysis to test transcript patterns and mRNA accumulation in wild-type and homoplasmic transformed Δycf3 plants. Total plant RNA was hybridized to probes specific for psaC (A), psaA (B), psaI (C), and psaJ (D). The major transcripts of ∼0.5 kb for psaC (18), 5.2 kb for psaA and psaB (17), 0.6 kb for psaI, and 0.5 kb for psaJ, respectively, are marked by arrows. No significant differences in mRNA accumulation between wild-type and mutant plants could be detected, thus excluding a pretranslational cause of the PSI-deficient phenotype. Note a difference in the size of a minor RNA species detected by the psaA-specific probe (asterisks; 6.9-kb transcript in mutant plants). This RNA species represents a polycistronic transcript initiating far upstream of psaA. The polymorphism thus reflects the size difference of ycf3 in wild type versus the chimeric aadA gene in mutant plastids. (The diffuse signal in the wild-type lane (wt) is due to the presence of splicing intermediates of the intron-containing ycf3 gene, which give rise to multiple bands.) Read-through transcription, as the cause of the appearance of these high molecular weight mRNA species, was verified by hybridizing the blot with an aadA-specific probe (B, right panel). This probe detects the same 6.9-kb transcript as the psaA-specific probe in Δycf3 plants and, in addition, the 1.0-kb monocistronic aadA transcript (and a 1.4-kb aadA transcript stabilized by the downstream 3′-UTR of the deleted ycf3 gene).
The psaA-specific probe detects a major RNA species of 5.2 kb (Fig. 6 B) spanning the cotranscribed psaA, psaB, and rps14 genes (17). The same transcript is also present in all of the Δycf3 mutant lines demonstrating that replacement of ycf3 with aadA does not interfere with transcription of the downstream psaA and psaB genes. This is in good agreement with the earlier finding that the tobacco psaA/B genes are independently transcribed from their own promoter as shown by capping analysis (17). However, wild-type and mutant lines differ in the size of a minor RNA species. This RNA species is the result of read-through transcription initiating upstream of ycf3 and aadA, respectively (Fig. 6 B). Thus the transcript-length polymorphism is merely caused by the size difference of the larger ycf3 in wild type versus the smaller aadA in mutant plastid genomes.
Transcription of the other two plastid-encoded PSI genes, psaI and psaJ, was also examined. Hybridization with a psaI-specific probe detects a complex transcript pattern (Fig. 6 C), suggesting that at least part of the mRNA population is synthesized by cotranscription with some of the adjacent reading frames (40). The most abundant mRNA species of ∼0.6 kb represents monocistronic psaI message. The psaJ hybridization probe detects a prominent transcript of ∼0.5 kb, in addition to a number of minor mRNA species of higher molecular weight (Fig. 6 D). The major band most likely represents monocistronic psaJ message since it is too small to also cover the downstream ribosomal protein gene rpl33 (29). Again, no difference in transcript pattern or mRNA accumulation could be observed between wild-type and Δycf3 plastids excluding a role of the ycf3 gene product in PSI mRNA synthesis or maturation.
These results suggest that translatable mRNAs of PSI genes accumulate in mutant plants. We therefore conclude that ycf3 is most likely not involved in any of the pretranslational steps in the expression of plastid-encoded PSI genes.
Transcripts of Photosystem I Genes Are Efficiently Loaded with Ribosomes in Δycf3 Plants
Since our Northern blot analyses suggest that no gene expression step before translation of PSI transcripts is blocked in Δycf3 plastids, formally two possibilities remain: (a) the ycf3 gene product plays a cotranslational role, i.e., ycf3 encodes an essential PSI gene-specific translation factor; or (b) the ycf3 gene product is posttranslationally involved in the assembly of PSI subunits into a stable complex. To distinguish between these two possibilities we set out to test whether or not transcripts of PSI genes are translated in Δycf3 plastids.
It has frequently been observed that unassembled subunits of PSI complexes are highly unstable (10, 25, 30). The rather lengthy pulse-labeling experiments may thus prevent the detection of PSI translation products in Δycf3 plastids by in organello translation assays. Analysis of the polysomal association of PSI mRNAs is therefore the method of choice to test for faithful translation initiation on PSI transcripts in Δycf3 plants. Wild-type and mutant leaf samples were lysed under conditions maintaining the integrity of polysomes (3). The lysates were then fractionated in sucrose gradients, and the distribution of chloroplast transcripts was analyzed by performing Northern hybridization experiments with RNA purified from gradient fractions. As a control, EDTA was added to a gradient containing lysate from mutant plants. EDTA treatment releases ribosomes from mRNAs. Comparison of EDTA-containing with EDTA-free gradient fractions thus allows for the identification of monosome- versus polysome-containing fractions (Fig. 7).
Figure 7.
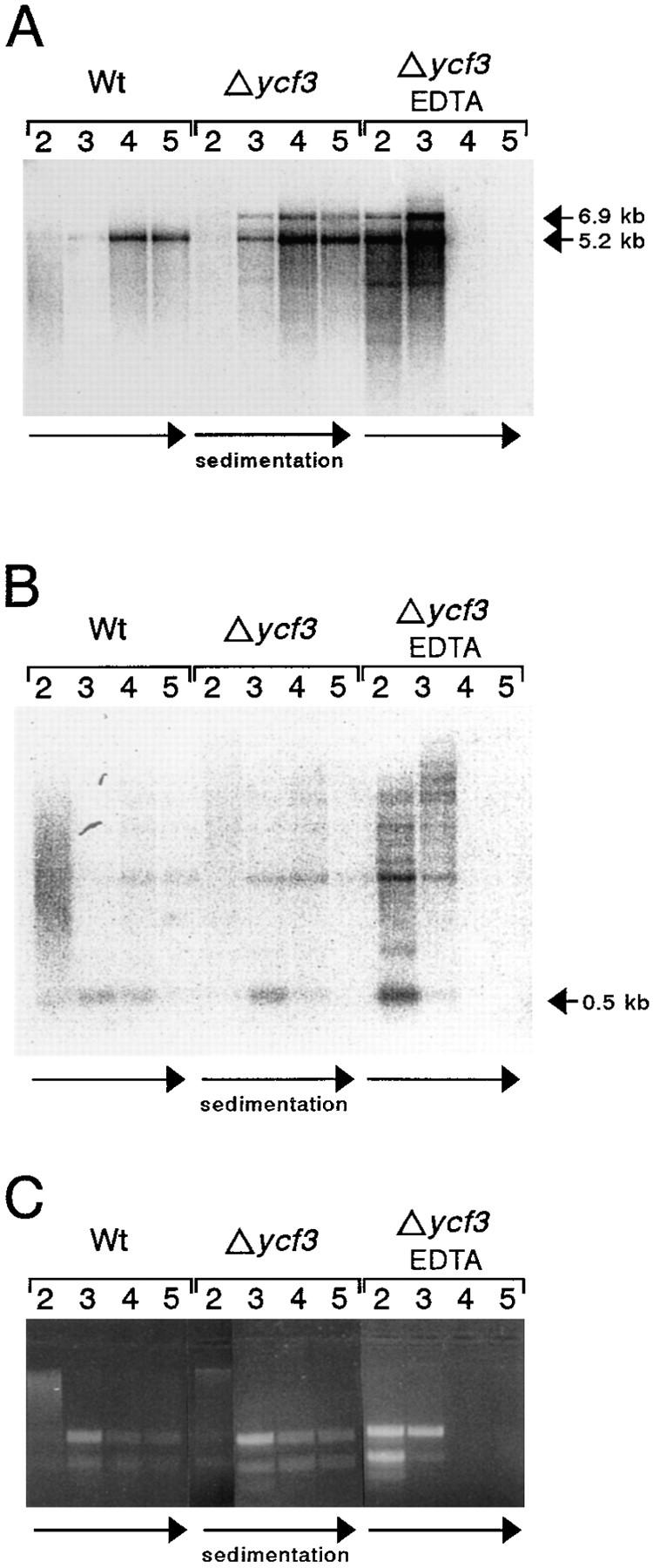
Test for association of PSI gene transcripts with polysomes in Δycf3 plants. (A) RNAs extracted from fractions 2–5 of analytical polysome isolation gradients were separated on 1% formaldehyde-containing agarose gels, transferred to nylon membranes, and hybridized to a psaA-specific probe mainly detecting the dicistronic psaA/B transcripts. Comparison of EDTA-free with EDTA-containing gradient fractions identifies fractions 2 and 3 as mainly monosome containing, and fractions 4 and 5 as polysome containing. The psaA/B transcripts in Δycf3 plastids are as efficiently associated with polysomes as in wild-type plastids (Wt). Note the prominent band for the read-through transcript initiating upstream of the aadA marker gene in mutant plastids (compare with Fig. 6 B), which is translated with extraordinarily high efficiency (most probably owing to the strong [rbcL-derived] Shine-Dalgarno sequence of the chimeric aadA). Transcript sizes are given at the right. The direction of polysome sedimentation is marked by horizontal arrows below the blot. (B) Analysis of polysome association for psaC transcripts. As psaA/B mRNAs, psaC transcripts are loaded with ribosomes with comparable efficiencies in wild-type and mutant plastids. The polysome-associated monocistronic psaC transcript (horizontal arrow) is predominantly present in fractions 3 and 4 for both wild-type and mutant plastids, but nearly exclusively in fraction 2 of the EDTA-containing gradient. (C) Ribosome content of the fractions collected. RNA aliquots of the fractions were separated under nondenaturing conditions on 2% agarose gels stained with ethidium bromide. The ribosome-containing fractions show prominent bands representing the ribosomal RNA species.
Hybridization using a psaA-specific probe (Fig. 7 A) detects RNA species identical with the ones identified in our mRNA accumulation analyses (Fig. 6 B). Mutant and wild-type plants again differ with respect to the read-through transcription product initiating upstream of ycf3 or aadA. The association of these read-through transcripts with polysomes demonstrates that they are efficient substrates for the chloroplast translation machinery. This finding is in accordance with the results of earlier studies showing that both monocistronic and polycistronic mRNAs are efficiently translated in higher plant chloroplasts (2, 34). No difference in polysome loading could be detected between mutant and wild-type plants suggesting that translation of psaA and psaB is initiated with comparable efficiencies in wild-type and Δycf3 plants.
We have also tested polysome association for psaC (Fig. 7 B) and, as a control, for a tetracistronic PSII transcript (psbE/F/L/J; data not shown). These analyses also failed to provide evidence for any defect in polysome loading in Δycf3 plastids.
DISCUSSION
The recent development of facile methods of transformation for higher plant chloroplasts has enabled us to address functional aspects of plastid open reading frames by reverse genetics. In the course of this work, we have performed the first targeted inactivation of a tobacco plastid open reading frame of unknown function by deleting the intron-containing ycf3 from the chloroplast genome. We have shown that homoplasmic Δycf3 plants display a pigment-deficient phenotype, most probably caused by the complete absence of PSI.
Several lines of evidence suggest that none of the general processes in plastid gene expression (i.e., transcription, RNA processing, translation) are impaired in Δycf3 plants. First, homoplasmic mutant plants display a high level of resistance to spectinomycin indicating that the chimeric aadA gene is highly expressed in the transgenic plastids. Second, the protein products of those plastid-encoded photosynthesis genes that are not related to PSI can be readily detected in mutant plastids. This finding also confirms that all of the chloroplast-encoded genes engaged in transcription or translation provide functional gene products in Δycf3 plastids. Third, transcripts of plastid-encoded PSI genes are faithfully synthesized, correctly processed, accumulate to wild-type levels in Δycf3 plants and are also efficiently loaded with ribosomes.
What then is the cause of the PSI-deficient phenotype? And consequently, what is the function of the ycf3 gene product? Our Northern blot and polysome association analyses suggest that neither transcription of PSI genes, nor transcript processing or translation initiation is impaired in Δycf3 plastids. Efficient loading with polysomes is suggestive of active translation of PSI mRNAs in mutant plastids. Identical distribution patterns of PSI transcripts across the polysome gradients also indicate that the numbers of ribosomes associated with PSI mRNAs do not significantly differ between wild-type and mutant plastids suggesting that translation elongation proceeds with comparable efficiencies. However, these data do not completely exclude a deficiency in a late step in PSI gene–specific translation elongation (or termination) in Δycf3 plastids.
The lack of evidence for a transcriptional or posttranscriptional role of the ycf3 gene product is consistent with the idea that the control of these steps in chloroplast gene expression is probably exclusively exerted by nuclear factors (for review see references 9, 11). We, therefore, propose that ycf3 encodes a factor involved in the assembly of a stable PSI unit in a posttranslational fashion. This could be the case if the Ycf3 protein is an integral part of PSI or alternatively, if it served as an auxiliary factor for the assembly or stability of the PSI complex in the thylakoid membrane. Both possibilities imply that the absence of virtually all PSI subunits from Δycf3 plastids is a secondary consequence of the destabilization of PSI caused by the missing ycf3 gene product.
Isolated cyanobacterial PSI complexes consist of 11 polypeptides (PsaA, B, C, D, E, F, I, J, K, L, and M) (for review see reference 21), which all are well characterized at the molecular level. In view of the presence of a cyanobacterial ycf3 homologue (39), it therefore appears unlikely that the ycf3 gene product is an integral component of the PSI complex. In this light, our results may be more consistent with the idea that the Ycf3 protein serves as an assembly or stability factor for PSI. However, at present we do not know the exact suborganellar localization of the ycf3 gene product since all our attempts to raise Ycf3-specific antibodies have failed.
Several PSI mutants have been described for cyanobacteria and Chlamydomonas reinhardtii. Insertional inactivation of psaC in C. reinhardtii was shown to result in destabilization of PSI and the concomitant loss of all PSI subunits (38). Given our failure to detect PsaC protein in Δycf3 plants, it is therefore not surprising that all the other PSI proteins tested by immunoblot analysis were not found either. A similar crucial role in PSI stability is attributed to the two large reaction center subunits PsaA and PsaB. In cyanobacteria (30) as well as in Chlamydomonas (10) and higher plants (25), a defective reaction center protein leads to a complete loss of the PSI complex and to a rapid turnover of all of its subunits.
In conclusion, our results indicate that the chloroplast ycf3 reading frame is indeed a functional gene. Its gene product is a heretofore unknown factor involved in the generation of functional PSI units. Future analyses will aim to determine the localization of the Ycf3 protein and to define the nature of its association or interaction with other components of photosystem I.
Acknowledgments
The authors wish to thank M. Hermann for excellent technical assistance, E. Schiefermayr for oligonucleotide synthesis and A. Herzfeld and M. Messerschmid for photographic work. We are grateful to P. Maliga and Z. Svab for making available ptDNA clones and the chimeric aadA gene construct and to K. Paal and P. Zeltz for valuable discussion. We are indebted to R. Oelmüller, R.B. Klösgen, and A. Barkan for generously providing antibodies. We wish to thank K. Biehler, W. Haehnel, and G. Buchholz for help with the physiological measurements, and G.L. Igloi for critical reading of this manuscript.
Abbreviations used in this paper
- PSI and PSII
photosystem I and II
- RFLP
restriction fragment–length polymorphism
Footnotes
This paper is dedicated to the memory of our late teacher Hans Kössel who passed away on December 24, 1995.
This research was supported by grants from the Deutsche Forschungsgemeinschaft (BO 1482/1-1); the SFB 388; the Land Baden-Württemberg (Graduiertenförderung); and the Human Frontiers Science Program Organization (RG-437/94 M).
Address all correspondence to Ralph Bock, Institut für Biologie III, Universität Freiburg, Schänzlestrasse 1, D-79104 Freiburg, Germany. Tel.: 49-761-2032721. Fax: 49-761-2032745. e-mail: bock@sun1.biologie.uni-freiburg.de
REFERENCES
- 1.Allison LA, Maliga P. Light-responsive and transcription- enhancing elements regulate the plastid psbDcore promoter. EMBO (Eur Mol Biol Organ) J. 1995;14:3721–3730. doi: 10.1002/j.1460-2075.1995.tb00042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO (Eur Mol Biol Organ) J. 1988;7:2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barkan A. Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell. 1993;5:389–402. doi: 10.1105/tpc.5.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock R, Maliga P. Correct splicing of a group II intron from a chimeric reporter gene transcript in tobacco plastids. Nucleic Acids Res. 1995;23:2544–2547. doi: 10.1093/nar/23.13.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bock R, Kössel H, Maliga P. Introduction of a heterologous editing site into the tobacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO (Eur Mol Biol Organ) J. 1994;13:4623–4628. doi: 10.1002/j.1460-2075.1994.tb06784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bock R, Hermann M, Kössel H. In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO (Eur Mol Biol Organ) J. 1996;15:5052–5059. [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhuri S, Carrer H, Maliga P. Site-specific factor involved in the editing of the psbLmRNA in tobacco plastids. EMBO (Eur Mol Biol Organ) J. 1995;14:2951–2957. doi: 10.1002/j.1460-2075.1995.tb07295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyle JJ, Doyle JL. Isolation of plant DNA form fresh tissue. Focus (Idaho) 1990;12:13–15. [Google Scholar]
- 9.Gillham NW, Boynton JE, Hauser CR. Translational regulation of gene expression in chloroplasts and mitochondria. Annu Rev Genet. 1994;28:71–93. doi: 10.1146/annurev.ge.28.120194.000443. [DOI] [PubMed] [Google Scholar]
- 10.Girard-Bascou J, Choquet Y, Schneider M, Delosme M, Dron M. Characterization of a chloroplast mutation in the psaA2 gene of Chlamydomonas reinhardtii. . Curr Genet. 1987;12:489–495. doi: 10.1007/BF00419557. [DOI] [PubMed] [Google Scholar]
- 11.Gruissem W, Tonkyn JC. Control mechanisms of plastid gene expression. Crit Rev Plant Sci. 1993;12:19–55. [Google Scholar]
- 12.Kanevski I, Maliga P. Relocation of the plastid rbcLgene to the nucleus yields functional ribulose-1,5-bisphosphate carboxylase in tobacco chloroplasts. Proc Natl Acad Sci USA. 1994;91:1969–1973. doi: 10.1073/pnas.91.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowallik KV, Stoebe B, Schaffran I, Kroth-Pancic P, Freier U. The chloroplast genome of a chlorophyll a+c-containing alga, Odontella sinensis. . Plant Mol Biol Rep. 1995;13:336–342. [Google Scholar]
- 14.Machold O, Simpson DJ, Møller BL. Chlorophyll-proteins of thylakoids from wild-type and mutants of barley (Hordeum vulgareL.) Carlsberg Res Commun. 1979;44:235–254. [Google Scholar]
- 15.Maier RM, Neckermann K, Igloi GL, Kössel H. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- 16.McCullough AJ, Kangasjärvi J, Gengenbach BG, Jones RJ. Plastid DNA in developing maize endosperm: genome structure, methylation, and transcript accumulation patterns. Plant Physiol. 1992;100:958–964. doi: 10.1104/pp.100.2.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng BY, Tanaka M, Wakasugi T, Ohme M, Shinozaki K, Sugiura M. Cotranscription of the genes encoding two P700 chlorophyll a apoproteins with the gene for ribosomal protein CS14: determination of the transcriptional initiation site by in vitrocapping. Curr Genet. 1988;14:395–400. doi: 10.1007/BF00419998. [DOI] [PubMed] [Google Scholar]
- 18.Meurer J, Berger A, Westhoff P. A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhCoperons. Plant Cell. 1996;8:1193–1207. doi: 10.1105/tpc.8.7.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 20.Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorphachloroplast DNA. Nature (Lond) 1986;322:572–574. [Google Scholar]
- 21.Pakrasi HB. Genetic analysis of the form and function of photosystem I and photosystem II. Annu Rev Genet. 1995;29:755–776. doi: 10.1146/annurev.ge.29.120195.003543. [DOI] [PubMed] [Google Scholar]
- 22.Reith M, Munholland J. Complete nucleotide sequence of the Porphyra purpureachloroplast genome. Plant Mol Biol Rep. 1995;13:333–335. [Google Scholar]
- 23.Ruf S, Zeltz P, Kössel H. Complete RNA editing of unspliced and dicistronic transcripts of the intron-containing reading frame IRF170 from maize chloroplasts. Proc Natl Acad Sci USA. 1994;91:2295–2299. doi: 10.1073/pnas.91.6.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 9.42–9.46.
- 25.Schaffner C, Laasch H, Hagemann R. Detection of point mutations in chloroplast genes of Antirrhinum majus L. I. Identification of a point mutation in the psaBgene of photosystem I plastome mutant. Mol Gen Genet. 1995;249:533–544. doi: 10.1007/BF00290579. [DOI] [PubMed] [Google Scholar]
- 26.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 27.Schreiber U, Schliwa U, Bilger W. Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth Res. 1986;10:51–62. doi: 10.1007/BF00024185. [DOI] [PubMed] [Google Scholar]
- 28.Shimada H, Sugiura M. Fine structural features of the chloroplast genome: comparison of the sequenced chloroplast genomes. Nucl Acids Res. 1991;19:983–995. doi: 10.1093/nar/19.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO (Eur Mol Biol Organ) J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smart LB, Anderson SL, McIntosh L. Targeted genetic inactivation of the photosystem I reaction center in the cyanobacterium Synechocystissp. PCC 6803. EMBO (Eur Mol Biol Organ) J. 1991;10:3289–3296. doi: 10.1002/j.1460-2075.1991.tb04893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staub JM, Maliga P. Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbAmRNA. EMBO (Eur Mol Biol Organ) J. 1993;12:601–606. doi: 10.1002/j.1460-2075.1993.tb05692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staub JM, Maliga P. Extrachromosomal elements in tobacco plastids. Proc Natl Acad Sci USA. 1994;91:7468–7472. doi: 10.1073/pnas.91.16.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staub JM, Maliga P. Translation of psbAmRNA is regulated by light via the 5′- untranslated region in tobacco plastids. Plant J. 1994;6:547–553. doi: 10.1046/j.1365-313x.1994.6040547.x. [DOI] [PubMed] [Google Scholar]
- 34.Staub JM, Maliga P. Expression of a chimeric uidAgene indicates that polycistronic mRNAs are efficiently translated in tobacco plastids. Plant J. 1995;7:845–848. doi: 10.1046/j.1365-313x.1995.07050845.x. [DOI] [PubMed] [Google Scholar]
- 35.Stirewalt VL, Michalowski CB, Löffelhardt W, Bohnert HJ, Bryant DA. Nucleotide sequence of the cyanelle genome from Cyanophora paradoxa. . Plant Mol Biol Rep. 1995;13:327–332. [Google Scholar]
- 36.Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadAgene. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svab Z, Hajdukiewicz P, Maliga P. Stable transformation of plastids in higher plants. Proc Natl Acad Sci USA. 1990;87:8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi Y, Goldschmidt-Clermont M, Soen S-Y, Franzén LG, Rochaix J-D. Directed chloroplast transformation in Chlamydomonas reinhardtii: insertional inactivation of the psaCgene encoding the iron sulfur protein destabilizes photosystem I. EMBO (Eur Mol Biol Organ) J. 1991;10:2033–2040. doi: 10.1002/j.1460-2075.1991.tb07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vörös K, Hübschmann T, Börner T. The cyanobacterium Synechocystis sp. PCC6803 contains a putative gene homologous to tobacco chloroplast open reading frame 168. Endocyt. . Cell Res. 1992;9:71–76. [Google Scholar]
- 40.Willey DL, Gray JC. An open reading frame encoding a putative haem-binding polypeptide is cotranscribed with the pea chloroplast gene for apocytochrome f. . Plant Mol Biol. 1990;15:347–356. doi: 10.1007/BF00036920. [DOI] [PubMed] [Google Scholar]
- 41.Wolfe KH, Sharp PM. Identification of functional open reading frames in chloroplast genomes. Gene (Amst) 1988;66:215–222. doi: 10.1016/0378-1119(88)90358-7. [DOI] [PubMed] [Google Scholar]