Abstract
Localization of yeast Kex2 protease to the TGN requires a signal (TLS1) in its cytosolic tail (C-tail). Mutation of TLS1 results in rapid transit of Kex2p to the vacuole. Isolation of suppressors of the Tyr713Ala mutation in TLS1 previously identified three SOI genes. SOI1, cloned by complementation of a sporulation defect, encodes a novel, hydrophilic 3,144-residue protein with homologues in Caenorhabditis elegans, Drosophila melanogaster, and humans. Epitope-tagged Soi1p existed in a detergent-insensitive, sedimentable form. Deletion of SOI1 impaired TGN localization of wild-type Kex2p and a fusion protein containing the C-tail of Ste13p, and also caused missorting of carboxypeptidase Y and accelerated vacuolar degradation of the Vps10p sorting receptor. Deletion of SOI1 improved retention of Tyr713Ala Kex2p in the pro-α-factor processing compartment but, unlike the original soi1 alleles, did not increase the half-life of Tyr713Ala Kex2p. These results suggested that Soi1p functions at two steps in the cycling of Kex2p and other proteins between the TGN and prevacuolar compartment (PVC). This hypothesis was confirmed in several ways. Soi1p was shown to be required for optimal function of TLS1. Suppression of the Tyr713Ala mutation by mutation of SOI1 was shown to be caused by activation of a second signal (TLS2) in the Kex2p C-tail. TLS2 delayed exit of Kex2p from the TGN, whereas TLS1 did not affect this step. We propose that Soi1p promotes cycling of TGN membrane proteins between the TGN and PVC by antagonizing a TGN retention signal (TLS2) and facilitating the function of a retrieval signal (TLS1) that acts at the PVC.
The yeast (Saccharomyces cerevisiae) Kex2 protease is required in MATα haploid cells for the production of the mating pheromone α-factor (19). A type 1 transmembrane protein, Kex2p, is localized to a late Golgi compartment that is analogous to the TGN in mammalian cells (15, 35, 55). Localization of Kex2p to the TGN requires a TGN localization signal (TLS)1 in the COOH-terminal, cytosolic tail (C-tail) of the protein. This signal includes important aromatic residues Tyr713 and Phe715 (36, 55). Substitution of alanine for either Tyr713 or Phe715 results in rapid transport of Kex2p to the vacuole without passing through the plasma membrane (34, 55). Truncation of Kex2p after Ile718 results in a form of the protein, I718tail Kex2p (called D719Amb in reference 34), which possesses only the first 19 residues of the 115-residue C-tail (34). I718tail Kex2p is localized efficiently to the TGN, suggesting that the TLS is sufficient for localization (34). A similar aromatic residue–containing TLS has been identified in the NH2-terminal, cytosolic tail of Ste13p (dipeptidyl aminopeptidase A), a type 2 transmembrane protein that is also required for α-factor biosynthesis (29, 30). Kex1p, a carboxypeptidase involved in α-factor maturation, and Vps10p, a vacuolar protein sorting receptor, also require sequences in their COOH-terminal, cytosolic tails for proper TGN localization (6, 8, 9). In each case, mutation or deletion of TLS sequences in these proteins results in mislocalization to the vacuole (6, 8, 9).
Several studies suggest that steady-state localization of membrane proteins to the TGN is mediated by a cycling pathway between the TGN and the prevacuolar compartment (PVC), the intersection between vacuolar biosynthetic traffic and endocytic traffic that corresponds to the mammalian late endosome (31, 32, 37, 51). In class E vps mutants, which accumulate active vacuolar proteases in an aberrant PVC (32), both Kex2p and Vps10p are degraded in a vacuolar protease–dependent manner (6, 31), consistent with continual cycling of these proteins between the TGN and PVC. Moreover, when class E vps27-ts mutant cells are shifted to the restrictive temperature, Vps10p is rapidly and reversibly redistributed from the TGN to the PVC (31).
Transport from the TGN to the PVC appears to depend on the clathrin heavy chain Chc1p (44, 45) and the dynamin/shibire homologue Vps1p (28, 41), and does not require cytosolic signals (36). Use of this TGN–PVC pathway by proteins devoid of C-tail sequences implies that membrane proteins enter this pathway by default (although a recent study suggests that the nature of the transmembrane domain may be a factor; see reference 33), and that TGN localization signals promote selective retrieval of transmembrane proteins from the PVC. Both the mannose-6-phosphate receptors (22) and furin (3) have been proposed to follow similar cycling pathways in mammalian cells.
To identify proteins involved in TLS function, we used a screen that depends on the essential role of Kex2p in mating of MATα haploids to isolate extragenic suppressors of the mislocalization defect of Y713A Kex2p (34). After a shift from galactose to glucose to repress expression of forms of Kex2p under GAL1 promoter control, MATα cells expressing Y713A Kex2p lose mating competence much more rapidly than do cells expressing wild-type (WT) Kex2p (34, 55). This reflects the more rapid loss of Y713A Kex2p from the pro-α-factor processing compartment. A subset of suppressors of this rapid “onset of impotence” (Soi+ phenotype) were allele specific, suppressing the effects of substitutions for Tyr713 but not the effects of deletions of the TLS or the entire C-tail (34). The allele specificity of these suppressor mutations, which mapped to the SOI1, SOI2, and SOI3 genes, suggested that the SOI gene products are involved in TLS-dependent events, possibly by interacting with the TLS or other C-tail sequences (34). These mutations also suppressed the effects of the F87A mutation in the Ste13p TLS, suggesting a role for the SOI genes in the localization of other TGN proteins as well (34).
The SOI1 gene was of particular interest because soi1 mutants also displayed defects in the TGN localization of WT Kex2p, suggesting a defect in the recognition of the Kex2p TLS (34). soi1 mutants were also defective for vacuolar protein sorting (Vps−), i.e., for sorting proCPY to the vacuole (34). All of the phenotypes of the soi1 mutants were recessive, suggesting that the phenotypes resulted from loss of function of the SOI1 gene product.
Here we describe the cloning and characterization of SOI1, which encodes a 358-kD protein that is conserved in metazoan eukaryotes. Soi1p is required for efficient TGN localization of not only Kex2p, but also of Ste13p and Vps10p. Analysis of soi1 null mutants revealed the existence of a second TGN localization signal (TLS2) in the C-tail of Kex2p. TLS2 has features of a “retention signal,” regulating the rate of exit of Kex2p from the TGN, whereas TLS1 (i.e., the Tyr713-based signal) appears to function in retrieval of Kex2p from the PVC. We propose that Soi1p promotes cycling of proteins between the TGN and the PVC by antagonizing TGN retention and promoting retrieval from the PVC to the TGN. This function is mediated through the modulation of the function of two TGN localization signals.
MATERIALS AND METHODS
Strains, Antibodies, Reagents, and Media
Yeast strains used appear in Table I. SPB227-5C was crossed to SEY6210 to produce JBY11 from which JBY11-r1 was a segregant (Table I). JBY96 was produced by a series of crosses between JBY11-r1, SEY6210, and YJM82. Synthetic minimal (SD) and synthetic complete containing glucose (SDC) or galactose (SGC), or rich medium (YPD; glucose) were as described (39), except that isoleucine, glutamate, aspartate, valine, and serine were omitted from SDC, and 50 μg/ml adenine sulfate was included in YPD. Dropout media are indicated as SDC-Ura, etc. Anti–alkaline phosphatase (ALP) serum was provided by Steve Nothwehr (University of Missouri, Columbia, MO) and by Nia Bryant and Tom Stevens (University of Oregon, Eugene, OR); anti-CPY and anti-Vps10p sera were furnished by Scott Emr (University of California, San Diego, CA). Monoclonal anti–carboxypeptidase Y (CPY) was from Molecular Probes, Inc. (Eugene, OR). Anti–hemagglutinin (HA) mAb 12CA5 was obtained from Boehringher Mannheim (Indianapolis, IN). Rabbit anti–mouse IgG and goat anti–rabbit IgG Texas red were from Jackson ImmunoResearch Laboratories (West Grove, PA). Unless otherwise noted, chemicals were from Sigma Chemical Co. (St. Louis, MO).
Table I.
Yeast Strains
| Strain | Genotype* | Parental strain | ||
|---|---|---|---|---|
| CRY1 | MAT a ade2-1 can1-100 his3-11,15 leu2-3,-112 trp1-1 ura3-1 | — | ||
| CRY2 | MATα ade 2-1 can1-100 his3-11,15 leu2-3,-112 trpl-1 ura3-1 | — | ||
| BFY106-4D | kex2Δ::HIS3 | CRY2 | ||
| KRY18-1A | kex2Δ::TRP1 | CRY2 | ||
| SEY6210‡ | MATα his3-Δ200 leu2-3,112 lys2-801 suc2-Δ9 trp1-Δ901 ura3-52 | — | ||
| 0472-28‡ | vps28 | SEY6210 | ||
| YJM82§ | MAT a ade2-101 cyh2 | — | ||
| CB018 | MAT a pep4::HIS3 prb1::hisG prc1::hisG | — | ||
| SPB227-5C | kex2Δ::TRP1 soi1-2 | CRY2 | ||
| JBY11-rl | MATα ade2-1 can1-100 his3 kex2Δ::TRP1 leu2-3,-112 lys2-801 soi1-2 trp1-1 ura3-1 | NA | ||
| JBY96 | a/α ade2-1/+cyh2/+ can1-100/+ his3/his3 kex2Δ::TRP1 leu2-3,-112/leu2-3,-112 lys2-801/lys2-801soi1-2/ soi1-2 suc2-Δ9/+ trp1/trp1 ura3-1/ura3-52 | NA | ||
| JBY134.1 | kex2Δ::HIS3 soi1Δ-1::kan r | CRY2 | ||
| JBY135-1A | soi1Δ-1::kan r | CRY2 | ||
| JBY135-2D | CRY2 | |||
| JBY154-1A | kex2Δ::TRP1 | CRY2 | ||
| JBY154-2A | kex2Δ::TRP1 soi1D-2::kan r | CRY2 | ||
| JBY154-1D | soi1Δ::kan r | CRY2 | ||
| JBY154-8B | CRY2 | |||
| CB018 | MAT a pep4::HIS3 prb1::hisG prc1::hisG | CRY1 | ||
| JBY171 | MAT a pep4::HIS3 prb1::hisG prc1::hisG SOI1::HA | CRY1 | ||
| JBY173 | MAT a pep4::HIS3 prb1::hisG prc1::hisG soi1Δ-2::kan r | CRY1 | ||
| JBY178 | soi1Δ-2::kan r vps 28 | SEY6210 |
Only those loci differing from the parental strain are described.
Provided by Dr. Scott Emr.
Provided by Dr. John McCusker (Duke University School of Medicine, Durham, NC).
SOI1 Cloning, Sequencing, Mapping, and Disruption
To create a selection for Spo+ transformants, we used a soi1-2/soi1-2 strain that was heterozygous at two recessive drug resistance loci, can1 and cyh2 (JBY96). After sporulation, one quarter of the haploid progeny of such a strain should be resistant to both drugs. Library (YPH1; LEU2 CEN yeast genomic library, ATCC No. 77162) transformants that were complemented for the Spo− phenotype were identified by selecting for haploid progeny on SDC-Arg containing 40 μg/ml canavanine sulfate and 10 μg/ml cycloheximide after incubation on sporulation medium. Transformants producing drug-resistant progeny were picked from the sporulation medium and examined microscopically for the production of asci. Plasmids were retrieved into Escherichia coli as described (53).
Oligonucleotide primers were synthesized and sequencing reactions were performed by the University of Michigan core facility. Both strands of SOI1 were sequenced, and sequence analysis was performed using software from Intelligenetics (Palo Alto, CA).
To determine whether the insert in pSOI1.1 was linked to the SOI1 locus, a 3.3-kb ClaI fragment from pSOI1.1 was cloned into pRS303 (HIS3 integrating vector) to create pSOI1.3. pSOI1.3, digested at a unique NcoI site in the insert, was used to transform CRY2. Four independent transformants were crossed to SPB227-5C, and 10 tetrads were dissected from the resulting diploids. All 39 complete tetrads displayed parental ditype segregation of these phenotypes (2 His+, Vps+: 2 His−, Vps−), indicating that the DNA insert from pSOI1.1 was tightly linked to the SOI1 locus.
SOI1 was replaced by the E. coli kan r gene by PCR amplification of the kan r gene using primers whose 5′ sequences were derived from SOI1 (52). Transformants were selected on YPAD + 400 mg/liter Geneticin (GIBCO BRL, Gaithersburg, MD). Two different disruptions were created: soi1Δ-1 was a partial deletion of SOI1 with kan r in place of bases 2,178–9,078 in the coding sequence of SOI1. Deletion soi1Δ-2 removed the entire SOI1 structural gene. No differences were observed between soi1Δ-1 and soi1Δ-2 strains (our unpublished data). The primers used to amplify the kan r gene from pFAkanMX2 were (5′ to 3′): soi1Δ-1, (upstream), CCCCTTGTGCGATCATTGATGCCGGTCATATGTCTATACTCAGTGATTTAA-AGCTTCGTACGCTGCAG; soi1Δ-1, (downstream), TCACGGTCTTGAGCCAGTATTGTCCTTGGGCCTCACGTAGATCATA A G G C G - GCCACTAGTGGATCTGA; soi1Δ-2, (upstream), TGACTACCAAAAGGGAAAAGGCAGAAAAAAGGAAAATTAAGAACAG T TA A - AAGCTTCGTACGCTGCAG; soi1Δ-2, (downstream), GAATTATAGCTACATAGTGTACAAAAGCGGGTATATACTTTCATATGTG- AGGCCACTAGTGGATCTGA. PCR products were introduced into diploid strains, and disruption was confirmed by three-primer PCR using genomic DNA from G418r transformants. The primers used to confirm disruptions were (5′ to 3′): kanmid, GACTCAGCTTTCGAGGCCGCG; soi1Δ-1 mid (antisense), ACATTGACGGCCTCCATTTT; soi1Δ-2 mid (antisense), GAATGGGGTCACGTAGCGGAG; soi1Δ-1 upstream (sense), TCAATTGCCAACCACGCTACC; soi1Δ-2 upstream (sense), ATTTAATTTGATGCGGATAGAA. Tetrads from diploid transformants were dissected, and the progeny were analyzed for the Vps phenotype and G418 resistance.
Indirect Immunofluorescence
Indirect immunofluorescence localization of Kex2p was performed as described (35), visualized using an Axioskop (Carl Zeiss, Inc., Thornwood, NY) microscope and photographed using Hypered Kodak Techpan 2415 film ASA 800 (Lumicon, Livermore, CA).
Phenotypic Assays and Complementation Analysis
The onset of impotence assay was performed as described (34). The Vps phenotype was analyzed essentially as described (40), using a monoclonal anti-CPY antibody followed by rabbit anti–mouse IgG and then donkey anti–rabbit IgG conjugated to HRP. Filters were processed for enhanced chemiluminescence according to the manufacturer's instructions (Amersham, Arlington Heights, IL). The Spo phenotype was scored microscopically as the production of asci. Resistance to G418 was scored as growth on YPAD + 400 mg/liter Geneticin.
The soi1 mutants were previously characterized as class A vps mutants (34), which display no alterations in vacuolar morphology (32). Complementation analysis with a complete collection of vps and pep mutants, provided by Dr. Bruce Horazdovsky and Dr. Scott Emr (University of California, San Diego, CA), indicated that all (1561-2, 0534-2, 1564-2) but one of the mutants designated as vps13 failed to complement the Vps− and Spo− phenotypes caused by the soi1-2 mutation (data not shown). The allele that did complement soi1 (0594-2) was not allelic with the other vps13 mutations (data not shown). The soi1-2 allele also failed to complement pep9 (18). These data suggest that SOI1, VPS13, and PEP9 are the same locus. This is consistent with the fact that vps13 mutations have also been characterized as class A vps mutants (32).
Subcellular Fractionation
Cells were grown and labeled with [35S]H2SO4 (54) for 30 min, 2 × 108 cells were harvested by centrifugation, washed twice in 100 mM Tris, pH 9.4, 10 mM DTT, resuspended in spheroplasting buffer (10 mM Tris, pH 7.4, 1 mM DTT, 0.7 M sorbitol, 625 μg/ml zymolyase 100T), and incubated at 30°C for 30 min. Spheroplasts were washed twice with 0.7 M sorbitol, 50 mM Hepes, pH 7, and resuspended in lysis buffer (0.3 M sorbitol, 10 mM Hepes, pH 7). DEAE dextran was then added slowly to a final concentration of 15 μg/ml (10), and the spheroplasts were incubated for 2 min on ice, at 37°C for 5 min, and then back on ice.
The supernatant from a 1,000-g clearing spin was spun at 13,000 g to produce a pellet (P13) and supernatant fraction (S13). The S13 was then added to either lysis buffer or Triton X-100 (0.1% final concentration), and was spun at 50,000 rpm for 90 min in a TLS55 rotor (Beckman Instruments, Palo Alto, CA). SDS was added to 1% to supernatant (S150) and P13 fractions, and the P150 pellets were resuspended in 8 M urea + 1% SDS. All samples were heated to 95°C for 3 min, diluted into 1 ml immunobuffer, and processed for immunoprecipitation (34).
Pulse-Chase/Immunoprecipitation/SDS-PAGE
Labeling with [35S]H2SO4 and chase for most immunoprecipitation experiments were performed as described (54, 55). Termination of chase, lysis, and immunoprecipitation of dipeptidyl aminopeptidase A–alkaline phosphatase fusion protein (A-ALP), ALP (34), CPY (20), and Vps10p (6) were performed as described. Soi1p was immunoprecipitated using both 12CA5 anti-HA and rabbit anti–mouse IgG with Pansorbin (Calbiochem-Novabiochem Corp., La Jolla, CA). Washes for Soi1p and Kex2p immunoprecipitation experiments were performed using a modified protocol (described in reference 34). Kex2p, Vps10p, A-ALP, CPY, and ALP immunoprecipitates were subjected to SDS-PAGE using 8% separating gels. 6% separating gels were used for Soi1p-HA immunoprecipitates.
For experiments in which the chase was longer than 100 min, or for the experiments performed at 23°C, cultures were diluted with 0.5 vol of fresh medium containing chase after 60 min (30°C) or 40 min (23°C), and the volume of cells harvested was increased from 1 to 1.5 ml for subsequent time points. The vps28 mutant was grown at 30°C in SDC-Met-Ura to OD600 of 0.4, labeled for 10 min with ∼300 μCi/ml EXPRE35S35S label (35S amino acid mix: New England Nuclear, Boston, MA), and chased by addition of cysteine and methionine to a final concentration of 2 mM each. Alternatively, cells were grown at 23°C in SD + Ade, His, Leu, Lys, Trp to OD600 of 1.0, labeled, and chased as described above. At various times after addition of chase, 10 mM NaF and 10 mM NaN3 were added to 107 cells, which were processed as described (54).
Radioactive band intensities were quantified using a Molecular Dynamics PhosphorImager (Sunnyvale, CA) and IPLabGel software (Signal Analytics, Vienna, VA), and t 1/2 values were determined by linear regression using data from five or more time points.
Site-directed Mutagenesis
DNA manipulations were performed as described (42). A NotI site was created at the 3′ end of SOI1 by mutagenesis of single-stranded, uracil-containing DNA (24) using the following primer: AGTACTGTGAAGGCGGCCGCTGATCACATATG. After subcloning this mutation into pUC19 (pUC19-NotI.1), the triple-HA tag from pGTEP1 (provided by Dennis Thiele, University of Michigan, Ann Arbor, MI) was introduced as a NotI fragment to create pUC19-HA1 (50). A 3.2-kb PstI fragment containing the 3′ 2,840 bp of SOI1 was then cloned into the pRS304 integrating vector (46). This plasmid was digested with NcoI, which cuts 1,293 bases upstream of the stop codon, and was transformed into BFY106-4D, CRY1, and CB018. Trp+ transformants were Vps+ and Spo+, indicating that Soi1p-HA was functional. Strains expressing Soi1p-HA specifically produced a band of >300 kD.
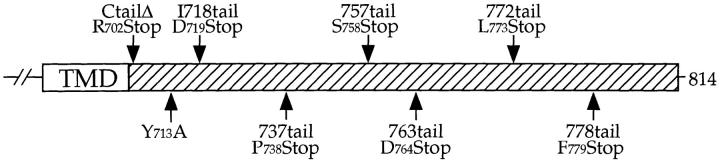
Mutations in the C-tail of Kex2p (shown schematically in Fig. 1) were introduced by two-step PCR using mutagenic primers (1). PCR-amplified mutant fragments and pCWKX20-D719Amb (34), which had been gapped with NarI and SnaBI, were cotransformed into yeast (4). Plasmids were retrieved from transformants into E. coli, and mutations were confirmed by DNA sequencing. The primers used were (number 1 = sense strand, number 2 = antisense strand): 737tail (P738Stop), 1) ATTACTGAGTGATCAGAGGTTGAG, 2) CTCAACCTCTGATCACTCAGTAAT; 757tail (S758Stop), 1) GCAAGTTTGTGATCATCAGAA, 2) TTCTGATGATCACAAACTTGC; 778tail (F779Stop), 1) AACGAAAATTCATGAAGTGACCCT, 2) AGGGTCACTTCATGAATTTTCGTT; Y713A I718tail, 1) AGAGCGGAAACGGCTGAGTTCGATATCATT, 2) AATGATATCGAACTCAGCCGTTTCCGCTCT; 763tail (D764Stop), 1) CATCAGAAAACTGAGATGCTGAAC, 2) GTTCAGCATCTCAGTTTTCTGATG; 772tail (L773Stop), 1) GATAGTGTATGAACAAACGAAAATCC, 2) GGATTTTCGTTTGTTCATACACTATC.
Figure 1.
Schematic of mutations in the Kex2p cytosolic tail (C-tail). The positions of mutations in various forms of Kex2p that were used in this study are indicated. The Y713A (55) and I718tail (34) mutations have been described. TMD, transmembrane domain.
RESULTS
Cloning of SOI1
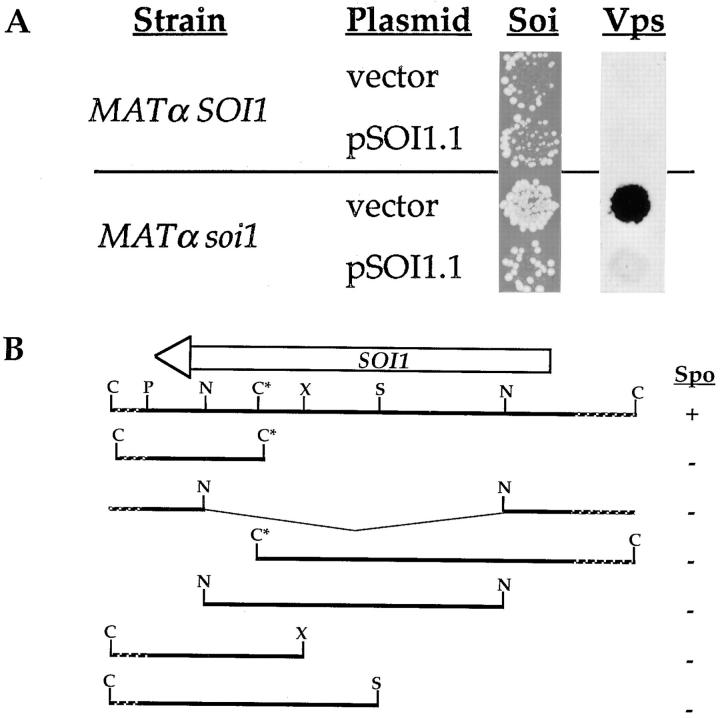
In addition to Soi+ and Vps− phenotypes, soi1 mutations also resulted in a severe sporulation defect. As measured by the production of viable spores (38), sporulation efficiency was reduced ∼2,500-fold in soi1 homozygous diploid strains (data not shown). However, asci were not identifiable by microscopic inspection of cultures of soi1 homozygous diploids. Because of the stringent nature of the sporulation phenotype, we exploited it to clone SOI1 by complementation, as described in Materials and Methods. Screening a single-copy yeast genomic library, only 2 of 20,000 transformants were Spo+. Plasmid loss and retransformation experiments confirmed that complementation of this phenotype was plasmid dependent (data not shown). These two Spo+ transformants contained the same plasmid, hereafter called pSOI1.1, that had a 12-kb insert. Transformation of a MATα soi1-2 kex2Δ strain with pSOI1.1 complemented both the Vps− and Soi+ phenotypes caused by the soi1-2 mutation (Fig. 2 A).
Figure 2.
Isolation of SOI1. (A) KRY18-1A (MATα kex2Δ) and JBY11-r1 (MATα kex2Δ soi1-2), expressing Y713A Kex2p under the control of the GAL1 promoter from plasmid pCWKX21 (55), were transformed with either pBS32 (vector control) or pSOI1.1, and were analyzed for their Soi and Vps phenotypes, as described in Materials and Methods. For the mating assay, strains were shifted to glucose for 6 h before testing mating competence. (B) Restriction map of pSOI1.1 and analysis of subcloned fragments. At the top is shown a restriction map of the insert from pSOI1.1. Indicated above the map is the extent and direction of the SOI1 open reading frame. Restriction fragments from pSOI1.1 were subcloned into either pBS32 or pRS313, transformed into JBY96 (MATa/MATα, soi1-2/soi1-2), and tested for complementation of the Spo− phenotype (scores are shown to the right of each subclone). Restriction sites are indicated as follows: C, ClaI; P, PstI; N, NcoI; S, SalI; X, XhoI. The ClaI site indicated by an asterisk is not unique, but is the only one within the insert not blocked by dam methylation.
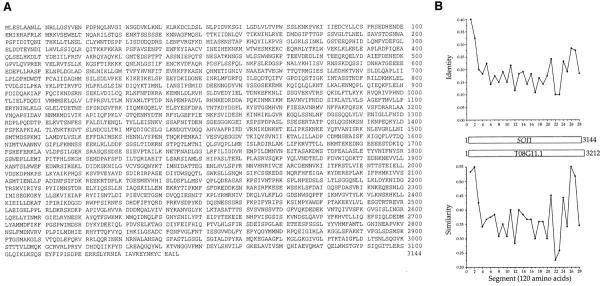
Genetic linkage between soi1-2 and the pSOI1.1 insert confirmed that pSOI1.1 contained the SOI1 gene (see Materials and Methods). Efforts to isolate smaller complementing fragments from pSOI1.1 were unsuccessful, suggesting that the length of the complementing gene was between 6 and 10 kb (Fig. 2 B). Sequencing of the insert from pSOI1.1 revealed a single long open reading frame of 9,432 bp (GenBank accession number AF001317; Fig. 2 B) predicted to encode a 3,144-residue polypeptide of 358 kD (Fig. 3 A). This open reading frame was flanked by fragments of the UBI4 and SDH2 genes, and comparison of this sequence to that from this region of chromosome XII (GenBank/EMBL/DDBJ accession number Z73145) revealed no significant differences. BLAST and FASTA similarity searches of the GenBank/EMBL/DDBJ database using the deduced amino acid sequence of Soi1p identified an open reading frame from Caenorhabditis elegans with significant similarity to SOI1 (T08G11.1; GenBank/EMBL/DDBJ accession number 1546759). This open reading frame would encode a protein of 3,212 amino acids that is 22% identical and 42% similar to Soi1p over its entire length, with the highest degree of similarity in the NH2 and COOH termini (Fig. 3 B). Given the strong conservation of NH2- and COOH-terminal domains along with nearly identical overall length and similar sequence composition, T08G11.1 represents a likely Soi1p homologue in C. elegans, suggesting conservation of the protein and its function between yeast and metazoans.
Figure 3.
Sequence of Soi1p and analysis of conserved sequences. (A) The deduced amino acid sequence of Soi1p (GenBank/EMBL/ DDBJ accession number AF001317). (B) Schematic representation of Soi1p and the deduced amino acid sequence from C. elegans open reading frame T08G11.1. Percent identity and similarity (using a PAM250 substitution matrix) were calculated using Clustal W (49) and plotted for segments of 120 amino acids (the COOH-terminal segment was 87 amino acids). Upper plot, percent identity; lower plot, percent similarity. A second potential C. elegans homologue of ∼3,200 codons (not shown) can be formed by joining two adjacent, predicted open reading frames from cosmid C25H3 (sequences C25H3.8 and C25H3.9, GenBank/EMBL/DDBJ accession No. U29535).
The sequence composition of Soi1p, as assessed by analysis of overlapping 100 residue segments, was surprisingly homogeneous throughout the length of the sequence. The sequence contained 25% charged residues (Glu, Asp, Arg, and Lys) with a predicted pI of 5.23. The sequence was rich in residues found in α-helical coiled coils (Leu + Ile + Val + Met = 26%). Levels of Ala (4.5%) and Gly (4.2%) in Soi1p were low. These features are suggestive of a protein with an extensive structural motif; however, no heptad repeats longer than two to three turns were predicted (using the program Coils; references 25, 26). Although the program TMpred (17) predicted three potential transmembrane domains in Soi1p, the interpretation of this result was uncertain because each potential transmembrane domain contained several strongly hydrophilic or charged residues. No other motifs, domains, or identifiable structural features were predicted through analysis of the Soi1p sequence with various search programs (data not shown).
SOI1 Encodes a High Molecular Weight Protein That Exists in a Sedimentable Complex
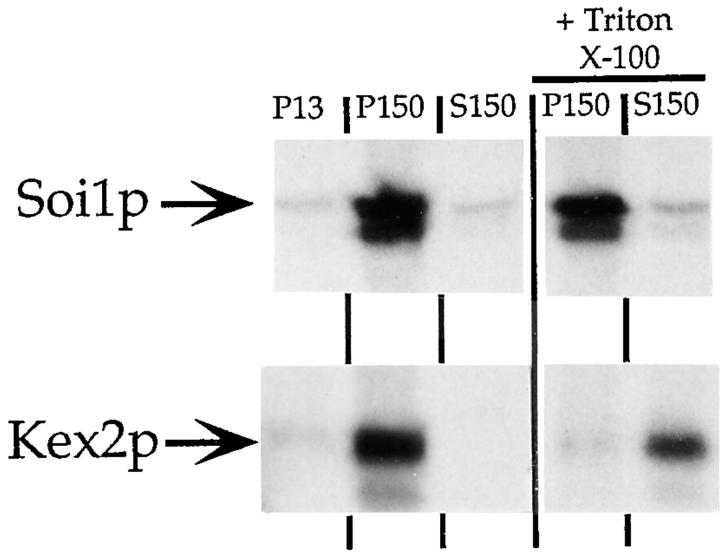
To identify the protein encoded by SOI1, we introduced three copies of the HA epitope at the carboxy terminus of Soi1p (see Materials and Methods). This allele of SOI1 (encoding Soi1p-HA) was integrated at the SOI1 locus and complemented both the Vps− and Spo− phenotypes of the soi1Δ strain (data not shown). Monoclonal anti-HA antibodies selectively immunoprecipitated a >300-kD polypeptide from strains expressing Soi1p-HA (data not shown). To assess the possible association of Soi1p with membranes, cells labeled with [35S]SO4 were converted to spheroplasts, and osmotic lysates were fractionated by differential centrifugation (51). In this fractionation, the low-speed pellet (P13) should contain ER and vacuolar membranes. The high-speed pellet (P150) should contain Golgi membranes, transport vesicles, and secretory vesicles, and the high speed supernatant fraction (S150) should contain soluble proteins (see Materials and Methods). Because Soi1p-HA was very sensitive to proteolytic degradation in lysates (data not shown), the experiments were performed in a strain devoid of vacuolar proteases. Similar to Kex2p, ∼90% of Soi1p-HA sedimented at 150,000 g, whereas very little (∼5%) sedimented at 13,000 g (Fig. 4). In contrast, the vacuolar ALP, as expected (51), was found in both the P13 and P150 fractions (data not shown). However, although addition of 0.1% Triton X-100 to the 13,000-g supernatant fraction before high speed centrifugation efficiently solubilized Kex2p, sedimentation of Soi1p-HA in the P150 fraction was unchanged (Fig. 4). Likewise, neither 1% deoxycholate nor 0.1 M Na2CO3, pH 11, had any effect on the sedimentation of Soi1p (data not shown). Thus, Soi1p-HA was associated with a sedimentable complex that was insensitive to detergent. Fractionation of lysates by discontinuous sucrose equilibrium density gradients revealed that >50% of Soi1p-HA cofractionated with a subpopulation of Kex2p-containing membranes (Sipos, G., J.H. Brickner, and R.S. Fuller, unpublished results). In Triton X-114 phase separation experiments, during which integral membrane proteins typically partition into the detergent phase (2), Soi1p-HA partitioned into the aqueous phase (data not shown). Therefore, Soi1p appears to exist in a high molecular weight heterooligomeric or homooligomeric complex that is peripherally associated with membranes.
Figure 4.
Soi1p is a high molecular weight protein that is found in a detergent-insensitive, sedimentable fraction. Labeled cell extracts were produced and processed as described in Materials and Methods. After a clearing spin, lysates were spun at ∼13,000 g to produce a pellet (P13). Triton X-100 (0.1%) was added to half of the supernatant fraction from this spin and the detergent-treated and untreated samples were then centrifuged at ∼150,000 g to produce a pellet (P150) and a supernatant fraction (S150). Kex2p and Soi1p-HA were immunoprecipitated from SDS-denatured samples and were separated by SDS-PAGE (see Materials and Methods).
Disruption of SOI1
Chromosomal deletions of SOI1 were created by gene replacement using the bacterial kan r gene (52), which confers resistance to G418. Diploid strains heterozygous for either of two SOI1 deletions (see Materials and Methods) were sporulated, and the tetrads were dissected. Nearly all tetrads produced four viable spores, indicating that SOI1 was not essential. In all complete tetrads, the Vps− and G418r phenotypes segregated as single loci (2:2) and displayed tight linkage. All G418r progeny were Vps− and all G418s progeny were Vps+ (data not shown). Thus, like the original soi1 mutant isolates (34), soi1Δ strains were Vps−.
The soi1 Null Mutation Affects Kex2p Localization, as Judged by Indirect Immunofluorescence
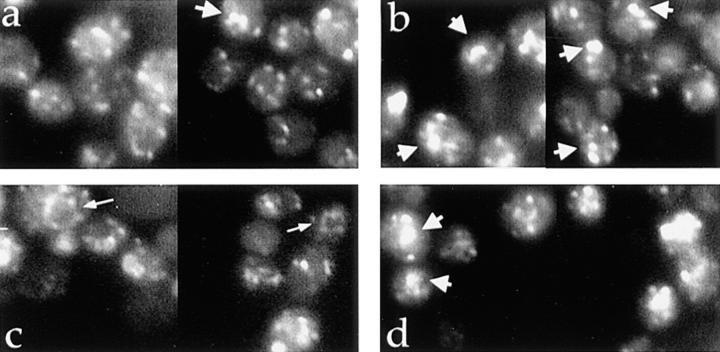
By indirect immunofluorescence, Kex2p displays a punctate cytoplasmic distribution (35) that is also observed for other Golgi proteins (12, 29, 31). We examined localization of both WT and Y713A Kex2p in SOI1 and soi1Δ strains by indirect immunofluorescence (Fig. 5). In the SOI1 strain (i.e., the soi1Δ strain transformed with pSOI1.1), WT Kex2p localized as expected to punctate structures throughout the cytoplasm (Fig. 5 a). WT Kex2p was also localized in a similar punctate distribution in the soi1Δ strain (Fig. 5 b); however, the Kex2p-containing structures were often larger and/or more elongated (see wide arrows in Fig. 5 b). Such structures were also observed in the SOI1 strain, but much less frequently (Fig. 5 a). Y713A Kex2p localized to faint punctate spots and occasionally to the vacuolar membrane in both the SOI1 and soi1Δ strains (Fig. 4, c and d). In the soi1Δ strain, Y713A Kex2p was also found associated with larger structures similar to those observed in the case of WT Kex2p in the soi1Δ strain (Fig. 5 d). Thus, the soi1Δ mutant exhibited a qualitative effect on the localization of both WT Kex2p and Y713A Kex2p that may reflect a change in either the structure of Kex2p-containing compartments or the organellar distribution of Kex2p.
Figure 5.
Indirect immunofluorescence analysis of Kex2p in SOI1 and soi1Δ-2 strains. JBY154-2A (kex2Δ soi1Δ-2) transformed with either pCWKX20 (a and b, WT Kex2p under GAL1 promoter control; reference 55) or pCWKX21 (c and d, Y713A Kex2p under GAL1 promoter control; reference 55), and either pSOI1.1 (a and c) or a vector control (b and d) were processed for immunofluorescence as described (35). Expression of Kex2p under GAL1 promoter control from a CEN plasmid results in ∼15-fold overexpression that is not thought to alter localization significantly (35). Vacuolar staining, apparent from bright field viewing, is indicated with narrow arrows. Wide arrows indicate larger structures present at a higher frequency in the soi1Δ strain.
TGN Localization of WT Kex2p, Vps10p, and Ste13p (A-ALP) Is Impaired in a soi1Δ Mutant
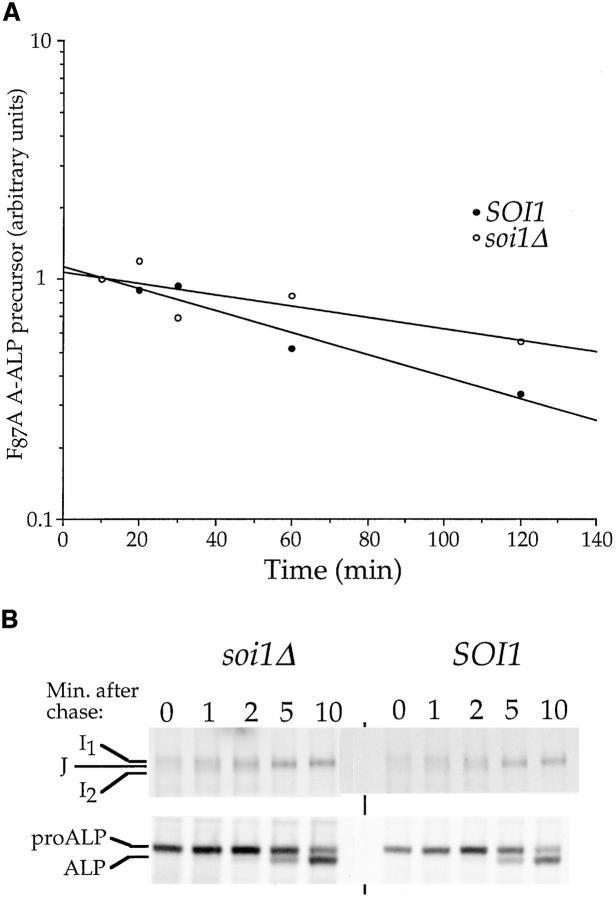
The original soi1 mutant alleles accelerated the degradation of WT Kex2p (34). To examine the effect of the soi1Δ mutation on the vacuolar degradation of WT Kex2p, we measured the t 1/2 of the protein in the soi1Δ strain. As in the case of the original soi1 alleles, WT Kex2p was degraded more rapidly in the soi1Δ strain (Fig. 6 A; t 1/2 = 50 min ± 4 min [SEM], n = 3) than in the wild-type strain (t 1/2 = 112 min ± 10 min, n = 3). Therefore, localization of WT Kex2p was perturbed by the soi1Δ mutation in the same way as by the original mutant alleles.
Figure 6.

SOI1 is required for TGN localization of Kex2p, Vps10p, and Ste13p, and for vacuolar targeting of proCPY. (A) Pulse-chase/immunoprecipitation of Kex2p. BFY106-4D (kex2Δ SOI1) and JBY134.1 (kex2Δ soi1Δ-1), transformed with CEN plasmids expressing either WT Kex2p (pCWKX10) or Y713A Kex2p (pCWKX11) from the KEX2 promoter (55), were pulse labeled with [35S]H2SO4 for 10 min before addition of chase. Cells were collected at the indicated times after addition of chase, lysed, and processed for immunoprecipitation of Kex2p. (B) BFY106-4D and JBY134.1 were labeled as described in A and chased. Samples were collected at the indicated times, separated into cells (I, intracellular) and medium (E, extracellular), and processed for immunoprecipitation of CPY. Forms of CPY are as follows: p1CPY, core-glycosylated ER form; p2CPY, Golgi form; mCPY, mature vacuolar form (47). (C and D) JBY135-1A (soi1Δ-1) and JBY135-2D (SOI1) were labeled with [35S]H2SO4 for 10 min and then chased. Samples were collected, lysed, and immunoprecipitated using anti-Vps10p (C) or anti-ALP (D) sera. Vps10p (C) and precursor A-ALP (D) were quantified after SDS-PAGE and plotted relative to initial band intensity. Half-life (C) or half-time of processing (D) were determined by linear regression (Table II).
The Vps− phenotype of the original soi1 mutants was evaluated qualitatively by a colony immunoblotting assay for CPY secretion (Fig. 2 A). To analyze the missorting of vacuolar proteins in greater detail, we examined the sorting and processing of CPY by pulse chase/immunoprecipitation. Transport of CPY through the secretory pathway can be followed by the modifications that occur during its transport to the vacuole. Core-glycosylated proCPY, p1CPY, exits the ER and is further glycosylated in the Golgi complex to produce p2CPY, which is proteolytically processed upon delivery to the vacuole to produce mature enzyme (mCPY; 47). Spheroplasts were labeled with [35S]H2SO4 for 10 min and then chased for 60 min. After 60 min of chase, all of the CPY in the SOI1 strain was found as intracellular mCPY (Fig. 6 B). In contrast, the soi1Δ strain missorted at least 50% of p2CPY to the cell surface after 60 min of chase (Fig. 6 B). A smaller fraction of proPrA, another vacuolar proenzyme (20), was also missorted to the cell surface in the soi1Δ mutant (data not shown).
In the late Golgi, p2CPY is actively sorted to the vacuole by the Vps10p sorting receptor (16, 27). Vps10p is localized to the TGN and requires sequences in its cytosolic tail for proper localization and sorting function (6, 9). Deletion of the cytosolic tail of Vps10p leads to rapid delivery of Vps10p to the vacuole, where it is degraded, and to poor sorting of p2CPY (6, 9). To determine whether missorting of p2CPY in soi1 mutants may result from a disruption of Vps10p localization, we examined the turnover rate of Vps10p by pulse-chase immunoprecipitation. In the SOI1 strain, Vps10p turned over slowly (t 1/2 = 122 min ± 9 min, n = 4; Fig. 6 C). Vps10p was degraded more quickly in the soi1Δ strain (t 1/2 = 70 min ± 6 min, n = 4). The behavior of Vps10p in the soi1Δ strain was similar to that of Kex2p. Both molecules were delivered to the vacuole approximately twofold more rapidly in the soi1Δ strain than in the wild-type strain.
We also assessed the function of the TGN localization signal of another TGN membrane protein, Ste13p, in the soi1Δ mutant by using a fusion protein consisting of the NH2-terminal cytosolic tail of Ste13p fused to the transmembrane and lumenal domains of ALP (A-ALP; 29). Delivery of this protein to the vacuole can be monitored by proteolytic maturation of proALP (21, 29). SOI1 and soi1Δ strains expressing A-ALP were labeled with [35S]H2SO4 for 10 min before adding chase. Samples were taken at regular intervals for up to 200 min. While A-ALP was very stable in the SOI1 strain (t 1/2 > 300 min; Fig. 6 B, closed circles), it was matured at an appreciable rate in the soi1Δ strain (t 1/2= 158 min ± 7 min, n = 2; Fig. 6 B, open circles). Thus, efficient TGN localization of three proteins, Kex2p, Ste13p, and Vps10p, requires Soi1p function.
Deletion of SOI1 Suppresses the Effect of the Y713A Substitution on Mating but Does Not Alter the Overall Rate of Delivery of Y713A Kex2p to the Vacuole
Mutations in SOI1 were originally isolated as suppressors of the rapid loss of mating competence exhibited by a MATα strain after shutting off expression of Y713A Kex2p (34). Deletion of SOI1 also resulted in suppression (Fig. 7 A). All three original soi1 alleles also decreased the rate of delivery of Y713A Kex2p to the vacuole (34). In contrast, deletion of SOI1 did not effect on the rate of vacuolar delivery of Y713A Kex2p. The rate of vacuolar degradation of Y713A Kex2p was the same in SOI1 and soi1Δ strains (Fig. 6 A; t 1/2 = 21 min ± 2 min [SEM] in soi1Δ strain, n = 4; t 1/2 = 23 min ± 3 min in SOI1 strain, n = 4). The onset of impotence assay reflects the level of Kex2p activity in the pro-α-factor processing compartment, presumably the TGN, whereas the t 1/2 of Kex2p may also reflect downstream events, e.g., the rate of Kex2p delivery from the PVC to the vacuole. Therefore, complete loss of Soi1p function apparently increased the concentration of Y713A Kex2p in the pro-α-factor processing compartment without measurably affecting the net rate of delivery of the protein to the vacuole. These results are consistent with a model in which Soi1p functions in two distinct steps in a pathway or cycle involved in Kex2p localization, one step governing the rate of transport of Kex2p from the PVC to the vacuole and the other affecting the concentration of the protein in the TGN.
Figure 7.
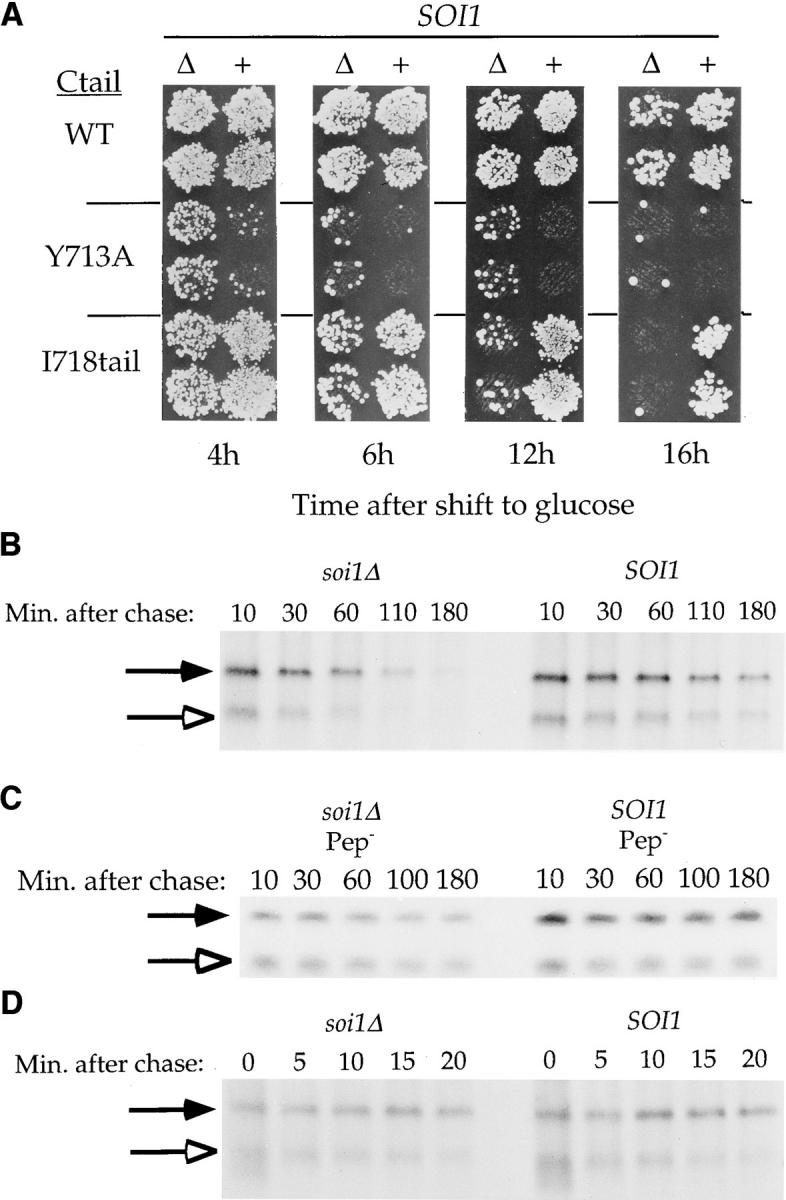
Deletion of SOI1 reveals a second TLS in the C-tail of Kex2p. (A) BFY106-4D (SOI1 kex2Δ) and JBY134.1 (soi1Δ-1 kex2Δ) expressing WT Kex2p, Y713A Kex2p or I718tail Kex2p, under the control of the GAL1 promoter, were shifted from galactose to glucose for the indicated times before testing mating competence. (B and D) JBY135-1A (soi1Δ-1) and JBY135-2D (SOI1) carried CEN plasmids expressing either I718tail Kex2p (pCWKX10-I718tail; B) or C-tailΔ Kex2p (pCWKX17; D) from the KEX2 promoter (34, 55). (C) JBY173 (soi1Δ-2 pep4 prb1 prc1) and CB018 (SOI1 pep4 prb1 prc1) carried pCWKX10-I718tail. Strains were labeled for 10 min (B and C) or 5 min (D), chased, and samples were collected at the indicated times after the addition of chase. Cells were lysed, and Kex2p was immunoprecipitated using antiserum raised against the Kex2p lumenal domain (55). WT Kex2p is indicated by closed arrows. I718tail Kex2p (B and C) and C-tailΔ Kex2p (D) are indicated by open arrows.
These data also demonstrate that the original three soi1 alleles, by their effects on the rate of transport of Y713A Kex2p to the vacuole, are phenotypically distinguishable from the soi1Δ allele. Therefore, the original alleles must encode partially functional proteins despite the fact that they are recessive to wild type. To determine whether this partial function also affected the degree of suppression of Y713A mutation in Kex2p in the onset of impotence assay, the mutant soi1 genes were deleted by one-step gene disruption in soi1-1, -2, and -3 strains. The resulting soi1Δ strains were compared side-by-side to the soi1-1, -2, and -3 strains in the onset of impotence assay for suppression of Y713A Kex2p. Suppression by the original soi1 allele and the deletion allele was identical in each case (data not shown). Therefore, suppression in this assay results from loss of Soi1p function, and the degree of suppression by the soi1 mutations does not correlate with the t 1/2 of the Y713A Kex2p.
Analysis of the soi1Δ Mutant Reveals the Existence of a Second TGN Localization Signal in the Kex2p C-tail, TLS2
Consistent with the decreased t 1/2 of WT Kex2p in soi1 mutant strains, the original soi1 alleles also exhibited a measurable effect on WT Kex2p in the onset of impotence assay (34). We therefore assessed the effect of the soi1Δ mutation on WT Kex2p using this assay. As can be seen in Fig. 7 A, 16 h after shutting off expression of WT Kex2p, the soi1Δ strain mated slightly less well than the SOI1 strain, though much better than the soi1Δ strain expressing Y713A Kex2p.
Also examined by the onset of impotence assay was I718tail Kex2p, which has only the first 19 amino acids of the tail. I718tail Kex2p behaved like WT Kex2p in both t 1/2 and in the onset of impotence assay, suggesting that it contains all sequences within the C-tail necessary for localization (34). Despite the presence of the Tyr713-based signal in I718tail Kex2p, loss of Soi1p function had a much more profound effect on I718tail Kex2p than on WT Kex2p. I718tail Kex2p was lost more quickly from the TGN (Fig. 7 A) and was delivered more rapidly to the vacuole (Fig. 7 B) in the soi1Δ strain than in the SOI1 strain. These results lead to two conclusions. First, Soi1p is required for the function of the TLS in I718tail Kex2p, presumably the Tyr713-based signal (Fig. 7 A, compare I718tail Kex2p in SOI1 and soi1Δ strains after 16 h on glucose). Second, to account for the superior TGN localization of WT Kex2p in the soi1Δ strain (Fig. 7 A, compare WT Kex2p to I718tail Kex2p in the soi1Δ strain after 12 or 16 h on glucose), a second localization signal must exist in the WT Kex2p C-tail that is not present in I718tail Kex2p. We have designated this second signal TLS2 and the Tyr713-based signal TLS1. The rates of degradation of both the WT and I718tail forms of Kex2p in the soi1Δ strain represent the rates of transport to the vacuole because these proteins were stable in strains lacking vacuolar proteases (Fig. 7 C).
Suppression of Y713A Kex2p by the soi1Δ Mutation Requires TLS2: Soi1p Antagonizes TLS2 Function
The suppression of the mating defect of Y713A Kex2p in soi1Δ strains could be explained in two ways. It might have resulted from loss of active discrimination against Ala at position 713, which might also explain the defect in the function of the WT TLS1. Alternatively, suppression might have resulted from the activation of TLS2 function by removal of Soi1p. The first model predicts that suppression of Tyr713Ala should be identical in the context of either the full-length tail or the I718tail, whereas the second model predicts that suppression should depend on TLS2. To distinguish between these models, we substituted Ala for Tyr713 in I718tail Kex2p and assessed the TGN localization of this protein in SOI1 and soi1Δ strains using the onset of impotence assay. Y713A I718tail Kex2p behaved identically in SOI1 and soi1Δ strains in the onset of impotence assay (Fig. 8, A and B). Suppression of Y713A was only observed when TLS2 was present, i.e., only in the context of the full-length tail. Therefore, suppression of Y713A Kex2p in soi1 mutants resulted from activation of TLS2, implying that TLS2 is normally antagonized by Soi1p. Y713A-I718tail Kex2p, lacking both TLS1 and TLS2, behaved like C-tailΔ Kex2p in this assay (Fig. 8 A).
Figure 8.

(A) Suppression of the localization defect of Y713A Kex2p in the soi1Δ strain requires TLS2. JBY154-1A (MATα soi1Δ-2 kex2Δ) and JBY154-2A (MATα SOI1 kex2Δ) expressing either Y713A Kex2p (pCWKX21), Y713A I718tail Kex2p (pCWKX21-I718), or C-tailΔ Kex2p (pCWKX27) under the control of the GAL1 promoter on CEN plasmids were shifted from galactose to glucose for 5 h before testing mating competence. (B) Optimal TLS1 function requires Soi1p, but TLS1 exhibits residual function in the absence of Soi1p. Strains JBY154-1A and JBY154-2A expressing either I718tail Kex2p (pCWKX20-I718) or Y713A I718tail Kex2p under the control of the GAL1 promoter on CEN plasmids were shifted from galactose to glucose medium for the indicated times before testing mating competence. Indicated to the left of each row of patches is the form of Kex2p expressed. The relevant SOI1 allele is indicated above the columns.
soi1 mutations were classified as allele-specific suppressors because they suppressed Ala and other substitutions at Tyr713, but not complete deletion of the C-tail in Kex2p (34). Examination of C-tailΔ Kex2p by both half-life determination (Fig. 7 D) and onset of impotence (Fig. 8 A) indicated that deletion of SOI1 had no effect on either the concentration of this protein in the TGN or its rate of delivery to the vacuole, consistent with results obtained with soi1-1, -2, and -3 strains (34). The basis for the allele-specific suppression of soi1 mutations, therefore, is the activation of TLS2, a signal that is absent from both Y713A-I718tail Kex2p and C-tailΔ Kex2p.
An obvious question was whether TLS2 functioned at all in the presence of Soi1p. If so, the TGN localization of Y713A Kex2p in the SOI1 strain would be measurably better than that of Y713A I718tail Kex2p in the same strain. Y713A I718tail Kex2p was delivered to the vacuole and degraded with a t 1/2 of 15 min in the wild-type SOI1 strain (Table II). The t 1/2 of Y713A Kex2p was 50% longer (t 1/2 = 23 ± 3 min; Table II) in the SOI1 strain. Thus, TLS2 appears to function to some degree even in a wild-type SOI1 strain.
Table II.
Rate of Delivery of TGN Membrane Proteins to the Vacuole in SOI1 and soi1Δ Strains
| Half-life or half-time of delivery to the vacuole | ||||
|---|---|---|---|---|
| Protein | SOI1 | soi1Δ | ||
| min | ||||
| WT Kex2p | 112 ± 10 | 50 ± 4 | ||
| I718tail Kex2p | 80 | 35 | ||
| Y713A Kex2p | 23 ± 3 | 21 ± 2 | ||
| Y713A I718tail Kex2p | 15 | 12 | ||
| C-tailΔ Kex2p | 8* | 11 | ||
| Vps10p | 122 ± 9 | 70 ± 6 | ||
| A-ALP | >300 | 158 ± 7 | ||
| F87A A-ALP | 60 | 125 | ||
The t1/2 for C-tailΔ Kex2p in the WT (SOI1) background was previously measured as 11 min (55).
Optimal TLS1 Function Requires Soi1p, but TLS1 Exhibits Residual Function in the Absence of Soi1p
As discussed previously, I718tail contains a TGN localization signal that requires Soi1p for optimal function (Fig. 7 A; see also Fig. 8 B). Substitution of Ala for Tyr713 in the I718tail Kex2p completely abrogated localization in both SOI1 and soi1Δ strains (Fig. 8 B). Therefore, the Soi1p- dependent signal responsible for TGN localization of I718tail Kex2p is TLS1. However, comparison of I718tail Kex2p with Y713A I718tail Kex2p in the soi1Δ strain at both the 7- and 11-h time points clearly indicated that TLS1 exhibited significant residual function in the absence of Soi1p (Fig. 8 B).
Soi1p Antagonizes a TGN Localization Signal in the Ste13p C-Tail That Is Distinct from the F-X-F87 Signal
Previously, we found that the original soi1 mutations suppressed the effects of the F87A substitution in the Ste13p C-tail, as measured by the t 1/2 of maturation of the A-ALP fusion protein (34). To determine conclusively whether suppression in this case resulted from loss of Soi1p function, we examined the t 1/2 for maturation of F87A A-ALP in the soi1Δ strain (Fig. 9 A). In the SOI1 strain, F87A A-ALP matured with a t 1/2 of ∼60 min (Fig. 9; references 29, 36). In the soi1Δ strain, maturation of F87A A-ALP was delayed, occurring with a t 1/2 of 125 min (Fig. 9 and Table II). Thus, deletion of SOI1 resulted in a slower rate of delivery of F87A A-ALP to the vacuole, suggesting that loss of Soi1p resulted in activation of a second TGN localization signal in the Ste13p C-tail.
Figure 9.
(A) Improved TGN localization of F87A A-ALP in a soi1 mutant strain. Strains JBY135-1A (soi1Δ-1) and JBY135-2D (SOI1), transformed with pSN98 (expressing F87A A-ALP; reference 29), were labeled for 10 min with [35S]H2SO4 and chased for 120 min. Samples were collected after 10, 20, 30, 60, and 120 min of chase and processed for immunoprecipitation using anti-ALP antiserum. Band intensity of pro-A-ALP was quantified after SDS-PAGE (see Materials and Methods) and plotted relative to initial band intensity. (B) Deletion of SOI1 does not affect transport through early secretory compartments. Strains JBY154-1D (KEX2 soi1Δ-2) and JBY154-8B (KEX2 SOI1) were labeled with EXPRE35S35S label for 2 min before addition of chase. Cells were collected at the indicated times after addition of chase. Indicated beside the top panel are precursor forms of Kex2p, I1 (pro-Kex2p possessing core glycosyl modifications) and I2 (mature, core glycosylated Kex2p), and Golgi-modified, mature Kex2p (J; reference 54). Indicated beside the bottom panel are proALP and mature ALP (21).
Deletion of SOI1 Does Not Affect Early Transport Steps in the Secretory Pathway
It was conceivable that the reduced rate of transport of F87A A-ALP to the vacuole and the suppression of Y713A Kex2p in the soi1Δ mutant might have resulted from slower transport steps proximal to the late Golgi. To address this possibility, we analyzed the rate of transport from the ER to the Golgi by following the rate of posttranslational modification of Kex2p. If transport from the ER to the Golgi were slowed in the soi1Δ mutant, then conversion of core-glycosylated Kex2p (I2; reference 54) to Golgi-glycosylated Kex2p (J) would be expected to be slowed. However, rapid pulse-chase immunoprecipiation indicated that the conversion from core-glycosylated pro-Kex2p (I1) to I2 and from I2 to J were identical in SOI1 and soi1Δ strains (Fig. 9 B). This result also shows that TLS2 does not express its function during the early steps of the secretory pathway.
We also analyzed the rate of transport of proALP through the Golgi and to the vacuole. If the rate of ER- to-Golgi and/or intra-Golgi transport steps were slowed in the soi1Δ strain, then the rate of delivery of ALP to the vacuole should have been slowed. Analysis of the proteolytic maturation of pro-ALP, which occurs upon delivery of this protein to the vacuole (21), indicated that deletion of SOI1 had no effect on the rate of delivery of ALP to the vacuole. Therefore, mutation of SOI1 had no effect on the transport of proteins from the ER to the Golgi or through the Golgi.
TLS2 Regulates the Rate of Delivery of Kex2p to the PVC
The fact that activation of TLS2 can suppress mutation of TLS1 (i.e., in Y713A Kex2p) suggested that TLS2 functions before TLS1. Specifically, the fact that deletion of SOI1 increased the amount of Y713A Kex2p in the pro-α-factor processing compartment suggested that TLS2 delays the exit of Kex2p from the TGN. Conversely, TLS1 might function to promote retrieval of Kex2p from the PVC. To test this hypothesis, we measured the rate of transport of Kex2p to the PVC directly using a class E Vps− mutant strain. We reasoned that, if TLS2 functioned to improve TGN retention of Kex2p, then it should slow the rate of degradation of Kex2p in class E PVC compartment. Furthermore, if TLS1 functioned in the retrieval of membrane proteins from the PVC to the TGN, then its integrity should have no effect on the rate of degradation of Kex2p in the class E vps mutant strains.
We analyzed the rate of turnover of WT Kex2p coexpressed with either I718tail Kex2p or Y713A I718tail Kex2p in a vps28 strain (Fig. 10). All three forms of Kex2p were degraded rapidly in the vps28 strain at 30°C (Fig. 10 A), with I718tail Kex2p and Y713A I718tail Kex2p exhibiting nearly identical t 1/2 values of 24 and 23 min, respectively (Fig. 10). This indicated that TLS1 has no effect on the rate of delivery to the PVC and likely functions to promote retrieval to the TGN. There was only a small difference between the rates of delivery of I718tail Kex2p and of WT Kex2p (i.e., full-length; t 1/2 = 27 min ± 1 min) to the PVC. To amplify the possible difference between WT Kex2p and I718tail Kex2p in this assay, we compared the t 1/2 values of the two proteins in the vps28 mutant at 23°C (see Materials and Methods). Under these conditions, WT Kex2p was degraded with a t 1/2 of 37 min ± 2 min, while I718tail Kex2p was degraded with a t 1/2 of 27 min ± 3 min (n = 2; Fig. 10 B). Thus, TLS2 slows the rate of delivery of Kex2p to the PVC. Together with the observation that activation of TLS2 leads to an increase in the concentration of Y713A Kex2p in the TGN (Fig. 8 A), these data suggest that TLS2 regulates the rate of exit of Kex2p from the TGN.
Figure 10.
TLS2 slows delivery of Kex2p to the PVC. (A) Mutation of TLS1 does not affect the rate of delivery to the PVC. Strain 0472-28 (vps28 KEX2) expressing either I718tail Kex2p (pCWKX10-I718tail; top panel) or Y713A I718tail Kex2p (pCWKX11-I718tail; bottom panel) on a CEN plasmid was pulse-labeled for 10 min at 30°C in SDC-Met-Ura and chased for 80 min (see Materials and Methods). At the indicated times after addition of chase, cells were collected, lysed, and immunoprecipitated using antisera against the Kex2p lumenal domain. After SDS PAGE, WT Kex2p, I718tail Kex2p, or Y713A I718tail Kex2p were quantified, and t 1/2 values were obtained by linear regression. Indicated are the positions of WT Kex2p (Full length; filled arrows), I718tail Kex2p, or Y713A I718tail Kex2p (open arrows). (B) Strain 0472-28 containing pCWKX10-I718tail was grown at 23°C in SD + Ade, His, Leu, Lys, and Trp, labeled, chased, and processed for immunoprecipitation as described in A. Note that under otherwise identical conditions, the rate of degradation of Kex2p in the vps28 mutant is slower in SDC-Met-Ura than in SD + Ade, His, Leu, Lys, and Trp (data not shown). This may be caused by increased proteolytic activity in the PVC caused by amino acid limitation.
We attempted to analyze the rates of transport of full-length and I718tail Kex2p to the PVC in the soi1 vps28 strain. However, deletion of SOI1 in the context of the vps28 mutation stabilized I718tail Kex2p as well as full-length Kex2p, suggesting that the combination of the two mutations reduced proteolytic activity in the PVC (data not shown).
Mapping of TLS2
To map the Kex2p TLS2, we examined a series of truncated forms of Kex2p in the soi1Δ strain using the onset of impotence assay. These truncation mutants were indistinguishable from full-length Kex2p in the SOI1 strain (Fig. 11). In the soi1Δ strain, however, mating behavior indicated that whereas 778tail Kex2p possessed TLS2, its function was lost by deletion of six additional residues (see 772tail Kex2p in Fig. 11). Thus, the COOH-terminal endpoint of TLS2 appears to be discrete.
Figure 11.

Mapping TLS2. A series of Kex2p truncation mutants (shown schematically in Fig. 1) was expressed under the control of the GAL1 promoter in JBY154-1A (MATα kex2Δ soi1Δ-2) and JBY154-2A (MATα kex2Δ SOI1), and was tested for mating competence in the onset of impotence assay after 10 h on glucose. Sequence analysis of the 778tail Kex2p used in this experiment revealed that this protein had a second mutation at the carboxy terminus (P778S). This second mutation was inconsequential in that other isolates of this truncation that lacked this second mutation behaved identically to the one shown (data not shown).
DISCUSSION
Analysis of the soi1Δ strain led to the identification of a second signal (TLS2) in the Kex2p C-tail whose function is antagonized by Soi1p. This signal appears to function at the TGN because it delays transport from the TGN to the PVC in the class E vps28 mutant, and because it increases the concentration of Kex2p in the pro-α-factor processing compartment. TLS1 does not appear to function at the TGN. Its presence does not affect the rate of transport from the TGN to the PVC in the class E vps28 mutant. This is consistent with the idea that transport of Kex2p from the TGN to the PVC does not depend on sequences in the C-tail (36). However, TLS1 function is important for proper TGN localization. Therefore, it must function in an event downstream from TGN to PVC transport. The most likely role of TLS1 is in promoting retrieval of Kex2p from the PVC. A similar conclusion has been reached concerning the role of the aromatic residue–containing signal, FXFXD, in the Ste13p C-tail (5). In contrast to TLS2, which is activated in the absence of Soi1p, TLS1 requires the presence of Soi1p for full function. Thus, Soi1p appears to function both at the TGN and the PVC.
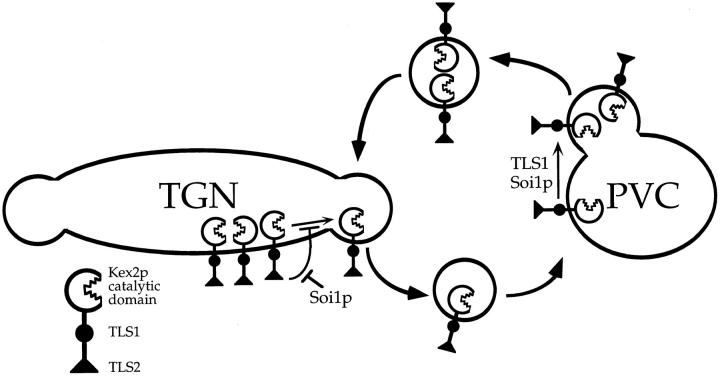
A model for Soi1p function and the localization of TGN transmembrane proteins such as Kex2p and Ste13p is presented in Fig. 12. In this model, Soi1p promotes cycling of TGN transmembrane proteins between the TGN and PVC. Soi1p antagonizes the function of “retention” signals (TLS2 in Kex2p) in the TGN, promoting entry of proteins into transport vesicles targeted to the PVC. At the PVC, Soi1p promotes entry of proteins containing aromatic residue–containing retrieval signals (TLS1 in Kex2p) into retrograde transport vesicles. The proposed function of Soi1p at two organelles and at two distinct steps in this cycling pathway is an unusual and important feature of this model. A second important feature is that Soi1p function is not required for transport of proteins between the TGN and PVC per se. Rather, Soi1p regulates the function of TLS1 and TLS2 which, in turn, regulate the entry of proteins into the cycling pathway. The localization of forms of Kex2p that lack both signals (e.g., Y713A I718tail Kex2p or CtailΔ Kex2p) is unaffected by the loss of Soi1p. In other words, the function of Soi1p in promoting cycling is achieved through regulating the function of localization information and not through the efficiency of the transport steps that constitute this pathway. The role proposed for Soi1p is different from that of other proteins shown to be involved in TGN-to-PVC transport, including clathrin heavy chain and Vps1p. These proteins appear to be required for transport of all proteins from the TGN to the PVC. Loss of their function blocks transport of proteins regardless of the presence of signals in their tails (28, 36).
Figure 12.
Model for role of Soi1p, TLS1, and TLS2 in the cycling of Kex2p between the TGN and PVC. TLS2 inhibits/delays entrance of Kex2p into the PVC transport vesicle at the TGN. Soi1p inhibits this function of TLS2. TLS1 and Soi1p together promote entry of Kex2p into newly forming TGN transport vesicle at PVC.
Although we have not characterized the role of Soi1p in localization of Ste13p in as much detail, it is likely to be similar to the role of Soi1p in Kex2p localization. Loss of Soi1p impaired the localization of WT A-ALP, but it improved the localization of F87A A-ALP, which lacks the aromatic residue–containing retrieval signal. These effects are consistent with a positive role for Soi1p function in the retrieval of Ste13p, which depends on the aromatic residue containing signal, and with Soi1p-dependent antagonism of a second signal in the Ste13p C-tail. Analysis of the Ste13p C-tail recently revealed the presence of a second signal, analogous to TLS2 in Kex2p, that is distinct from the aromatic residue–containing TLS, and that appears to prevent exit of Ste13p from the TGN (5). It seems likely that this signal is the one that is activated in the soi1Δ mutant, although this has not been tested. Analysis of residues 758–778 in Kex2p revealed two sequence blocks (E761-G763 and T768-D770) that had potential counterparts in the Ste13p C-tail. Moreover, truncation of the Kex2p C-tail between or downstream of these blocks disrupted TLS2 function (Fig. 11, 763tail and 772tail). However, mutant forms of Kex2p in which three alanines were substituted for either of these blocks behaved like full-length Kex2p in both SOI1 and soi1Δ strains, indicating that neither of these blocks was required for TLS2 function (data not shown). Conversely, deletions that inactivated the retention signal in the Ste13p C-tail did not remove the counterparts of the E761-G763 and T768-D770 sequences in the Ste13p C-tail (5). Thus, although they may fulfill analogous roles, the TGN retention signals in Kex2p and in Ste13p lack obvious sequence similarity.
Several observations suggest that this second TGN localization signal in Ste13p is a stronger signal than TLS2 in Kex2p. In SOI1 strains, the difference in the rate of delivery to the PVC between full-length Kex2p and I718tail Kex2p was small, indicating that TLS2 is ordinarily a weak signal (Fig. 10). In an analogous experiment using a class E vps27 strain, there was a substantial difference in the rate of delivery to the PVC between forms of A-ALP having the retention signal (t 1/2 = 60 min) and forms lacking it (t 1/2 = 15 min; reference 5). Deletion of SOI1, and, hence activation of TLS2, had no effect on the net rate of delivery of Y713A Kex2p to the vacuole. In contrast, deletion of SOI1 resulted in a significant decrease in the t 1/2 of delivery of F87A A-ALP to the vacuole. Moreover, mutation of the aromatic residue–containing retrieval signal of Ste13p in F87A A-ALP resulted in a relatively modest defect in TGN localization (29). Therefore, it seems likely that the retention signal of Ste13p is more effective than the Kex2p TLS2 and, conversely, that the aromatic residue–based TLS1 is more important in the localization of Kex2p.
The difference between the Kex2p TLS2 and the Ste13p retention signal provides a reasonable explanation for the large difference in t 1/2 between the two proteins (Figs. 5 and 6; references 29, 55). Why might different proteins have localization signals of different strengths? Variation in the strength of a TGN retention signal offers a mechanism to adjust both the distribution of membrane proteins between the TGN and the PVC and the relative rates of turnover of TGN membrane proteins. The weak TLS2 in Kex2p might result in a more balanced distribution of the protein between the TGN and the PVC. This could have physiological significance if, for example, Kex2p processing activity were required in both the TGN and in the PVC. Some proteins may use one or the other kind of localization signal exclusively. Kex1p carboxypeptidase, for example, depends on sequences in its C-tail for proper TGN localization, but lacks an obvious aromatic residue– containing TLS (8). It is possible that the Kex1p C-tail contains only a TGN retention signal. Conversely, because the role of Vps10p in vacuolar protein sorting requires that it cycle efficiently between the TGN and PVC, Vps10p might not be expected to have a TGN “retention” signal. Localization of Vps10p might be mediated exclusively by signal- and Soi1p-dependent retrieval from the PVC.
Although the deduced amino acid sequence of Soi1p is unlike that of any proteins of known function, it does appear to be conserved between yeast and C. elegans (Fig. 3 B). We have also identified human (cDNA clone 727106, GenBank/EMBL/DDBJ accession numbers AA292831 and AA398770) and Drosophila melanogaster (cDNA clone CK01879, GenBank/EMBL/DDBJ accession number AA141511) expressed sequence tags that represent other likely homologues. The existence of these probable homologues argues that mechanisms of TGN membrane protein localization are conserved between yeast and higher eukaryotes, including mammals. The cycling of Kex2p, Ste13p and Vps10p between the TGN and PVC is similar to that of mannose-6 phosphate receptors between the TGN and the late endosome in mammalian cells (22). In addition, net TGN localization of the mammalian Kex2p homologue furin is achieved by cycling between the TGN, the cell surface, and endosomal compartments (3, 7, 48).
The biochemical role of Soi1p is still largely unknown. Even though extensive heptad repeat regions are not found, the highly conserved domains at the NH2 and COOH termini, combined with a monotonous amino acid composition rich in aliphatic and charged residues in between, are reminiscent of intermediate filament proteins (13). Models of Soi1p function must accomodate roles at both the TGN and PVC. Soi1p might be associated with the TGN and PVC independently or might itself cycle between the two organelles in association with transport vesicles. An interesting possibility is that Soi1p forms a cytoskeletal element that physically links the TGN and PVC. Unfortunately, efforts to visualize Soi1p tagged with the HA epitope or with green fluorescent protein have been unsuccessful thus far, most likely due to the fact that Soi1p appears to be present at very low levels in cells (data not shown). On the basis of comparing the efficiency of labeling with [35S]methionine, we estimate Soi1p to be 5–10-fold less abundant than Kex2p, which is present at only a few hundred molecules per cell (reference 14 and data not shown).
It remains to be determined whether Soi1p physically interacts with TLS1 or TLS2. The sorting of TLS1-containing proteins back to the TGN is impaired in soi1Δ strains, which is consistent with a requirement for Soi1p in effective TLS1-dependent retrieval. Soi1p is not absolutely required for the recognition of TLS1 in that substitution of Ala for Tyr713 in the Kex2p C-tail has a small but measurable effect in a soi1Δ strain (Fig. 8 B). The defect in retrieval exhibited by the soi1Δ strain may result from either a sorting defect or a subtle defect in transport from the PVC to the TGN. A unifying role for Soi1p that is consistent with all the current data is in the recruitment of TGN membrane proteins into transport vesicles leaving both the TGN and the PVC. Such a function would necessarily antagonize a TGN retention signal and would also facilitate the use of a retrieval signal. This “recruitment factor” role for Soi1p is analogous to roles proposed for proteins such as Emp24p (43), Bst2p (11), and Shr3p (23), which are thought to promote sorting of cargo molecules into COPII-coated vesicles at the ER. Given the apparent disposition of Soi1p as a cytosolic peripheral membrane protein, we think Soi1p must function through interactions with other cytosolic factors or with cytosolic domains of transmembrane proteins. Identification of the interacting partners of Soi1p should help in dissecting the signal- mediated events in the localization of TGN membrane proteins.
Acknowledgments
We thank Nia Bryant, Scott Emr, Bruce Horazdovsky, Eric Marcusson, John McCusker, Steve Nothwehr, Tom Stevens, and Dennis Thiele for generously providing plasmids, antibodies, and strains. We also thank Susan Brown, N. Bryant, Amy Chang, S. Emr, and T. Stevens for helpful discussions and/or for communicating results before publication. We thank Kevin Redding, Andrea Stoddard, and György Sipos for technical and intellectual input; Kevin Morano, D. Thiele, and the Fuller Laboratory members for helpful discussions and comments on the manuscript.
Abbreviations used in this paper
- A-ALP
dipeptidyl aminopeptidase A–alkaline phosphatase fusion protein
- ALP
alkaline phosphatase
- C-tail
cytosolic tail
- CPY
carboxypeptidase Y
- HA
hemagglutinin
- PVC
prevacuolar compartment
- Soi
suppression of the onset of impotence
- Spo
sporulation competence
- TLS
trans-Golgi network localization sequence
- Vps
vacuolar protein sorting
- WT
wild type
Note Added in Proof
While this paper was in press, a paper was published that described the identification of an insertion mutation in VPS13 (SOI1) as a suppressor of mutation in plasma membrane ATPase that diverts Pma1p to the vacuole (Luo, W., and Chang, A. 1997. Novel genes involved in endosomal traffic in yeast revealed by suppression of a targeting-defective plasma membrane ATPase mutant. J. Cell Biol. 138:731–746).
Footnotes
Address all correspondence to Robert S. Fuller, Department of Biological Chemistry, Room 5413 Med. Sci. I, 1301 Catherine Road, University of Michigan Medical School, Ann Arbor, MI 48109-0606. Tel.: (313) 936-9764. Fax: (313) 763-7799. e-mail: bfuller@umich.edu
This work supported in part by National Institutes of Health (NIH) training grant GM07599 (J.H. Brickner) and NIH grant GM50915 (R.S. Fuller).
REFERENCES
- 1.Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1989. Mutagenesis of cloned DNA. In Current Protocols in Molecular Biology. Vol. 1. John Wiley and Sons, New York. 8.0.1–8.5.9.
- 2.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 3.Bosshart H, Humphrey J, Deignan E, Davidson J, Drazba J, Yuan L, Oorschot V, Peters PJ, Bonifacino JS. The cytoplasmic domain mediates localization of furin to the trans-Golgi network en route to the endosomal/lysosomal system. J Cell Biol. 1994;126:1157–1172. doi: 10.1083/jcb.126.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner C, Bevan A, Fuller RS. One-step site-directed mutagenesis of the Kex2 protease oxyanion hole. Curr Biol. 1993;3:498–506. doi: 10.1016/0960-9822(93)90040-u. [DOI] [PubMed] [Google Scholar]
- 5.Bryant NJ, Stevens TH. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPSgene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman RE, Munro S. Retrieval of TGN proteins from the cell surface requires endosomal acidification. EMBO (Eur Mol Biol Organ) J. 1994;13:2305–2312. doi: 10.1002/j.1460-2075.1994.tb06514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper A, Bussey H. Yeast Kex1p is a Golgi-associated membrane protein: deletions in a cytoplasmic targeting domain result in mislocalization to the vacuolar membrane. J Cell Biol. 1992;119:1459–1468. doi: 10.1083/jcb.119.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durr M, Boller T, Wiemken A. Polybase-induced lysis of yeast spheroplasts. Arch Microbiol. 1975;105:319–327. doi: 10.1007/BF00447152. [DOI] [PubMed] [Google Scholar]
- 11.Elrod-Erickson MJ, Kaiser CA. Genes that control the fidelity of endoplasmic reticulum to Golgi transport identified as suppressors of vesicle budding mutations. Mol Biol Cell. 1996;7:1043–1058. doi: 10.1091/mbc.7.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzusoff A, Redding K, Crosby J, Fuller RS, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function and disease. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 14.Fuller RS, Brake A, Thorner J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc Natl Acad Sci USA. 1989;86:1434–1438. doi: 10.1073/pnas.86.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuller RS, Sterne RE, Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- 16.Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18(NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann K, Stoffel W. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 18.Jones EW. Proteinase mutants of Saccharomyces cerevisiae. . Genetics. 1977;85:23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julius D, Brake A, Blair L, Kunisawa R, Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- 20.Klionsky DJ, Banta LM, Emr SD. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol Cell Biol. 1988;8:2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO (Eur Mol Biol Organ) J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornfeld S. Structure and function of the mannose 6-phosphate/insulinlike growth factor II receptors. Annu Rev Biochem. 1992;61:307–330. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- 23.Kuehn MJ, Schekman R, Ljungdahl PO. Amino acid permeases require COPII components and the ER resident membrane protein Shr3p for packaging into transport vesicles in vitro. J Cell Biol. 1996;135:585–595. doi: 10.1083/jcb.135.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;100:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 25.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 26.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science (Wash DC) 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 27.Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 28.Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nothwehr SF, Stevens TH. Sorting of membrane proteins in the yeast secretory pathway. J Biol Chem. 1994;269:10185–10188. [PubMed] [Google Scholar]
- 31.Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. . J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raymond CK, Howald SI, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vpsmutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rayner JC, Pelham HRB. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO (Eur Mol Biol Organ) J. 1997;16:1832–1841. doi: 10.1093/emboj/16.8.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redding K, Brickner JH, Marschall LG, Nichols JW, Fuller RS. Allele-specific suppression of a defective trans-Golgi network (TGN) localization signal in Kex2p identifies three genes involved in localization of TGN transmembrane proteins. Mol Cell Biol. 1996;16:6208–6217. doi: 10.1128/mcb.16.11.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. . J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redding KE, Seeger M, Payne GS, Fuller RS. The effects of clathrin inactivation on localization of Kex2 protease are independent of the TGN localization signal in the cytosolic tail of Kex2p. Mol Biol Cell. 1996;7:1667–1677. doi: 10.1091/mbc.7.11.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rieder SE, Banta LM, Koher K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment acculumates in the yeast vps28vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rockmill B, Lambie EJ, Roeder GS. Spore enrichment. Methods Enzymol. 1991;194:146–149. doi: 10.1016/0076-6879(91)94012-2. [DOI] [PubMed] [Google Scholar]
- 39.Rose, M.D., F. Winston, and P. Heiter. 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Vol. 198. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 198 pp.
- 40.Rothman JH, Hunter CP, Valls LA, Stevens TH. Overproduction-induced mislocalization of a yeast vacuolar protein allows isolation of its structural gene. Proc Natl Acad Sci USA. 1986;83:3248–3252. doi: 10.1073/pnas.83.10.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothman JH, Raymond CK, Gilbert T, O'Hara PJ, Stevens TH. A putative GTP binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell. 1990;61:1063–1074. doi: 10.1016/0092-8674(90)90070-u. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning. C. Nolan, editor. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Schimmoller F, Singer-Kruger B, Schroder S, Kruger U, Barlowe C, Riezman H. The absence of Emp24p, a component of ER- derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO (Eur Mol Biol Organ) J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seeger M, Payne GS. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO (Eur Mol Biol Organ) J. 1992;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seeger M, Payne GS. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. . J Cell Biol. 1992;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silkorsky RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi S, Nakagawa T, Banno T, Watanabe T, Murakami K, Nakayama K. Localization of furin to the trans-Golgi network and recycling from the cell surface involves Ser and Tyr residues within the cytoplasmic domain. J Biol Chem. 1995;270:28397–28401. doi: 10.1074/jbc.270.47.28397. [DOI] [PubMed] [Google Scholar]
- 49.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiaeis regulated by proteolysis and phosphorylation. EMBO (Eur Mol Biol Organ) J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vida TA, Huyer G, Emr SD. Yeast vacuolar proenzymes are sorted in the late Golgi complex and transported to the vacuole via a prevacuolar endosome-like compartment. J Cell Biol. 1993;121:1245–1256. doi: 10.1083/jcb.121.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. . Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 53.Ward AC. Single-step purification of shuttle vectors from yeast for high frequency back-transformation into E. coli. . Nucleic Acids Res. 1990;18:5319. doi: 10.1093/nar/18.17.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilcox CA, Fuller RS. Posttranslational processing of the prohormone-cleaving Kex2 protease in the Saccharomyces cerevisiaesecretory pathway. J Cell Biol. 1991;115:297–307. doi: 10.1083/jcb.115.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]