Abstract
We have recently shown that two proteins related to two of the adaptor subunits of clathrincoated vesicles, p47 (μ3) and β-NAP (β3B), are part of an adaptor-like complex not associated with clathrin (Simpson, F., N.A. Bright, M.A. West, L.S. Newman, R.B. Darnell, and M.S. Robinson, 1996. J. Cell Biol. 133:749–760). In the present study we have searched the EST database and have identified, cloned, and sequenced a ubiquitously expressed homologue of β-NAP, β3A, as well as homologues of the α/γ and σ adaptor subunits, δ and σ3, which are also ubiquitously expressed. Antibodies raised against recombinant δ and σ3 show that they are the other two subunits of the adaptor-like complex. We are calling this complex AP-3, a name that has also been used for the neuronalspecific phosphoprotein AP180, but we feel that it is a more appropriate designation for an adaptor-related heterotetramer. Immunofluorescence using anti-δ antibodies reveals that the AP-3 complex is associated with the Golgi region of the cell as well as with more peripheral structures. These peripheral structures show only limited colocalization with endosomal markers and may correspond to a postTGN biosynthetic compartment. The δ subunit is closely related to the protein product of the Drosophila garnet gene, which when mutated results in reduced pigmentation of the eyes and other tissues. Because pigment granules are believed to be similar to lysosomes, this suggests either that the AP-3 complex may be directly involved in trafficking to lysosomes or alternatively that it may be involved in another pathway, but that missorting in that pathway may indirectly lead to defects in pigment granules.
The first step in the trafficking of proteins from a donor to an acceptor membrane compartment is the formation of a transport vesicle. This process is mediated by coat proteins, which are recruited from the cytosol onto the donor membrane. Here they form a scaffold that drives vesicle budding while at the same time choosing the vesicle cargo by interacting with the cytoplasmic domains of selected transmembrane proteins. The first coated vesicles to be described were the clathrin-coated vesicles. These are coated with clathrin and adaptor (or AP) complexes, and they bud from the TGN and the plasma membrane where they concentrate cargo proteins for delivery to endosomal compartments. The clathrin associated with these two membranes is the same, but the adaptors are different: the AP-1 complex is associated with the TGN, while the AP-2 complex is associated with the plasma membrane. More recently, two other types of coated vesicles have been identified: COPI-coated vesicles, which bud from the Golgi stack and the intermediate compartment; and COPII-coated vesicles, which bud from the ER. Although clathrin, COPI, and COPII coats are assembled from different components, there are certain features that are shared by all coated vesicles. In all cases the membrane needs to be primed by the binding of a small GTP-binding protein (ARF or Sar1p) before the coat proteins can be recruited, and in all cases the coat appears to be required not only for vesicle budding but also for cargo selection (for review see Schekman and Orci, 1996).
There are many other membrane traffic pathways in the cell in addition to those mediated by the three well characterized types of coats, and this has led to the suggestion that there must be additional coat proteins to carry out these pathways (Robinson, 1991). This possibility is supported by the observation that tyrosine-based sorting signals, which promote internalization of plasma membrane proteins by binding to AP-2 adaptor complexes, can function in other pathways as well, suggesting that the coats associated with such pathways may contain components related to adaptors (Matter and Mellman, 1994; Ohno et al., 1995). Recently we described a novel adaptor-related protein complex that has many of the properties expected for a component of a new type of coat (Simpson et al., 1996). Two of the subunits of the complex are proteins that have previously been identified: p47, a homologue of the adaptor medium chains, or μ subunits (Pevsner et al., 1994); and β-NAP, a homologue of the adaptor β subunits (Newman et al., 1995). p47 exists as two isoforms: p47A, which is expressed ubiquitously, and p47B, which is specifically expressed in neuronal tissues (Pevsner et al., 1994). β-NAP is also neuronal specific (Newman et al., 1995), which suggests that in nonneuronal cells there may be another, ubiquitously expressed isoform of β-NAP to form complexes with p47A. Using a heterologous in vitro system we showed that the p47/β-NAP complex can be recruited from brain cytosol onto membranes prepared from tissue culture cells or liver fractions. Recruitment is enhanced by GTPγS and inhibited by brefeldin A, indicating that it is ARF dependent. Cell fractionation, immunofluorescence, and immunogold electron microscopy suggested that the complex binds to a subcompartment of the TGN, and this was confirmed by EM labeling of endogenous complex in primary cultures of neuronal cells. Both the newly recruited exogenous complex and the endogenous complex were found to be associated with buds and vesicles that were coated on the cytoplasmic side, but these coats were thinner than a clathrin coat. In addition, the complex did not colocalize with clathrin at either the light or the electron microscope level and did not copurify with clathrin-coated vesicles. Thus, we proposed that the complex is required for a nonclathrin-mediated budding event from the TGN (Simpson et al., 1996).
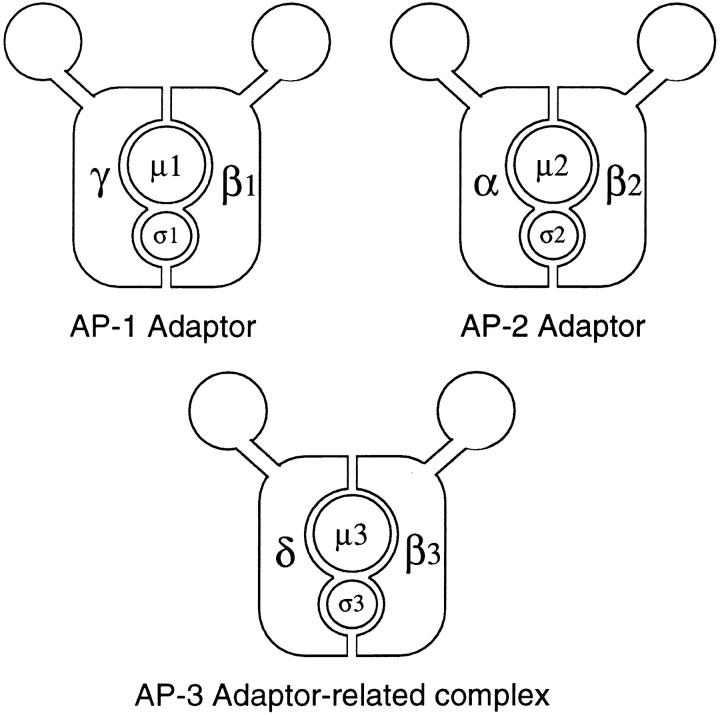
Immunoprecipitation experiments indicated that the complex consists not only of β-NAP and p47, but also of two other proteins of ∼160 (p160) and ∼25 kD (p25), making it a heterotetramer like an adaptor complex. A model for this complex is shown in Fig. 1, together with models of the AP-1 and AP-2 complexes. We are calling this complex AP-3, by analogy with AP-1 and AP-2. The name AP-3 has also been used for AP180, a clathrin assembly–promoting phosphoprotein specifically expressed in neurons (Keen, 1987; Morris et al., 1993). However, AP180 is a monomer that does not show any sequence homology with the adaptor subunits, so it seems more appropriate to use this name for an adaptor-related heterotetrameric complex. Both AP-1 and AP-2 consist of four subunits: γ- or α-adaptin; β1- or β2-adaptin (also called β′ and β); μ1 (AP47) or μ2 (AP50); and σ1 (AP19) or σ2 (AP17). The two subunits of AP-3 that have already been identified, p47 and β-NAP, are labeled μ3 and β3 on the diagram. The other two proteins that coimmunoprecipitate with μ3 and β3, p160 and p25, are labeled δ and σ3, respectively. We predicted that δ would turn out to be a homologue of the α- and γ-adaptins and that σ3 would turn out to be a homologue of σ1 and σ2 (Simpson et al., 1996). In the EST database, there are candidates for both of these proteins as well as a candidate for a ubiquitously expressed isoform of β-NAP. We have now raised antibodies against the δ and σ3 candidates expressed as fusion proteins, and here we show by coimmunoprecipitation that they are indeed the missing subunits of the complex. To learn more about the function of the AP-3 complex we have used the antibodies to localize endogenous AP-3 in nonneuronal cells and to compare its distribution with that of other proteins. In addition, we have found a close homologue of the δ subunit in Drosophila, which is encoded by the garnet gene. Studying the mutant phenotype of the gene provides additional insights into the role of the complex.
Figure 1.
Diagrams of the two conventional adaptor complexes, together with the adaptor-related complex, AP-3. The AP-1 complex is associated with the TGN, the AP-2 complex is associated with the plasma membrane, and the AP-3 complex also appears to be associated with the TGN as well as with more peripheral membranes. Each complex consists of four subunits, belonging to four different families. γ, α, and δ are related; β1 (β′), β2 (β), and β3 (β-NAP/β3B and β3A) are related; μ1 (AP47), μ2 (AP50), and μ3 (p47A/μ3A and p47B/μ3B) are related; and σ1 (AP19), σ2 (AP17), and σ3 (A and B) are related. EM studies of the AP-2 complex have revealed that it has a structure resembling a head flanked by two ears connected by flexible hinges (Heuser and Keen, 1988), and although such studies have not yet been carried out on AP-1 or AP-3, the sequence homologies suggest that all three complexes have a similar structure.
Materials and Methods
Cloning and Sequencing
The EST database was searched for proteins with homology to the α- and γ-adaptins, to β-NAP, and to the adaptor small chains σ1 and σ2. Clones were then obtained from the appropriate source, sequenced, and expressed as recombinant fusion proteins for antibody production. Most molecular biology procedures were carried out as described by Sambrook et al. (1989).
The δ subunit of the complex was first identified as GenBank/EMBL/ DDBJ clone T30164. This is a cDNA cloned from human uterus encoding a protein with homology to both α- and γ-adaptins, available from the American Type Tissue Culture (Rockville, MD). Sequencing of the clone indicated that both the 5′ and 3′ ends of the cDNA were missing (the clone encodes amino acids 22–756 of the full length δ subunit; Fig. 2). The 3′ end of the clone was found to be the human homologue of a bovine cDNA previously cloned as a candidate bovine leukemia virus receptor (Ban et al., 1993). Although it is not clear why the δ subunit was cloned in this way, it is possible that its unusually high content of charged amino acids caused it to bind the probe nonspecifically (the screen was carried out using an expression library). We then searched the EST database with the bovine sequence and identified another human EST (GenBank/EMBL/ DDBJ clone R54523; Image consortium clone ID 39286) encoding the COOH-terminal end of the δ subunit (amino acids 865–1112). To obtain the 5′ end and the middle part of the cDNA, two library screens were carried out using a human heart cDNA library (Clontech Laboratories, Inc., Palo Alto, CA). A 60-mer oligonucleotide (antisense for amino acids 22– 81) was used for the 5′ end, while two restriction fragments, one from clone T30164 encoding amino acids 485–756 and one from clone R54523 encoding amino acids 865–925, were used on duplicate filters to isolate a clone overlapping both of the database clones. Sequencing was carried out by John Lester (University of Cambridge, Cambridge, UK) on an automated ABI sequencer using oligonucleotide primers to “walk out” along the DNA. The entire coding sequence was read in both directions.
Figure 2.

Sequences of novel components of the AP-3 complex. cDNAs encoding the four components were originally identified as ESTs encoding homologues of the known AP subunits. The complete sequences of the δ and β3A subunits were obtained by library screening. Both the cDNA and the protein sequence data are available from GenBank/EMBL/ DDBJ under accession numbers U91930 (δ), U91931 (β3A), U91932 (σ3A), and U91931 (σ3B).
A similar strategy was used in the cloning of the β-NAP homologue, β3A. Two cDNAs were initially found in the EST database (GenBank/ EMBL/DDBJ clones R02669 and T98538; Image consortium clone ID 124011 and 123092), encoding a protein or proteins homologous to β-NAP but without the neuronal specificity (both were cloned from a human liver and spleen cDNA library). Sequencing revealed that they did not overlap, but a preliminary Northern blot indicated that they were derived from the same mRNA. Thus, duplicate filter lifts were taken from the human heart cDNA library and screened with probes prepared from each of the two database clones. One of the clones that hybridized to both probes contained an insert of 3.6 kb, and when sequenced in its entirety it was found to encode the complete open reading frame of the protein.
Two small chain homologues were initially identified in the EST database (GenBank/EMBL/DDBJ clones R23892 and R87391; Image consortium clone ID 131033 and 166044), cloned from human placenta and human brain, respectively, which encoded closely related but distinct proteins, and these were used for initial sequencing and for antibody production. Although neither clone is full length (R23892 encodes amino acids 79–193 of σ3A, and R87391 encodes amino acids 85–193 of σ3B), subsequent database searches identified full length clones for both (GenBank/ EMBL/DDBJ clones N30960 and W97614; Image consortium clone ID 266095 and 422534), from human melanocytes for σ3A and from mouse embryo for σ3B, respectively. The mouse and human σ3B protein sequences are 100% identical in the region of overlap. Moreover, while this work was in progress, Dell'Angelica et al. (1997) independently isolated a full length human clone encoding σ3B that also shows 100% identity at the protein level to mouse σ3B.
Northern Blotting
Human multiple tissue Northern blots were purchased from Clontech Laboratories, Inc. and probed according to the manufacturer's instructions, except that higher stringency washing conditions were used. Three of the probes consisted of 60-mer antisense oligonucleotides that had been end labeled with 32P. The δ probe was the same oligonucleotide used for library screening; the two σ3 probes both corresponded to the last five amino acids, the stop codon, and 42 bases of 3′ UTR. The probe used for β3A was the insert from clone T98538, encoding amino acids 946–1093, plus ∼500 bp of 3′ UTR, labeled with 32P by random priming.
Antibody Production and Coimmunoprecipitation
Antibodies were raised in rabbits against GST fusion proteins using PCR to amplify the appropriate sequences that were then cloned into the expression vector pGEX-3X (Pharmacia Biotech, Piscataway, NJ). For the δ subunit, the entire insert of clone T30164 was expressed, resulting in a fusion protein containing amino acids 22–756. This construct was insoluble and was purified from inclusion bodies (Page and Robinson, 1995). For the two σ subunits, only their COOH-terminal domains, amino acids 113– 193 were expressed, since previous studies on σ1 and σ2 indicated that this region was likely to be the most antigenic part of the protein (Page and Robinson, 1995). These constructs were soluble and were purified by glutathione–Sepharose affinity chromatography. Immunization and affinity purification of the resulting antisera were carried out as described by Page and Robinson (1995). After affinity purification, the antisera were tested on Western blots of whole brain homogenate. The δ antiserum gave a very clean signal, strongly labeling an ∼160-kD band and much more weakly labeling an ∼120-kD band, possibly a breakdown product. The two σ antisera were less specific, both labeling high molecular weight bands as well as bands of ∼20–25 kD. However, when these antisera were used for immunoprecipitations (see below) and then tested on blots of the proteins they brought down, the low molecular weight bands were found to be specifically precipitated.
Immunoprecipitations of both AP-3 and AP-1 complexes were carried out under nondenaturing conditions. Pig brain cytosol was prepared in PBS (Seaman et al., 1993) and precleared by the addition of 100 μl of 50% protein A Sepharose for each ml of cytosol. 10 μl of affinity-purified antibody were then added to 100 μl aliquots of the precleared cytosol, and the samples were incubated for 1 h at room temperature. The antibodies that were used were the δ, σ3A, and σ3B antibodies described above, anti–βNAP (Simpson et al., 1996), and anti–γ-adaptin (Seaman et al., 1996). After the antibody incubations, 70 μl of 50% protein A Sepharose were added and the samples incubated for an additional h at room temperature, after which the Sepharose was collected by centrifugation and washed five times with PBS containing 0.1% NP-40. The samples were then boiled in sample buffer, run on SDS polyacrylamide gels, and subjected to Western blotting. The δ subunit was found to elute very poorly in the absence of carrier proteins, so in some experiments yeast cytosol was added to the immunoprecipitates to improve the blotting efficiency. Blots were probed with the various antibodies followed by 125I-protein A, as previously described (Robinson and Pearse, 1986).
Immunofluorescence
NRK cells and MDBK cells were grown on multiwell test slides, fixed with either 2% paraformaldehyde followed by 0.1% NP-40 or with methanol and acetone, and prepared for immunofluorescence as previously described (Robinson, 1987). For some experiments the cells were allowed to internalize rhodamine-conjugated wheat germ agglutinin before fixation (Seaman et al., 1993). Other experiments involved treating the cells with 5 μg/ml brefeldin A (Sigma Chemical Co., St. Louis, MO) for 2 min before fixation. The cells were then labeled with rabbit anti-δ, either alone or together with a mouse monoclonal antibody. The monoclonal antibodies included anti-transferrin receptor (Chemicon International Inc., Temecula, CA), anti-lgp120 (Grimaldi et al., 1987; generously provided by Paul Luzio, University of Cambridge, Cambridge, UK), anti-p200 (Narula et al., 1992; generously provided by Jenny Stow, University of Brisbane, Brisbane, Australia), anti-γ-adaptin (Sigma Chemical Co.), and anti-clathrin heavy chain (X22; generously provided by Frances Brodsky, University of California at San Francisco, CA; Brodsky, 1985). Secondary antibodies were fluorescein-conjugated donkey anti–rabbit IgG and Texas red–conjugated sheep anti–mouse IgG, both obtained from Amersham Life Science (Arlington Heights, IL). The cells were viewed either using an Axioplan fluorescence microscope (Zeiss, Inc., Thornwood, NY) or using a confocal microscope (MRC1000; BioRad Laboratories, Hercules, CA).
Preparation of Drosphila Eye Sections
Wild-type (Barton strain) and mutant Drosophila were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN). Their eyes were dissected out and fixed in glutaraldehyde followed by osmium tetroxide as described by Seaman et al. (1993). The tissue was infiltrated and embedded in Spurrs resin, and sections were cut for both light and electron microscopy. The pigment granules tended to lose their contents in thin sections, so most studies were carried out on thick (1-μm) sections, using phase contrast and bright field microscopy.
Results
Identification of New Adaptor-related Proteins
To find the missing subunits of the AP-3 complex we searched the EST database for likely candidates. Both μ3 and β3 are more distantly related to μ1/μ2 and β1/β2 than μ1 and μ2 or β1 and β2 are to each other (Fig. 1 and see Fig. 3); thus, we looked for proteins with a similar type of relationship to α- and γ-adaptin and to σ1 and σ2, as candidates for the δ and σ3 subunits. We also looked for a nonneuronal-specific homologue of β-NAP. cDNAs with the appropriate properties were then obtained and sequenced. In some cases the cDNAs were not full length, and it was necessary to carry out library screens to obtain the complete coding sequence.
Figure 3.
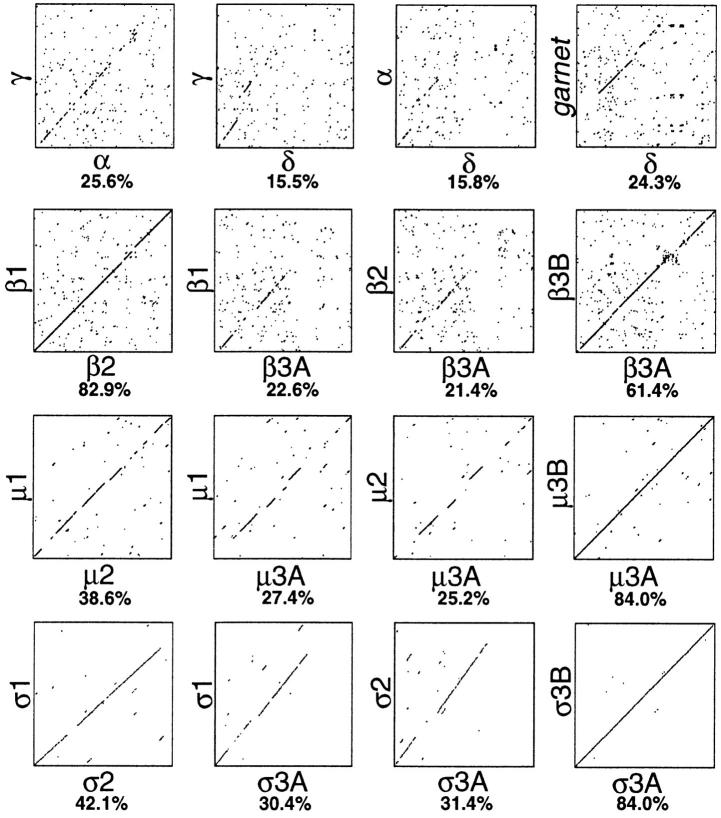
Diagon plots comparing related proteins in the AP-1, AP-2, and AP-3 complexes. The sequences of all four types of subunits were compared using the “SIP” program (Staden, 1990), which was also used to calculate the percent identity. The δ subunit shows the least similarity with its counterparts in the AP-1 and AP-2 complexes, while the σ subunits (σ3A and σ3B) show the most. The protein product of the Drosophila garnet gene is also shown, compared with the δ subunit.
Fig. 2 shows the sequences of the candidate δ subunit, a homologue of β-NAP (β3A), and two closely related σ1/ σ2 homologues (σ3A and σ3B), while Fig. 3 shows diagon plots illustrating the relationship between these proteins and other members of the same families. The δ subunit has a predicted size of 125 kD, which is less than the apparent molecular weight (∼160 kD) of the large protein that coimmunoprecipitates with μ3 and β3, but its unusual amino acid content (see below) may make it run anomalously on SDS polyacrylamide gels. It shares the least sequence identity of any of the novel sequences with its counterparts in the AP-1 and AP-2 complexes, being only ∼15% identical to α- and γ-adaptins, and this homology is mainly restricted to the extreme NH2-terminal portion of the protein. However, it does have a “WIIGEY” consensus sequence, WICGEF, at the appropriate position (amino acids 396–401). This is a motif of unknown function found not only in the α- and γ-adaptins (Robinson, 1989, 1990) but also in all three types of β subunits (Kirchhausen et al., 1989; Ponnambalam et al., 1990; Newman et al., 1995) and even in the very distantly related β-COP, a subunit of the COPI complex (Duden et al., 1991). The protein is also adaptin like in that it appears to have a three domain structure consisting of an NH2-terminal domain of ∼650 amino acids, a highly hydrophilic linker or hinge domain (normally particularly rich in acidic residues, prolines, and alanines, although the δ subunit has a high proportion of basic residues as well), and a COOH-terminal (“ear”) domain.
The β-NAP homologue, β3A, is much more closely related to β3B (β-NAP; 61.4% identity) than to either β1 or β2 (22.6% and 21.4% identity, respectively). Its homology to β1 and β2 is restricted to its NH2-terminal domain of ∼600 amino acids, while its hinge and ear domains appear to be unrelated. Similarly, β3B only appears to be related to β1 and β2 in its NH2-terminal domain (Newman et al., 1995). The major difference between β3A and β3B lies within their hinge domains, including amino acids 644– 814. Both have a similar amino acid composition in this region, with a high concentration of acidic residues and serines, but their sequences diverge considerably. Interestingly, this is precisely the domain that was used for raising β3B antibodies (Newman et al., 1995), which probably explains why the antibodies are neuronal specific. While this paper was under review we raised a similar antibody against the hinge domain of β3A, which immunoprecipitates the other AP-3 subunits in nonneuronal cells (data not shown).
The σ3A and σ3B subunits are the most similar to their counterparts in the AP-1 and AP-2 complexes, with over 30% identity. They mainly differ from the other members of the σ family in that they extend farther at their COOHterminal ends, giving them predicted molecular weights of 21.7 and 22 kD, as compared with 19 kD for σ1 and 17 kD for σ2. These subunits have been independently cloned and sequenced by Dell'Angelica et al. (1997), using the same approach of searching the EST database for σ1/σ2 homologues.
Both μ3 and β3 have neuronal-specific isoforms, so we probed multiple tissue Northern blots to find out whether any of the novel sequences also show tissue-specific expression. Fig. 4 shows that δ, β3A, σ3A, and σ3B are all expressed ubiquitously. Thus, β3A is indeed a nonneuronal-specific isoform of β3; the only known isoform of δ is expressed in all tissues examined; and although there are two isoforms of σ3, they have similar expression patterns.
Figure 4.
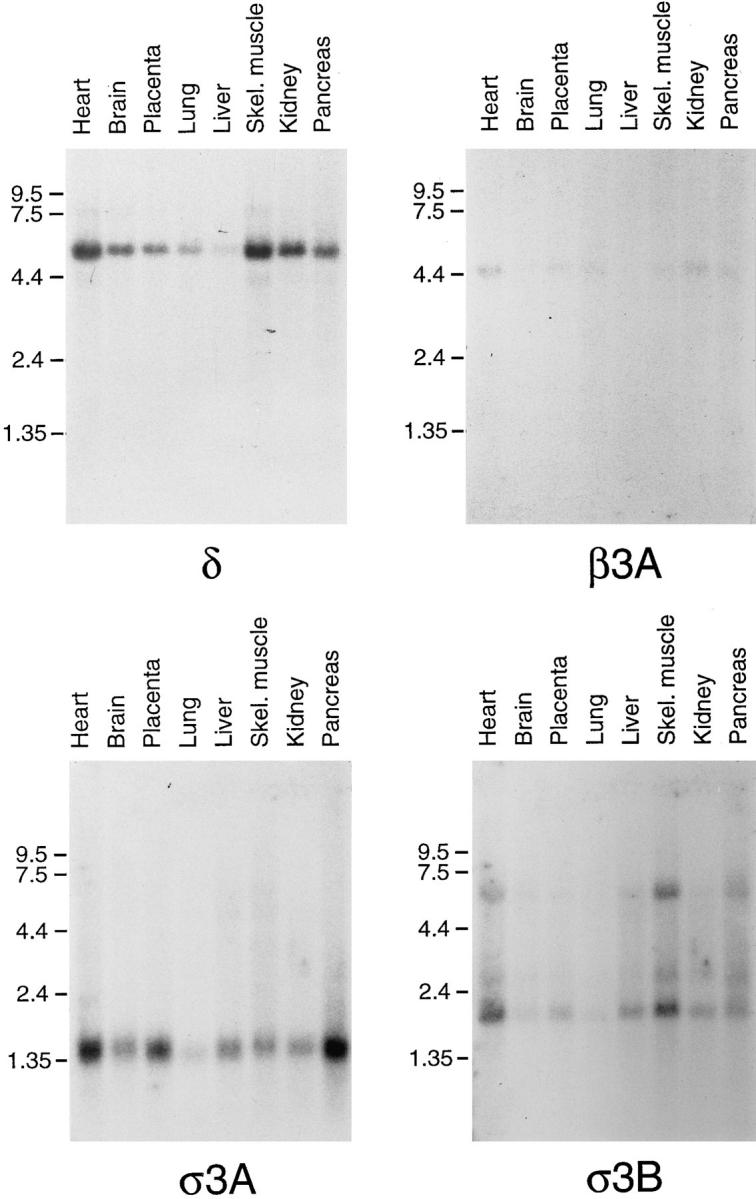
Expression patterns of δ, β3A, σ3A, and σ3B. Northern blots were probed with either oligonucleotides or cDNAs specific for each of the four sequences. All four genes are expressed ubiquitously. The relative weakness of the signal obtained with β3A probe is probably a consequence of the blot already having been probed several times.
Composition of the Adaptor-related Protein Complex, AP-3
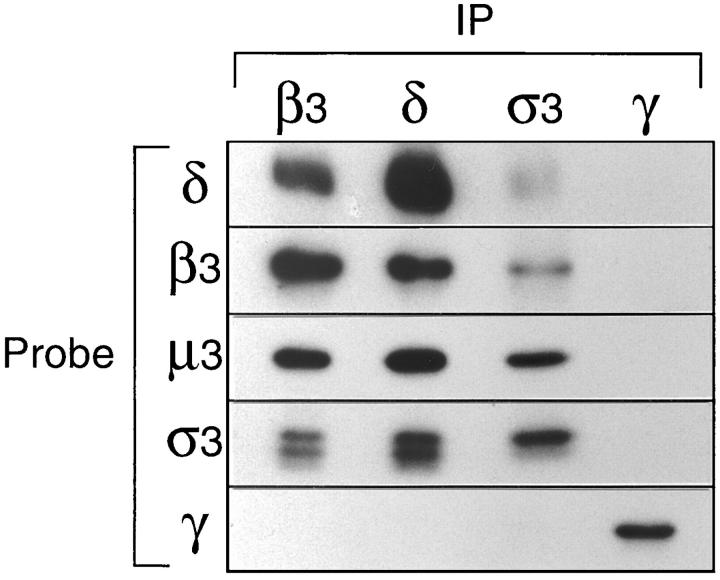
To characterize δ and σ3 further and to find out whether they are in fact associated with μ3 and β3, portions of them were expressed as fusion proteins and used to raise antibodies in rabbits. The antibodies were then used in immunoprecipitation and Western blotting experiments, together with the previously described antibodies against μ3 and β3. Fig. 5 shows the results of one such experiment. Pig brain cytosol was immunoprecipitated under nondenaturing conditions with anti-β3, anti-δ, and anti-σ3 (using pooled antibodies raised against both the A and B isoforms). As a control, cytosol was also immunoprecipitated with anti–γ-adaptin to bring down the AP-1 adaptor complex. Strips were then cut from Western blots and probed with antibodies against δ, β3, μ3, σ3, and γ.
Figure 5.
Coimmunoprecipitation of AP-3 subunits. Pig brain cytosol was immunoprecipitated under nondenaturing conditions with affinity-purified polyclonal antibodies against β3B, δ, σ3 (crossreacting with both the A and B isoforms), and γ. Gels were blotted, and the appropriate region was cut out and probed with each of the above antibodies, as well as with anti-μ3 (which does not recognize the native complex). The four subunits of the AP-3 complex, δ, β3, μ3, and σ3, all coimmunoprecipitate; the γ subunit of the AP-1 complex does not coimmunoprecipitate with antibodies against the AP-3 components, and antibodies against γ do not bring down any of these components. The doublet labeled with anti-σ3 presumably corresponds to the A and B isoforms, one of which appears to be preferentially immunoprecipitated with this antibody.
The β3 antibody brings down not only β3 and μ3, as has been previously reported, but also δ and σ3 (which appears as a doublet, presumably because there are two isoforms of the protein). It does not bring down γ, as expected, since γ is a component of a different complex. Similarly, the new δ antibody brings down δ, β3, μ3, and σ3, but not γ. The σ3 antibodies bring down these four subunits as well, although the signal from δ, β3, and μ3 is weaker than with the other two antibodies, and one of the σ3 isoforms appears to be preferentially immunoprecipitated. The control antibody, anti–γ-adaptin, brings down γ, but not δ, β3, μ3, or σ3.
Thus, we have correctly identified the two missing subunits of the adaptor-related complex. Like a conventional adaptor, the complex consists of an α/γ-like subunit (δ), a β subunit, a μ subunit, and a σ subunit. We propose that by analogy with AP-1 and AP-2, the complex should be called AP-3.
Localization of AP-3 in Nonneuronal Cells
In our previous study we localized the AP-3 complex by immunofluorescence and immunogold EM using antibodies against β3B. However, because of the tissue specificity of this antibody, we were only able to examine the distribution of the complex in neurons, which are not well suited for immunofluorescence, or in our heterologous system using permeabilized cells incubated with brain cytosol (Simpson et al., 1996). The δ antibody also works well for immunofluorescence, so we are now able to localize the endogenous complex in nonneuronal cells.
Fig. 6 shows the normal distribution of the complex in NRK cells (a–c, and g) and in MDBK cells (h). Like the newly recruited exogenous complex, endogenous AP-3 is mainly perinuclear, with punctate labeling extending out towards the cell periphery. Such punctate labeling is reminiscent of the pattern seen with markers for endosomal compartments, so we carried out double labeling with antibodies against the transferrin receptor (Fig. 6 d), as a marker for the early and/or recycling endosomal compartment, and against lgp120 (Fig. 6 e), as a marker for late endosomes and lysosomes. Both antibodies produced similar types of patterns to those seen with the δ antibody (Fig. 6 a and b), but close examination of the more peripheral structures revealed little if any colocalization. As a more general marker for the endosomal system we allowed cells to endocytose rhodamine-conjugated wheat germ agglutinin (r-WGA) for various lengths of time. The cells in Fig. 6 f had been incubated with r-WGA for 5 min and show a small but significant amount of colocalization of r-WGApositive structures with the δ antibody (Fig. 6, c and f). Longer incubations in r-WGA did not increase the amount of colocalization (data not shown), indicating that endocytosed proteins have access to compartments that can bind AP-3 but only at early time points.
Figure 6.
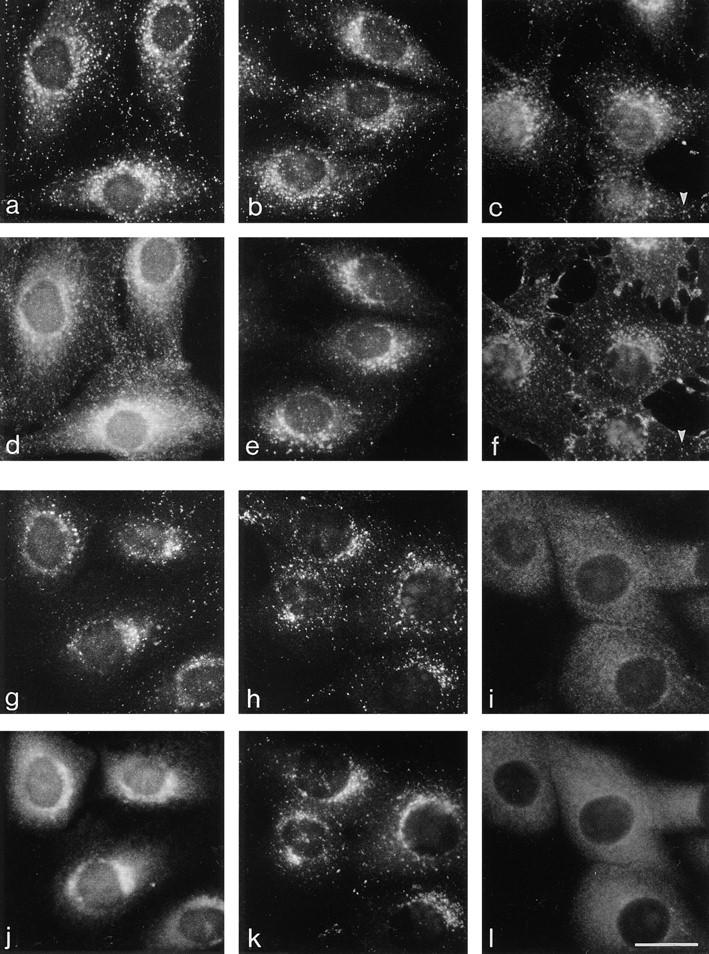
Immunofluorescence localization of the δ subunit of the AP-3 complex. (a, b, d, and e) NRK cells were fixed with methanol/acetone and double labeled with anti-δ (a and b) and anti-transferrin receptor (d) or anti-lgp120 (e). Although all three antibodies show perinuclear labeling, many organelles in the cell are concentrated in this region, and there is little colocalization of the more peripheral elements. (c and f) Cells were allowed to endocytose rhodamine-conjugated wheat germ agglutinin (f) for 5 min before fixation in methanol/ acetone and were then labeled with anti-δ (c). Again, there is little colocalization of the more peripheral elements, although some structures do coincide (arrowheads). (g and j) NRK cells were fixed with paraformaldehyde and permeabilized with NP-40 and then double labeled with anti-δ (g) and antip200 (j), a putative TGN coat. Both antigens have a perinuclear distribution, but the fine details are different, indicating that they are not components of the same coat. (h, i, k, and l) MDBK cells were fixed with methanol/acetone, either with (i and l) or without (h and k) prior incubation for 2 min in 5 μg/ ml brefeldin A and then double labeled with anti-δ (h and i) and anti-γ (k and l). Both antigens are brefeldin A sensitive, and both have a similar perinuclear distribution in control cells, but little if any colocalization of more peripheral structures. Bar, 20 μm.
Cells were also double labeled with the δ antibody and antibodies against TGN markers, since our earlier study indicated that at least some of the AP-3 complex is associated with the TGN. The cells in Fig. 6, g and j, were double labeled with anti-δ (g) and anti-p200 (j), a TGN-associated protein that may be a component of a nonclathrin coat (Narula et al., 1992; Narula and Stow, 1995). Although both show the same type of perinuclear localization, the two patterns are quite distinct, indicating that the two proteins are not associated with each other and thus are unlikely to be components of the same coat. The cells in Fig. 6, h and k, were double labeled with anti-δ (h) and anti–γadaptin (k), a component of the AP-1–containing coat that is also associated with the TGN. Again, although they are concentrated in the same perinuclear region they have distinct labeling patterns. In the final pair of panels the cells were treated with 5 μg/ml brefeldin A for 2 min before double labeling with anti-δ (Fig. 6 i) and anti–γ-adaptin (Fig. 6 l). Both proteins can be seen to have completely redistributed to the cytoplasm. Thus, the membrane localization of the AP-3 complex, like that of the AP-1 complex, appears to be ARF dependent, not only in our in vitro system but in vivo as well.
We also compared the distribution of δ with that of clathrin heavy chain. Because of the large number of clathrincoated pits and vesicles in the cell, it was necessary to use confocal microscopy to assess the degree of colocalization. Fig. 7 shows δ labeled in green and clathrin labeled in red. Although occasionally the labeling coincides, especially in the perinuclear region, for the most part the two patterns are distinct. The small amount of colocalization at the light microscope level is consistent with our earlier observations using immunogold electron microscopy, which revealed that β3B and clathrin were often in close proximity, although they were found on different budding profiles (Simpson et al., 1996). Thus, these results, together with the finding that AP-3 subunits are not enriched in clathrincoated vesicles prepared from brain (Simpson et al., 1996) or from liver (data not shown), indicate that the AP-3 complex is a component of a novel, nonclathrin coat.
Figure 7.
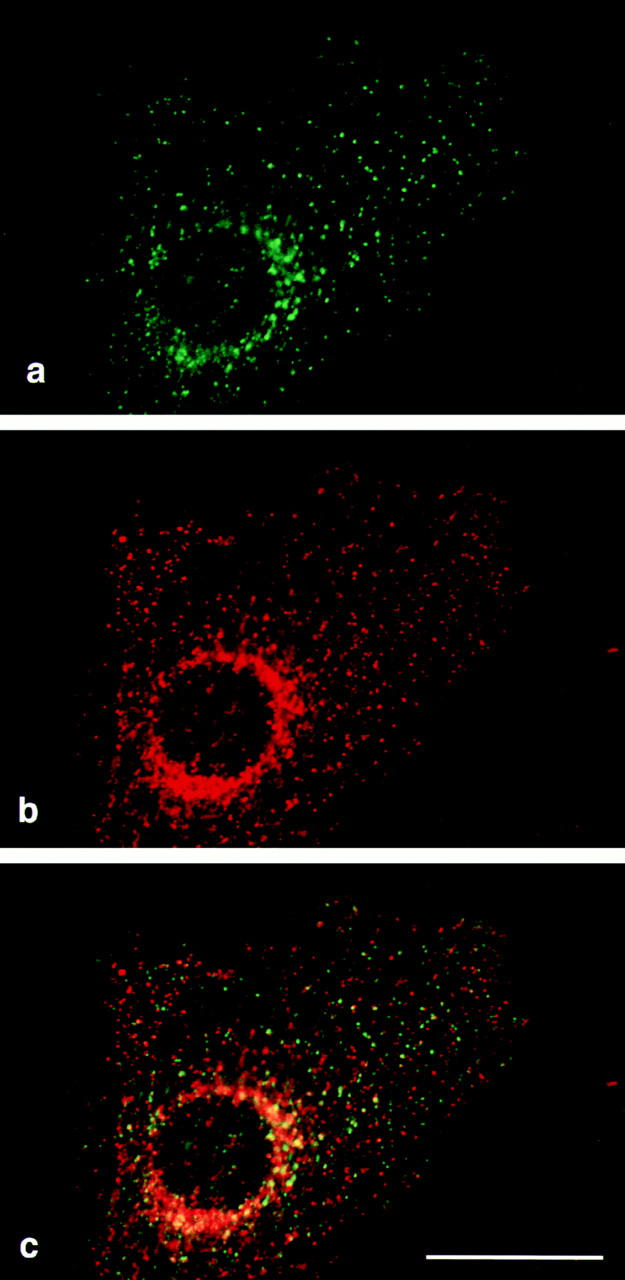
Confocal micrographs of an NRK cell double labeled for δ and clathrin. NRK cells were fixed with methanol/acetone and labeled with anti-δ (a, shown in green) and a monoclonal antibody against clathrin heavy chain (b, shown in red). The merged images (c) show numerous dots that are positive for δ but not for clathrin, and the limited amount of overlap is consistent with the two being found on the same membranes but different populations of coated buds. Bar, 20 μm.
An AP-3 Mutant in Drosophila
When the database was searched for homologues of the AP-3 subunits, we found that δ is closely related to the protein product of the Drosophila garnet gene (GenBank/ EMBL/DDBJ DMU31351). The garnet (g) locus was first described in 1916 as an eye color gene (Bridges, 1916) and was cloned by P element tagging and partially sequenced by V. Lloyd in 1995. The homology between the two proteins is strongest near their NH2-terminal ends, including a stretch of >100 amino acids (178–287 in δ) where human δ and Drosophila garnet are >96% identical (Fig. 3). Thus, garnet is almost certainly Drosophila δ, and an analysis of the garnet mutant phenotype should help to establish the function of the AP-3 complex.
Fig. 8 shows sections through the eyes of both wild type (a and d) and mutant (b, c, e, and f) flies. Two different garnet alleles are illustrated: g3 (Fig. 8, b and e), which results in reduced pigmentation throughout the fly's body, including dull brownish eyes; and g53d (Fig. 8, c and f), which appears to be tissue specific, giving rise to pale orange eyes and colorless Malpighian tubules but normal pigmentation of the testis sheath. Both phase contrast (Fig. 8, a–c) and bright field (Fig. 8, d–f) views are shown.
Figure 8.
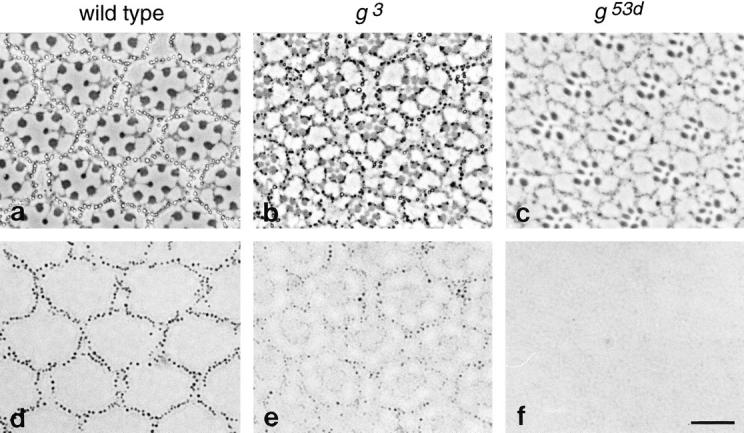
Phenotype of Drosophila with mutations in the garnet gene, which encodes the δ subunit. Sections were cut from the eyes of both wild type (a and d) and mutant (b, c, e, and f) flies. Two alleles were examined: g3 (b and e) and g53d (c and f). a–c are phase contrast micrographs; d–f show brightfield views of the same images. Each ommatidium consists of photoreceptor cells surrounded by pigment cells. In wild-type flies the pigment granules in the pigment cells can be readily seen in brightfield as well as phase contrast micrographs (a and d). The granules are still visible in the g3 flies but contain less pigment and thus are less prominent when viewed by brightfield (b and e). They are essentially undetectable in the g53d flies (c and f). Bar, 10 μm.
The repeating units, or ommatidia, of the compound eye consist of photoreceptor cells surrounded by pigment cells containing prominent pigment granules. In phase contrast micrographs of eyes from wild type flies (Fig. 8 a), the pigment granules appear as bright structures surrounded by dark rims. They are also easily visible in bright field micrographs (Fig. 8 d) because of their intense color. In g3 flies, the pigment granules tend to be dark in phase contrast micrographs (Fig. 8 b) and are much less visible in bright field micrographs (Fig. 8 e). This is consistent with reports that both red and brown pigments are reduced in flies with this allele (Nolte, 1954, 1959). However, the granules are of normal size and shape, and the number of granules surrounding each ommatidium is not appreciably different in g3 and wild-type flies. A much more severe phenotype is seen in the g53d flies. In phase contrast micrographs (Fig. 8 c), the ommatidia can be seen to be well developed, implying that the pigment cells are present; but no pigment granules are detectable, and the sections are nearly invisible in bright field micrographs (Fig. 8 f).
These observations indicate that the garnet gene is required for pigment granule biogenesis. However, it is likely to be involved in other pathways as well, since in situ hybridization indicates that it is also expressed in nonpigment cells (Lloyd, V., personal communication), and its mammalian homologue, δ, is also ubiquitously expressed, as shown in Fig. 4. Thus, the AP-3 complex may play a fundamental role in all types of cells, but mutations in the δ subunit appear to have a particularly strong effect on pigment granules.
Discussion
There are many membrane traffic pathways for which no coats have yet been identified, and this is what prompted us initially to look for adaptor-related proteins that might be components of novel types of coats. In a previous report we showed that two recently described proteins, p47 (μ3) and β-NAP (β3B), are associated with each other in an adaptor-like complex (Simpson et al., 1996). We have now identified the remaining subunits of the complex and have further investigated its function. We have named the complex AP-3, by analogy with AP-1 and AP-2.
The identification of the missing subunits of the AP-3 complex relied upon the availability of random cDNA sequences in the EST database. Based on the degree of homology between μ3 and μ1/μ2 and between β3 and β1/β2 we predicted that the δ and σ3 subunits would be relatively distant homologues of α/γ and of σ1/σ2, respectively. Because the complex is fairly abundant, it was easy to find entries in the database for both types of subunits; indeed, a recent search revealed over 50 mammalian ESTs for the δ subunit alone. Interestingly, there are now additional homologues of the β, μ, and σ subunits in the database, suggesting that there is likely to be at least one more type of AP complex.
Both the μ and the β subunit have neuronal-specific as well as ubiquitously expressed isoforms; however, no neuronal-specific isoforms have been found for either of the other two subunits, δ and σ. Although we cannot rule out the possibility that such isoforms may exist, candidates for them do not appear to be present in the EST database, in spite of the fact that many of the entries are from brain and that both μ3B and β3B can be found as multiple “hits.” Why are there neuronal-specific isoforms of two of the subunits? Is μ3B always associated with β3B and μ3A with β3A, or can the subunits “mix and match”? We do not yet know the answers to either of these questions, but studies on the μ and β subunits of the AP-1 and AP-2 complexes may provide some clues. μ1 and μ2 have been shown to bind to tyrosine-based sorting signals (Ohno et al., 1995), and recently μ3 has also been shown to bind such sequences (Dell Angelica et al., 1997). Possibly μ3A and μ3B recognize different repertoires of signals. β1 and β2 have been shown to bind to clathrin, an activity that involves the hinge domain of the protein (Gallusser and Kirchhausen, 1993; Shih et al., 1995). The AP-3 complex is not clathrin associated (Simpson et al., 1996) but may interact with another type of scaffolding protein, and such an interaction would be likely to involve the β3 subunit, with the two different β3 isoforms possibly interacting with different types of scaffolds. Alternatively, it may be that only one of the two proteins has a neuronal-specific role. The μ and β subunits of the AP-1 and AP-2 complexes show a strong interactaction with each other (Page and Robinson, 1995; Seaman et al., 1996), and it is possible that neuronal-specific isoforms of μ3 and β3 may have coevolved because there is an obligatory coupling between them.
In our previous study we were able to localize the neuronal-specific complex using antibodies against β3B (Simpson et al., 1996). The present study confirms and extends our earlier findings by localizing the endogenous AP-3 complex in nonneuronal cells. We have shown that the complex is brefeldin A–sensitive in vivo as well as in vitro and have confirmed that it is associated with both perinuclear and more peripheral membranes. What do these peripheral membranes correspond to? There is only very limited colocalization of either δ or β3B with endosomal markers, suggesting that the complex is more likely to be involved in a biosynthetic pathway than in an endocytic one. Our earlier study indicated that some of the complex is associated with the TGN (Simpson et al., 1996), and the more peripheral labeling may represent a post-TGN compartment. This compartment may also be able to receive endocytosed proteins without being a conventional type of endosome. The presence of a μ subunit in the complex indicates that it plays a role in the sorting of proteins containing tyrosine-based signals (Ohno et al., 1995; Dell'Angelica et al., 1997). Such signals have been shown to function in the post-Golgi biosynthetic pathway as well as in the endocytic pathway and may be used to send proteins to any one of several destinations: the (basolateral) plasma membrane, endosomes, lysosomes, or back to the TGN (Humphrey et al., 1993; Matter and Mellman, 1994). Which of these pathways might the complex mediate?
An important clue is provided by the finding that the δ subunit is the mammalian homologue of the Drosophila garnet gene product. The garnet mutant alleles that have been described only appear to affect pigmentation, yet the gene is expressed ubiquitously, not just in pigment cells. In addition, although a number of alleles have been identified, so far none of those tested by Northern blotting have been found to be nulls; thus, the protein may be essential (Lloyd, V., personal communication). Of the two alleles shown in Fig. 8, g53d is tissue specific, suggesting that the mutation is in the 5′ upstream regulatory region, while g3 contains an insertion within the coding portion of the gene (Lloyd, V., personal communication). Thus, the g3 mutation is likely to affect all the cells in the flies' bodies, yet the animals are viable and in particular do not appear to have any neurological problems. This is in contrast to flies with conditional mutations in two other genes encoding membrane traffic proteins, dynamin and NSF, where incubation at a nonpermissive temperature causes a block in neurotransmission (Chen et al., 1991; Van der Bliek and Meyerowitz, 1991; Pallanck et al., 1995). Although more information is needed about the activity of the mutant protein, the g3 phenotype appears to be inconsistent with a role previously proposed for both the β3 and μ3 subunits in synaptic vesicle biogenesis (Pevsner et al., 1994; Newman et al., 1995).
The most likely explanation for the garnet phenotype is a defect in the delivery of proteins to pigment granules. The pigments themselves are small molecules, but the granules must also contain a specific set of proteins for synthesizing, transporting, and/or storing the pigments, and it is possible that the sorting of these proteins may be impaired in the garnet flies. But what role might the complex play in nonpigment cells? In mammalian cells, pigment granules have been shown to be like modified lysosomes. Thus, patients with Chediak Higashi syndrome, or mice with the beige mutation, have not only giant lysosomes but also giant melanosomes (Burkhardt et al., 1993). Less is known about the formation of pigment granules in flies, but it seems likely that they too are lysosome-like in origin. This possibility is supported by the discovery that VPS18, a yeast gene involved in the sorting of proteins to the lysosome-like vacuole (Robinson et al., 1991), is homologous to another fly eye color gene, deep orange (Reider, S., and S. Emr, personal communication).
Taken together, these observations suggest that the AP-3 complex may play a role in trafficking from the TGN to the lysosome. However, a coat already exists for this pathway: the AP-1 and clathrin-containing coat. One possibility is that both coats are involved in this pathway but that they act at different stages or sort different types of cargo molecules. Alternatively, the role of the AP-3 coat in lysosomal biogenesis may be less direct. For instance, it may be involved in a different pathway, such as trafficking to the plasma membrane or recycling back to the TGN, but defective sorting may result in a certain amount of “scrambling” of proteins and interfere with other pathways as well. Specialized organelles such as pigment granules may be particularly susceptible to such missorting and may also be especially sensitive to problems at earlier stages of the secretory pathway (e.g., ER to Golgi), as has been shown for the specialized secretory granule-like trichocysts of Paramecium (Gautier et al., 1994). There are numerous Drosophila eye color genes, many of which have not yet been cloned, and it seems likely that some of these other genes will also be found to encode proteins involved in membrane traffic.
Although the garnet phenotype provides important information about the role of the AP-3 complex, further studies will be required to establish this role definitively. One promising lead comes from the observation that another genetically tractable organism, Sarcharomyces cerevisiae, contains genes encoding homologues of all four types of AP subunits, including some (e.g., SCYPL195W [δ] and SCYJL024C [σ3]) whose protein products are much more closely related to components of the AP-3 complex than to components of either the AP-1 or the AP-2 complexes. Targeted gene disruptions may indicate whether the complex is involved in trafficking to the vacuole, to the plasma membrane, or in some other pathway. Microinjection and/or immunodepletion experiments, using the antibodies we have generated that recognize the nonneuronal complex in its native form, may also help to define the role of the complex. AP-3 is not the only novel coat component that has recently been identified. Database searches indicate that there is at least one other AP-type complex, and there may be more. Monoclonal antibodies have identified p200 as yet another possible coat component (Narula and Stow, 1995), and electron microscopy has revealed the existence of lace-like coats associated with the TGN (Ladinsky et al., 1994). Thus, there are pathways in the cell that require coats and coats that require pathways, and the next step will be to fit the two together.
Acknowledgments
We are grateful to Vett Lloyd for communicating unpublished information about the garnet gene, to Matthew Seaman, Scott Emr, and Stephanie Reider for telling us about the VPS18-deep orange connection, to Paul Luzio, Jenny Stow, and Frances Brodsky for antibodies, and to Matthew Freeman for introducing us to Drosophila. We also thank Rainer Duden, Matthew Freeman, John Kilmartin, Vett Lloyd, Paul Luzio, and members of the Robinson lab for comments on the manuscript and for helpful discussions.
This work was supported by grants from the Medical Research Council, the Wellcome Trust, and the Human Frontier Science Program.
Footnotes
Fiona Simpson and Andrew Peden contributed equally to this paper.
Please address all correspondence to Margaret Robinson, University of Cambridge, Department of Clinical Biochemistry, Cambridge CB2 2QR, UK. Tel.: (44) 1223-330163; Fax: (44) 1223-330598.
Fiona Simpson's current address is Department of Cell Biology, Scripps Research Institute, La Jolla, CA 92037-1027.
References
- Ban J, Portetelle D, Altaner C, Horizon B, Milan D, Krchnak V, Burny A, Kettmann R. Isolation and characterization of a 2.3-kilobase-pair cDNA fragment encoding the binding domain of the bovine leukemia virus receptor. J Virol. 1993;67:1050–1057. doi: 10.1128/jvi.67.2.1050-1057.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB. Non-disjunction as proof of the chromosome theory of heredity. Genetics. 1916;1:1–52. doi: 10.1093/genetics/1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky FM. Clathrin structure characterized with monoclonal antibodies. I. Analysis of multiple antigenic sites. J Cell Biol. 1985;101:2047–2054. doi: 10.1083/jcb.101.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt JK, Wiebel FA, Hester S, Argon Y. The giant organelles in beige and Chediak-Higashi fibroblasts are derived from late endosomes and mature lysosomes. J Exp Med. 1993;178:1845–1856. doi: 10.1084/jem.178.6.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Obar RA, Schroeder CC, Austin TW, Poodry CA, Wadsworth SA, Vallee RB. Multiple forms of dynamin are encoded by shibire, a Drosophilagene involved in endocytosis. Nature (Lond) 1991;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Ohno H, Ooi CE, Rabinovich E, Roche KW, Bonifacino JS. An adaptor-like protein complex with ubiquitous expression. EMBO (Eur Mol Biol Organ) J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duden R, Griffiths G, Frank R, Argos P, Kreis TE. β-COP, a 100 kD protein associated with non-clathrin-coated vesicles and the Golgi complex, shows homology to β-adaptin. Cell. 1991;64:649–665. doi: 10.1016/0092-8674(91)90248-w. [DOI] [PubMed] [Google Scholar]
- Gallusser A, Kirchhausen T. The β1 and β2 subunits of the AP complexes are the clathrin coat assembly components. EMBO (Eur Mol Biol Organ) J. 1993;12:5237–5244. doi: 10.1002/j.1460-2075.1993.tb06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier MC, De Loubresse NG, Modeddu L, Sperling L. Evidence for defects in membrane traffic in Parameciumsecretory mutants unable to produce functional storage granules. J Cell Biol. 1994;124:893–902. doi: 10.1083/jcb.124.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi KA, Hutton JC, Siddle K. Production and characterisation of monoclonal antibodies to insulin secretory granule membranes. Biochem J. 1987;245:557–566. doi: 10.1042/bj2450557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE, Keen JH. Deep-etch visualization of proteins involved in clathrin assembly. J Cell Biol. 1988;107:877–886. doi: 10.1083/jcb.107.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey JS, Peters PJ, Yuan LC, Bonifacino JS. Localization of TGN38 to the trans-Golgi network: involvement of a cytoplasmic tyrosine-containing sequence. J Cell Biol. 1993;120:1123–1136. doi: 10.1083/jcb.120.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen JH. Clathrin assembly proteins: affinity purification and a model for coat assembly. J Cell Biol. 1987;105:1989–1998. doi: 10.1083/jcb.105.5.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Nathanson KL, Matsui W, Vaisberg A, Chow EP, Burne C, Keen JH, Davis AE. Structural and functional division into two domains of the large (100- to 115-kDa) chains of the clathrin-associated protein complex AP-2. Proc Natl Acad Sci USA. 1989;86:2612–2616. doi: 10.1073/pnas.86.8.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky MS, Kremer JR, Furcinitti PS, McIntosh RJ, Howell KE. HVEM tomography of the trans-Golgi network: structural insights and identification of a lace-like vesicle coat. J Cell Biol. 1994;127:29–38. doi: 10.1083/jcb.127.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter K, Mellman I. Mechanisms of cell polarity: sorting and transport in epithelial cells. Curr Opin Cell Biol. 1994;6:545–554. doi: 10.1016/0955-0674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- Morris SA, Schroder S, Plessman U, Weber K, Ungewickell E. Clathrin assembly protein 180: primary structure, domain organization, and identification of a clathrin binding site. EMBO (Eur Mol Biol Organ) J. 1993;12:667–675. doi: 10.1002/j.1460-2075.1993.tb05700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narula N, Stow JL. Distinct coated vesicles labeled for p200 bud from trans-Golgi network membranes. Proc Natl Acad Sci USA. 1995;92:2874–2878. doi: 10.1073/pnas.92.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narula N, McMorrow I, Plopper G, Doherty J, Matlin KS, Burke B, Stow JL. Identification of a 200–kD, brefeldin–sensitive protein on Golgi membranes. J Cell Biol. 1992;117:27–38. doi: 10.1083/jcb.117.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LS, McKeever MO, Hirotaka OJ, Darnell RB. β-NAP, a cerebellar degeneration antigen, is a neuron-specific vesicle coat protein. Cell. 1995;82:773–783. doi: 10.1016/0092-8674(95)90474-3. [DOI] [PubMed] [Google Scholar]
- Nolte DJ. The eye pigmentary system of Drosophila: V. The pigments of the light and dark groups of mutants. J Genet. 1954;52:127–135. [Google Scholar]
- Nolte DJ. The eye pigmentary system of Drosophila: VIII. Series of multiple alleles. Heredity. 1959;13:233–241. [Google Scholar]
- Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science (Wash DC) 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Page LJ, Robinson MS. Targeting signals and subunit interactions in coated vesicle adaptor complexes. J Cell Biol. 1995;131:619–630. doi: 10.1083/jcb.131.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallanck L, Ordway RW, Ganetzky B. A DrosophilaNSF mutant. Nature (Lond) 1995;376:25. doi: 10.1038/376025a0. [DOI] [PubMed] [Google Scholar]
- Pevsner J, Volknandt W, Wong BR, Scheller RH. Two rat homologs of clathrin-associated adaptor proteins. Gene (Amst) 1994;146:279–283. doi: 10.1016/0378-1119(94)90306-9. [DOI] [PubMed] [Google Scholar]
- Ponnambalam S, Robinson MS, Jackson AP, Peiperl L, Parham P. Conservation and diversity in families of coated vesicle adaptins. J Biol Chem. 1990;265:4814–4820. [PubMed] [Google Scholar]
- Robinson JS, Graham TR, Emr SD. A putative zinc finger protein, Saccharomyces cerevisiaeVps18p, affects late Golgi functions required for vacuolar protein sorting and efficient alpha-factor prohormone maturation. Mol Cell Biol. 1991;11:5813–5824. doi: 10.1128/mcb.11.12.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. 100-kD coated vesicle proteins: molecular heterogeneity and intracellular distribution studied with monoclonal antibodies. J Cell Biol. 1987;104:887–895. doi: 10.1083/jcb.104.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Cloning of cDNAs encoding two related 100-kD coated vesicle proteins (α-adaptins) J Cell Biol. 1989;108:833–842. doi: 10.1083/jcb.108.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Cloning and expression of γ-adaptin, a component of clathrin-coated vesicles associated with the Golgi apparatus. J Cell Biol. 1990;111:2319–2326. doi: 10.1083/jcb.111.6.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Cell Biology. Membrane traffic COPs. Nature (Lond) 1991;349:343–344. doi: 10.1038/349743a0. [DOI] [PubMed] [Google Scholar]
- Robinson MS, Pearse BMF. Immunofluorescent localization of 100K coated vesicle proteins. J Cell Biol. 1986;102:48–54. doi: 10.1083/jcb.102.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Schekman R, Orci L. Coat proteins and vesicle budding. Science (Wash DC) 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Seaman MNJ, Ball CL, Robinson MS. Targeting and mistargeting of plasma membrane adaptors in vitro. J Cell Biol. 1993;123:1093–1105. doi: 10.1083/jcb.123.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, Sowerby PJ, Robinson MS. Cytosolic and membrane-associated proteins involved in the recruitment of AP-1 adaptors onto the trans-Golgi network. J Biol Chem. 1996;271:25446–25451. doi: 10.1074/jbc.271.41.25446. [DOI] [PubMed] [Google Scholar]
- Shih W, Gallusser A, Kirchhausen T. A clathrin binding site in the hinge of the β2 chain of mammalian AP-2 complexes. J Biol Chem. 1995;270:31083–31090. doi: 10.1074/jbc.270.52.31083. [DOI] [PubMed] [Google Scholar]
- Simpson F, Bright NA, West MA, Newman LS, Darnell RB, Robinson MS. A novel adaptor-related protein complex. J Cell Biol. 1996;133:749–760. doi: 10.1083/jcb.133.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Searching for patterns in protein and nucleic acid sequences. Method Enzymol. 1990;183:193–211. doi: 10.1016/0076-6879(90)83014-z. [DOI] [PubMed] [Google Scholar]
- Van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibiregene associated with vesicular traffic. Nature (Lond) 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]