Abstract
Heat stress is an obvious hazard, and mechanisms to recover from thermal damage, largely unknown as of yet, have evolved in all organisms. We have recently shown that a marker protein in the ER of Saccharomyces cerevisiae, denatured by exposure of cells to 50°C after preconditioning at 37°C, was reactivated by an ATP-dependent machinery, when the cells were returned to physiological temperature 24°C. Here we show that refolding of the marker enzyme Hsp150Δ–β-lactamase, inactivated and aggregated by the 50°C treatment, required a novel ER-located homologue of the Hsp70 family, Lhs1p. In the absence of Lhs1p, Hsp150Δ–β-lactamase failed to be solubilized and reactivated and was slowly degraded. Coimmunoprecipitation experiments suggested that Lhs1p was somehow associated with heat-denatured Hsp150Δ– β-lactamase, whereas no association with native marker protein molecules could be detected. Similar findings were obtained for a natural glycoprotein of S. cerevisiae, pro-carboxypeptidase Y (pro-CPY). Lhs1p had no significant role in folding or secretion of newly synthesized Hsp150Δ–β-lactamase or pro-CPY, suggesting that the machinery repairing heat-damaged proteins may have specific features as compared to chaperones assisting de novo folding. After preconditioning and 50°C treatment, cells lacking Lhs1p remained capable of protein synthesis and secretion for several hours at 24°C, but only 10% were able to form colonies, as compared to wild-type cells. We suggest that Lhs1p is involved in a novel function operating in the yeast ER, refolding and stabilization against proteolysis of heatdenatured protein. Lhs1p may be part of a fundamental heat-resistant survival machinery needed for recovery of yeast cells from severe heat stress.
When Saccharomyces cerevisiae cells are shifted from physiological temperature (24°C) to 50°C, they die. However, if preconditioned at the temperature where heat shock genes are activated, 37°C, they survive a subsequent thermal insult at 50°C and form colonies at 24°C. All organisms initiate a protective response to heat stress at characteristic temperatures in a similar way (1, 24, 28). Though the primary lesions causing cell death from thermal insult are not known, denaturation of proteins in various subcellular locations can be lethal, unless it is reversible (28, 30, 32, 34). Heat shock proteins, which are upregulated during the preconditioning period, have been assumed to function in the acquisition of thermotolerance (28, 30). However, little is known of how they protect or repair heat-damaged target proteins, though a wealth of information is available on their roles in folding of newly synthesized proteins under physiological conditions (12). Moreover, the Hsp26, Hsp70, and Hsp90 classes of heat shock proteins do not significantly affect thermotolerance in yeast (3, 31, 50). In contrast, Hsp104, an ATPbinding member of the Hsp100 (Clp) family of stress proteins, is essential for acquisition of thermotolerance in S. cerevisiae (35). In its absence, cytoplasmic protein aggregates generated by thermal insult to the cells at 50°C fail to be solubilized, and heat-inactivated mRNA splicing is not reactivated, resulting in cell death (29, 47).
The ER is the folding compartment for proteins destined to secretory organelles and to the exterior of the cell (14). When a newly synthesized polypeptide chain emerges in the ER lumen, it adopts a conformation dictated by its amino acid sequence. Folding is thought to be assisted by chaperones like the Hsp70 family member BiP/Kar2p, apparently by prevention of incorrect associations between nonnative chains (10). We showed recently that a fusion protein with β-lactamase activity, Hsp150Δ–β-lactamase, was inactivated in the yeast ER upon shift of the cells to 50°C after preconditioning at 37°C. When the cells were returned to 24°C, the enzyme was reactivated in an ATPdependent way, even in the absence of de novo protein synthesis (21). In these studies the marker enzyme was accumulated in the ER of the temperature-sensitive secretion mutants sec23 or sec18 before the thermal insult at 50°C. When the marker enzyme was preaccumulated in the Golgi in sec7 or sec14 mutants, it failed to be reactivated, demonstrating that the refolding apparatus was confined to the ER (21). We show here that conformational repair of heat-damaged protein in the ER involves a novel member of the Hsp70 family, Lhs1p. Lhs1p was needed for reactivation, solubilization, and stabilization against proteolysis of the marker enzyme Hsp150Δ–β-lactamase, which was denatured and aggregated by thermal insult to living cells. Lhs1p appeared to have no significant role in folding and secretion of newly synthesized Hsp150Δ–β-lactamase molecules.
Materials and Methods
Yeast Strains and Media
The following yeast strains were grown at 24°C in shake flasks overnight to early logarithmic phase in YPD medium or synthetic complete (SC)1 medium lacking histidine (H645, H647, H656, and H660): H393 (Matα sec18-1 ura3-52 trp1-289 leu2-3,112 URA3::HSP150Δ–β-lactamase [43]), H454 (Mat a ura3-1 his3-11,15 leu2-3,112 trp1-2 ade2-1 can1-100 hsp104:: LEU2 [35]), H486 (Mat a sec23-1 leu2-3,11 ura3-52 URA3::HSP150Δ–β-lactamase [12]), H503 (RCY127 Mat a sec18-1 trp1-1 ade2-1 his3 ura3-52 lhs1:: TRP1[6]), H540 (RCY104 Mat a ura3-52 ade2-1 trp1-1 lys2 his3 lhs1::TRP1 [6]), H541 (TR2 Mat a ura3-52 ade2-1 trp1-1 lys2 his3 [27]), H602 (Mat a ura3-52 ade2-1 trp1-1 lys2 his3 lhs1::TRP1 URA3::HSP150Δ–β-lactamase), H604 (Mat a ura3-52 ade2-1 trp1-1 lys2 his3 URA3::HSP150Δ–β-lactamase), H621 (Mat a sec18-1 ura3-52 ade2-1 trp1-1 his3 lhs1::TRP1 URA3:: HSP150Δ–β-lactamase), H645 (Mat a sec18-1 ura3-52 ade2-1 trp1-1 his3 lhs1::TRP1 URA3::HSP150Δ–β-lactamase [pRC42]), H647 (Mat a sec18-1 ura3-52 ade2-1 trp1-1 his3 lhs1::TRP1 URA3::HSP150Δ–β-lactamase [pRC51]), H656 (Mat a ura3-52 ade2-1 trp1-1 lys2 his3 lhs1::TRP1 [pRC51]), and H660 (Mat a sec18-1 trp1-1 ade2-1 his3 ura3-52 lhs1::TRP1 [pRC51]). Strains H602, H604, and H621 were created by integrating the HSP150Δ– β-lactamase gene in plasmid pKTH4544 (43), into the genome of strains H540, H541, and H503, respectively. Centromeric plasmid pRC42 containing the LHS1 gene (6) was introduced into strain H621, creating strain H645, and centromeric plasmid pRC51 containing the c-myc–LHS1 gene (6) was introduced into strains H621, H540, and H503 to create strains H647, H656, and H660, respectively. Transformations were with the lithium acetate method (15). Strain H534 (Matα sec18-1 ade2-1 his3-11,15 trp1 hsp104::LEU2 URA3::HSP150Δ–β-lactamase) was created by mating strains H393 and H454 and dissecting the spores of the resulting diploid. The 4-kb BamHI/SacI fragment containing the LHS1 gene was removed from pRC40 (6) and ligated into BamHI/SacI-digested pRS313 (CEN HIS3 [42]), to create pRC42. A 4.1-kb BamHI/SacI fragment containing a modified copy of the LHS1 gene encoding Lhs1p tagged with the c-myc epitope was removed from pRC45 (6) and ligated into pRS313 (CEN HIS3) to create pRC51.
Metabolic Labeling and Immunoprecipitation
Metabolic labeling of cells (108/200 μl) was with 20 μCi of [35S]methionine/cysteine (1,000 Ci/mmol; Amersham International, Buckinghamshire, UK) in SC medium lacking methionine and cysteine. The labelings were terminated with NaN3, and the culture medium and cell lysate samples were immunoprecipitated as described (19). Briefly, cells were lysed mechanically with glass beads in NET buffer (0.05 M Tris-HCl, pH 8.0, containing 0.4 M NaCl, 5 mM EDTA, 1% NP-40, and 100 U/ml of aprotinin) in the presence of 2 mM PMSF. The lysates were boiled and precleared for 1 h at 4°C with protein A–Sepharose (Pharmacia LKB Biotechnology, Inc., Piscataway, NJ). The samples were immunoprecipitated with anti–β-lactamase antiserum (1:100) or anti–carboxypeptidase Y (CPY) antiserum (1:100) and protein A–Sepharose for 2 h at 4°C. After washing with diluted NET buffer (1:1), wash buffer (0.1 M Tris-HCl, pH 7.5, containing 0.2 M NaCl, 2 M urea, and 0.5% Tween-20), and with 0.1% SDS, the precipitates were analyzed by SDS-PAGE.
Coimmunoprecipitation and Western Analysis
Cells were lysed with glass beads in the presence of protease inhibitors as above, but in the presence of Triton X-100 and absence of SDS and boiling. The lysates were precleared with preimmune serum (1:100) and protein A–Sepharose and incubated overnight with anti–β-lactamase antiserum (1:100), anti-CPY antiserum (1:100), or preimmune serum (1:100) at 4°C, followed by a 3-h incubation with protein A–Sepharose at 4°C. The precipitates were washed twice with NET buffer containing 10 mM CaCl2 and lacking EDTA, and twice with the same buffer diluted 1:1 and subjected to SDS-PAGE. Proteins were blotted from gel onto nitrocellulose membrane (Hybond-C Extra; Amersham International) and immunostained with anti–β-lactamase antiserum (1:6,000) or anti-CPY antiserum (1:1,000) and alkaline-phosphatase–conjugated anti–rabbit antibody (1:7,500; Promega Corp., Madison, WI), or with monoclonal anti–c-myc antibody (1:1,000; Chemicon International, Temecula, CA) and alkaline phosphatase– conjugated anti–mouse antibody (1:3,000; Promega Corp.).
Sucrose Gradient Centrifugation
Cells were lysed under nondenaturing conditions (see above) and loaded on 10–40% (wt/vol) linear sucrose gradients and subjected to velocity sedimentation centrifugation (44). Fractions of 0.5 ml were collected from top to bottom. The pellet was dissolved in 10 μl of 20% SDS and diluted in 0.5 ml of NET buffer. The fractions were immunoprecipitated under nondenaturing conditions.
Other Methods and Materials
For determination of β-lactamase activity, duplicate cell samples (25 × 106/0.5 ml of YPD or SC medium) were incubated at indicated temperatures. After termination of the incubation with NaN3, the cells were separated from the media, and cell-associated and secreted β-lactamase activity was assayed using nitrocefin as a substrate as described (43). Invertase synthesis was induced by incubating cells (5 × 107/0.5 ml) in YPD medium containing 0.1% glucose, and intracellular and cell wall–bound invertase activity of duplicate samples was measured as described in (19). Thermotolerance was assayed by incubating duplicate cell samples (3 × 106/ml) in YPD medium (except strain H647 in SC medium lacking histidine) in Wassermann tubes successively at 37 and 50°C (35). After cooling on ice, the cells were diluted and plated for 3–4 d at 24°C. Cycloheximide (CHX), DTT, and NaN3 were from Sigma Chemical Co. (St. Louis, MO) and used in final concentrations of 100 μg/ml, 20 mM, and 10 mM, respectively. BSA and porcine thyroglobulin were from Sigma Chemical Co. SDSPAGE was in 8% reducing gels.
Results
Effect of Lhs1p on Reactivation of Heat-denatured Hsp150Δ–β-lactamase In Vivo
Our assay to study heat-denaturation and refolding of ERlocated proteins in living S. cerevisiae cells is presented at the top of Fig. 1. Cells grown at 24°C are incubated in liquid medium for an h at 37°C. The marker protein Hsp150Δ–β-lactamase is expressed under the heat-activated HSP150 promoter in a temperature-sensitive mutant (sec18), where ER-derived transport vesicles are unable to fuse with Golgi membranes at 37°C. The 37°C incubation thus preconditions the cells to survive a subsequent thermal insult at 50°C, upregulates the synthesis of the marker protein, and causes its retention in the preGolgi compartment. The cells are then incubated for 20 min at 50°C, resulting in inactivation of the β-lactamase fusion protein, and shifted to 24°C to monitor reactivation of the marker enzyme. In tens of experiments, 20–70% of the original β-lactamase activity was resumed in 3–6 h at 24°C in the presence of CHX (21). The variation in the reactivation yields evidently reflects the vulnerability of the cells to extreme temperature. Hsp150Δ–β-lactamase consists of Escherichia coli β-lactamase fused to the COOH terminus of the Hsp150Δ-carrier, an NH2-terminal 321–amino acid fragment of the yeast secretory glycoprotein Hsp150 (20). The carrier is needed for translocation and to promote proper folding of foreign proteins that by themselves are not secretion-competent in yeast (25, 43).
Figure 1.

Dependence of reactivation of heat-denatured Hsp150Δ– β-lactamase on Lhs1p. (Top) Scheme of the renaturation assay. After growth at 24°C, cells were incubated for 60–70 min at 37°C (preconditioning), thereafter for 20 min at 50°C (thermal insult), and then for 6 h at 24°C (recovery). Strains (A) H621 (lhs1− sec18), (B) H393 (sec18), and (C) H645 (lhs1− LHS1 + sec18) were preincubated for 10 min at 37°C, pelleted, resuspended in prewarmed medium, and incubated at 37°C for 1 h and then at 50°C for 20 min. CHX (closed circles) or NaN3 (open circles) was added, and the cells were incubated for the indicated times at 24°C. The β-lactamase activity of lysed cells was determined and plotted against time. The absence (lhs1−) and presence (WT) of the LHS1 gene is indicated. The designation wild-type (WT) refers only to the LHS1 gene.
To study whether Lhs1p has a role in refolding of proteins denatured by thermal insult to living cells, Hsp150Δ–β-lactamase was expressed in S. cerevisiae sec18 cells lacking a functional LHS1 gene because of targeted gene disruption (strain H621, lhs1− sec18). When H621 cells were incubated at 37°C, β-lactamase activity accumulated inside of the cells (Fig. 1 A), to a somewhat lower level than in wildtype cells (Fig. 1 B). Hsp150Δ–β-lactamase adopts an enzymatically active conformation upon translocation into the ER and concomitant disulfide formation. The ER form of Hsp150Δ–β-lactamase is primary O-glycosylated at many serines and threonines of the carrier portion and migrates in SDS-PAGE like a protein of ∼110 kD, whereas the mature protein with extended O-glycans migrates like a 145-kD protein (21, 43). Shift of the cells from 37 to 50°C abolished the β-lactamase activity. When CHX was added to inhibit protein synthesis and the cells were shifted to 24°C for recovery, very little of the initial β-lactamase activity was resumed in 6 h (Fig. 1 A, closed circles). Parallel cells, which received NaN3 at the time of shift to 24°C, served as a negative control: no reactivation was observed in the absence of ATP (open circles). H393 cells, which harbored the wild-type genomic LHS1 gene and the sec18 mutation, served as a positive control: 38% of the initial β-lactamase activity, which had accumulated in the ER before inactivation at 50°C, was recovered in 6 h (Fig. 1 B, closed circles), and NaN3 inhibited the reactivation (open circles). We returned one copy of the wild-type LHS1 gene on a centromeric plasmid to strain H621, creating strain H645 (lhs1 − LHS1 + sec18). Reactivation of heat-denatured Hsp150Δ– β-lactamase was rescued to the level observed in the wildtype strain H393, again in an ATP-dependent fashion (Fig. 1 C). In none of the strains was β-lactamase activity found in the culture medium. These experiments show that Lhs1p did not protect the marker protein from thermal inactivation but was involved in reactivation of the heatdenatured marker protein.
Aggregation and Solubilization of Heat-denatured Hsp150Δ–β-lactamase
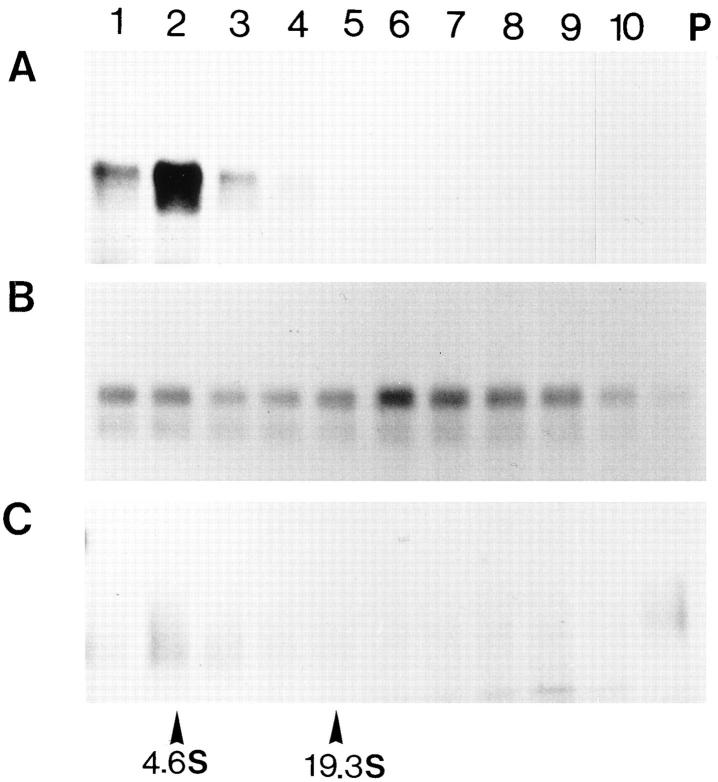
Next, we studied whether the thermal treatments caused aggregation of Hsp150Δ–β-lactamase. H393 cells (sec18) were 35S-labeled for 1 h at 37°C (Fig. 2 A), a parallel cell sample was incubated thereafter for 20 min at 50°C (Fig. 2 B), and a third sample was allowed to recover for 6 h at 24°C in the presence of CHX (Fig. 2 C). A fourth sample received NaN3 for the time of recovery (Fig. 2 D). The cells were lysed under nondenaturing conditions and subjected to velocity sedimentation centrifugation in sucrose gradients. The fractions plus the pellet material (lane P) were subjected to immunoprecipitation with anti–β-lactamase antiserum and SDS-PAGE analysis. After labeling at 37°C, Hsp150Δ–β-lactamase was detected in the top fractions, indicating that it was soluble (Fig. 2 A). When immunoprecipitated from the gradient fractions, the protein migrated in SDS-PAGE as a doublet of ∼95 and 110 kD. Thermal insult to the cells caused Hsp150Δ–β-lactamase to form aggregates since it was detected across the whole gradient and in the pellet fraction (Fig. 2 B). After the recovery period at 24°C, it sedimented again mainly in the top fractions, showing that it was largely solubilized. Solubilization required metabolic energy, since it was inhibited in the presence of NaN3 (Fig. 2 D). Since the sec18-1 mutation blocks fusion of ER-derived vesicles with the Golgi membranes, at least part of our marker protein may have resided in the vesicles in addition to the ER proper. Thus, we repeated the aggregation assay described above using sec23-1 cells (H486), where the marker is blocked quantitatively in the ER proper. Similar results were obtained as shown in Fig. 2, A–D, for the sec18-1 mutant. We have shown before that resumption of enzymatic activity of heat-inactivated Hsp150Δ–β-lactamase occurred equally efficiently in the sec23-1 and sec18-1 mutants (21).
Figure 2.
Reversible heat-induced aggregation of Hsp150Δ– β-lactamase. (A) Strain H393 (sec18) was labeled, after a preincubation for 10 min at 37°C, with [35S]methionine/cysteine for 1 h at 37°C. (B) Parallel cells were incubated for 20 min at 50°C after the labeling, or incubated thereafter for 6 h at 24°C in the presence of CHX (C) or NaN3 (D). The cells were lysed under nondenaturing conditions and centrifuged in linear 10–40% sucrose gradients. 10 fractions from top to bottom plus the pellet (lane P) were collected and immunoprecipitated with anti–β-lactamase, followed by SDS-PAGE and fluorography. Molecular mass markers of 97 and 69 kD are indicated on the left of each panel, and the sedimentation of bovine serum albumin (4.6 S, 66 kD) and thyroglobulin (19.3 S, 660 kD) are indicated at the bottom.
Next we assayed the β-lactamase activity after fractionation. After incubation of H393 cells at 37°C, β-lactamase activity coincided with 35S-Hsp150Δ–β-lactamase in fractions 1–3 of the gradient (Fig. 3 A). After the thermal insult, no activity could be detected in the fractions (Fig. 3 B), whereas after 6 h at 24°C in the presence of CHX, ∼22% of the original β-lactamase activity reappeared in the top fractions (Fig. 3 C). We conclude that our marker protein was soluble when accumulated in the ER, aggregated by thermal insult to the living cells, and resolubilized in an ATP-dependent fashion, even in the absence of de novo protein synthesis, during recovery of the cells at 24°C.
Figure 3.
β-Lactamase activity of the gradient fractions. Strain H393 (sec18) was incubated at 37°C (A), then at 50°C (B), and thereafter for 6 h at 24°C with CHX (C). The cells were lysed and subjected to sucrose gradient centrifugation as described in Fig. 2, A–C. The fractions were assayed for β-lactamase activity.
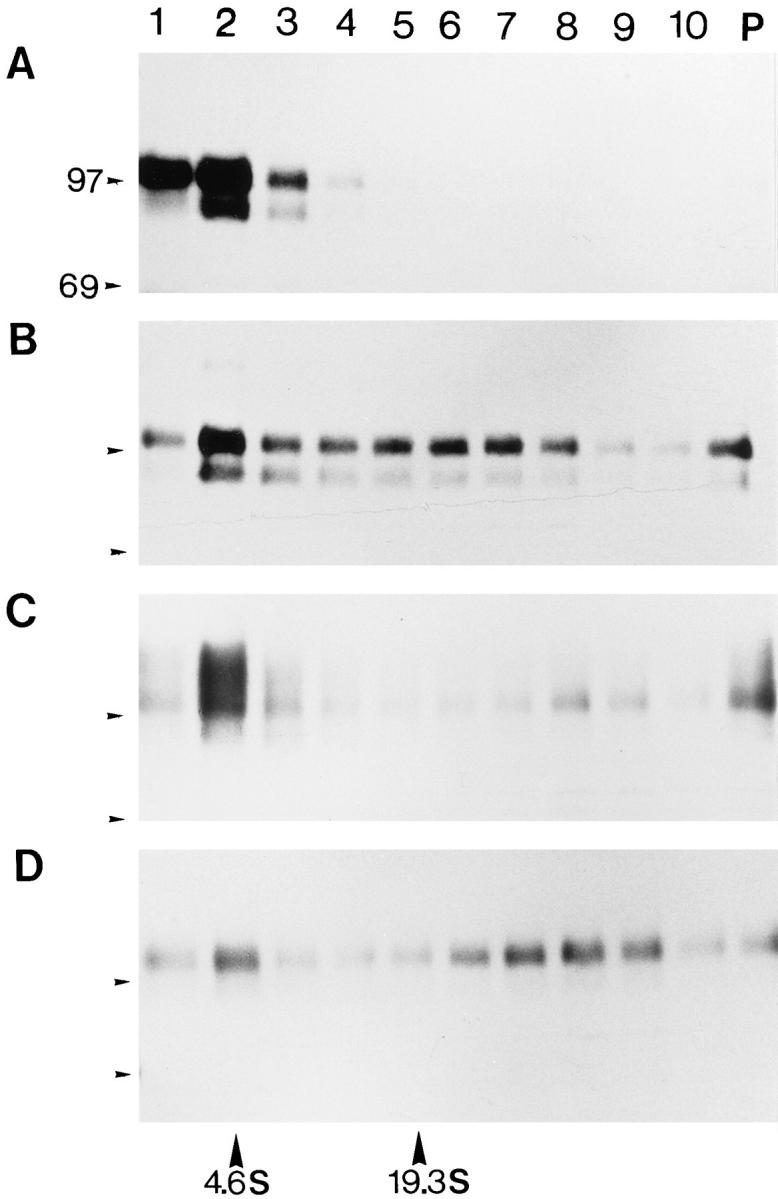
In the absence of the LHS1 gene (strain H621, lhs1− sec18), 35S-Hsp150Δ–β-lactamase labeled at 37°C was soluble and migrated in SDS-PAGE again as a doublet of 95 and 110 kD (Fig. 4 A). After the 50°C treatment, it was found in aggregates (Fig. 4 B), and after the recovery period it was barely detectable (Fig. 4 C). After the thermal insult and the recovery period, very little of the marker protein could be immunoprecipitated. Exposure time for B and C was five times longer than for A. Fractions 1–3 contained β-lactamase activity when the cells had been incubated at 37°C, whereas after thermal insult and recovery, no activity could be detected in the gradients (not shown). In the rescue strain H645 (lhs1− LHS1 + sec18), ATP-dependent solubilization of the Hsp150Δ–β-lactamase– containing aggregates occurred similarly as in strain H393 (see Fig. 2; not shown). Thus, heat-denatured Hsp150Δ–βlactamase remained in aggregates and was susceptible for proteolysis in the absence of Lhs1p. Degradation in this experiment occurred mainly by cellular proteases after cell lysis since less degradation occurred in vivo, as shown below. We conclude that thermal insult caused aggregation of ERlocated Hsp150Δ–β-lactamase, whether Lhs1p was present or not. In the absence of Lhs1p, the Hsp150Δ–β-lactamase–containing aggregates failed to be solubilized and were susceptible to proteolytic degradation in vitro.
Figure 4.
Aggregation and degradation of Hsp150Δ–β-lactamase in the absence of Lhs1p. The same experiments as described in the legend of Fig. 2, A–C, were performed using strain H621 (lhs1− sec18). B and C were exposed five times longer than A. The sedimentation of BSA (4.6 S) and thyroglobulin (19.3 S) is indicated at the bottom.
Coimmunoprecipitation of Lhs1p with Heat-denatured Hsp150Δ–β-lactamase and Pro-CPY
Coimmunoprecipitation experiments were performed to study whether Lhs1p was associated with Hsp150Δ–β-lactamase. H647 cells (lhs1− c-myc–LHS1+ sec18) were incubated successively for 1 h at 37°C (Fig. 5, A and B, lanes 1), 20 min at 50°C (lanes 2), and with CHX at 24°C for 3 h (lanes 3) or 6 h (lanes 4). The cells were lysed under nondenaturing conditions and divided in two. One lysate set was immunoprecipitated with anti–β-lactamase antiserum (Fig. 5, A and B, lanes 1–4) and the other with preimmune serum (lanes 6–9). Both sets were divided in two and subjected to SDS-PAGE and Western analysis using anti– c-myc antibody (Fig. 5 A) or anti–β-lactamase antiserum (Fig. 5 B). After incubation of the cells at 37°C, no precipitation of c-myc–Lhs1p with anti–β-lactamase antiserum could be detected (Fig. 5 A, lane 1). However, after thermal insult at 50°C and recovery at 24°C, c-myc–Lhs1p was coimmunoprecipitated with anti–β-lactamase antiserum (lanes 2–4). The total cell lysate samples (Fig. 5 A, lanes 5 and 10) show that a minor portion of Lhs1p was coprecipitated with Hsp150Δ–β-lactamase. Anti–β-lactamase antiserum precipitated Hsp150Δ–β-lactamase of ∼110 kD from all samples (Fig. 5 B, lanes 1–4). Neither Lhs1p (Fig. 5 A, lanes 6–9) nor the marker protein (Fig. 5 B, lanes 6–9) were precipitated with preimmune serum. Thus, Lhs1p appeared to occur in association predominantly with heatdenatured marker protein molecules. Whether the association was direct or mediated by other components is not known.
Figure 5.
Coimmunoprecipitation of Lhs1p with Hsp150Δ–β-lactamase. (A and B) H647 (lhs1− c-myc–LHS1 + sec18) cells were incubated for 1 h at 37°C (lanes 1 and 6), 20 min at 50°C (lanes 2 and 7), and at 24°C with CHX for 3 h (lanes 3 and 8) or 6 h (lanes 4 and 9). The samples were lysed under mild detergent conditions and divided in two. One set of samples was subjected under nondenaturing conditions to immunoprecipitation (IP) with anti– β-lactamase antiserum (α-bla; A and B, lanes 1–4) and the other with preimmune serum (pim; A and B, lanes 6–9). The precipitates were separated by SDS-PAGE, blotted, and immunostained (IS) using anti–c-myc antibody (α-c-myc; A, lanes 1–10) or anti– β-lactamase antiserum (B, lanes 1–10). Lanes 5 and 10 in both panels contained total lysate sample. c-myc–Lhs1p (lhs1p) and Hsp150Δ–β-lactamase (bla) are indicated by arrowheads. Molecular mass markers of 212, 170, 116, and 76 kD are indicated in lane M. (C) A cell sample incubated at 37° and 50°C, like above in lane 2, was lysed like above and centrifuged in a 10–40% sucrose gradient as described in Fig. 2. The fractions (lanes 1–10) and pellet material (lane P) were immunoprecipitated with anti– β-lactamase antiserum and subjected to blotting and immunostaining with anti–c-myc antibody, as in A. T, total cell lysate sample stained with anti–c-myc antibody. Arrowhead indicates c-myc–Lhs1p.
When a 50°C-treated sample, like the one in Fig. 5 A, lane 2, was fractionated in a sucrose gradient before immunoprecipitation with anti–β-lactamase antiserum in nondenaturing conditions, c-myc–Lhs1p was detected by anti– c-myc antibody in Western analysis mostly in fractions 7–9 and in the pellet fraction P (Fig. 5 C), i.e., in aggregates. We showed earlier that BiP/Kar2p was found in association with heat-treated as well as native Hsp150Δ–β-lactamase (21). Thus, the aggregates immunoprecipitated with anti–β-lactamase antiserum contained at least Hsp150Δ– β-lactamase, Lhs1p, and BiP/Kar2p.
To study whether Lhs1p could be found in association even with an authentic yeast glycoprotein, we repeated the above coimmunoprecipitation experiment using anti-CPY antiserum instead of anti–β-lactamase antiserum (Fig. 6). Normally, CPY is synthesized as a precursor, which is N-glycosylated in the ER (pro-CPY of 67 kD), and then transported to the vacuole, where it is processed to mature CPY of 62 kD. Similar results were obtained for pro-CPY as for Hsp150Δ–β-lactamase. After incubation of H660 cells (lhs1− c-myc–LHS1+ sec18) at 37°C, very little c-myc– Lhs1p (Fig. 6 A, lane 1) was immunoprecipitated with proCPY (Fig. 6 B, lane 1). After treatment of the cells for 20 min at 50°C and incubation for 3 or 6 h at 24°C, much more c-myc–Lhs1p (Fig. 6 A, lanes 2–4) was precipitated with pro-CPY (Fig. 6 B, lanes 2–4). Immunoprecipitation with preimmune serum served as a negative control, as before (Fig. 6, A and B, lanes 5–8). Lanes 9 contain total cell lysate. In Fig. 6 B, IgG is shown because of close migration with pro-CPY. Thus, even in the case of pro-CPY did Lhs1p appear to be predominantly associated with molecules that had undergone the thermal insult.
Figure 6.
Coimmunoprecipitation of Lhs1p with pro-CPY. (A and B) H660 (lhs1− c-myc–LHS1 + sec18) cells were incubated as in Fig. 5 for 1 h at 37°C (lanes 1 and 5), 20 min at 50°C (lanes 2 and 6), and at 24°C with CHX for 3 h (lanes 3 and 7) or 6 h (lanes 4 and 8). After lysis of the cells under nondenaturing conditions, the samples were divided in two. One set of samples was immunoprecipitated (IP) with anti-CPY antiserum (α-CPY, A and B, lanes 1–4) and the other with preimmune serum (pim; A and B, lanes 5–8). Lanes 9 contained total lysate sample. After SDSPAGE and blotting, the proteins were immunostained (IS) using anti–c-myc antibody (α-c-myc; A, lanes 1–9) or anti-CPY antiserum (B, lanes 1–9). c-myc–Lhs1p (Lhs1p), pro-CPY (CPY), and IgG are indicated. Molecular mass markers (kD) are indicated on the left.
Stability of Heat-denatured Marker Protein
To study the stability of heat-aggregated Hsp150Δ–β-lactamase in the absence of Lhs1p in vivo, strain H621 (lhs1− sec18) was labeled with [35S]methionine/cysteine at 37°C (Fig. 7 A, lane 1). Parallel cells were then incubated at 50°C (Fig. 7, lane 2), or thereafter at 24°C for 2–8 h (lanes 3–6). The cell lysates were immunoprecipitated with anti– β-lactamase antiserum, followed by SDS-PAGE analysis. Hsp150Δ–β-lactamase persisted for about 4 h at 24°C, growing in size apparently because of some increase in O-glycosylation. During the next 4 h, it was largely degraded. In the rescue strain H647 (lhs1− c-myc–LHS1 + sec18), the amount of the marker protein remained virtually unchanged (Fig. 7 B), as shown before for strain H393 harboring the genomic LHS1 gene (21). Degradation of the marker in the absence of Lhs1p was not due to abrupt cell death, since heat-treated H621 cells were capable of protein synthesis for hours after shift to 24°C (see below). To confirm this, we studied the stability of Hsp150Δ–β-lactamase in a strain lacking a functional HSP104 gene (H534), which does not acquire thermotolerance at 50°C in spite of preconditioning at 37°C (35) (see below). In this strain, the thermal treatments inhibited protein synthesis irreversibly (see below). Hsp150Δ–β-lactamase 35S-labeled at 37°C remained after the 50°C treatment apparently undegraded at 24°C at least for 8 h (Fig. 7 C). These data show that Lhs1p suppressed in vivo degradation of the heat-denatured marker protein.
Figure 7.

In vivo degradation of heat-denatured Hsp150Δ–β-lactamase. (A) H621 (lhs1− sec18), (B) H647 (lhs1− c-myc–LHS1 + sec18), and (C) H534 (Hsp104− sec18) cells were preincubated for 15 min at 37°C, labeled in SC medium lacking methionine and cysteine with [35S]methionine/cysteine for 20 min, diluted with excess SC medium containing unlabeled methionine and cysteine, and incubated further at 37°C for 40 min (lanes 1). Parallel cells were incubated thereafter for 20 min at 50°C (lanes 2), or after this at 24°C for 2 h (lanes 3), 4 h (lanes 4), 6 h (lanes 5), or 8 h (lanes 6). The cells were lysed and immunoprecipitated with anti– β-lactamase antiserum, followed by SDS-PAGE analysis.
Role of Lhs1p in De Novo Folding and Secretion of Hsp150Δ–β-lactamase and Pro-CPY
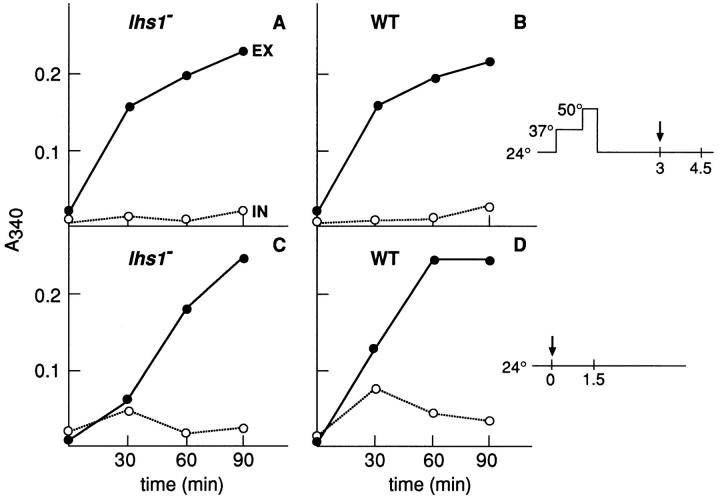
Lhs1p is required for translocation into the ER of a subset of proteins (2, 6). Thus, we studied the role of Lhs1p in translocation, folding, and secretion of de novo–synthesized Hsp150Δ–β-lactamase in more detail. Strain H602 (lhs1 −) (Fig. 8 A) and its parental strain H604 (WT) (Fig. 8 B), both wild-type for secretion, were incubated at 37°C, and the β-lactamase activities of the culture medium samples (Fig. 8, closed circles, EX) and lysed cell samples (open circles, IN) were determined. In both strains, β-lactamase activity externalized to the culture medium increased linearly, a little faster in the parental strain than in the lhs1− mutant. In the parental strain, little intracellular activity accumulated (0.12 U/ml in 90 min), indicating efficient secretion of the marker enzyme. In the lhs1− mutant, twice as much intracellular activity accumulated during the incubation, and its level in the beginning of the experiment was high. This suggests that a fraction of the newly synthesized marker protein remained cell associated in active form and gradually built up an intracellular pool in the absence of a functional LHS1 gene.
Figure 8.
Secretion of β-lactamase activity. Strains (A) H602 (lhs1−) and (B) H604 (WT) were incubated at 37°C. The β-lactamase activity of the culture medium (closed circles, EX) and lysed cells (open circles, IN) was determined and plotted against incubation time.
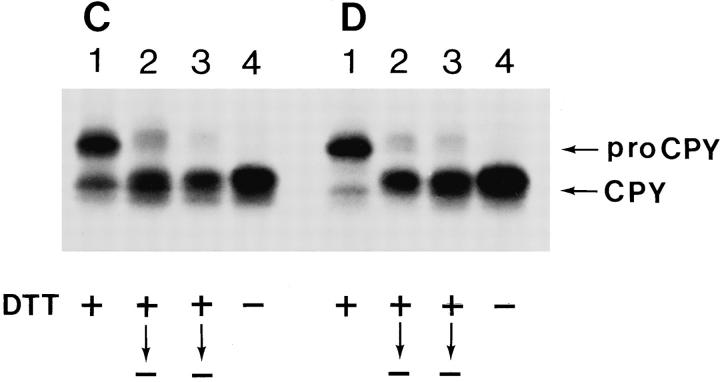
Proteins that normally acquire disulfide bonds are retained in the yeast ER if cotranslocational disulfide formation is prevented by incubating cells with a reducing agent like DTT, though the secretory machinery remains functional. When oxidizing conditions are reestablished by removing DTT, the sulfhydryls are oxidized, and the proteins undergo conformational maturation resulting in resumption of secretion (19, 44). To study whether Lhs1p was involved in conformational maturation of newly synthesized proteins, H602 (lhs1−) cells were 35S-labeled in the presence of DTT to allow translocation, while folding of Hsp150Δ–β-lactamase was inhibited. Immunoprecipitation with anti–β-lactamase antiserum of the culture medium sample showed that very little of the marker protein was secreted (Fig. 9 A, lane 3). In the absence of DTT, mature Hsp150Δ–β-lactamase of 145 kD was immunoprecipitated from the culture medium (Fig. 9 A, lane 1), and some 66- and 145-kD protein from the cell lysate (lane 2). The intracellular reduced molecules were difficult to detect (Fig. 9 A, lane 4), as noted before (43). When cells labeled under reducing conditions were washed and chased in the absence of DTT and presence of CHX, secretion of the marker protein was resumed, since it could be detected in the culture medium (Fig. 9 A, lane 5). Some 66-, 110-, and 145-kD proteins remained cell associated (Fig. 9 A, lane 6). Similar results were obtained for the parental strain H604: DTT prevented reversibly the secretion of the marker protein (Fig. 9 B), but less intracellular proteins were detected in the cell lysates as compared to H602 (Fig. 9 B, lanes 2, 4, and 6).
Figure 9.
Conformational maturation of Hsp150Δ–β-lactamase and pro-CPY. (A) Strains H602 (lhs1−) and (B) H604 (WT) were preincubated for 10 min in the absence or presence of DTT and labeled with [35S]methionine/cysteine for 1 h in the absence (lanes 1 and 2) or presence of the reducing agent (lanes 3 and 4). CHX was added and the incubations continued for 20 min. In lanes 5 and 6, the labeling was performed in the presence of DTT as above, after which DTT was removed and the cells incubated further in fresh medium with CHX for 1 h. All incubations were at 37°C. The culture medium (m; lanes 1, 3, and 5) and cell lysate samples (c; lanes 2, 4, and 6) were immunoprecipitated with anti–β-lactamase antiserum before SDS-PAGE analysis. Apparent molecular masses of Hsp150Δ–β-lactamase–related proteins (kD) are indicated on the right, and the molecular mass markers (kD) on the left. (C) Strains H602 (lhs1−) and (D) H604 (WT) were 35S-labeled for 1 h after a preincubation of 10 min in the presence of DTT (lanes 1–3), washed, and incubated with CHX for 45 min (lanes 2) or 90 min (lanes 3). In lanes 4, the labeling was as in lanes 1, but in the absence of DTT. All incubations were at 24°C. The cell lysates were precipitated with anti-CPY antiserum, followed by SDS-PAGE analysis.
The role of Lhs1p in conformational maturation of proCPY was then studied. Both in strains H602 (lhs1−; Fig. 9 C) and H604 (WT; Fig. 9 D), pro-CPY was accumulated in the ER in the presence of DTT (lanes 1), and converted similarly to CPY after DTT was replaced by CHX (lanes 2 and 3). In lanes 4, the cells were labeled in the absence of DTT, and thus mature CPY could be detected because of its rapid transport to the vacuole. From these data we conclude that Lhs1p had no significant role in translocation or conformational maturation of Hsp150Δ–β-lactamase and pro-CPY under our experimental conditions.
Acquisition of Thermotolerance in the Absence of Lhs1p
Next we studied whether Lhs1p affected the ability of yeast cells to acquire thermotolerance. Strain H540 (lhs1−), the parental strain H541 (WT), and strain H656 (lhs1− c-myc–LHS1 +) were incubated for different times at 37°C and thereafter for 20 min at 50°C and plated at 24°C (Fig. 10 A). More than 50% of H541 (Fig. 10 A, open circles) and H656 cells (closed circles) formed colonies when preconditioned for 45 min at 37°C before the 50°C treatment, whereas only 4% of H540 cells (Fig. 10 A, squares) survived. Strain H454 lacking a functional HSP104 gene (Fig. 10 A, triangles) served as a negative control: only 0.1% of them were able to form colonies after the thermal treatments. When the assay was performed by preincubating strains H540 (Fig. 10 B, lhs1−; squares) and H656 (lhs1− LHS1 +; circles) for 45 min at 37°C followed by different times at 50°C, lack of LHS1 again caused a 10-fold decrease in survival.
Figure 10.
Effect of Lhs1p on acquisition of thermotolerance. (A) Strains H541 (WT, open circles), H540 (lhs1−, squares), H656 (lhs1− c-myc–LHS1 +, closed circles), and H454 (hsp104−, triangles) were incubated for different times at 37°C and then for 20 min at 50°C, and plated at 24°C. (B) Strains H656 (closed circles) and H540 (squares) were incubated for 45 min at 37°C and then for different times at 50°C, and plated at 24°C. The colonies were counted to determine the percentage of survival, which was plotted against incubation times.
Though only 10% of lhs1− cells, as compared to wild-type cells, were able to form colonies after the thermal insult, most of them remained biochemically active for several hours after the 50°C treatment as shown by de novo synthesis of invertase during the recovery period. H621 cells (lhs1− sec18; Fig. 11 A) and H393 cells (sec18; Fig. 11 B) were incubated successively for 1 h at 37°C, 20 min at 50°C, and 3 h at 24°C. The cells were then shifted to low glucose medium to derepress the synthesis of invertase (Fig. 11 B, arrow), which is normally destined to the cell wall. Determination of invertase activity in the cell wall (Fig. 11, A and B, closed circles, EX) and inside of the cells (open circles, IN) showed that similar amounts of the enzyme were synthesized and secreted in the presence and absence of Lhs1p. Both strains secreted similar amounts of active invertase also at 24°C in the absence of any thermal treatments (Fig. 11, C and D). In contrast, no invertase synthesis during a recovery period of 6 h could be detected in strain H454 (hsp104−) (not shown). We conclude that synthesis and secretion, but not repair of the heat-damaged marker protein, functioned for many hours during the recovery period at 24°C in the absence of Lhs1p. Decreased thermotolerance of lhs1− cells, as measured by plating efficiency, was thus not due to abrupt cell death soon after the thermal insult, but to later events resulting finally in failure of colony formation in >90% of the cells.
Figure 11.
Invertase synthesis during recovery at 24°C. (A) Strains H621 (lhs1− sec18) and (B) H393 (sec18, indicated WT) were incubated successively for 1 h at 37°C, 20 min at 50°C, and 3 h at 24°C in YPD medium containing 2% glucose. The cells were shifted to YPD medium containing 0.1% glucose (arrow in upper scheme of experimental design) and incubated further at 24°C. In C and D, H621 and H393 cells, respectively, were incubated at 24°C in YPD medium containing 0.1% glucose (arrow in lower scheme). Cell samples were withdrawn for determination of intracellular (open circles, IN) and externalized (closed circles, EX) invertase activity. The relative activities are plotted against incubation time.
Discussion
The open reading frame of the LHS1 gene (YKL 073) was discovered in the Yeast Genome Sequencing Project. The translated amino acid sequence begins with a potential NH2terminal signal peptide and terminates with an ER-retrieval signal HDEL. The ER-located Hsp70 cognate BiP/Kar2p has 25% identical and 50% similar amino acids with Lhs1p (33). LHS1 encodes a glycoprotein of 113–119 kD, is located in an ER-like compartment, and is required for translocation of a subset of proteins (2, 6). The LHS1 gene and Lhs1p are also called SSI1 and Ssi1p, respectively (2). We show here that Lhs1p is involved in conformational repair and stability of heat-damaged protein in the yeast ER. In the absence of a functional LHS1 gene, Hsp150Δ–β-lactamase, aggregated and denatured by thermal insult to the cells, failed to be solubilized and reactivated like in wildtype cells.
According to coimmunoprecipitation experiments, Lhs1p was associated with heat-denatured Hsp150Δ–β-lactamase, whereas association with native marker protein molecules could not be detected. Lhs1p could be immunoprecipitated even with a natural glycoprotein of S. cerevisiae, proCPY, predominantly after thermal insult. Also, pro-CPY was damaged by the thermal insult to the cells since it became secretion incompetent (Saris, N., and M. Makarow, to be published). Whether association of the heat-damaged marker proteins with Lhs1p was mediated by other components is not known. Similar studies demonstrated earlier that BiP/Kar2p also could be coimmunoprecipitated with heat-denatured Hsp150Δ–β-lactamase, and to a lesser extent with the native marker protein, whereas no association with protein disulfide isomerase could be detected (21). Heat-denatured Hsp150Δ–β-lactamase thus occurred in aggregates containing at least Lhs1p and BiP/ Kar2p. The role of BiP/Kar2p in refolding of heat-damaged Hsp150Δ–β-lactamase could not be studied in vivo because it is required for translocation. Under conditions where BiP/Kar2p is nonfunctional, the marker protein is unable to translocate into the ER, and under normal conditions the marker protein escapes from the ER immediately after translocation (not shown). An ER-located hsp70like stress protein grp170 has been found in association with BiP and immunoglobulin in CHO cells (5, 23). Sequence comparisons suggest that grp170 could be the mammalian homologue of yeast Lhs1p, raising the possibility that this novel molecular chaperone is ubiquitous amongst eukaryotes.
Though heat-damaged aggregated Hsp150Δ–β-lactamase was solubilized and part of its enzymatic activity resumed, it remained in the ER at least for 6 h of recovery at 24°C (21). We have found that different secretory proteins, damaged by the thermal insult to the cells while residing in the ER, resume secretion competence with widely varying kinetics and efficiency (Saris, N., and M. Makarow, manuscript in preparation). Apparently, the structural features of Hsp150Δ–β-lactamase were not compatible with secretion competence at the time points examined.
In the presence of Lhs1p, Hsp150Δ–β-lactamase remained stable during the recovery period at 24°C, whereas it was slowly degraded in vivo in the absence of Lhs1p. Lately, degradation of mutant pro-CPY was shown to be retrotranslocated from the ER back to the cytosol for degradation by the proteasome complex (16). Since retrotranslocation of protein aggregates is unlikely, degradation of the heatdenatured marker protein probably occurred by some other mechanism, perhaps in the ER lumen or by autophagy (22, 46). Recently, a misfolded soluble protein was shown to be targeted to the vacuole for degradation (18). It has been suggested that heat shock proteins dissolve protein aggregates and facilitate protein degradation (13, 17). Newly synthesized misfolded proteins are degraded in the mitochondrial matrix by the PIM1 protease, if they are maintained in a soluble state by the chaperones mt-Hsp70 and Mdj1p (49). Lhs1p did not promote degradation of heatdamaged Hsp150Δ–β-lactamase but suppressed its degradation. Exposed internal domains of pro-insulin were shown to display signals for degradation in the ER of mammalian cells. BiP has been suggested to mask these domains, suppressing the degradation of unfolded proinsulin molecules (39).
Lhs1p thus appears to be involved both in translocation and in conformational repair of heat-damaged protein. Yeast BiP/Kar2p also has been assigned two functions. It is generally needed for translocation (36, 48) and for conformational maturation of newly synthesized pro-CPY (44) and Hsp150Δ–β-lactamase (Holkeri, H., E. Paunola, E. Jämsä, and M. Makarow, manuscript submitted for publication). In the latter studies, the marker proteins were translocated into the ER in reduced form in the presence of DTT and did not adopt a transport-competent conformation after reestablishment of oxidizing conditions unless BiP/Kar2p was functional. To compare the functions of BiP/Kar2p and Lhs1p, we studied the role of Lhs1p in folding of newly synthesized protein. Lack of Lhs1p did not inhibit translocation, conformational maturation, or intracellular transport of Hsp150Δ–β-lactamase, pro-CPY, and invertase. Thus, Lhs1p differed from BiP/Kar2p in that it had no significant role in folding or secretion competence of our newly synthesized marker protein, but instead was required for its conformational repair and stability after heat denaturation. This suggests that the repair machinery in the ER may have distinct features as compared to the chaperone apparatus involved in folding of newly synthesized polypeptides. Newly synthesized unfolded polypeptides are fed to the chaperone machinery during translocation domain by domain, whereas the repair machinery deals mostly with full-length protein molecules that have been folded before stress-induced unfolding and aggregation. Frydman and Hartl (8) found that molecular chaperones interact differently with newly translated polypeptides and their chemically denatured counterparts in the eukaryotic cytosol. The in vivo kinetics of folding and conformational repair of Hsp150Δ–β-lactamase differed strikingly: de novo folding occurs apparently in seconds, whereas conformational repair took several hours. Although recovery of yeast cells from thermal insult is efficient, it is remarkably slow. We found bulk protein synthesis to be inhibited for 2–3 h and fully recovered only after 6 h (21). Inhibition of protein synthesis for hours after severe heat stress has been noted also in mammalian cells (26). Evidently, in the absence of protein synthesis, survival requires refolding of heat-denatured proteins that perform vital functions.
Chaperone-dependent refolding of heat-denatured proteins has been shown before to occur in vivo in the cytosol of yeast and mammalian cells, and in E. coli. Heat-denatured firefly luciferase was reactivated in S. cerevisiae in an Hsp104-dependent manner (29). In E. coli, refolding of λ repressor protein and luciferase were dependent on the chaperones DnaK (Hsp70), DnaJ (Hsp40), and GrpE (9, 40). In vitro, the DnaK/DnaJ/GrpE chaperone system and the chaperonins GroEL/GroES (Hsp60) are able to reactivate heat-damaged or chemically denatured protein (11, 45, 51). The human cytosolic chaperone Hsp90 was shown to convert chemically denatured β-galactosidase to a folding-competent state, whereafter the substrate protein was refolded by Hsp70 and hdj-1. It was suggested that Hsp90 maintains nonnative proteins in a soluble state in heatstressed cells, whereafter Hsp70 and hdj-1 refold them at normal temperature (7). As BiP/Kar2p and other Hsp70 chaperones require DnaJ/Hsp40 chaperones for activity (4, 37, 41), Lhs1p may need accessory proteins as well.
In the absence of Lhs1p, the ability of the yeast cells to form colonies after thermal insult was reduced 10-fold, whereas in the absence of Hsp104, thermotolerance is reduced 1,000-fold (35). Thermal insult did not result in abrupt death of the lhs1− mutant cells since they were capable of protein synthesis and secretion for many hours thereafter, and thus cell death must have occurred because of later events. In contrast, in the hsp104− mutant protein synthesis was completely and irreversibly inactivated by the 50°C treatment. The upstream sequence of LHS1 contains an unfolded protein response element. Indeed, LHS1 was upregulated under conditions where unfolded proteins accumulated in the ER, and moreover, the absence of Lhs1p caused induction of the unfolded protein response (2, 6). However, LHS1 is not upregulated at 37°C, whereas HSP104 is strongly induced by elevated temperature (35). A mitochondrial member of the Clp/Hsp100 family of S. cerevisiae, Hsp78, was recently found to be needed for maintenance of respiratory competence and resumption of mitochondrial protein synthesis after thermal insult at 50°C; however, it did not affect acquisition of cellular thermotolerance (38). Chaperones that repair vital heat-damaged proteins evidently occur in all compartments of the cell. Lhs1p located in the ER may be part of this fundamental survival system, which is needed for cells to recover from heat stress causing protein denaturation.
Acknowledgments
We thank Dr. Neil Bulleid (Manchester University, UK) for drawing our attention to YKL 073. Drs. Randy Schekman (University of California, Berkeley, CA), Susan Lindquist (Howard Hughes Medical Institute, Chicago, IL), and Sören W. Rasmussen (Carlsberg Laboratory, Denmark) generously gave us yeast strains and plasmids. Drs. Leevi Kääriäinen and Eija Jämsä from our institute had valuable suggestions on the manuscript, and Ms. Anna Liisa Nyfors provided excellent technical assistance.
Footnotes
1. Abbreviations used in this paper: CHX, cycloheximide; CPY, carboxypeptidase Y; SC, synthetic complete.
Support from the Academy of Finland and the Foundation of the University of Helsinki is gratefully acknowledged. M. Makarow is a Biocentrum Helsinki fellow. C.J. Stirling is a Lister Institute Jenner Research Fellow.
Address all correspondence to Marja Makarow, Institute of Biotechnology, P.O. Box 56, 00014 University of Helsinki, Finland. Tel.: 358-970859419. Fax: 358-9-70859366. E-mail: Makarow@operoni.helsinki.fi
References
- 1.Ang D, Liberek K, Skowyra D, Zylicz M, Georgopoulos C. Biological role and regulation of the universally conserved heat shock proteins. J Biol Chem. 1991;266:24233–24236. [PubMed] [Google Scholar]
- 2.Baxter, B.K., P. James, T. Evans, and E.A. Craig. 1996. SSI1 encodes a novel Hsp70 of the Saccharomyces cerevisiae endoplasmic reticulum. [DOI] [PMC free article] [PubMed]
- 3.Borkovich KA, Farelly FW, Finkelstein DB, Taulien J, Lindquist S. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodsky JL, Schekman R. A sec63-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Easton D, Oh H, Lee-Yoon D, Liu X, Subjek JR. The 170 kDa glucose regulated stress protein is a large HSP70-HSP110like protein of the endoplasmic reticulum. FEBS (Fed Exp Biol Soc) Lett. 1996;380:68–72. doi: 10.1016/0014-5793(96)00011-7. [DOI] [PubMed] [Google Scholar]
- 6.Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO (Eur Mol Biol Organ) J. 1996;15:2640–2650. [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman BC, Morimoto R. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO (Eur Mol Biol Organ) J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- 8.Frydman J, Hartl FU. Principles of chaperone-assisted protein folding: differences between in vitro and in vivomechanisms. Science (Wash DC) 1996;272:1497–1502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- 9.Gaitanaris GA, Papavassiliou AG, Rubock P, Silverstein SJ, Gottesman ME. Renaturation of denatured lambda repressor requires heat shock proteins. Cell. 1990;61:1013–1020. doi: 10.1016/0092-8674(90)90066-n. [DOI] [PubMed] [Google Scholar]
- 10.Gething MJ, Sambrook JF. Protein folding in the cell. Nature (Lond) 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 11.Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and Mg-ATP. Nature (Lond) 1989;342:884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- 12.Hartl FU. Molecular chaperones in cellular protein folding. Nature (Lond) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 13.Hayes SA, Dice JF. Roles of molecular chaperones in protein degradation. J Cell Biol. 1996;132:255–258. doi: 10.1083/jcb.132.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helenius A, Marquardt T, Braakman I. The endoplasmic reticulum as a protein-folding compartment. Trends Cell Biol. 1992;2:227–231. doi: 10.1016/0962-8924(92)90309-b. [DOI] [PubMed] [Google Scholar]
- 15.Hill J, Donald KAIG, Griffiths DE. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiller MM, Finger A, Schweiger M, Wolf D. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science (Wash DC) 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 17.Hilt W, Wolf D. Stress-induced proteolysis in yeast. Mol Microbiol. 1992;6:2437–2442. doi: 10.1111/j.1365-2958.1992.tb01419.x. [DOI] [PubMed] [Google Scholar]
- 18.Hong E, Davidson AR, Kaiser C. A pathway for targeting soluble misfolded proteins to the yeast vacuole. J Cell Biol. 1996;135:623–633. doi: 10.1083/jcb.135.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jämsä E, Simonen M, Makarow M. Selective retention of secretory protein in the yeast endoplasmic reticulum by treatment of cells with a reducing agent. Yeast. 1994;10:355–370. doi: 10.1002/yea.320100308. [DOI] [PubMed] [Google Scholar]
- 20.Jämsä E, Holkeri H, Vihinen H, Wikström M, Simonen M, Walse B, Kalkkinen N, Paakkola J, Makarow M. Structural features of a polypeptide carrier promoting secretion of a β-lactamase fusion protein in yeast. Yeast. 1995;11:1381–1391. doi: 10.1002/yea.320111406. [DOI] [PubMed] [Google Scholar]
- 21.Jämsä E, Vakula N, Arffman A, Kilpeläinen I, Makarow M. In vivoreactivation of heat-denatured protein in the endoplasmic reticulum of yeast. EMBO (Eur Mol Biol Organ) J. 1995;14:6028–6033. doi: 10.1002/j.1460-2075.1995.tb00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knop M, Finger A, Braun T, Hellmuth K, Wolf D. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO (Eur Mol Biol Organ) J. 1996;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- 23.Lin H, Masso-Welch P, Cai J, Shen J, Subjek JR. The 170kDa glucose-regulated stress protein is an endoplasmic reticulum protein that binds immunoglobulin. Mol Biol Cell. 1993;4:1109–1119. doi: 10.1091/mbc.4.11.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindquist S, Craig E. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 25.Mattila P, Joutsjoki V, Kaitera E, Majuri ML, Niittymäki J, Maaheimo H, Renkonen O, Renkonen R, Makarow M. Targeting of active rat α2,3-sialyltransferase to the yeast cell wall by the aid of the Hsp150Δ-carrier—towards synthesis of sLex-decorated L-selectin ligands. Glycobiology. 1996;6:851–859. doi: 10.1093/glycob/6.8.851. [DOI] [PubMed] [Google Scholar]
- 26.Mizzen LA, Welch WJ. Characterization of the thermotolerant cell. I. Effects on protein synthesis activity and the regulation of heatshock protein 70 expression. J Cell Biol. 1988;106:1105–1116. doi: 10.1083/jcb.106.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker R, Simmons T, Shuter EO, Siliciano PG, Guthrie C. Genetic analysis of small nuclear RNAs in Saccharomyces cerevisiae: viable sextuple mutant. Mol Cell Biol. 1988;8:3150–3159. doi: 10.1128/mcb.8.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 29.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature (Lond) 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 30.Pelham HRB. Hsp70 accelerates the recovery of nucleolar morphology after heat shock. EMBO (Eur Mol Biol Organ) J. 1984;3:3095–3100. doi: 10.1002/j.1460-2075.1984.tb02264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petko L, Lindquist S. Hsp26 is not required for growth at higher temperatures nor for thermotolerance, spore development or germination. Cell. 1986;54:885–894. doi: 10.1016/0092-8674(86)90563-5. [DOI] [PubMed] [Google Scholar]
- 32.Pinto M, Moranges M, Bensaude O. Denaturation of proteins during heat shock. J Biol Chem. 1991;266:13941–13946. [PubMed] [Google Scholar]
- 33.Rasmussen SW. Sequence of a 20.7 kb region of yeast chromosome XI includes the NUP100gene, an open reading frame (ORF) possibly representing a nucleoside diphosphate kinase gene, tRNA for His, Val and Trp in addition to seven ORFs with weak or no significant similarity to known proteins. Yeast. 1994;10:S69–S74. doi: 10.1002/yea.320100009. [DOI] [PubMed] [Google Scholar]
- 34.Rothman JE. Polypeptide chain binding proteins: catalysts of protein folding and related processes in cells. Cell. 1989;59:591–601. doi: 10.1016/0092-8674(89)90005-6. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez Y, Lindquist S. Hsp104 is required for induced thermotolerance. Science (Wash DC) 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 36.Sanders SL, Whitfield KM, Vogel JP, Rose MD, Schekman RW. Sec61 and BiP directly facilitate polypeptide translocation into the ER. Cell. 1992;69:353–365. doi: 10.1016/0092-8674(92)90415-9. [DOI] [PubMed] [Google Scholar]
- 37.Schlenstedt G, Harris S, Risse B, Lill R, Silver PA. A yeast DnaJ homologue Scj1p can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with Hsp70s. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt M, Neupert W, Langer T. The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J Cell Biol. 1996;134:1375–1386. doi: 10.1083/jcb.134.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitz A, Maintz M, Kehle K, Herzog V. In vivoiodination of a misfolded proinsulin reveals co-localized signals for BiP binding and for degradation in the ER. EMBO (Eur Mol Biol Organ) J. 1995;14:1091–1098. doi: 10.1002/j.1460-2075.1995.tb07092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schröder H, Langer T, Hartl FU, Bukau B. DnaK, DnaJ and GrpE from a cellular chaperone machinery capable of repairing heat- induced protein damage. EMBO (Eur Mol Biol Organ) J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scidmore MA, Okamura HH, Rose MD. Genetic interactions between KAR2 and SEC63, encoding eukaryotic homologues of DnaK and DnaJ in the endoplasmic reticulum. Mol Biol Cell. 1993;4:1145–1159. doi: 10.1091/mbc.4.11.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simonen M, Jämsä E, Makarow M. The role of the carrier protein and disulfide formation in the folding of β-lactamase fusion proteins in the endoplasmic reticulum of yeast. J Biol Chem. 1994;269:13889–13892. [PubMed] [Google Scholar]
- 44.Simons JF, Ferro-Novick S, Rose MD, Helenius A. BiP/ Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J Cell Biol. 1995;130:41–49. doi: 10.1083/jcb.130.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skowyra D, Georgopoulos C, Zylicz M. The E. coli dnaKgene product, the hsp70 homolog, can reactivate heat-inactivated RNA polymerase in an ATP hydrolysis-dependent manner. Cell. 1990;62:939–944. doi: 10.1016/0092-8674(90)90268-j. [DOI] [PubMed] [Google Scholar]
- 46.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogel JL, Parsell DA, Lindquist S. Heat-shock proteins Hsp104 and Hsp70 reactivate mRNA splicing after heat inactivation. Curr Biol. 1995;5:306–317. doi: 10.1016/s0960-9822(95)00061-3. [DOI] [PubMed] [Google Scholar]
- 48.Vogel JP, Misra MD, Rose MD. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner I, Arlt H, van Dyck L, Langer T, Neupert W. Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO (Eur Mol Biol Organ) J. 1994;13:5135–5145. doi: 10.1002/j.1460-2075.1994.tb06843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Werner-Walshburne M, Stone DE, Craig E. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. . Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziemienowicz A, Skowyra D, Zeilstra-Ryalls J, Fayet O, Georgopoulos C, Zylicz M. Both the Escherichia colichaperone systems, GroEL/GroES and DnaK/DnaJ/GrpE, can reactivate heat-treated RNA polymerase. J Biol Chem. 1993;268:25425–25431. [PubMed] [Google Scholar]