Abstract
In Saccharomyces cerevisiae, the growing bud inherits a portion of the mitochondrial network from the mother cell soon after it emerges. Although this polarized transport of mitochondria is thought to require functions of the cytoskeleton, there are conflicting reports concerning the nature of the cytoskeletal element involved. Here we report the isolation of a yeast mutant, mdm20, in which both mitochondrial inheritance and actin cables (bundles of actin filaments) are disrupted. The MDM20 gene encodes a 93-kD polypeptide with no homology to other characterized proteins. Extra copies of TPM1, a gene encoding the actin filament–binding protein tropomyosin, suppress mitochondrial inheritance defects and partially restore actin cables in mdm20Δ cells. Synthetic lethality is also observed between mdm20 and tpm1 mutant strains. Overexpression of a second yeast tropomyosin, Tpm2p, rescues mutant phenotypes in the mdm20 strain to a lesser extent. Together, these results provide compelling evidence that mitochondrial inheritance in yeast is an actin-mediated process. MDM20 and TPM1 also exhibit the same pattern of genetic interactions; mutations in MDM20 are synthetically lethal with mutations in BEM2 and MYO2 but not SAC6. Although MDM20 and TPM1 are both required for the formation and/or stabilization of actin cables, mutations in these genes disrupt mitochondrial inheritance and nuclear segregation to different extents. Thus, Mdm20p and Tpm1p may act in vivo to establish molecular and functional heterogeneity of the actin cytoskeleton.
Mitochondria are essential organelles whose morphology, copy number, and distribution can vary dramatically in different eukaryotic cell types (Bereiter-Hahn, 1990; Yaffe, 1991; Thorsness, 1992; Bereiter-Hahn and Voth, 1994; Warren and Wickner, 1996). Like all organelles that cannot be synthesized de novo, the mitochondrial compartment must grow, divide, and be delivered to newly formed daughter cells during division. While substantial progress has been made in identifying the molecular components that mediate mitochondrial biogenesis (Hannavy et al., 1993; Schwarz and Neupert, 1994; Kubrich et al., 1995; Ryan and Jensen, 1995; Lill and Neupert, 1996), a detailed picture of the mechanisms that control mitochondrial morphology, distribution, and motility throughout the cell cycle has not yet emerged.
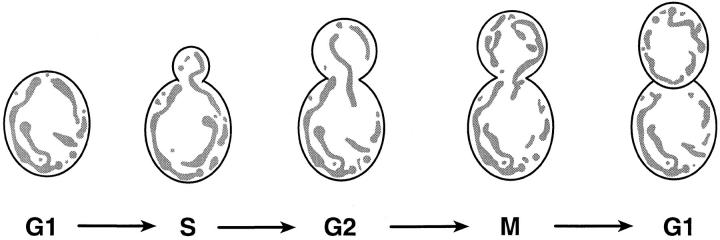
In the budding yeast Saccharomyces cerevisiae, respiring mitochondria form an elaborate network of tubular membranes located near the cell periphery (Hoffman and Avers, 1973). Very early in the cell cycle (early S phase), a portion of this mitochondrial network is transported from the mother cell into the growing bud (Fig. 1) (Stevens, 1981). Recent genetic studies have identified a few proteins that control mitochondrial morphology and distribution in cells and mitochondrial transmission during cell division. These include Mdm10p and Mmm1p, two proteins of the outer mitochondrial membrane required for the maintenance of normal organelle morphology (Sogo and Yaffe, 1994, mdm [mitochondrial distribution and morphology]; Burgess et al., 1994, mmm [maintenance of mitochondrial morphology]; Mdm2p/Ole1p, a fatty acid desaturase that may regulate the level of unsaturated fatty acids in the mitochondrial membrane (Stukey et al., 1989, 1990; Stewart and Yaffe, 1991, ole [oleic acid requiring]); and Mdm1p, a protein with characteristics of the mammalian intermediate filament–like proteins keratin and vimentin (McConnell and Yaffe, 1992, 1993). In addition, there are several proteins that are required for the normal maintenance or segregation of mitochondrial DNA. Mgm1p is a member of the dynamin family of GTP binding proteins (Jones and Fangman, 1992; Guan et al., 1993; Backer, 1995; mgm [mitochondrial genome maintenance]); ILV5 encodes a protein that plays an as yet unidentified role in mitochondrial genome maintenance (Zelenaya-Troitskaya et al., 1995; ilv [isoleucine-plus-valine requiring]); and Abf2p is an HMG1-like DNA binding protein (Diffley and Stillman, 1991, 1992; abf [ARS binding factor]). Studies are in progress in a number of laboratories to determine how the activities of these proteins control the behavior of the mitochondrial compartment and the replication and segregation of the mitochondrial genome.
Figure 1.
Mitochondrial inheritance during mitotic cell division. Mitochondria are localized near the cortex of unbudded and budding cells. Early in the cell cycle (S phase) a portion of the maternal mitochondrial network extends into the developing bud. As the bud grows (S phase to G2 phase), mitochondria continue to accumulate in the daughter cell. At cytokinesis (M phase/G1 phase boundary), the mitochondrial content of the daughter cell is often greater than that of the mother cell.
Most forms of organelle motility are directed by cytoskeletal structures and their associated motor proteins. Mitochondria have been reported to colocalize with actin filaments (Drubin et al., 1993), microtubules (Heggeness et al., 1978; Ball and Singer, 1982; Summerhayes et al., 1983), and intermediate filaments (Mose-Larsen et al., 1982; Summerhayes et al., 1983; Stromer and Bendayan, 1990) in different cell types, and recent studies indicate that mitochondria can be transported in a polarized fashion using both microtubule- (Nangaku et al., 1994; Morris and Hollenbeck, 1995; Hollenbeck, 1996) and actin-based (Kuznetsov et al., 1992; Morris and Hollenbeck, 1995; Simon et al., 1995; Hollenbeck, 1996) motors. In yeast, two different cytoskeletal elements, Mdm1p and actin, have been proposed to direct the transfer of mitochondria into yeast buds. Mdm1p shares structural features with mammalian intermediate filament proteins and will form 10-nm filaments in vitro (McConnell and Yaffe, 1992, 1993). Mutations in MDM1 cause defects in mitochondrial morphology and also in both mitochondrial and nuclear inheritance, suggesting that this protein is important for organelle positioning and/or segregation.
Recent studies have also established a role for the yeast actin cytoskeleton in mitochondrial distribution and inheritance. Yeast cells contain a single, essential, conventional actin gene (ACT1) (Shortle et al., 1982; Welch et al., 1994). In vivo, actin is organized into two different types of structures: (a) cortical actin patches (invaginations of the plasma membrane associated with F-actin) and (b) actin cables (bundles of F-actin; Adams and Pringle, 1984; Mulholland et al., 1994). In mitotically dividing cells, cortical actin patches cluster in the growing bud, and actin cables aligned along the mother-bud axis are well positioned to deliver mitochondria to the daughter cell (Kilmartin and Adams, 1984). Mitochondria have been shown to colocalize with actin cables in yeast, and some mutations in actin result in abnormal mitochondrial morphology, distribution, and motility (Drubin et al., 1993; Lazzarino et al., 1994; Simon et al., 1995). In addition, recent studies established that isolated yeast mitochondria bind reversibly to phalloidin-stabilized actin filaments in vitro (Lazzarino et al., 1994). These mitochondria were shown in a related assay to contain a myosin-like actin-dependent motor activity on their surface (Simon et al., 1995). Based on these results, it has been suggested that yeast mitochondria are transported from the mother cell into the emerging bud using actin filaments as “tracks” and a myosin-like motor activity on the surface of the organelle.
To learn more about molecules that control mitochondrial inheritance, we screened a collection of yeast mutants for strains that failed to transport mitochondria into growing buds. In one of these mutants, mdm20, both mitochondrial inheritance and the organization of actin cables were disrupted. The MDM20 gene encodes a novel 93-kD protein with no sequence similarities to proteins in the database. Extra copies of TPM1, a gene encoding the actin filament–binding protein tropomyosin, partially restored actin cables and suppressed mitochondrial inheritance defects in the mdm20 strain. Furthermore, the combination of null mutations in MDM20 and TPM1 resulted in synthetic lethality. Overexpression of a second yeast tropomyosin, Tpm2p, also rescued actin organization defects and mitochondrial inheritance defects in mdm20 cells, but to a lesser extent. Together, these results provide compelling evidence that mitochondrial inheritance in yeast is an actin-mediated process. Like tpm1, mdm20 mutations are synthetically lethal with mutations in BEM2 (Rho-GAP) and MYO2 (type V myosin) but not SAC6 (fimbrin). Thus, TPM1 and MDM20 exhibit the same pattern of genetic interactions. Despite the fact that mutations in MDM20 and TPM1 both result in the loss of observable actin cables in cells, we found that mitochondrial inheritance and nuclear segregation were disrupted to different extents in mdm20 and tpm1 mutant strains. Our results suggest that these two proteins act in vivo to establish molecular and functional heterogeneity of the actin cytoskeleton.
Materials and Methods
Strains, Media, and Genetic Techniques
Yeast strains used in this study are listed in Table I. Rich medium (yeast/ peptone/dextrose [YPD] and yeast/peptone/glycerol [YPG]), synthetic medium (synthetic dextrose [SD] and synthetic galactose [SGal]), and media containing 5-fluoro-orotic acid (5-FOA)1 were prepared as described by Sherman et al. (1986). Yeast mating, sporulation, tetrad dissection, complementation studies, and meiotic segregation analysis were performed using standard genetic techniques (Sherman et al., 1986). The lithium acetate method was used for all yeast transformations (Ito et al., 1983) and gene disruption experiments (Rothstein, 1991; Baudin et al., 1993). Escherichia coli strains DH5α and JM109 (Promega, Madison, WI) were used for bacterial manipulations of clones and plasmids. Molecular cloning techniques were performed as described by Maniatis et al. (1982).
Table I.
Yeast Strains Used in This Study
| Name | Genotype* | Source | ||
|---|---|---|---|---|
| FY10 | MATα leu2Δ1 ura3-52 | F. Winston | ||
| FY22 | MATa his3Δ200 ura3-52 | F. Winston | ||
| GHY1 | MATα leu2Δ1 his3Δ200 ura3-52 mdm20-1 | This study | ||
| JSY707 | MATa his3Δ200 ura3-52 tpm1Δ::HIS3 | This study | ||
| JSY948 | MATa/α leu2Δ1/leu2Δ1 ura3-52/ura3-52 | This study | ||
| JSY999 | MATα leu2Δ1 his3Δ200 ura3-52 | This study | ||
| JSY1065 | MATα leu2Δ1 his3Δ200 ura3-52 mdm20Δ::LEU2 | This study | ||
| JSY1084 | MATa leu2Δ1 his3Δ200 ura3-52 tpm1Δ::HIS3 | This study | ||
| JSY1138 | MATa/α leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 ura3-52/ura3-52 tpm1Δ::HIS3/+ mdm20Δ::LEU2/+ | This study | ||
| JSY1285 | MATα leu2Δ1 his3Δ200 ura3-52 tpm2Δ::HIS3 | This study | ||
| JSY1340 | MATa leu2Δ1 his3Δ200 ura3-52 mdm20Δ::LEU2 | This study | ||
| JSY1374 | MATa/α leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 ura3-52/ura3-52 tpm2Δ::HIS3/+ mdm20Δ::LEU2/+ | This study | ||
| ABY1249 | MATa leu2-3,112 ura3-52 lys2-801 ade2-101 ade3 bem2-10 | A. Bretscher | ||
| IGY4 | MATα leu2-3,112 his3Δ200 ura3-52 lys2-801 ade2 sac6Δ::LEU2 | A. Adams | ||
| SLY63 | MATα leu2-3,112 ura3-52 trp1-Δ1 his6 myo2-66 | S. Brown |
All of the GHY and JSY strains used in this study are isogenic to FY10 (Winston et al., 1995).
Identification of mdm Mutants
The wild-type strain FY10 (Winston et al., 1995) was UV mutagenized (25% survival), and a collection of 4,000 temperature-sensitive yeast strains was generated. To identify mitochondrial distribution and morphology (mdm) mutants in this collection, individual strains were grown for 16 h at the permissive temperature (25°C) in 500 μl of YPD, diluted 1:10 into 500 μl fresh YPD, and shifted to the nonpermissive temperature (37°C) for 3 h. 100 μl of the culture was stained with a final concentration of 100 nM 3,3′ dihexyloxacarbocyanine (DiOC6) (Molecular Probes, Eugene, OR), and mitochondria were visualized by fluorescence microscopy. 18 mdm mutant strains were identified that produced large buds lacking DiOC6-stained mitochondria. Standard genetic methods were used to show that the mdm phenotype in each strain was due to a single recessive mutation. In all but one case, the mdm phenotype was linked to the temperature-sensitive growth defect in three or more backcrosses to the wild-type parent. The exception was mdm29, which exhibited only a mitochondrial morphology defect. Complementation analysis was performed between the 18 mdm mutants identified in this screen and the existing mdm and mmm mutants generously provided by M. Yaffe (University of California, San Diego) (McConnell et al., 1990; Sogo and Yaffe, 1994) and R. Jensen (Johns Hopkins University, Baltimore, MD) (Burgess et al., 1994). One mutation was allelic to the previously identified mdm2/ole1 gene (Stukey et al., 1989, 1990; Stewart and Yaffe, 1991). The remaining 17 mdm mutants define 10 new complementation groups designated mdm20-mdm29.
Cloning, Sequencing, and Disruption of MDM20 and Identification of TPM1 as an Unlinked Suppressor
An mdm20-1 strain (GHY1) that had been backcrossed three times to the wild-type parent strains FY10 and FY22 was transformed with a yeast genomic library contained within the low copy plasmids YCp50 (Rose et al., 1987) (obtained from the American Type Culture Collection, Rockville, MD) and p366 (kindly provided by T. Formosa, University of Utah, Salt Lake City). 12,000 Ura+ transformants containing the YCp50 library were tested for complementation of the temperature-sensitive growth defect in the mdm20-1 strain. 14 colonies contained rescuing plasmids. Restriction digest and sequence analysis showed that these strains contained six different overlapping genomic inserts. The plasmid A6-2p, containing a 9-kb genomic insert, was selected for further analysis. A 1.6-kb BsaBI/KpnI fragment from A6-2p inserted into pRS416 (low copy CEN plasmid) or pRS426 (multicopy 2μ plasmid) (Stratagene, La Jolla, CA) digested with KpnI and SmaI was sufficient for complementation of the temperaturesensitive growth defect in mdm20-1 cells. The sequence of a large genomic region containing this fragment was released as part of the Yeast Genome Sequencing Project (GenBank accession number X86470), and the 1.6-kb fragment was shown to contain only the TPM1 gene. Integrative mapping (Rose and Broach, 1991) revealed that mdm20 is not linked to TPM1. Thus, TPM1 is an unlinked extra copy suppressor of the mdm20 mutation.
The MDM20 gene was isolated by transforming the mdm20-1 strain GHY1 with the p366 genomic library. Of the 23,000 Leu+ transformants screened, 24 contained plasmids that could rescue the temperature-sensitive growth defect in mdm20-1 cells. Of these 24 plasmids, restriction digestion and Southern analysis revealed that seven contained TPM1. The remaining 17 plasmids contained four different genomic inserts that shared common overlapping restriction fragments. Plasmid A15-5p contained a 5.2-kb SphI fragment that, when cloned into YCp50, was sufficient to fully complement the mdm20-1 mutation. The DNA carried by this plasmid was sequenced on both strands using an ABI373 automated sequencer at the University of Utah Health Sciences Sequencing Facility (supported by National Cancer Institute grant 5-P30CA42014). Deletion mapping and transposon mutagenesis (Sedgwick and Morgan, 1994) revealed that a 2,388-bp open reading frame was responsible for complementation. A Pfu polymerase (Stratagene) generated a PCR fragment containing the open reading frame, and 500 bp of 5′ and 3′ flanking sequence was subcloned into the EagI and SalI sites of pRS416 and pRS426. The two resulting plasmids complemented both the temperature-sensitive growth defects and mitochondrial inheritance defects in mdm20-1 cells. The wild-type sequence of the MDM20 gene inserted into both plasmids was verified by sequence analysis. Meiotic segregation analysis showed that the cloned open reading frame (MDM20) mapped to the mdm20-1 mutation. The predicted Mdm20 protein sequence was compared with all available databases using the FASTA (Pearson and Lipman, 1988), BLAST (Altschul et al., 1990), and Prosite (Bairoch et al., 1995) programs. No significant homologies were detected. The putative coiled-coil domains in the Mdm20p were found using the COILS 2.1 program (Lupas et al., 1991). The MDM20 GenBank accession number is U54799.
To generate the mdm20 null mutation (mdm20Δ), a 1.9-kb LEU2 PCR fragment was subcloned into pRS406-MDM20 digested with HindIII and SpeI, replacing 2,219 bp (codon 12–751) of the MDM20 open reading frame with LEU2. Digesting this plasmid with SalI and EagI released a 2.9-kb fragment containing the LEU2 gene bordered by MDM20-flanking sequences that was used to transform the diploid yeast strain JSY948. The mdm20Δ::LEU2 disruption was confirmed by Southern blotting of genomic DNA isolated from a Leu+ transformant. Tetrad analysis of sporulated diploids confirmed that the LEU2 marker cosegregated 2:2 with mdm20 phenotypes (temperature sensitivity and defective mitochondrial inheritance) and that cells harboring the mdm20Δ mutation were viable at 25°C. To document the mitochondrial inheritance defect in the mdm20Δ strain, wild-type (JSY999) and isogenic mdm20Δ (JSY1065) cells were stained with DiOC6 and examined microscopically as described below.
Suppression of the Temperature-sensitive and Organelle Inheritance Defects in mdm20Δ Cells
To examine the suppression of the mdm20Δ temperature-sensitive growth defect by TPM1, the wild-type (JSY999) or isogenic mdm20Δ (JSY1065) strains harboring pRS426, pRS426-MDM20, or pRS426-TPM1 were grown to log phase at 25°C in SD minus uracil (SD − Ura) liquid medium. Equal numbers of cells were then serially diluted and spotted on SD − Ura solid media and grown at 37°C for 3 d. To examine the suppression of the mdm20Δ temperature-sensitive growth defect by TPM2, the wild-type (JSY999) or isogenic mdm20Δ (JSY1065) strains harboring pRS416, pRS416-MDM20, or GAL1-TPM2 (pEH006; a generous gift from E. Harsay and A. Bretscher, Cornell University, Ithaca, NY) were grown to log phase at 25°C in SGal − Ura liquid medium. Equal numbers of cells were then serially diluted and spotted on SGal − Ura solid media and grown at 37°C for 3 d.
To determine the effects of extra copies of TPM1 and TPM2 on the mitochondrial and nuclear inheritance defects in mdm20Δ cells, the wildtype (JSY999) or mdm20Δ (JSY1065) strains harboring pRS416 or pRS426, pRS426-TPM1, or GAL1-TPM2 were grown in SGal − Ura or SD − Ura liquid media for 16 h at 25°C, diluted, and grown in log phase at 25° or 37°C for 3 h. Mitochondrial inheritance was evaluated by DiOC6 staining as described above. Nuclear segregation was analyzed by 4′,6-diamidino2-phenylindole staining (DAPI) as described previously (Pringle et al., 1991). In each experiment, at least 400 cells were counted. tpm1Δ (JSY707) cells were examined for defects in mitochondrial and nuclear inheritance using the same protocol.
Characterization of Mdm20p-HA Expression
To monitor Mdm20p levels, the protein was tagged by the addition of three hemagglutinin (HA) epitopes at the amino and carboxy termini. A 3.1-kb SalI/HpaI fragment from pRS416-MDM20 was inserted into p-ALTER-1 (Promega) digested with SalI and SmaI to create p-ALTER1-MDM20. Site-directed mutagenesis (Altered Sites II; Promega) was used to insert NotI sites immediately downstream from the initiating Met or the carboxy-terminal Lys of MDM20. An NotI fragment from plasmid GTEP1 containing three tandem HA epitopes was inserted into the NotI sites at the 5′ or 3′ ends of MDM20. The correct orientation was verified by sequence analysis. MDM20-N-HA and MDM20-C-HA SalI/ KpnI fragments were inserted into pRS416 and pRS426 digested with SalI and KpnI to create pRS416-MDM20-N-HA, pRS426-MDM20-N-HA, pRS416-MDM20-C-HA, and pRS426-MDM20-C-HA. All of the MDM20HA constructs fully rescued the temperature-sensitive growth defect, the mitochondrial inheritance defect, and the loss of actin cables in mdm20Δ cells (JSY1065), indicating that the fusion proteins are functional.
To detect the Mdm20p–HA fusion proteins, 108 cells from log phase cultures were harvested and lysed using 0.2-mm glass beads in 500 μl of SDS-PAGE sample buffer. 25 μl of each lysate was loaded onto an 8% SDS-PAGE gel, and, after electrophoresis, the proteins were transferred to nitrocellulose. Western blot analysis was performed using standard protocols (Harlow and Lane, 1988). Mdm20p–HA fusion proteins were detected with polyclonal rabbit antibodies against HA (1:1,000) (BAbCO, Richmond, CA). Detection was carried out using the ECL kit (New England Nuclear, Boston, MA). The fusion proteins encoded by the MDM20-N-HA and MDM20-C-HA constructs have a predicted molecular mass of 97 kD but migrated with an apparent molecular mass of 90 kD on SDS-PAGE gels. Since the amino- and carboxy-terminal HA-tagged Mdm20p gave identical results, the aberrant size is likely not due to proteolysis, but instead may result from some structural feature or posttranslational modification of the fusion proteins. Comparison of Mdm20p–HA levels was performed using NIH Image 1.60 (National Institutes of Health, Bethesda, MD).
Synthetic Interactions Involving mdm20
The effects of combining mdm20Δ with tpm1Δ or tpm2Δ mutations were determined by constructing diploids heterozygous for mdm20Δ and tpm1Δ (JSY1138) or mdm20Δ and tpm2Δ (JSY1374). The tpm1Δ mutation was generated by transforming the wild-type strain FY22 with a PCR fragment containing 50 bp of TPM1 flanking sequence interrupted by the HIS3 gene using the technique described by Baudin et al. (1993). The disruption removes all but 42 bp of the 3′ TPM1 coding region. The disruption in JSY707 was verified by Southern analysis. The tpm2Δ disruption was constructed in JSY999 using the TPM2 disruption plasmid pBD103 (a gift from A. Bretscher) as described previously (Drees et al., 1995) to produce the tpm2Δ::HIS3 strain JSY1285. The tpm2Δ::HIS3 disruption was verified by Southern analysis. The heterozygous diploids JSY1138 and JSY1374 were sporulated, and tetrads were dissected. The tpm1Δ::HIS3 mdm20Δ::LEU2 mutants germinated and gave rise to microcolonies containing 50–100 cells. In separate experiments, tpm1Δ::HIS3 mdm20Δ:: LEU2 double mutants were recovered when JSY1138 diploids containing the wild-type MDM20 gene on a URA3 plasmid were sporulated and dissected. The synthetic growth defect of mdm20Δ tpm1Δ double mutants was verified by showing that these cells were unable to lose the plasmid when grown on 5-FOA–containing medium. The JSY1374 diploid gave rise to viable mdm20Δ::LEU2 tpm2Δ::HIS3 double mutants with no obvious synthetic growth defects. Crosses to test possible synthetic interactions between mdm20Δ::LEU2 and bem2-10 (Wang and Bretscher, 1995), myo2-66 (Johnston et al., 1991), and sac6Δ::LEU2 (Adams et al., 1991) were performed essentially as described above.
Analysis of the Actin Cytoskeleton in mdm20Δ and tpm1Δ Cells
To visualize F-actin, the wild-type (JSY999) or mdm20Δ (JSY1065) strains containing pRS426 were grown 16 h at 25°C in SD − Ura liquid medium. The cultures were then diluted (to 2 × 106 cells per ml) into fresh SD − Ura and grown at 25° or 37°C for 3 h. F-actin was detected with rhodamine-phalloidin (Molecular Probes) as described previously (Adams and Pringle, 1991). To determine the effects of overexpressing TPM1 and TPM2 on the actin cytoskeleton in mdm20Δ mutants, the mdm20Δ strain JSY1065 carrying pRS426-TPM1 or GAL1-TPM2 was grown in SD − Ura or SGal − Ura at 25° or 37°C as described above, and then stained with rhodamine-phalloidin. To analyze the effects of Mdm20p–HA overexpression on actin organization in tpm1Δ cells, the tpm1Δ strain JSY707 carrying pRS426, pRS426-MDM20, or pRS426-MDM20-C-HA was grown and stained as described above.
Microscopy
Cells were viewed on an Axioplan microscope using a ×100 Plan-neofluor (NA 1.3) objective (Carl Zeiss, Inc., Thornwood, NY) using an optivar setting of ×1.6, a 200-W mercury lamp, and the following excitation wavelengths: DiOC6 (450–490 nm), DAPI (365 nm), and rhodamine (546 nm). Differential interference contrast (DIC) and fluorescence digital images were captured and assembled into figures as described previously (Roeder and Shaw, 1996). Images were printed on a Tektronix Phaser IIsdx dyesublimation printer (Wilsonville, OR).
Results
mdm20 Mutants Exhibit Defects in Mitochondrial Inheritance
To identify yeast genes required for mitochondrial inheritance, we generated and screened a collection of temperature-sensitive mutants for strains that failed to transport mitochondria into growing buds. Individual strains grown at 25°C were shifted to 37°C for 3 h and stained with the lipophilic, fluorescent dye, DiOC6, under conditions that preferentially label yeast mitochondria (Pringle et al., 1989; Shaw and Wickner, 1991; Koning et al., 1993). 18 of the 4,000 mutants examined exhibited defects in mitochondrial distribution and/or morphology. The mutations in these strains were recessive in heterozygous diploids and segregated 2:2 in backcrosses with the wild-type parent strain. 17 of these strains fell into ten complementation groups designated mdm20-mdm29; the remaining mutation was a new allele of the previously characterized gene MDM2/OLE1 (Stukey et al., 1989, 1990; Stewart and Yaffe, 1991). In all but one mutant (mdm29), the temperaturesensitive growth defects were linked to the defects in mitochondrial distribution and morphology. The MDM20 complementation group, defined by seven mutant alleles, was chosen for further study.
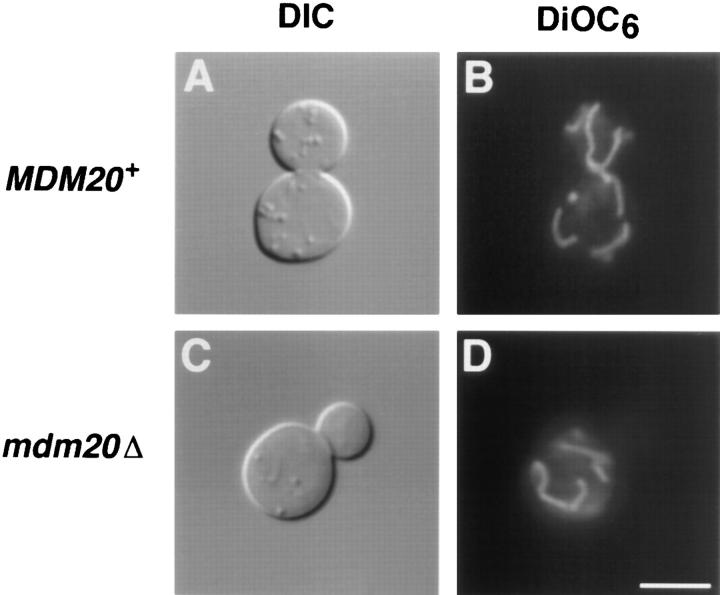
Wild-type strains grown at 25° or 37°C (Fig. 2, A and B) segregate a portion of the mitochondrial network into buds almost as soon as they can be detected on the surface of the mother cell. In contrast, mdm20 mutants retained their branched mitochondrial networks but produced large buds that lack this organelle (Fig. 2, C and D). This mitochondrial inheritance defect was observed at the permissive temperature and became more pronounced at 37°C (Table II). Both transmission EM and DAPI staining of mitochondrial genomes confirmed that mitochondria were absent from mdm20 buds (data not shown). In addition, organelles labeled with a mitochondrially targeted form of the green fluorescent protein (kindly provided by R. Jensen) also failed to be transported into buds in the mdm20 strain (data not shown). Although the mdm20 mutation caused a striking mitochondrial inheritance defect, it did not severely affect metabolic functions of mitochondria since they could still be efficiently labeled with the potential-dependent dye DiOC6, which only stains actively respiring organelles (Pringle et al., 1989).
Figure 2.
Cells lacking MDM20 have normal mitochondrial morphology but do not segregate mitochondria into buds at 37°C. DIC images (A and C) and DiOC6 mitochondrial staining (B and D) of MDM20 + (JSY999) (A and B) and mdm20Δ (JSY1065) (C and D) cells grown at 37°C for 3 h. Bar, 5 μm.
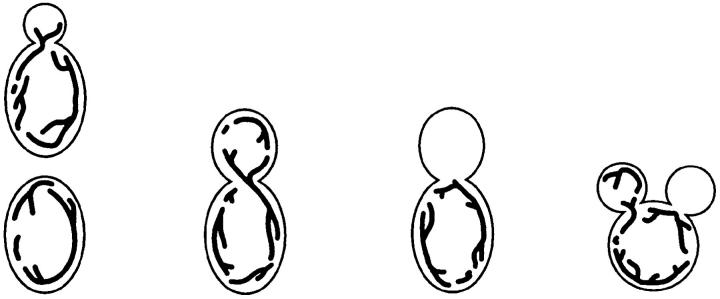
Table II.
Mitochondrial Inheritance in mdm20 and tpm1 Mutants
| Genotype/(Plasmid)* | °C |
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | 25 | 48 | 52 | 0 | 0 | ||||||
| Wild type | 37 | 55 | 45 | <1 | 0 | ||||||
| mdm20Δ | 25 | 37 | 49 | 14 | <1 | ||||||
| mdm20Δ | 37 | 36 | 6 | 53 | 5 | ||||||
| mdm20Δ + (pRS426- TPM1) | 25 | 46 | 52 | 2 | 0 | ||||||
| mdm20Δ + (pRS426- TPM1) | 37 | 41 | 42 | 17 | 0 | ||||||
| mdm20Δ + (GAL1- TPM2)‡ | 25 | 39 | 52 | 9 | 0 | ||||||
| mdm20Δ + (GAL1- TPM2) | 37 | 46 | 20 | 34 | 0 | ||||||
| tpm1Δ | 25 | 42 | 57 | 1 | 0 | ||||||
| tpm1Δ | 37 | 45 | 50 | 4 | 1 | ||||||
Mitochondrial inheritance scored in log-phase cells grown at 25° or 37°C for 3 h. The cartoons depict classes scored and include unbudded/small-budded, large-budded, and multiply budded cells with or without mitochondria in buds. The percentage of cells in each class is shown (n > 400).
The strains used were JSY999 (wild type), JSY1065 (mdm20Δ), and JSY707 (tpm1Δ).
mdm20Δ cells carrying the GAL1-TPM2 plasmid were grown in SGal − Ura medium. The mitochondrial inheritance defect was not affected by growth in galactose media (not shown).
Isolation and Sequence Analysis of the MDM20 Gene
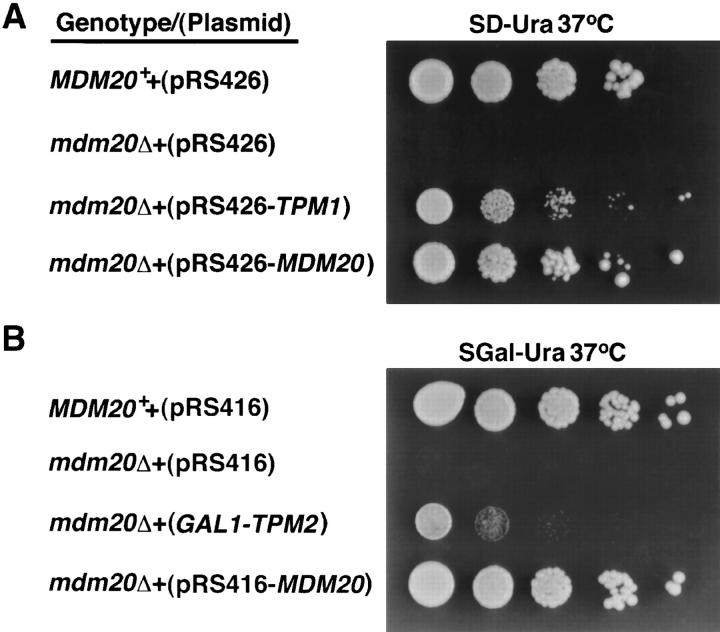
The MDM20 gene was isolated by complementing the temperature-sensitive growth defect of the mdm20-1 mutant allele (see Materials and Methods). Integrative mapping studies indicated that the cloned DNA contained the wild-type MDM20 gene. Transposon mutagenesis (Sedgwick and Morgan, 1994) and DNA sequence analysis revealed that a 3.2-kb fragment (Fig. 3 A) was sufficient to rescue both the temperature-sensitive growth defect (Fig. 4) and the mitochondrial inheritance defect (not shown) in the mdm20 mutant strain.
Figure 3.
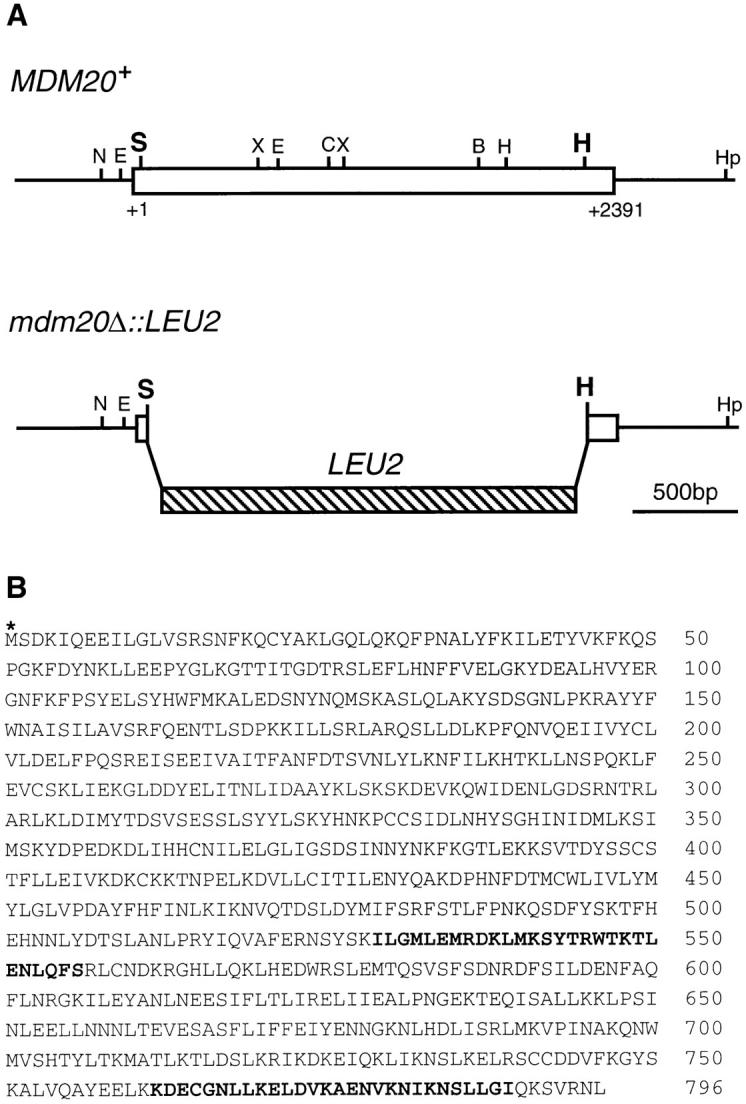
MDM20 encodes a novel 93-kD protein. (A, top) Restriction map of the MDM20 gene (open box). (A, bottom) The LEU2 construct used to generate the MDM20 disruption that replaces 2,219 bp (codons 12–751) of the MDM20 coding region. All restriction enzyme sites in the MDM20 gene are indicated: B, BglII; C, ClaI; E, EcoRI; H, HindIII; Hp, HpaI; S, SpeI; and X, XbaI. (B) Predicted amino acid sequence (single letter code) of the Mdm20p. The sequence begins at the first potential initiating methionine (asterisk). Indicated in boldface are two putative heptad repeats predicted by the COILS 2.1 program (Lupas et al., 1991). These sequence data are available from GenBank/EMBL/ DDBJ under accession number U54799.
Figure 4.
Extra copies of TPM1 and TPM2 can suppress the temperature-sensitive growth defect in mdm20Δ cells. (A) MDM20 + (JSY999) or isogenic mdm20Δ (JSY1065) strains harboring pRS426, pRS426-MDM20, or pRS426-TPM1 plasmids were serially diluted and spotted on SD minus uracil (SD − Ura) solid medium and grown at 37°C for 3 d. (B) MDM20 + (JSY999) or isogenic mdm20Δ (JSY1065) strains harboring pRS416, pRS416- MDM20, or GAL1-TPM2 plasmids were serially diluted and spotted on SGal minus uracil (SGal − Ura) solid medium and grown at 37°C for 3 d.
The MDM20 gene predicts a novel, hydrophilic protein that contains 796 amino acids with a calculated molecular mass of 92,821 daltons (Fig. 3 B). The sequences adjacent to the first potential initiating methionine (Fig. 3 B, asterisk) fit the consensus for a translational initiation site in yeast (Cigan and Donahue, 1987). Moreover, we demonstrated that an in-frame epitope tag (HA) inserted immediately downstream of this methionine is translated and can be detected on a Western blot (data not shown). Comparison of the Mdm20 protein sequence with all available databases using the FASTA (Pearson and Lipman, 1988), BLAST (Altschul et al., 1990), and Prosite (Bairoch et al., 1995) algorithms revealed no significant homologies with other proteins. However, two potential heptad repeat– containing regions located in the carboxy-terminal portion of the polypeptide are predicted to adopt an α-helical coiled-coil structure (Fig. 3 B, boldface amino acids). Heptad repeats have been implicated in the homo- and heterodimerization of a variety of proteins with diverse functions (Cohen and Parry, 1986, 1990).
Mdm20p Is Required for Mitochondrial Inheritance during Budding but Does Not Alter Mitochondrial Morphology
Yeast cells harboring mdm20-1 through mdm20-7 mutations exhibited defects in mitochondrial inheritance, grew slowly compared to wild type at 25°C, and were dead at 37°C. To further evaluate the role of MDM20 in vivo, we generated a null allele by replacing codons 12–751 with the LEU2 gene (Fig. 3 A). When diploids heterozygous for the mdm20::LEU2 null allele were sporulated, dissected into tetrads, and incubated at 25°C, all four spores in each tetrad germinated. However, spores lacking MDM20 grew slowly and formed small colonies relative to wild type. Further analysis indicated that spores containing the mdm20:: LEU2 disruption did not grow on fermentable (dextrose) or nonfermentable (glycerol) carbon sources at 37°C.
The mitochondrial inheritance defect in the mdm20:: LEU2 cells (mdm20Δ) was similar to that observed for the original seven mdm20 mutant alleles. In log phase cultures grown at 25°C, 14% of mdm20Δ mother cells remained attached to buds that failed to inherit mitochondria (Table II). In contrast, mitochondrial segregation defects were not observed in wild-type cultures grown at 25°C (Table II). Incubation at 37°C for 3 h resulted in an increased fraction of mdm20Δ cells exhibiting mitochondrial inheritance defects (58%), whereas mitochondrial partitioning in wild-type cells did not change significantly (<1%) (Table II). Moreover, when mdm20Δ cells arrested in the unbudded stage were released upon temperature shift, >90% of the buds that formed at 37°C did not inherit mitochondria (data not shown). Despite the severe defect in mitochondrial transmission to buds, DiOC6-stained mitochondrial networks appeared wild type in the mdm20Δ cells at both 25°C (98% wild type) and 37°C (95% wild type) (see Fig. 2 D). Thus, MDM20 is required for normal mitochondrial inheritance in yeast but does not appear to play an essential role in maintaining the reticulated network of mitochondrial membranes.
Although nuclear inheritance was normal in mdm20Δ cells grown at 25°C, a small number (5%) of binucleate cells was observed at 37°C (Table III; discussed below). This defect in nuclear segregation, while significant, is much less severe than the defect in mitochondrial inheritance in the mdm20 mutant. We also examined the distribution of the vacuole in the mdm20 mutant. We found that, at both 25° and 37°C, new mdm20Δ buds contained vacuolar material even when mitochondria were absent from the buds (data not shown). Thus, mutations in the mdm20 gene appear to have the most dramatic affect on inheritance of the mitochondrial compartment.
Table III.
Nuclear Segregation in mdm20 and tpm1 Mutants
| Genotype/(Plasmid)* | °C |
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | 25 | 56 | 20 | 7 | 17 | 0 | 0 | 0 | |||||||||
| Wild type | 37 | 52 | 26 | 10 | 12 | 0 | <1 | 0 | |||||||||
| mdm20Δ | 25 | 36 | 23 | 6 | 35 | 0 | <1 | 0 | |||||||||
| mdm20Δ | 37 | 28 | 37 | 2 | 27 | 1 | 5 | <1 | |||||||||
| mdm20Δ + (pRS426-TPM1) | 25 | 51 | 25 | 6 | 18 | 0 | 0 | 0 | |||||||||
| mdm20Δ + (pRS426-TPM1) | 37 | 40 | 34 | 8 | 18 | 0 | <1 | 0 | |||||||||
| mdm20Δ + (GAL1-TPM2)‡ | 25 | 46 | 31 | 8 | 15 | 0 | 0 | 0 | |||||||||
| mdm20Δ + (GAL1-TPM2) | 37 | 44 | 27 | 9 | 17 | 0 | 3 | 0 | |||||||||
| tpm1Δ | 25 | 35 | 24 | 6 | 22 | 0 | 13 | 0 | |||||||||
| tpm1Δ | 37 | 36 | 26 | 9 | 13 | 0 | 16 | 0 | |||||||||
Nuclear inheritance scored in log-phase cells grown at 25° or 37°C for 3 h. The cartoons depict classes scored. The percentage of cells in each class is shown (n > 400).
The strains used were JSY999 (wild type), JSY1065 (mdm20Δ), and JSY707 (tpm1Δ).
mdm20Δ cells carrying the GAL1-TPM2 plasmid were grown in SGal − Ura media. The nuclear inheritance defect was not affected by growth in galactose medium.
Extra Copies of Genes Encoding the Actin Binding Proteins Tpm1p and Tpm2p Partially Rescue Mitochondrial Inheritance Defects and Temperature-sensitive Growth Defects in the mdm20 Mutant
To identify loci that interact genetically with MDM20 and to learn more about the role of the Mdm20 protein, we analyzed multicopy suppressors of the mdm20-1 temperature-sensitive growth defect. The multicopy suppressors were isolated from a genomic library in a centromerebased yeast vector at the same time the MDM20 gene was cloned (see Materials and Methods). One of the clones we isolated contained TPM1, a previously identified gene that encodes the actin binding protein tropomyosin (Liu and Bretscher, 1989). Tropomyosins are rod-shaped coiled-coil dimers that bind along the groove of the actin helix. Skeletal tropomyosin modulates the interaction between actin and myosin during muscle contraction. In yeast, however, tropomyosins are thought to function in the assembly and/ or stabilization of actin cables (Liu and Bretscher, 1989; Bretscher et al., 1994). Although TPM1 is not an essential gene, cells lacking TPM1 grow slowly, are heterogeneous in size, and are often multinucleate due to defects in aligning the mitotic spindle (Liu and Bretscher, 1992; Palmer et al., 1992; Wang and Bretscher, 1995). Loss of TPM1 also results in severe defects in actin cytoskeletal organization, most notably the loss of actin cables (see below) (Liu and Bretscher, 1989). In addition to TPM1, yeast contain a second tropomyosin encoded by the TPM2 gene (Drees et al., 1995). Disruption of the TPM2 gene does not visibly affect the organization of the actin cytoskeleton and does not hamper cell growth (Drees et al., 1995). However, the genetic and biochemical behavior of Tpm2p and the observed synthetic lethality between tpm1 and tpm2 mutations indicate that cellular functions of these two proteins overlap (Drees et al., 1995).
Extra copies of the TPM1 gene provided on a low copy number (not shown) or high copy number plasmid partially rescued the temperature-sensitive lethality in mdm20Δ cells grown at 37°C (Fig. 4). In contrast, the conditional lethality of the mdm20Δ mutation was completely suppressed by a wild-type copy of the MDM20 gene (Fig. 4). Extra copies of TPM1 also suppressed mitochondrial inheritance defects in the mdm20Δ strain. In a culture of mdm20Δ cells harboring TPM1 on a multicopy plasmid, the fraction of cells lacking mitochondria in buds was reduced from 14% to 2% at 25°C and from 58% to 17% at 37°C (Table II). These results indicated that the Tpm1 protein could functionally substitute for the loss of the Mdm20 protein.
To determine whether functional overlaps also existed between Mdm20p and Tpm2p, we asked whether extra copies of TPM2 could rescue mutant phenotypes in mdm20Δ cells. As shown in Fig. 4, the temperature-sensitive growth defect of mdm20Δ cells was partially suppressed when Tpm2p was overexpressed from the GAL1 promoter (Fig. 4 B). Overexpression of Tpm2 was also sufficient to reduce the mitochondrial inheritance defect in mdm20Δ from 14% to 9% at 25°C and from 58% to 34% at 37°C (Table II). Thus, Mdm20p and Tpm2p apparently have some overlapping functions as well.
MDM20 Is Essential for the Integrity of Actin Cables in Yeast
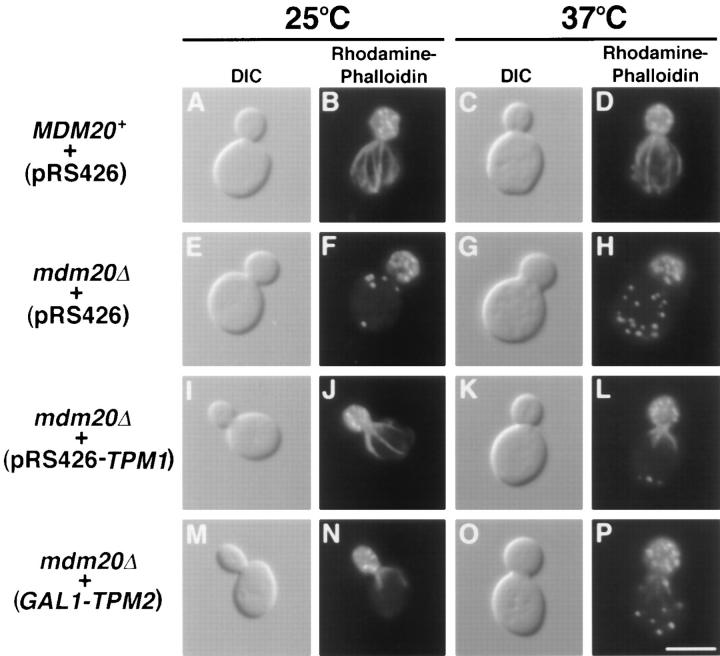
Taken together, the data presented above suggested that (a) MDM20 and TPM1 participate in overlapping, essential cellular functions; and (b) Tpm2p can partially substitute for Mdm20p when overexpressed. Given what is known about the function of tropomyosins (Pittenger et al., 1994), these results also suggested that the integrity of the actin cytoskeleton is important for mitochondrial inheritance during yeast budding, and that the mdm20 mutation affects the assembly or organization of F-actin. To further investigate this possibility, we examined the actin cytoskeleton in mdm20 null cells by rhodamine-phalloidin staining. At both 25° and 37°C, wild-type cells contained well- defined actin cables and cortical actin patches concentrated in growing buds (Fig. 5, A–D). In contrast, actin cables were absent in 100% of the mdm20 null cells examined at both temperatures (Fig. 5, E–H). mdm20Δ cells were able to divide, however, and contained brightly staining cortical patches that were predominantly clustered in buds at 25°C (Fig. 5, E and F; >95% of cells) and partially delocalized at 37°C (Fig. 5, G and H; >50% of cells). The fact that mitochondrial inheritance defects are already present at 25°C in the mdm20 mutant (at a temperature where patches are still properly localized) suggests that it is the disruption of actin cables that is primarily responsible for the lack of mitochondrial transmission.
Figure 5.
Actin cables lacking in mdm20Δ cells are partially restored by introducing extra copies of TPM1 and TPM2. MDM20 + cells (JSY999) carrying pRS426 (A–D) and mdm20Δ cells (JSY1065) carrying pRS426 (E–H), pRS426-TPM1 (I–L), or GAL1-TPM2 (M–P) were grown in SD − Ura or SGal − Ura at 25°C, and then shifted to 25° and 37°C for 3 h. Cells were stained with rhodamine-phalloidin and visualized by DIC or fluorescence microscopy as indicated. Representative cells are shown. Bar, 5 μm.
Rhodamine-phalloidin staining also revealed that multiple copies of TPM1 restored actin cable formation in the mdm20 mutant. At 25°C, >95% of the mdm20 cells containing TPM1 on a multicopy plasmid had actin cables that were robust and extended the full length of the mother cell (Fig. 5, I and J). However, these restored cables appeared fewer in number than those observed in wild type. At 37°C, >95% of mdm20 cells also contained the TPM1- restored cables. However, these cables were shorter than normal, and, while visible near the mother-bud neck, they rarely extended the full length of the mother cell (Fig. 5, K and L). The delocalization of actin cortical patches in mdm20Δ cells at 37°C was also rescued by introducing extra copies of TPM1 (Fig. 5, K and L; >95% of cells). Abbreviated actin cables were also observed in >90% of the mdm20Δ cells overexpressing Tpm2p from the GAL1 promoter at 25°C (Fig. 5, M and N). At 37°C, only faint cables were observed near the mother-bud neck in mdm20Δ cells containing GAL1-TPM2 (Fig. 5, O and P; >90% of cells). Finally, the wild-type MDM20 gene on a low copy number plasmid was able to completely restore cables in >98% of mdm20Δ cells (data not shown). This partial restoration of actin cables and mitochondrial inheritance by TPM1 and TPM2 was striking and suggested that the defect in mitochondrial inheritance observed in mdm20 is linked to the loss of actin cables in these cells.
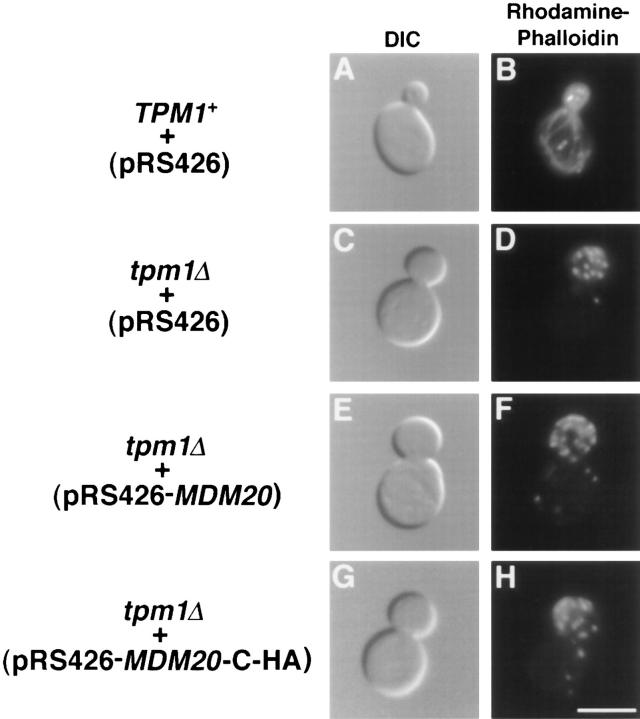
MDM20 Fails to Complement Mutant Phenotypes in tpm1Δ Cells
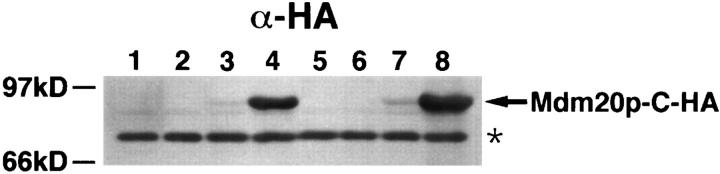
Although overexpression of TPM1 suppressed mitochondrial inheritance defects and restored actin cables in the mdm20 mutant, the functions of the Tpm1 and Mdm20 proteins were not completely interchangeable. As shown in Fig. 6, C and D, and reported previously, actin cables are absent in tpm1Δ mutant cells (Liu and Bretscher, 1989). However, actin cables could not be restored by introducing extra copies of the MDM20 gene into tpm1Δ cells on a high copy number plasmid (Fig. 6, E and F) or by overexpressing epitope-tagged versions of the Mdm20p (Fig. 6, G and H; data not shown). In the latter experiments, Mdm20p overexpression was monitored using amino- and carboxyterminal HA-tagged Mdm20 proteins (see Materials and Methods). Wild-type cells containing Mdm20p-C-HA on a centromere-based plasmid expressed relatively low levels of the tagged protein (Fig. 7, lane 3). The steady state protein level increased more than eightfold when Mdm20-CHA protein was expressed from a multicopy plasmid (Fig. 7, lane 4). Identical results were obtained when expression of the Mdm20p-N-HA was examined in wild-type cells (data not shown). Fusion of the HA tag to the Mdm20 protein had no obvious effects on its function, as these constructs completely rescued temperature-sensitive growth defects, mitochondrial inheritance defects, and loss of actin cables in mdm20 mutant strains (data not shown). Furthermore, overexpression of HA-tagged Mdm20p constructs did not induce additional phenotypes in wild-type cells (data not shown). We found that tpm1Δ cells overexpressing Mdm20p (Fig. 7, lane 8) did not grow significantly better than tpm1Δ cells alone. In addition, overexpression of Mdm20p did not rescue the lethality observed in tpm1Δ tpm2Δ double mutants. Together, these findings suggest that MDM20 cannot bypass the requirement for Tpm1p and Tpm2p in cells. Furthermore, although expression of Tpm1p in wildtype cells results in thicker actin cables (Liu and Bretscher, 1989), the actin cytoskeleton was not affected by overexpressing Mdm20p in a wild-type strain (data not shown). Thus, Mdm20p and Tpm1p appear to perform some distinct functions in cells.
Figure 6.
Actin cables lacking in tpm1Δ cells are not restored by overexpression of Mdm20p. TPM1 + cells (FY22) containing pRS426 (A and B) and tpm1Δ cells (JSY707) containing pRS426 (C and D), pRS426-MDM20 (E and F), or pRS426-MDM20-CHA (G and H) were grown at 25°C, stained with rhodamine-phalloidin, and visualized by DIC or fluorescence microscopy as indicated. Representative cells are shown. Bar, 5 μm.
Figure 7.
Expression of Mdm20p-C-HA in wild-type and tpm1Δ cells. Extracts were prepared from wild-type cells (JSY999) containing pRS416 (lane 1), pRS426 (lane 2), pRS416-MDM20-CHA (lane 3), and pRS426-MDM20-C-HA (lane 4), or tpm1Δ cells (JSY707) containing pRS416 (lane 5), pRS426 (lane 6), pRS416- MDM20-C-HA (lane 7), and pRS426-MDM20-C-HA (lane 8). The cells were harvested after growth to mid log phase at 25°C, and equal amounts (equivalent OD units) of the cell extracts were subjected to SDS-PAGE and transferred to nitrocellulose. The nitrocellulose was probed with antibodies to the HA epitope. The arrowhead identifies the Mdm20-C-HA protein, and the asterisk identifies a background band that was used to normalize the amount of protein present in each lane.
Loss of Actin Cables in mdm20Δ and tpm1Δ Cells Disrupts Mitochondrial Inheritance and Nuclear Segregation to Different Extents
To further evaluate functional overlaps between Mdm20p and Tpm1p, we examined mitochondrial inheritance and morphology in the tpm1Δ mutant. Although actin cables are also absent in tpm1Δ cells, we were surprised to find that tpm1 null strains did not exhibit significant defects in mitochondrial inheritance at 25° (<1%) or 37°C (5%) (Table II). In addition, mitochondrial morphology appeared normal in tpm1Δ cells at both temperatures (99% wild type at 25°C and 96% wild type at 37°C). The mitochondrial inheritance phenotype of tpm1Δ contrasted with that of mdm20 null cells, which displayed more severe defects in mitochondrial inheritance at both temperatures (14% at 25°C and 58% at 37°C; Table II). These phenotypic differences, together with the results described above, supported the notion that the Mdm20 and Tpm1 proteins perform some distinct roles in cells. To extend these observations, we examined nuclear segregation (a process known to be defective in tpm1 mutants) in isogenic tpm1 and mdm20 strains.
During division, the mitotic spindle is oriented by interactions of astral microtubules with the actin cytoskeleton. Disruption of this interaction in act1 mutants results in misoriented spindles that give rise to binucleate cells (Palmer et al., 1992). Consistent with previous reports (Liu and Bretscher, 1989; Wang and Bretscher, 1995), we found that tpm1Δ mutants lacking actin cables exhibited nuclear segregation defects and produced a significant proportion of multinucleate cells at 25°C (13%) and 37°C (16%) (Table III). Conversely, nuclear segregation was wild type in mdm20Δ cells grown at 25°C, although a small number (5%) of binucleate cells was observed at 37°C (Table III). Extra copies of TPM1 (but not overexpression of Tpm2p) suppressed the formation of binucleate cells at 37°C in the mdm20Δ strain (Table III). Thus, tpm1 mutants displayed more severe defects in nuclear segregation than mdm20 mutants. Together, these data indicate that, although TPM1 and MDM20 are both required for the formation and/or stabilization of actin cables, their roles in cellular processes such as mitochondrial inheritance and nuclear segregation differ significantly.
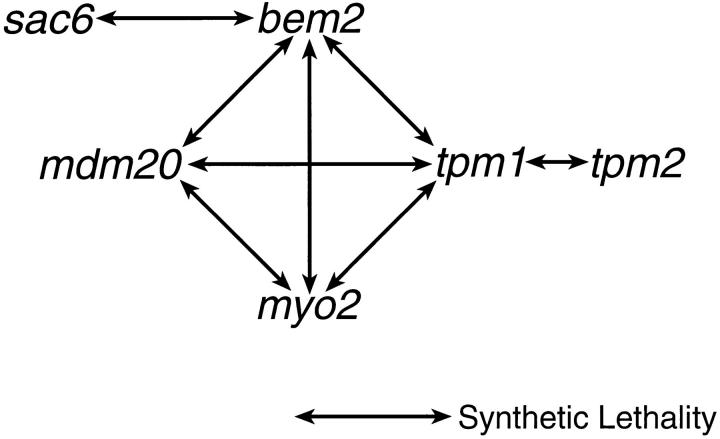
Null Mutations in mdm20 are Synthetically Lethal With Mutations in tpm1, myo2, and bem2
Although TPM1 and TPM2 null mutants are alive at 25°C, the combinatorial loss of these two genes in a haploid strain results in cell death (Drees et al., 1995). This synthetic lethality is thought to arise because Tpm1p and Tpm2p carry out some redundant functions required for an essential cellular process (Drees et al., 1995). The similar actin phenotypes we observed in tpm1 and mdm20 mutants prompted us to genetically test the possibility that MDM20 acts in a common pathway with tropomyosins. The phenotypes of haploid mdm20Δ tpm1Δ and mdm20Δ tpm2Δ double mutants were examined after dissecting tetrads generated from diploid cells heterozygous for null mutations in both MDM20 and TPM1. In 33 tetrads analyzed, double mdm20Δ tpm1Δ mutants were never observed (30 expected; Table IV). Although mdm20Δ tpm1Δ spores germinated, they were defective in mitotic growth and gave rise to microcolonies of 50–100 cells that could not be analyzed further. These results were confirmed by repeating the cross with an mdm20Δ strain carrying the wild-type MDM20 gene on a URA3-marked plasmid. In this experiment, mdm20Δ tpm1Δ cells harboring the MDM20 URA3 plasmid were recovered and were viable unless forced to lose the plasmid on 5-FOA plates (data not shown). Synthetic lethal interactions were not observed between MDM20 and TPM2 in similar crosses (Table IV), and mitochondrial inheritance and morphology defects were not observed in tpm2Δ cells (data not shown).
Table IV.
Synthetic Lethal Relationship Involving mdm20
| Segregation pattern* | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cross | PD | TT | NPD | Double mutants | ||||
| alive:dead | ||||||||
| mdm20Δ::LEU2(JSY1065) × tpm1Δ::HIS3(JSY1084) | 8 | 20 | 5 | 0:30‡ | ||||
| mdm20Δ::LEU2(JSY1340) × tpm2Δ::HIS3(JSY1285 | 11 | 48 | 8 | 64:0 | ||||
| mdm20Δ::LEU2(JSY1065) × bem2-10(ABY249) | 8 | 26 | 3 | 0:32§ | ||||
| mdm20Δ::LEU2(JSY1340) × myo2-66(SLY63) | 12 | 35 | 18 | 0:71‖ | ||||
| mdm20Δ::LEU2(JSY1340) × sac6Δ::LEU2(IGY4) | 2 | 5 | 2 | 9:0 | ||||
The segregation patterns for mdm20Δ::LEU2, tpm1Δ::HIS3, tpm2Δ::HIS3, bem2-10 (enlarged cells), myo2-66 (t.s.), and sac6Δ::LEU2 are shown (PD, parental ditype; TT, tetratype; NPD, nonparental ditype). In cases where synthetic lethality was observed, the segregation pattern was inferred from other viable members of the tetrad.
mdm20Δtpm1Δ double mutants form terminal microcolonies containing 50–100 cells.
mdm20Δbem2-10 double mutants form terminal microcolonies containing 20–40 cells.
mdm20Δmyo2-66 double mutants form terminal microcolonies containing 100–500 cells.
Previous studies indicated that tpm1 also exhibits synthetic growth defects with mutations in BEM2 (Wang and Bretscher, 1995) and MYO2 (Liu and Bretscher, 1992). The BEM2 gene encodes a GTPase-activating protein required for bud emergence, a process that involves polarization of actin cables and patches to the site of bud growth (Kim et al., 1994; Peterson et al., 1994). MYO2 encodes a type V myosin family member that forms a cap at or near the plasma membrane on unbudded cells and small buds (Johnston et al., 1991; Lillie and Brown, 1994). In crosses similar to those described above, we determined that mutations in BEM2 and MYO2 are also synthetically lethal with mdm20 (Table IV). We also examined synthetic interactions between mutations in MDM20 and the SAC6 gene that encodes the actin cross-linking protein, fimbrin (Adams et al., 1991). Although Sac6p, like Tpm1p, localizes to actin cables in wild-type yeast cells, synthetic growth defects were not detected for tpm1 sac6 double mutants (Adams et al., 1993) and were not observed in crosses we performed with mdm20 and sac6 mutant strains (Table IV). Thus, mdm20 and tpm1 mutants not only exhibit similar actin organization defects but also show the same pattern of genetic interactions.
Discussion
We have demonstrated that the MDM20 gene encodes a novel protein required for proper mitochondrial segregation in yeast. Cells lacking Mdm20p are temperature sensitive for growth and fail to transport mitochondria into newly formed buds. Our results indicate that the defect in the mdm20 mutant specifically disrupts mitochondrial transport but not mitochondrial morphology in these cells. In addition, the mdm20 mutation does not severely interrupt the division and segregation of the nucleus, and other cytoplasmic organelles, such as vacuoles, are always detected in mdm20 daughter cells that fail to inherit the mitochondrial compartment. Together, our findings establish an essential role for MDM20 in mitochondrial partitioning during mitotic division.
Two different cytoskeletal elements in yeast, actin (Drubin et al., 1993; Lazzarino et al., 1994) and an intermediate filament–like protein (McConnell and Yaffe, 1992), have been proposed to play a central role in mitochondrial inheritance. In this study we present two lines of in vivo evidence that mitochondrial inheritance in yeast is an actin-mediated process. First, two major phenotypic consequences of disrupting MDM20 appear to be the cessation of mitochondrial movement into buds during division and the loss of observable actin cables in cells. Second, overexpression of two well-characterized actin binding proteins, Tpm1p and Tpm2p, suppresses temperature-sensitive growth and mitochondrial inheritance defects and partially restores actin cables in the mdm20 mutant. Although it is possible that Mdm20p fulfills distinct functions in mitochondrial inheritance and actin cable organization, the observation that overexpression of Tpm1p and Tpm2p can partially rescue defects in both processes in mdm20 mutants suggests that these two phenotypes cannot be separated from one another. Moreover, a direct link between mitochondrial segregation and the actin cytoskeleton is consistent with the recent observations that yeast mitochondria colocalize with actin cables in vivo (Drubin et al., 1993), can bind to phalloidin-stabilized actin filaments in vitro (Lazzarino et al., 1994), and contain an actin-dependent myosin-like motor activity on their surface (Simon et al., 1995). Thus, our data strongly support a model in which mitochondria are transported into yeast buds along polarized actin filaments or cables.
The organization and stabilization of the actin cytoskeleton in yeast depends on the activities of a large number of actin binding proteins and regulatory factors. Mdm20p is a new member of this important group of proteins required for the integrity of actin cables in cells. Previous studies indicate that defects in actin organization cause a variety of phenotypes including temperature-sensitive lethality, osmotic sensitivity, and defects in cell morphology, mitochondrial dynamics, secretion, endocytosis, and nuclear segregation (for review see Bretscher et al., 1994; Welch et al., 1994). In addition to the phenotypes we have reported here for the mdm20 mutant, our preliminary studies suggest that mdm20 cells are rounder than wild type and heterogeneous in size, exhibit defects in chitin localization, and accumulate small vesicles (unpublished observations). These data provide further support that MDM20 is required for normal functions of the actin cytoskeleton. Most importantly, MDM20 defines a new class of genes that control both actin organization and mitochondrial transport during division.
Previous studies of the actin–myosin complex identified a myosin “footprint” on the actin monomer (Rayment et al., 1993; Schroder et al., 1993). Charge-to-alanine substitutions located under the equivalent myosin footprint in the yeast actin monomer have been shown to cause defects in mitochondrial morphology, specifically mitochondrial aggregation (Drubin et al., 1993). The mutants with aberrant mitochondrial morphology exhibit severe actin organization defects that are most pronounced at 37°C, including loss of actin cables and/or patches, delocalized patches, and reduced or faint cables. In contrast, we find that mdm20 mutants lacking actin cables exhibit disrupted mitochondrial transport but not mitochondrial morphology. Similarly, we do not observe defects in mitochondrial morphology in isogenic tpm1 mutant cells. If the actin cytoskeleton plays a role in regulating mitochondrial morphology, then the structures required must still be present in mdm20 and tpm1 mutant cells. Alternatively, other cytoplasmic structures or cytoskeletal proteins may be responsible for controlling yeast mitochondrial morphology.
Our observation that mitochondrial morphology and inheritance is normal in the tpm1Δ mutant is interesting in light of previous reports that tpm1Δ strains generate petites (lose mitochondrial genome function) at a higher frequency than wild type (Liu and Bretscher, 1992). We also find that mdm20Δ and tpm1Δ cells generate petites at a high frequency when maintained on dextrose. One possible explanation for these results is that tpm1 daughter cells frequently receive DNA-free mitochondrial compartments. In our experiments, mitochondrial morphology and distribution were measured by labeling the mitochondrial compartment with the potential dependent dye, DiOC6, and labeling mtDNA nucleoids with DAPI. We observed that tpm1Δ cells segregated both the mitochondrial compartment (Table II) and mitochondrial nucleoids (data not shown) into daughter cells. Furthermore, we did not detect a large number of tpm1 cells lacking mt nucleoids. Thus, the increased petite formation in tpm1 cells is not simply the result of a block in mtDNA transfer to buds. Further experiments are required to determine the mechanism by which these strains become petite.
Some of our studies suggest that Tpm1p and Mdm20p might perform overlapping functions required for the integrity of the actin cytoskeleton. The disruption of both genes causes a similar loss of actin cables in cells and combinations of mutations in MDM20 and TPM1 result in synthetic lethality. In addition, like tpm1, mdm20 exhibits synthetic growth defects in combination with mutations in either BEM2 or MYO2 but not SAC6 (Fig. 8). Furthermore, we observe that overexpression of Tpm1p restores actin cables and rescues mitochondrial inheritance defects in mdm20. However, this suppression is not reciprocal; overexpression of Mdm20p does not rescue mutant phenotypes in tpm1 cells, suggesting that their roles in organizing actin differ. Mdm20p and Tpm1p do not appear to be structurally homologous since their amino acid sequences and sizes are quite different. If Mdm20p and Tpm1p do not have redundant functions, then an alternative explanation for the rescue of mdm20 mutant phenotypes by overexpressed Tpm1p is required. One possibility is that the overexpression of Tpm1p leads to the global stabilization of all actin filaments and cables in mdm20 cells, including those necessary for mitochondrial transport and inheritance. In support of this model, overexpression of Tpm1p is reported to cause more prominent cables and actin networks in wild-type cells and can restore actin cables in act1-2 mutant strains (Liu and Bretscher, 1989). Although overexpression of Tpm2p also suppresses mutant phenotypes in mdm20, we did not observe synthetic lethality in mdm20 tpm2 double mutants. Previous studies showed that the level of Tpm2p expression in wild-type cells is much lower than that of Tpm1p, and that cells lacking TPM2 do not exhibit defects in actin organization or growth (Drees et al., 1995). Thus, mdm20Δ tpm2Δ cells may contain enough Tpm1p to compensate for the loss of Tpm2p and prevent the cell death observed in mdm20Δ tpm1Δ double mutants.
Figure 8.
Summary of the synthetic interactions that involve mdm20. The synthetic interactions between tpm1 and bem2 (Wang and Bretscher, 1995), myo2 (Liu and Bretscher, 1992), and tpm2 (Drees et al., 1995) were previously reported.
Interestingly, although mutations in MDM20 and TPM1 cause similar defects in actin organization, the mutant phenotypes in these two strains are distinct. mdm20Δ mutant cells exhibit severe defects in mitochondrial inheritance (58%) and only slight defects in nuclear segregation (5%). Conversely, tpm1Δ strains exhibit weak defects in mitochondrial inheritance (5%) and stronger defects in nuclear segregation (16%). As discussed above, while overexpression of Tpm1p can rescue mutant phenotypes in mdm20, the converse is not true. These observations suggest that Mdm20p and Tpm1p perform different roles in vivo and support the hypothesis that there are functionally distinct classes of actin-containing structures in cells. It is important to note that mdm20 and tpm1 mutant cells lacking actin cables may contain individual actin filaments that are not detected by rhodamine-phalloidin staining. Thus, actin filaments that play a role in mitochondrial inheritance may be present in the tpm1 mutant but absent in the mdm20 mutant. By the same reasoning, actin-containing structures required for nuclear segregation may be more severely disrupted in the tpm1 mutant than in the mdm20 mutant. In this way, MDM20 and TPM1 could be acting to establish molecular and functional heterogeneity in the actin cytoskeleton.
As illustrated in Fig. 8, our synthetic lethal studies suggest that MDM20 functions in a cellular process that also requires TPM1, BEM2, and MYO2. Assembly and/or stabilization of the actin cytoskeleton is an obvious candidate for this process since defects in actin organization have been reported for mutations in all of these genes (Liu and Bretscher, 1989; Johnston et al., 1991; Kim et al., 1994; Wang and Bretscher, 1995). The synthetic growth defects we observe between mdm20 and tpm1, bem2, and myo2 may simply reflect the fact that partial defects in actin organization can be tolerated at 25°C, but combinations of defects cannot. However, this interpretation is not supported by the findings that double mutants lacking both MDM20 and SAC6 (Table IV and Fig. 8) or SAC6 and TPM1 (Adams et al., 1993) are viable. An alternative possibility is that synthetic lethality between mutations in these genes results from a failure to coordinate bud site assembly with the actin polarization required for directed membrane transport and bud growth. Lillie and Brown (1994) have suggested that Myo2p localized at the incipient bud site might function to anchor actin filaments that are required for the polarized delivery of vesicles to the bud. In support of this model, both myo2 and tpm1 mutants accumulate small vesicles, and we have observed similar vesicles in a small number of mdm20 cells (data not shown). Our studies of the mdm20 mutant indicate that polarized actin cables are also required for the delivery of mitochondria to buds. Physical and genetic interactions reported for BEM2 suggest that the Bem2p protein may also localize to the bud site where it is postulated to regulate Tpm1p-containing actin filaments (Wang and Bretscher, 1995). Based on the synthetic lethal interactions observed to date, we speculate that viable mdm20 and tpm1 cells lacking actin cables still contain some undetected actin filaments that are properly polarized to the bud site. However, when mdm20 and tpm1 are combined with mutations in genes that control actin filament polarization to the bud site, synthetic lethality results.
Although MDM20 encodes a novel protein required for the organization of the actin cytoskeleton, analysis of the predicted Mdm20 amino acid sequence does not reveal any known actin binding domains. Mdm20p does contain two potential heptad repeats, however, which could modulate its homo- or heterodimerization through the formation of α-helical coiled-coils. While the specific biochemical activity of Mdm20p is unknown, it is likely to participate in the assembly or function of actin-containing structures necessary for mitochondrial segregation. In this regard, Mdm20p could bind directly to filamentous actin or might act in a signaling pathway that directs the assembly or stabilization of actin filaments or cables that transport mitochondria during cell division. We think it is unlikely that Mdm20p is a major structural component of actin filaments or cables for two reasons. First, our studies with the HA-tagged Mdm20p suggest that relatively low levels of the protein are able to completely rescue mutant phenotypes in the mdm20 strain (see Fig. 7). Second, we have been unable to localize the low abundance of HA-tagged Mdm20p in wild-type or mdm20 cells (data not shown). In cells overexpressing HA-tagged Mdm20p, the protein is found dispersed throughout the cytoplasm and does not appear to be concentrated along actin cables or at actin patches (data not shown). Based on these results, we suggest that Mdm20p serves a regulatory function in cells. We are currently studying the function of this interesting protein in organizing actin and in directing mitochondrial transport and inheritance.
Acknowledgments
We are grateful to Drs. Alison Adams, Anthony Bretscher, Susan Brown, John Cooper, Tim Formosa, Edina Harsay, Rob Jensen, Sue Lillie, David Stillman, Fred Winston, and Michael Yaffe for providing strains, antibodies, and plasmids. We especially thank Jason Singer and William Bleazard for the mdm20/sac6 and mdm20/bem2 synthetic lethality data. We also thank David Nix for technical advice, and Drs. Mary Beckerle, Tim Formosa, David Gard, and members of the Shaw laboratory for stimulating discussions and careful review of the manuscript.
This work was supported by grants from the American Cancer Society (CB-97) and the National Institutes of Health (NIH) (GM53466) to J. Shaw. G. Hermann was supported in part by an NIH Predoctoral Genetics training grant and a University of Utah Graduate Research fellowship.
Abbreviations used in this paper
- DAPI
4′,6-diamidino-2-phenylindole
- DIC
differential interference contrast
- DiOC6
3,3′ dihexyloxacarbocyanine
- 5-FOA
5-fluoro-orotic acid
- HA
hemagglutinin
- SD
synthetic dextrose
- SGal
synthetic galactose
- Ura
uracil
Footnotes
Please address all correspondence to Janet M. Shaw, Department of Biology, University of Utah, Salt Lake City, UT 84112. Tel.: (801) 585-6205. Fax: (801) 581-4668.
References
- Adams AEM, Pringle JR. Relationship of actin and tubulin distribution in wild-type and morphogenetic mutant Saccharomyces cerevisiae. . J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AEM, Pringle JR. Staining of actin with fluorochromeconjugated phalloidin. Methods Enzymol. 1991;194:729–731. doi: 10.1016/0076-6879(91)94054-g. [DOI] [PubMed] [Google Scholar]
- Adams AEM, Botstein D, Drubin DG. Requirement of yeast fimbrin for actin organization and morphogenesis in vivo. . Nature (Lond) 1991;354:404–408. doi: 10.1038/354404a0. [DOI] [PubMed] [Google Scholar]
- Adams AEM, Cooper JA, Drubin DG. Unexpected combinations of null mutations in genes encoding the actin cytoskeleton are lethal in yeast. Mol Biol Cell. 1993;4:459–468. doi: 10.1091/mbc.4.5.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Backer JS. New alleles of mgm1: a gene encoding a protein with a GTPbinding domain related to dynamin. Curr Genet. 1995;28:499–501. doi: 10.1007/BF00310822. [DOI] [PubMed] [Google Scholar]
- Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1995. Nucleic Acids Res. 1995;24:189–196. doi: 10.1093/nar/24.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball EH, Singer SJ. Mitochondria are associated with microtubules and not with intermediate filaments in cultured fibroblasts. Proc Natl Acad Sci USA. 1982;79:123–126. doi: 10.1073/pnas.79.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. . Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–63. doi: 10.1016/s0074-7696(08)61205-x. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- Bretscher A, Drees B, Harsay E, Schott D, Wang T. What are the basic functions of microfilaments? Insights from studies in budding yeast. J Cell Biol. 1994;126:821–825. doi: 10.1083/jcb.126.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Delannoy M, Jensen RE. MMM1encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan AM, Donahue TF. Sequence and structural features associated with translational initiator regions in yeast: a review. Gene (Amst) 1987;59:1–18. doi: 10.1016/0378-1119(87)90261-7. [DOI] [PubMed] [Google Scholar]
- Cohen C, Parry DAD. α-helical coiled coils: a widespread motif in proteins. Trends Biochem Sci. 1986;11:245–248. [Google Scholar]
- Cohen C, Parry DAD. α-helical coiled coils and bundles: how to design an α-helical protein. Proteins Struct Funct Genet. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Stillman B. DNA binding properties of an HMG1- related protein from yeast mitochondria. J Biol Chem. 1992;267:3368–3374. [PubMed] [Google Scholar]
- Drees B, Brown C, Barrell BG, Bretscher A. Tropomyosin is essential in yeast, yet the TPM1 and TPM2products perform distinct functions. J Cell Biol. 1995;128:383–392. doi: 10.1083/jcb.128.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Jones HD, Wertman KF. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1227–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K, Farh L, Marshall TK, Deschenes RJ. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1gene. Curr Genet. 1993;24:141–148. doi: 10.1007/BF00324678. [DOI] [PubMed] [Google Scholar]
- Hannavy K, Rospert S, Schatz G. Protein import into mitochondria: a paradigm for the translocation of polypeptides across membranes. Curr Opin Cell Biol. 1993;5:694–700. doi: 10.1016/0955-0674(93)90142-d. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 726 pp.
- Heggeness MH, Simon M, Singer SJ. Association of mitochondria with microtubules in cultured cells. Proc Natl Acad Sci USA. 1978;75:3863–3866. doi: 10.1073/pnas.75.8.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman H, Avers CJ. Mitochondrion of yeast: ultrastructural evidence for one giant, branched organelle per cell. Science (Wash DC) 1973;181:749–751. doi: 10.1126/science.181.4101.749. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ. The pattern and mechanism of mitochondrial transport in axons. Front Biosci. 1996;1:91–102. doi: 10.2741/a118. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GC, Prendergast JA, Singer RA. The Saccharomyces cerevisiae MYO2gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BA, Fangman WL. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes & Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- Kilmartin J, Adams AEM. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. . J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Francisco L, Chen G, Marcotte E, Chan CSM. Control of cellular morphogenesis by the Ipl2/Bem2 GTPase-activating protein: possible role of protein phosphorylation. J Cell Biol. 1994;127:1381–1394. doi: 10.1083/jcb.127.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning AJ, Lum PY, Williams JM, Wright R. DiOC6staining reveals organelle structure and dynamics in living yeast cells. Cell Motil Cytoskeleton. 1993;25:111–128. doi: 10.1002/cm.970250202. [DOI] [PubMed] [Google Scholar]
- Kubrich M, Dietmeier K, Pfanner N. Genetic and biochemical dissection of the mitochondrial protein-import machinery. Curr Genet. 1995;27:393–403. doi: 10.1007/BF00311207. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SA, Langford GM, Weiss DG. Actin-dependent organelle movement in squid axoplasm. Nature (Lond) 1992;356:722–725. doi: 10.1038/356722a0. [DOI] [PubMed] [Google Scholar]
- Lazzarino DA, Boldogh I, Smith MG, Rosand J, Pon LA. Yeast mitochondria contain ATP-sensitive, reversible actin-binding activity. Mol Biol Cell. 1994;5:807–818. doi: 10.1091/mbc.5.7.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Neupert W. Mechanisms of protein import across the mitochondrial outer membrane. Trends Cell Biol. 1996;6:56–61. doi: 10.1016/0962-8924(96)81015-4. [DOI] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. . J Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57:233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- Liu H, Bretscher A. Characterization of TPM1disrupted yeast cells indicates an involvement of tropomyosin in directed vesicular transport. J Cell Biol. 1992;118:285–299. doi: 10.1083/jcb.118.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science (Wash DC) 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., E.F. Fritsch, and J. Sambrook. 1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 545 pp.
- McConnell SJ, Yaffe MP. Nuclear and mitochondrial inheritance in yeast depends on novel cytoplasmic structures defined by the MDM1 protein. J Cell Biol. 1992;118:385–395. doi: 10.1083/jcb.118.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SJ, Yaffe MP. Intermediate filament formation by a yeast protein essential for organelle inheritance. Science (Wash DC) 1993;260:687–689. doi: 10.1126/science.8480179. [DOI] [PubMed] [Google Scholar]
- McConnell SJ, Stewart LC, Talin A, Yaffe MP. Temperaturesensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol. 1990;111:967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mose-Larsen P, Bravo R, Fey SJ, Small JV, Celis JE. Putative association of mitochondria with a subpopulation of intermediate-sized filaments in cultured human skin fibroblasts. Cell. 1982;31:681–692. doi: 10.1016/0092-8674(82)90323-3. [DOI] [PubMed] [Google Scholar]
- Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end- directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence analysis. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. Interactions between the bud emergence proteins Bem1p and Bem2p and the Rho-type GTPases in yeast. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger MF, Kazzaz JA, Helfman DM. Functional properties of nonmuscle tropomyosin isoforms. Curr Opin Cell Biol. 1994;6:96–104. doi: 10.1016/0955-0674(94)90122-8. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Rayment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC, Milligan RA. Structure of the actin-myosin complex and its implications for muscle contraction. Science (Wash DC) 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- Roeder AD, Shaw JM. Vacuole partitioning during meiotic division in yeast. Genetics. 1996;144:445–458. doi: 10.1093/genetics/144.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Broach JR. Cloning genes by complemenation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiaegenomic plasmid bank based on a centromere-containing shuttle vector. Gene (Amst) 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Ryan KR, Jensen RE. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- Schroder RR, Manstein DJ, Jahn W, Holden H, Rayment I, Holmes KC, Spudich JA. Three-dimensional atomic model of F-actin decorated with Dictyosteliummyosin S1. Nature (Lond) 1993;364:171–174. doi: 10.1038/364171a0. [DOI] [PubMed] [Google Scholar]
- Schwarz E, Neupert W. Mitochondrial protein import: mechanisms, components and energetics. Biochim Biophys Acta. 1994;1187:270–274. doi: 10.1016/0005-2728(94)90125-2. [DOI] [PubMed] [Google Scholar]
- Sedgwick SG, Morgan BA. Locating, DNA sequencing, and disrupting yeast genes using tagged Tn1000. . Methods Mol Genet. 1994;3:131–140. [Google Scholar]
- Shaw JM, Wickner WT. vac2: a yeast mutant which distinguishes vacuole segregation from Golgi-to-vacuole protein targeting. EMBO (Eur Mol Biol Organ) J. 1991;10:1741–1748. doi: 10.1002/j.1460-2075.1991.tb07698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Methods in Yeast Genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY. 186 pp.
- Shortle D, Haber JE, Botstein D. Lethal disruption of the yeast actin gene by integrative DNA transformation. Science (Wash DC) 1982;217:371–373. doi: 10.1126/science.7046050. [DOI] [PubMed] [Google Scholar]
- Simon VR, Swayne TC, Pon LA. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J Cell Biol. 1995;130:345–354. doi: 10.1083/jcb.130.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, B. 1981. Mitochondrial structure. In Molecular Biology of the Yeast Saccharomyces. J.M. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Press, Cold Spring Harbor, NY. 471–504.
- Stewart LC, Yaffe MP. A role for unsaturated fatty acids in mitochondrial movement and inheritance. J Cell Biol. 1991;115:1249–1257. doi: 10.1083/jcb.115.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromer MH, Bendayan M. Immunocytochemical identification of cytoskeletal linkages to smooth muscle cell nuclei and mitochondria. Cell Motil Cytoskeleton. 1990;17:11–18. doi: 10.1002/cm.970170104. [DOI] [PubMed] [Google Scholar]
- Stukey JE, McDonough VM, Martin CE. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. . J Biol Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- Stukey JE, McDonough VM, Martin CE. The OLE1 gene of Saccharomyces cerevisiaeencodes the Δ9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- Summerhayes IC, Wong D, Chen LB. Effect of microtubules and intermediate filaments on mitochondrial distribution. J Cell Sci. 1983;61:87–105. doi: 10.1242/jcs.61.1.87. [DOI] [PubMed] [Google Scholar]
- Thorsness PE. Structural dynamics of the mitochondrial compartment. Mutat Res. 1992;275:237–241. doi: 10.1016/0921-8734(92)90027-m. [DOI] [PubMed] [Google Scholar]
- Wang T, Bretscher A. The rho-GAP encoded by BEM2regulates cytoskeletal structure in budding yeast. Mol Biol Cell. 1995;6:1011–1024. doi: 10.1091/mbc.6.8.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, Wickner W. Organelle inheritance. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- Welch MD, Holtzman DA, Drubin DG. The yeast actin cytoskeleton. Curr Opin Cell Biol. 1994;6:110–119. doi: 10.1016/0955-0674(94)90124-4. [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiaestrains that are isogenic to S228C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Yaffe MP. Organelle inheritance in the yeast cell cycle. Trends Cell Biol. 1991;1:160–163. doi: 10.1016/0962-8924(91)90017-4. [DOI] [PubMed] [Google Scholar]
- Zelenaya-Troitskaya O, Perlman PS, Butow RA. An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO (Eur Mol Biol Organ) J. 1995;14:3268–3276. doi: 10.1002/j.1460-2075.1995.tb07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]