Abstract
During development, the lens of the eye becomes transparent, in part because of the elimination of nuclei and other organelles from the central lens fiber cells by an apoptotic-like mechanism. Using confocal microscopy we showed that, at the border of the organelle-free zone (OFZ), fiber cell nuclei became suddenly irregular in shape, with marginalized chromatin. Subsequently, holes appeared in the nuclear envelope and underlying laminae, and the nuclei collapsed into condensed, spherical structures. Nuclear remnants, containing DNA, histones, lamin B2, and fragments of nuclear membrane, were detected deep in the OFZ. We used in situ electrophoresis to demonstrate that fragmented DNA was present only in cells bordering the OFZ. Confocal microscopy of terminal deoxynucleotidyl transferase (TdT)–labeled lens slices confirmed that DNA fragmentation was a relatively late event in fiber differentiation, occurring after the loss of the nuclear membrane. DNA fragments with 3′-OH or 3′-PO4 ends were not observed elsewhere in the lens under normal conditions, although they could be produced by pretreatment with DNase I or micrococcal nuclease, respectively. Dual labeling with TdT and an antibody against protein disulfide isomerase, an ER-resident protein, revealed a distinct spatial and temporal gap between the disappearance of ER and nuclear membranes and the onset of DNA degradation. Thus, fiber cell chromatin disassembly differs significantly from classical apoptosis, in both the sequence of events and the time course of the process. The fact that DNA degradation occurs only after the disappearance of mitochondrial, ER, and nuclear membranes suggests that damage to intracellular membranes may be an initiating event in nuclear breakdown.
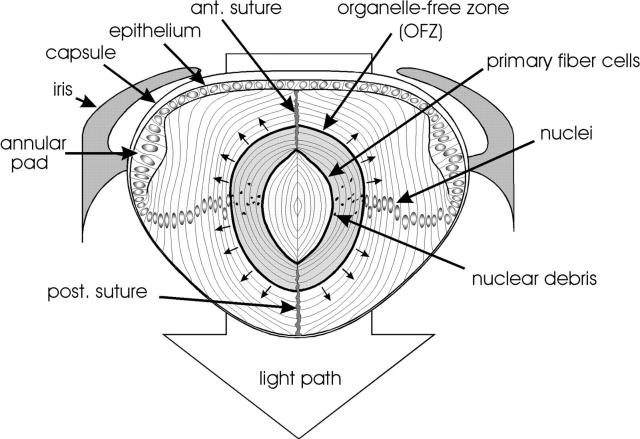
The lens of the eye is a transparent cellular structure that focuses light on the retina. The bulk of the lens consists of concentric layers of fiber cells that are formed throughout life by the differentiation of cells at the equatorial margin of the lens epithelium (see Fig. 1). Fiber cell differentiation is characterized by cellular elongation, the synthesis of certain crystallin proteins, and the degradation of all membrane-bound organelles, including nuclei (Piatigorsky, 1981). Because there is no cell turnover in the lens, all cells are retained within the tissue, those nearest the center being the oldest and those nearest the surface being the youngest.
Figure 1.
Diagram of a midsagittal slice of a chicken lens at E15. The lens is bounded by an acellular collagenous capsule. An epithelial monolayer covers the anterior surface of the lens and thickens at the periphery to form the annular pad. The bulk of the lens consists of concentric layers of highly elongated lens fiber cells derived from the edges of the epithelium. Primary fiber cells, formed early in development, are situated in the center of the lens. The rest of the fiber mass is composed of secondary fiber cells, formed throughout life by the differentiation of epithelial cells at the lens equator. The tips of secondary fiber cells make contact with fibers from the opposite hemisphere of the lens at the sutures. The outer fiber cells contain a normal complement of organelles, including nuclei. However, organelles are absent from cells in the center of the lens, giving rise to the OFZ. The OFZ increases steadily in size throughout embryonic development (small arrows).
During development, optical clarity is ensured by the degradation of intracellular organelles in fiber cells near the center of the lens (i.e., those cells directly in the light path). The mechanism by which organelles are disassembled is not known, but recent work has demonstrated that this process is under strict temporal and spatial control and that, in any given cell, the endoplasmic reticulum, mitochondria, and nuclei are rapidly and synchronously degraded (Bassnett, 1992, 1995; Bassnett and Beebe, 1992).
In the embryonic lens, organelles are present initially throughout the volume of the tissue, but, beginning on embryonic day 12 (E12)1 in the chicken, they are eliminated from cells in the center of the lens (Bassnett and Beebe, 1992). This results in the formation of an organelle-free zone (OFZ; Fig. 1). Once formed, the OFZ expands in diameter at a rate of 80 μm/d during embryonic development (Bassnett and Beebe, 1992), engulfing ∼10,000 cells per day (Bassnett, 1995). In adult lenses, those organelles that remain are concentrated in the periphery of the lens, lying largely in the shadow of the iris (Bassnett, 1992). In certain pathological conditions, however, organelles persist in the central fiber cells. For example, the failure of fiber cells to properly degrade their nuclei is a common feature of human congenital cataract (Zimmerman and Font, 1966).
Because both processes involve the systematic degradation of chromatin, several authors have pointed out the parallels between lens fiber cell denucleation and classical apoptosis (Chaudun et al., 1994; Arruti et al., 1995; Pan and Griep, 1995; Torriglia et al., 1995). Apoptosis was first described by Kerr et al. (1972) as a morphologically distinct form of cell death that involves (among other things) cell shrinkage, membrane blebbing, and chromatin condensation. One characteristic early feature of apoptotic cell death is internucleosomal DNA cleavage (Wyllie, 1980), which has been variously ascribed to the activity of DNase I (Peitsch et al., 1993), DNase II (Barry and Eastman, 1993), and NUC 18 (Montague et al., 1994). Interestingly, low– molecular weight fractions of DNA have been isolated from anucleate fiber cells in the center of the lens (Appleby and Modak, 1977). In fact, the lens was one of the first tissues in which such “DNA ladders” were observed. However, an obvious difference between apoptosis and lens fiber cell denucleation is that, in the latter, the cells are not phagocytosed by their neighbors, and the anucleate fiber cells persist throughout the life of the organism. One of the purposes of the present study was to determine the extent to which lens fiber cell denucleation resembles classical apoptosis in its time course and in its biochemical and morphological characteristics.
It has been known since the turn of the century that differentiation of lens fiber cells involves the loss of cell nuclei (Rabl, 1899), and, consequently, this aspect of lens fiber differentiation has received the most attention. The morphological changes that precede denucleation have been described in detail for a number of species (see for example Modak and Perdue, 1970; Kuwabara and Imaizumi, 1974; Sanwal et al., 1986; Vrensen et al., 1991). During fiber cell maturation, the cell nuclei undergo striking changes in shape, accompanied by progressive alterations in chromatin structure and eventual loss of integrity of the nuclear membrane and infiltration of cytosolic proteins (Sandilands et al., 1995).
Although there is a general consensus on the morphological aspects of lens cell denucleation, there is considerable disagreement regarding the underlying biochemical mechanism(s) and, importantly, the timing of the process. In a series of experiments in the early 1970s, the then newly discovered enzyme terminal deoxynucleotidyl transferase (TdT) was used to radiolabel the nuclei of sectioned embryonic chicken lenses (Modak and Bollum, 1970, 1971, 1972). TdT catalyzes the addition of dNTPs to the 3′-hydroxyl (3′-OH) termini of DNA molecules in a template-independent fashion. The fact that fiber cells contained initiation sites for the addition of radiolabeled nucleotides was taken as evidence for the presence of significant numbers of single-strand breaks in the DNA. Quantitative analysis showed that the nuclei of fiber cells in the deep cortex of the lens were more heavily labeled than in the superficial cells, suggesting that DNA damage (in the form of free 3′-OH ends) accumulated steadily during fiber cell differentiation. Taken together, these studies support the notion that fiber cell denucleation is a gradual process, beginning in the superficial cells and characterized by the steady accumulation of single-strand breaks in DNA. According to this view, the DNA is ultimately cleaved into small, nucleosome-sized fragments, either by extensive overlapping single-strand scissions or direct double-strand breaks.
However, in marked contrast to the work described above, recent studies have been unable to detect the presence of DNA strand breaks in differentiating chicken embryo fiber cells (Chaudun et al., 1994) or in mouse lenses (Fromm et al., 1994; Morgenbesser et al., 1994; Pan and Griep, 1994; Chow et al., 1995; Robinson et al., 1995), and DNase I transcripts are not detectable in the lens by reverse transcriptase–PCR (Hess and Fitzgerald, 1996). The involvement of a DNase II–like enzyme (which generates 3′-PO4 ends that would not be labeled by the conventional TdT technique) in the fiber denucleation process in chicken lenses has been invoked to explain this discrepancy (Torriglia et al., 1995). The present experiments were also designed to resolve this issue, using a combination of enzymatic labeling and electrophoretic techniques.
Materials and Methods
Animals
Fertilized White Leghorn chicken eggs were obtained from Truslow Farms (Chestertown, MD) and incubated at 38°C in a forced-draft incubator. Embryonic lenses, with their attached vitreous humor, were removed through an incision in the posterior globe.
Preparation of Lens Slices
Fixed lenses were embedded in 4% agar/PBS and sectioned in the midsagittal plane using a Vibratome (model 3000; Technical Products International, Inc., St. Louis, MO) as previously described (Bassnett, 1995). Fixed lenses were routinely sectioned at 200 μm.
DNA and Membrane Staining
In some preparations, it was necessary to colocalize DNA and cellular membranes. This was done by washing fixed lens slices in TBS (10 mM Tris, 150 mM NaCl, pH 7.4) for 5 min and then incubating the slices for 40 min in TBS containing 0.005% Triton X-100, 1% BSA, 1 μM SYTO 17 (Molecular Probes, Eugene, OR), and 1 μg/ml 3,3′-dihexyloxacarbocyanine iodide (DiOC6; Molecular Probes). SYTO 17 is a fluorescent DNA probe with spectral properties similar to Texas red. DiOC6 is a lipophilic membrane probe with fluorescein-like fluorescence. We have used DiOC6 previously to visualize membrane systems in the embryonic lens (Bassnett, 1995). After staining, the lens slices were washed in TBS for 30 min and viewed with the confocal microscope using the 488-nm laser line and a 515–565-nm band-pass filter for DiOC6 fluorescence and the 568-nm laser line and a 590-nm long pass filter for SYTO 17 fluorescence.
Immunofluorescence
E18 lenses were fixed for 1 h in 4% paraformaldehyde/PBS, pH 7.4, and sliced at 200 μm. Slices were permeabilized with 0.1% Triton X-100 for 30 min at room temperature and incubated in blocking solution (10% normal goat serum/1% BSA/PBS) for 1 h. Slices were incubated overnight at 4°C in primary antibody diluted 1:500 in blocking solution. After a 1-h wash in PBS, slices were incubated for 2 h in fluorescein-conjugated goat anti– mouse IgG antibody (Jackson ImmunoResearch Labs., West Grove, PA) diluted 1:500 in blocking solution. Slices were then washed for 1 h in PBS, mounted, and viewed.
To visualize the fate of histone proteins during fiber differentiation, we used a monoclonal antibody that recognizes an epitope present in histone H1, H2A, H2B, H3, and H4 (clone H11-4; Boehringer Mannheim Corp., Indianapolis, IN). To visualize the distribution of the ER in lens slices, we used a monoclonal antibody (clone ID3; StressGen Biotechnologies Corp., Sidney, BC, Canada) against protein disulfide isomerase (PDI), an ER-resident protein, as described previously (Bassnett, 1995; Bassnett and Shiels, 1996). The nuclear lamina was visualized using a monoclonal antibody raised against human lamin B2 (clone NCL-LAM-B2; Novocastra Laboratories Ltd., Newcastle upon Tyne, England). This antibody was only effective on fixed lens slices following the high-temperature antigen retrieval technique (Shi et al., 1991). This involved immersing lens slices in 0.01 M citrate buffer, pH 6.0, and heating at full power for 10 min in a 600-W domestic microwave oven. The lens slices were then allowed to cool, rinsed briefly in PBS, and processed for immunofluorescence as described above.
In Situ TdT Labeling
Lenses were fixed in 4% paraformaldehyde/PBS, pH 7.4, for 1 h at room temperature. Lens slices were permeabilized for 30 min in 0.1% Triton X-100/PBS at room temperature and washed for 20 min in PBS. The TdT labeling reaction was performed using an in situ cell death detection kit (Boehringer Mannheim Corp.) according to the manufacturer's instructions. In this assay, a homopolymeric tail of fluorescein-labeled nucleotides is incorporated at the 3′-OH ends of DNA molecules by the enzyme TdT. Preliminary experiments revealed significant nonspecific labeling of nuclei in the absence of the TdT enzyme. It was necessary to include a washing step (gentle agitation in 0.1% SDS at 60°C for 30 min) to remove these unincorporated fluorescent nucleotides. The washing procedure reduced nonspecific labeling to low levels. In each experiment, some lens slices were incubated in labeling mixture lacking TdT, as a negative control.
Permeabilized slices were pretreated with DNase I (Boehringer Mannheim Corp., 50 U/ml in 10 mM Tris, pH 7.6) for 30 min at 37°C, as a positive control. DNase I cleaves DNA into fragments with 3′-OH termini. Because there is evidence for DNase II–like activity in the chicken lens (Torriglia et al., 1995), we pretreated some slices with calf intestinal alkaline phosphatase (CIAP; GIBCO BRL, Gaithersburg, MD). CIAP treatment should convert any 3′-PO4 termini (produced by DNase II digestion) to 3′-OH termini, the latter being detectable by the TdT assay. Permeabilized slices were incubated for 1 h at 50°C in 100 μl of dephosphorylation buffer (supplied with the enzyme) containing 1 μl (26 U) of CIAP. As a positive control, some lens slices were first incubated for 30 min at 37°C with 0.5 U/μl micrococcal nuclease (Boehringer Mannheim Corp.), an enzyme that produces DNA fragments with 3′-PO4 termini.
In Situ Electrophoresis
The location of nuclei with degraded DNA was visualized by an in situ electrophoretic technique under alkaline conditions. This method was modified from that used previously to measure DNA damage in single cells (Ostling and Johanson, 1984; Singh et al., 1988), including lens epithelial cells (Kleiman and Spector, 1993). Single-strand scissions are 5- to 2,000-fold more numerous than double-strand breaks in damaged DNA (Bradley and Kohn, 1979) and are readily detected under denaturing alkaline conditions. Lenses were fixed in 0.5% paraformaldehyde/PBS, pH 7.4, for 30 min at room temperature, embedded in 4% agar/PBS, and sliced at 100 μm. Slices were reembedded in 0.3% agarose. The agarose block was trimmed to a cube with a volume of <1 cm3. The agarose cube containing the lens slice was immersed in permeabilizing buffer (1% sodium sarcosinate, 1% Triton X-100, 2.5 M NaCl, 100 mM EDTA, 10 mM Tris, pH 10) for 1 h and then soaked in electrophoresis buffer (10 mM EDTA, 0.4 M NaOH) for an additional hour. Some slices were incubated in DNase I (50 U/ml) for 2 h at 37°C after the permeabilization step as a positive control. The agarose cube was transferred to a horizontal gel electrophoresis apparatus and oriented so that the lens slice was perpendicular to the applied electric field. Electrophoresis was performed for 15 min at 1.5 V/cm. After electrophoresis, the agarose cubes were neutralized in 0.5 M Tris, pH 7.5, for 30 min, stained with ethidium bromide (1 μg/ml) for 45 min, and destained in distilled water. Blocks were transferred to the stage of the confocal microscope and viewed using the 488-nm line of an argon/krypton laser and a 515-nm long-pass dichroic filter.
Confocal Microscopy
Slices were viewed using a confocal microscope equipped with an Argon/ Krypton laser (model LSM 410; Carl Zeiss Inc., Thornwood, NY).
Results
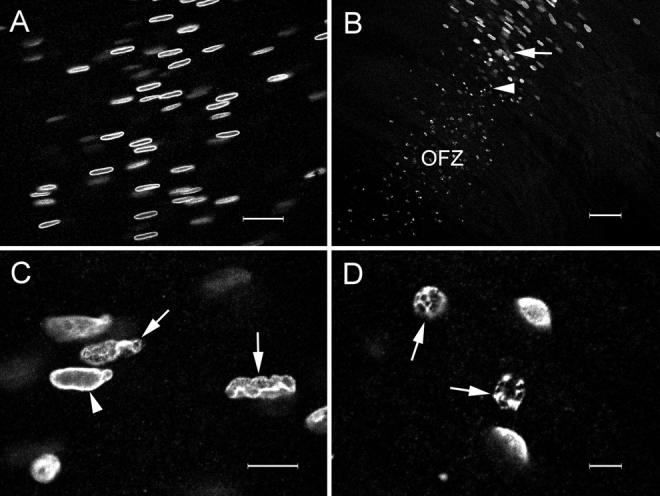
Cut open along the optic axis, an E15 embryonic chicken lens has the cellular organization shown in diagrammatic form in Fig. 1. Confocal microscopic images of the changes in nuclear organization in cells near the border of the OFZ are shown in Fig. 2. Lens slices were stained with both DiOC6 and SYTO 17, enabling the simultaneous visualization of the fiber cell membrane systems (Fig. 2, B, E, H, and K) and DNA (Fig. 2, C, F, I, and L), respectively, in addition to differential interference contrast (DIC) images (Fig. 2, A, D, G, and J). In the outer cortex of the lens, the leaf-shaped fiber cell nuclei were the only prominent subcellular feature of the DIC images (Fig. 2 A). The corresponding confocal image obtained with DiOC6 (Fig. 2 C) showed labeling of the fiber cell membrane systems, including the plasma membrane and the nuclear envelope. The distribution of DNA was visualized simultaneously using SYTO 17. The fiber cell nuclei were evenly stained by SYTO 17, with prominent nucleoli. In fiber cells near the border of the OFZ, a change in the organization of the nuclei was apparent (Fig. 2, D–F). In the space of a few cell layers, nuclei passed through distinct stages. In the first stage, the leaf-shaped nuclei characteristic of the outer cell layers became irregular in shape with an undulating nuclear membrane. At this point, the DNA staining was uneven, with marginalization of the chromatin. Subsequently, the nuclei collapsed into small spherical structures that were visible under DIC. Although the collapsed nuclei were intensely stained by the SYTO 17, the chromatin now appeared “naked” (i.e., no longer contained within a nuclear membrane). The last nucleus with an intact nuclear membrane is indicated by the arrow in Fig. 2 E. Fiber cells containing collapsed nuclei extend for 100–200 μm into the OFZ. Even in cells at the very center of the lens, small, positively stained nuclear remnants could be discerned (Fig. 2 L) in the otherwise featureless fiber cell cytoplasm.
Figure 2.

The fate of the nuclear membrane and DNA in fiber cells near the border of the OFZ. The lens slice was stained with DiOC6 (for membranes) and SYTO 17 (for DNA) and viewed with a confocal microscope. Cortical fiber cells are shown in A–C, cells just outside the border of the OFZ are shown in D–F, cells immediately within the OFZ are shown in G–I, and cells in the center of the lens are shown in J–L. A, D, G, and J are differential interference contrast images; B, E, H, and K, are confocal images of the DiOC6 fluorescence; and C, F, I, and L are the corresponding confocal images of the SYTO 17 fluorescence. Note the sudden change in nuclear morphology at the border of the OFZ and the loss of the nuclear membrane (D–F). The last nucleus to possess a nuclear membrane is indicated by the arrow in D–F. Collapsed nuclei are evident in cells within the OFZ and SYTO 17–stained nuclear remnants (L, arrows) can be discerned, even in the most central fiber cells. Bar, 25 μm.
Confocal microscopy and immunofluorescence were used to visualize the fate of two nuclear components, lamin B2 (Fig. 3) and histone proteins (Fig. 4), in fiber cells near the border of the OFZ. Lamin B2 is a member of a family of intermediate filament proteins that together form the karyoskeleton and, as such, are usually distributed immediately beneath the nuclear membrane. In the superficial cortex of the lens, the lamin B2 immunofluorescence specifically delineates the fiber cell nuclear membrane (Fig. 3 A). In these experiments, omission of the primary antibody, or its substitution by ascites fluid, resulted in complete absence of nuclear fluorescence, demonstrating the specificity of the antibody (data not shown). At the border of the OFZ, the nuclei collapsed and lamin B2–positive nuclear debris persisted for hundreds of cell widths into the OFZ (Fig. 3 B). Immediately before nuclear collapse (Fig. 3 B, arrow), lamin B2 immunofluorescence became uneven and the nuclear profile was irregular (Fig. 3 C). Two nuclei in the early stages of nuclear collapse are indicated by arrows in Fig. 3 C. An adjacent nucleus, with a relatively intact lamina, is indicated by an arrowhead. Soon after the collapse of the nuclei (Fig. 3, B, arrowhead), rents were evident in the nuclear lamina (Fig. 3, D).
Figure 3.
Confocal micrographs of lamin B2 immunofluorescence in an E17 lens slice after high-temperature antigen retrieval (see text for details). (A) Fiber cells in the peripheral cortex. The immunofluorescence is restricted to the lamina of the fiber cell nuclei. (B) Lowmagnification view of cells near the border of the OFZ. Note that even nuclear remnants (arrowhead) are positive for lamin B2 immunofluorescence. (C) High-magnification view of cells immediately adjacent to the OFZ (B, arrow). In many of these cells, the nuclear lamina has become distorted, and the profiles of the nuclei are irregular (arrows), although some nuclei still retain a more normal appearance (arrowhead). (D) High-magnification view of cells immediately within the OFZ (B, arrowhead). At this point in the denucleation process, the fiber nuclei have collapsed into small spherical structures (arrows), and rents are apparent in the nuclear lamina. Bars: (A) 25 μm; (B) 50 μm; (C) 10 μm; (D) 5 μm.
Figure 4.

DIC and corresponding confocal immunofluorescence images of histone distribution in an E17 lens slice. The antibody recognizes an epitope common to histones H1, H2A, H2B, H3, and H4. (A) DIC image of the equatorial region of a lens slice showing the annular pad (ap) and superficial fibers. (B) Immunofluorescence image of the region shown in A. Note the strong labeling of nuclei in the annular pad and superficial fibers. (C) DIC image of fiber cells at the border of the OFZ. Fiber cell nuclei at various stages of disassembly are present, including those with marginalized (arrow) or condensed chromatin (arrowhead). (D) Immunofluorescence image of the region shown in C. During fiber cell denucleation, the distribution of histone proteins parallels that of the DNA (compare with Fig. 2 F ), becoming first marginalized (arrow) and then collapsing into condensed residual structures (arrowhead). Bars, (A and B) 50 μm; (C and D) 10 μm.
The histones are the major protein components of the chromatin. The nuclei of epithelial cells and superficial fiber cells were readily labeled by the histone antibody (Fig. 4, A and B). As the denucleation process commenced, at the border of the OFZ, the change in the pattern of nuclear histone immunofluorescence paralleled that of the nuclear DNA (compare Fig. 4 D with Fig. 2 F). Within a few cell widths, the histone immunofluorescence became heterogeneous and marginalized, and it ultimately collapsed into brightly stained residual structures (Fig. 4, C and D).
As the karyoskeleton was disassembled at the border of the OFZ, the integrity of the nuclear envelope was also lost. Once the nuclei collapsed, only ragged fragments of membrane remained, adhering to the naked chromatin (Fig. 5). The condition of nuclear DNA was also visualized during various stages of fiber cell differentiation using an in situ TdT assay. In this assay, a tail of fluorescein-labeled dUTP molecules is incorporated at 3′-OH ends of DNA fragments. Labeled nuclei were rarely observed in the epithelial cell layer and never in the superficial fiber cells. However, strongly labeled nuclei were always detected at the border of the OFZ in lenses from embryos E15 and older (Fig. 6 A). At earlier developmental stages, labeled nuclei were not observed in the fiber cells. If the labeling reaction was performed in the absence of the TdT enzyme, no labeling was observed (data not shown). Surprisingly, only fully condensed nuclei were labeled. Irregularly shaped nuclei with heterogeneous or marginalized chromatin were not labeled by this technique. End-labeled DNA fragments were seen to persist in the cytoplasm of fiber cells hundreds of cell widths inside the border of the OFZ. The lack of labeling in the more superficial fibers could have been due to the presence of an inhibitor of the TdT assay in these cells. To control for this, some slices were preincubated in DNase I to cause extensive DNA fragmentation in vitro. In DNase I–treated tissue, all of the nuclei were labeled by the TdT assay, including those of the epithelial cells and annular pad (data not shown) and superficial fiber cells (Fig. 6 B). The pattern of DNA damage induced by the DNase I treatment was qualitatively different from that occurring naturally at the border of the OFZ. In DNase I–treated cells, the labeling was heterogeneous and strongest immediately beneath the nuclear membrane (Fig. 6 B). In contrast, the collapsed nuclei of cells at the border of the OFZ were homogeneously stained. The fact that all nuclei in the lens were labeled after the DNase I treatment suggests that the lack of label in the outer cells of untreated lenses was not due to the presence of an inhibitor of the TdT assay in these cells.
Figure 5.
Merged confocal image of fiber cells immediately inside the border of the OFZ after staining with DiOC6 (green) and SYTO 17 (red). The combination of these fluorescent probes allows the simultaneous visualization of the fiber cell membranes (DiOC6) and nuclear DNA (SYTO 17). At this point in fiber cell differentiation, the nuclei have collapsed into condensed spherical structures. Remnants of the nuclear membrane are still visible (arrowheads), attached to the naked chromatin. The highly interdigitated lateral membranes of the fiber cells are also evident. Bar, 5 μm.
Figure 6.
Merged confocal and DIC images of lens slices after TdT labeling with fluorescein-dUTP. The DIC images are shown in green and positively labeled nuclei (containing fragmented DNA) are shown in red. (A) At the border of the OFZ, the nuclei lose their regular shape (arrows) and collapse into condensed structures that are strongly labeled by the TdT assay (arrowheads). Positively labeled debris, resulting presumably from the disintegration of labeled nuclei, extends deep into the OFZ. (B) Cortical fiber cells from a lens slice that was pretreated for 30 min with 50 U/ml DNase I. Note that after DNase I treatment, all nuclei are labeled by the TdT assay and that the labeling is strongest immediately beneath the nuclear membrane. (C) Equatorial region of a lens slice that had been incubated with CIAP before TdT labeling. None of the nuclei are labeled, indicating that the superficial fibers do not contain fragmented DNA with 3′PO4 termini (see text for details). (D) Equatorial region of a lens slice that was treated sequentially with micrococcal nuclease and CIAP before TdT labeling. All the nuclei are labeled, demonstrating the efficacy of the CIAP technique for detecting fragmented DNA with 3′-PO4 termini. Bars: (A) 50 μm; (B) 10 μm; (C) 50 μm; (D) 50 μm.
It has been suggested that a DNase II–like enzyme may play a role in lens nuclear degradation (Chaudun et al., 1994; Torriglia et al., 1995). DNA damage caused by DNase II would not be detected by a conventional TdT technique because TdT only catalyzes the addition of labeled nucleotides to 3′-OH termini. DNase II–like enzymes, however, would be expected to generate 3′-PO4 termini. We modified the TdT assay by incorporating a pretreatment with calf intestinal alkaline phosphatase (CIAP) to allow the detection of 3′-PO4 termini. A CIAP-treated lens slice is shown in Fig. 6 C. There were no labeled nuclei in the epithelium or superficial fiber cells of CIAP-treated lens slices. Positively labeled nuclei were observed at the border of the OFZ in CIAP-treated slices, although no more were observed than in untreated lens slices (data not shown). To verify the efficacy of the CIAP treatment, some lens slices were first incubated in micrococcal nuclease to induce the formation of DNA fragments with 3′-PO4 termini. In lens slices treated in this fashion, all the nuclei were positively labeled by the TdT assay. This indicates that lack of labeling in CIAP-treated lenses reflected the absence of 3′-PO4 DNA termini in the nuclei of superficial fibers and epithelial cells.
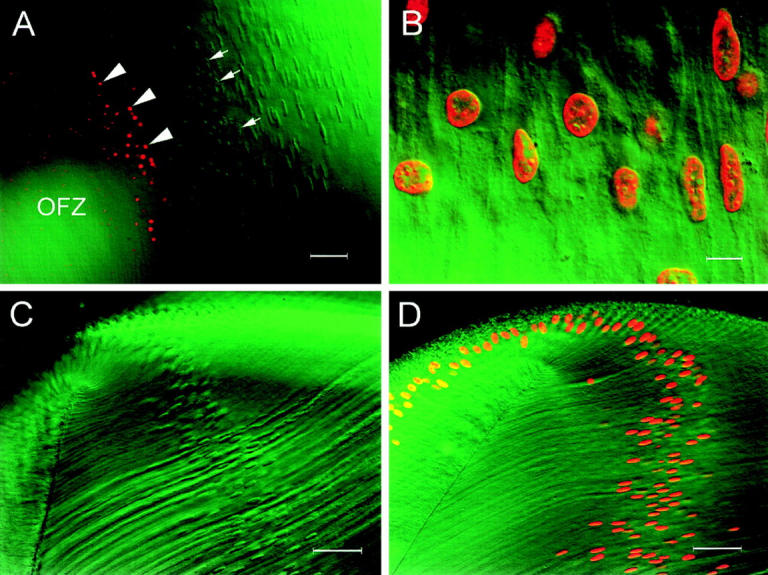
From studies such as those shown in Fig. 6 A, it appeared that positively labeled nuclei (i.e., those containing degraded DNA) were only detected some distance within the OFZ. If this were true, it would suggest that extensive DNA cleavage occurs some time after the disappearance of mitochondria, endoplasmic reticulum, and other organelles. To verify that this is the correct sequence of events, we performed immunofluorescence labeling of PDI (an ER marker) on E19 lens slices that had previously been labeled with TdT. In lens slices labeled with both TdT and antiPDI, a clear gap was revealed between the border of the OFZ (delineated by the PDI immunofluorescence) and the first TdT-positive nuclei (Fig. 7 A). At E19, cells that were unstained by either treatment formed a region ∼90 μm wide. We also noted an interesting developmental sequence in slices labeled with both TdT and anti-PDI. At E12, the OFZ was already clearly delineated, as described previously (Bassnett and Beebe, 1992). At this stage of development, the OFZ contained many condensed nuclei, but there was no evidence of TdT labeling. Only 3 d later, at E15, did the condensed nuclei become labeled by the TdT assay. Thus, the spatial gap observed at E19, between the disappearance of PDI immunofluorescence and the onset of DNA degradation (Fig. 7 A), paralleled the temporal gap observed at earlier stages.
Figure 7.
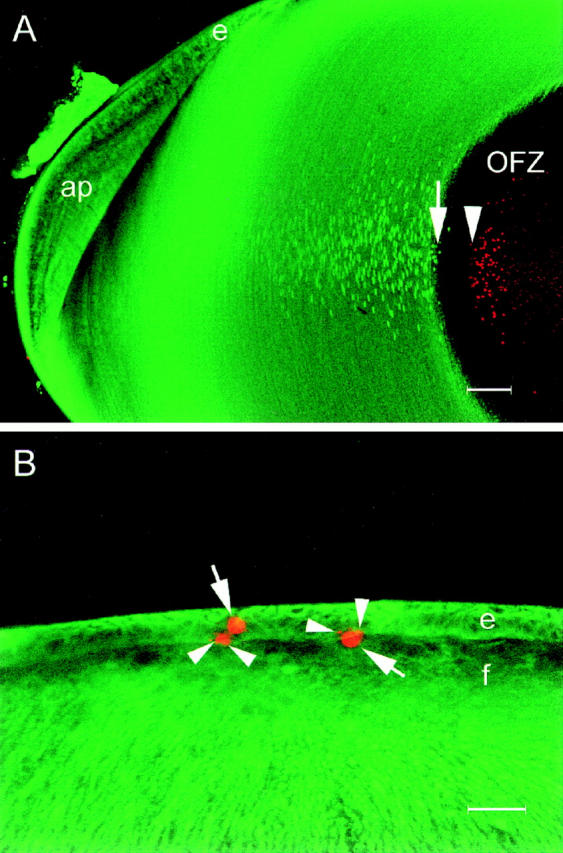
Confocal images of lens slices showing the distribution of ER (green) and degraded DNA (red). (A) The ER (visualized by immunofluorescence with a protein disulfide isomerase antibody) is abundant in the superficial layers of the lens but completely absent from the well-defined central OFZ. The fiber cell nuclear membranes are also labeled by this antibody. Degraded DNA is localized in condensed nuclear remnants, scattered throughout the cytoplasm of fiber cells in the OFZ. Note the gap of ∼90 μm between the last fiber cell to contain ER (arrow) and the first to contain degraded DNA (arrowhead). e, epithelium; ap, annular pad. (B) Apoptotic cells are occasionally detected in the anterior epithelium of the E9 lens. At this stage of development, the epithelial cells (e) and all of the fiber cells ( f ) contain abundant ER. Apoptotic nuclei (arrows) are strongly labeled by the TdT assay, and small apoptotic bodies are often observed in the adjacent tissue (arrowheads). Note that, in contrast to the central fiber cells shown in A, the apoptotic nuclei of the epithelial cells are found in cells in which the ER is still present. Bars: (A) 100 μm; (B) 25 μm.
In contrast to the fiber cells, all the epithelial cells contained abundant ER (as evidenced by strong PDI immunofluorescence), including those rare cells with TdT-labeled nuclei. Fig. 7 B shows a merged confocal micrograph of the central region of an E9 lens epithelium. The embryonic lens epithelium contained a few scattered apoptotic cells, and these were strongly labeled by the TdT assay. The labeled nuclei had a characteristic condensed morphology with small, positively labeled apoptotic bodies often found in close association (Fig. 7 B, arrows).
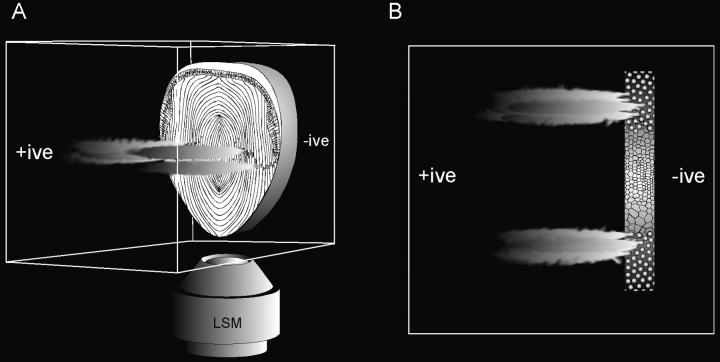
We also used a novel nonenzymatic technique for visualizing the condition of the chromatin during fiber cell differentiation. In our in situ electrophoresis assay, a lens slice was embedded in agarose, permeabilized, and oriented perpendicular to a weak electric field. Fragments of DNA that were sufficiently small to be electrophoretically mobile were driven from the lens cells into the agarose, where they were stained with ethidium bromide. A diagram showing the orientation of the lens slice and the expected appearance of the tissue under the confocal microscope is shown in Fig. 8. The optical-sectioning capability of the confocal microscope enabled us to position the focal plane at the level of the fiber cell nuclei. With this optical arrangement, the OFZ appeared as a dark gap between two arms of strongly fluorescent nuclei (Fig. 9 A). If the lens slice was pretreated with DNase I before electrophoresis, two diffuse clouds of ethidium-stained material were observed emanating from the fiber cells (Fig. 9 B). The clouds of DNA appeared to come from all the annular pad and fiber cells. In contrast, in non–DNase I–treated slices, ethidium-stained material was only observed emanating from cells at the immediate border of the OFZ (Fig. 9, C and D). We assume that, in each case, the ethidium-stained material emanating from the lens slices represents low–molecular weight DNA because cellular RNA (which would also be weakly stained by ethidium bromide) is degraded under the extremely alkaline conditions of the assay.
Figure 8.
Diagram showing the principle of in situ electrophoresis. (A) A permeabilized, alkali-treated lens slice is embedded in a block of 0.3% agarose and placed in a weak electric field. Electrophoretically mobile, fragmented DNA is driven from the slice into the gel. The preparation is stained with ethidium bromide and transferred to the stage of a laser scanning confocal microscope (LSM). The focal plane of the microscope is positioned midway up the slice at the level of the fiber cell nuclei. A hypothetical view down the microscope is shown in B, the OFZ appearing as a dark region in a strip of brightly stained nuclei. Fragmented DNA is visualized as streams of positively stained material emanating from regions of the lens where the DNA was sufficiently degraded to be rendered electrophoretically mobile.
Figure 9.

In situ electrophoresis of an E15 lens slice. (A) Low-magnification image of an ethidium-stained lens slice after electrophoresis. The slice is embedded in a block of 0.3% agarose. The dark space between the two bright arms (arrowheads) represents the OFZ. The arms are formed by the ethidium-stained fiber cell nuclei and are continuous with the band of annular pad and epithelial nuclei seen dimly in the background. The uneven bright strip extending across the image is a reflection from the edge of the agarose block. The arrow depicts the direction of the electrical field. (B) Intermediate-magnification view of a lens slice that was pretreated with DNase I before electrophoresis to cause fragmentation of fiber cell DNA. Note the diffuse clouds of ethidium-stained material emanating from all of the fiber cell nuclei. (C) Untreated lens slice showing two streams of ethidium-stained material emanating from nuclei immediately adjacent to the OFZ. (D) High-magnification image of cells at the border of the OFZ. Brightly stained individual nuclei are visible. A stream of ethidium-stained material can be seen issuing from cells immediately adjacent to the OFZ. Bars: (A) 500 μm; (B) 250 μm; (C) 100 μm; (D) 25 μm.
Discussion
The programmed loss of lens cell organelles is one of the most characteristic features of lens fiber cell differentiation and also one of the least well understood. Modak and Bollum suggested that lens cell denucleation is a gradual process characterized by the steady accumulation of DNA damage (1970, 1971, 1972). Such damage might result from the impairment of DNA repair mechanisms in the lens fiber cells (Counis et al., 1981) or the activation of lens endonucleases. However, recent studies have been unable to reproduce the original observation that DNA fragments with 3′-OH ends accumulate during fiber cell differentiation (Chaudun et al., 1994). Furthermore, transcripts for DNase I (the enzyme most likely to produce such 3′-OH ends) have not been detected in the lens (Hess and Fitzgerald, 1996). This has led to the possibility that lens nuclei may be degraded through the action of a DNase II– like enzyme. A DNase II–like enzyme has been identified in the lens on the basis of activity assays and immunological approaches (Torriglia et al., 1995).
In previous studies, we noted that the loss of organelles from lens fiber cells occurs over a time course of a few hours and within the space of a few cells (Bassnett and Beebe, 1992; Bassnett, 1995). This led us to ask whether nuclear disassembly is also a rapid event. We now report that nuclear breakdown is indeed a rapid event that begins shortly before cells reach the border of the OFZ. Over the space of 20–30 cells and a time frame of a few days, nuclei undergo a dramatic transformation, involving marginalization and subsequent collapse of the chromatin, loss of integrity of the nuclear lamina and nuclear envelope, and, finally, fragmentation of the DNA. Both the TdT labeling and in situ electrophoresis data suggest that DNA damage occurs only near the border of the OFZ, in cells that have already lost their nuclear membranes. Thus, our results contradict those of Modak and Bollum (1972), who reported DNA damage throughout the lens, albeit with increasing severity towards the center. The DNA damage that we detected was in the form of 3′-OH termini. These observations are, therefore, at odds with those of Chaudun et al. (1994), who did not observe 3′-OH termini in any region of the embryonic chicken lens using a nick translation technique to detect fragmented DNA in situ. Interestingly, Chaudun et al. (1994) did observe DNA fragmentation in the central fiber cells in tissue samples that were not fixed immediately after dissection. This raises the concern that the TdT-positive nuclei detected in our experiments could be a tissue preparation artifact due, perhaps, to postmortem activation of endogenous nucleases in the inner fiber cells. However, in the present study, tissue samples were always fixed immediately, the location of labeled cells was reproducible (the border of the OFZ), and positively labeled cells were never observed in lenses younger than E15. Thus, it seems unlikely that the TdT labeling of cells at the border of the OFZ that we have observed represents an artifact of tissue preparation. It is possible that discrepancies in the TdT-labeling results reported by various groups simply reflect species variation or subtle methodological differences. As our present results demonstrate, to observe positively labeled nuclei, it is important to examine cells in the appropriate region of the lens and at the appropriate stage of development.
In the present experiments, we used CIAP treatment to determine whether 3′-PO4 termini accumulated during lens cell differentiation, as would be the case if a DNase II–like enzyme was activated (Liao, 1985). We were unable to detect 3′-PO4 termini in lens fiber nuclei under assay conditions where 3′-PO4 termini induced by micrococcal nuclease treatment were readily observed. Thus, we found no evidence for the activity of a DNase II–like enzyme in the fiber cell denucleation process. We cannot rule out, however, the possibility that the positively labeled nuclei in cells at the border of the OFZ contained 3′-PO4 termini in addition to 3′-OH termini, as would be the case if both DNase I– and II–like enzymes were activated simultaneously at the border of the OFZ.
Lens nuclear breakdown is associated with the appearance of 3′-OH ends and, ultimately, the release of nucleosome-sized fragments of DNA into the cytoplasm (Appleby and Modak, 1977). This kind of DNA fragmentation is often found in cells undergoing apoptosis, and, indeed, some authors have suggested that lens denucleation represents a form of apoptosis (Chaudun et al., 1994). However, despite some superficial similarities, lens denucleation appears to be distinctly different from classical apoptosis. In mature lens fibers, the other cytoplasmic organelles have already disappeared 2–3 d before DNA fragmentation occurs. In contrast, apoptosis in many cell types (including the lens epithelium; Fig. 7 B) is characterized by the presence of organelles in the cytoplasm of cells in which the DNA has already been extensively degraded (Cossarizza et al., 1994; Watt et al., 1994; Weis et al., 1995). Furthermore, there is no evidence of membrane blebbing or the formation of apoptotic bodies in lens fiber cells, although fiber nuclei can sometimes have a lobulated appearance as they collapse (Fig. 5). We have also shown that enhanced binding of annexin V to the cell membranes (a marker for apoptosis in many cell types) does not occur at the time of fiber cell denucleation (data not shown). The time course of lens denucleation (3-4 d from the onset of nuclear changes to the complete disappearance of the DNA), although shown here to be more rapid than previously suspected, is nevertheless far slower than classical apoptosis. Apoptosis in normal hepatic cells, for example, is completed in ∼3 h (Bursch et al., 1990) and overt DNA damage can be detected within 30 min of the application of the apoptotic stimulus (Schmid et al., 1986). Taken together, these data suggest that lens denucleation is morphologically and biochemically distinct from apoptosis. It is likely, however, that these processes share fundamental regulatory mechanisms that determine whether the cells undergo proliferation, apoptosis, or nuclear degradation. For example, expression of the viral oncoprotein, E7, in the developing lenses of transgenic mice causes inappropriate proliferation and triggers apoptosis in the fiber cells (Pan and Griep, 1994). These effects can be inhibited by another oncoprotein, E6, possibly via its interaction with the tumor suppressor protein, p53. Interestingly, nuclear degradation is inhibited by expression of the E6 transgene, although by a p53–independent mechanism (Pan and Griep, 1995).
Lens transparency depends on the maintenance of a highly regular optically homogeneous cytoplasm. If fibers near the center of the lens were to undergo classical apoptosis, the transparency of the tissue would be compromised, and, presumably, a nuclear cataract would result. Perhaps the denucleation process in the lens has evolved in response to twin evolutionary pressures; the need to eliminate light-scattering organelles without disturbing the optical properties of the tissue.
The enzyme(s) responsible for digesting nuclear DNA in cells at the border of the OFZ remains to be identified. The apparent rapidity of the denucleation process suggests that a failure of DNA repair mechanisms (Counis et al., 1981) alone is unlikely to account for the sudden degradation of DNA. Active degradation of DNA by fiber cell endonucleases remains the most plausible explanation for fiber cell denucleation. A variety of candidate nuclease activities have been identified in embryonic chicken lens tissue. Muel et al. (1986) have demonstrated the presence of a Ca2+-dependent endonuclease in epithelial and fiber cell nuclei. This activity may correspond to a DNase I–like, 30-kD protein identified in lens tissue by DNase activity gels and immunoblots (Counis et al., 1991; Arruti et al., 1995; Torriglia et al., 1995). A 40-kD DNase, inhibited by high concentrations of divalent ions but with an absolute requirement for trace amounts of Ca2+ and Mg2+, has also been demonstrated in both epithelial and fiber cells (Counis et al., 1991) in addition to a cation-independent DNase active at neutral pH (Chaudun et al., 1994). Three DNase I isoforms have been identified in the lens by immunoblot. All three isoforms (18-, 32-, and 60-kD bands) are present in the epithelium, whereas the fiber cells contain the 32-kD isoform only (Torriglia et al., 1995). A DNase II–like activity has also been described in the embryonic lens (Torriglia et al., 1995), where it is found in the cytoplasm of the epithelial cells and the fiber cell nuclei and, surprisingly, in the lens capsule, a cell-free extracellular matrix. This enzyme activity is evident only at low pH (5.5), but, as intracellular pH is known to decrease during fiber differentiation (Bassnett and Duncan, 1986), a physiological role for such an enzyme is not inconceivable. Immunoblot studies have identified four different isoforms of DNase II in the lens. The fiber cells contain bands of 18, 23, and 60 kD, whereas two isoforms of 18 and 100 kD are found in the epithelium (Torriglia et al., 1995).
From the data reported here and previous studies (Bassnett and Beebe, 1992; Bassnett, 1995), it is now possible to describe the events that lead to lens fiber denucleation and offer a hypothesis regarding the mechanism of denucleation. It seems likely that a triggering event occurs in the center of the lens on or about E12. The triggering event could be related to the increasing size of the lens because the distance from the border of the OFZ to the lens surface remains approximately constant through embryonic development. As the volume of the tissue increases, diffusion-generated gradients of metabolites will be established within the lens as described by Bassnett et al. (1987). A drop in cytoplasmic pH, a fall in oxygen tension, or the build up of metabolic waste products are all candidates for the triggering event. We can speculate that the trigger may act at the level of mitochondrial or ER membranes, as these appear to be lost first. The loss of these major Ca2+ stores is expected to lead to an increase in cytosolic Ca2+ that may, in turn, mediate later events in the denucleation program. At this point, the nuclear lamina is broken down, perhaps after phosphorylation of lamin protein substrates. The breakdown of the nuclear envelope that occurs during denucleation may be similar to that which occurs during mitosis, where the disassembly of the nuclear lamina is known to result from the direct phosphorylation of lamins by the cdc2/cyclin B complex (Nigg, 1995). In this regard, it is interesting to note that Gao et al. (1995) have demonstrated the presence of both cyclin B and the cyclin-dependent kinase, p34cdc2, in E15 lens fiber cells. After the breakdown of the nuclear envelope in the lens fiber cells, there is a delay before overt DNA breakdown occurs. From a knowledge of the growth rate of the OFZ (Bassnett and Beebe, 1992), we can estimate that this delay is about 2 or 3 d. There are no obvious changes in nuclear morphology during this period, but perhaps the structure of the chromatin is undergoing transformation because, after this time, extensive DNA damage is apparent. The fact that DNA degradation was not observed until after the dissolution of the nuclear membrane raises the possibility that the nucleus may be susceptible to invasion by cytoplasmic nucleases or factors that can activate previously quiescent nuclear enzymes. Further experiments, where nuclei isolated from one region of the lens are mixed with cytoplasmic extracts from another, may help clarify this point.
Acknowledgments
We thank Drs. Andley, Beebe, and Petrash for their comments on the manuscript.
These studies were supported in part by National Institutes of Health grants RO1 EY09852 and EY02687 (Core Grant for Vision Research) and an unrestricted grant to the Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness, Inc.
Abbreviations used in this paper
- CIAP
calf intestinal alkaline phosphatase
- DIC
differential interference contrast
- DiOC6
3,3′-dihexyloxacarbocyanine iodide
- E
embryonic day
- OFZ
organelle-free zone
- PDI
protein disulfide isomerase
- TdT
terminal deoxynucleotidyl transferase
Footnotes
Address all correspondence to Steven Bassnett, Ph.D., Department of Ophthalmology and Visual Sciences, Washington University Medical School, 660 S. Euclid Avenue, Campus Box 8096, St. Louis, MO 63110-1093. Tel.: (314) 362-1604. Fax: (314) 747-1405. E-mail: Bassnetts@am.seer.wustl.edu
References
- Appleby DW, Modak SP. DNA degradation in terminally differentiating lens fiber cells from chick embryos. Proc Natl Acad Sci USA. 1977;4:5579–5583. doi: 10.1073/pnas.74.12.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruti C, Chaudun E, De Maria A, Courtois Y, Counis M-F. Characterization of eye-lens DNases: long term persistence of activity in post apoptotic lens fiber cells. Cell Death Differ. 1995;2:47–56. [PubMed] [Google Scholar]
- Barry MA, Eastman A. Identification of deoxyribonuclease II as an endonuclease involved in apoptosis. Arch Biochem Biophys. 1993;300:440–450. doi: 10.1006/abbi.1993.1060. [DOI] [PubMed] [Google Scholar]
- Bassnett S. Mitochondrial dynamics in differentiating fiber cells of the mammalian lens. Curr Eye Res. 1992;11:1227–1232. doi: 10.3109/02713689208999548. [DOI] [PubMed] [Google Scholar]
- Bassnett S. The fate of the Golgi apparatus and the endoplasmic reticulum during lens fiber cell differentiation. Invest Ophthalmol Visual Sci. 1995;36:1783–1803. [PubMed] [Google Scholar]
- Bassnett S, Beebe DC. Coincident loss of mitochondria and nuclei during lens fiber cell differentiation. Dev Dyn. 1992;195:85–93. doi: 10.1002/aja.1001940202. [DOI] [PubMed] [Google Scholar]
- Bassnett, S., and G. Duncan. 1986. Variation of pH with depth in the rat lens measured by double-barrelled ion sensitive microelectrodes. In The Lens: Transparency and Cataract: Proceedings of the EURAGE/BBS Symposium. G. Duncan, editor. Eurage, Rijswijk, The Netherlands. 77–85.
- Bassnett S, Shiels A. Mutations in the founder of the MIP gene family underlie cataract development in the mouse. Nat Genet. 1996;12:212–215. doi: 10.1038/ng0296-212. [DOI] [PubMed] [Google Scholar]
- Bassnett S, Croghan PC, Duncan G. Diffusion of lactate and its role in determining intracellular pH in the lens of the eye. Exp Eye Res. 1987;44:143–147. doi: 10.1016/s0014-4835(87)80032-5. [DOI] [PubMed] [Google Scholar]
- Bradley MO, Kohn KW. X-ray induced DNA double strand break production and repair in mammalian cells as measured by neutral filter elution. Nucl Acids Res. 1979;7:793–804. doi: 10.1093/nar/7.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch W, Paffe S, Putz B, Barthel G, Schulte-Hermann R. Determination of the length of the histological stages of apoptosis in normal liver and in altered hepatic foci of rats. Carcinogenesis. 1990;11:847–853. doi: 10.1093/carcin/11.5.847. [DOI] [PubMed] [Google Scholar]
- Chaudun E, Arruti C, Courtois Y, Ferrag F, Jeanny JC, Patel BN, Skidmore C, Torriglia A, Counis MF. DNA strand breakage during physiological apoptosis of the embryonic chick lens: free 3′OH end single strand breaks do not accumulate even in the presence of a cation-independent deoxyribonuclease. J Cell Physiol. 1994;158:354–364. doi: 10.1002/jcp.1041580218. [DOI] [PubMed] [Google Scholar]
- Chow RL, Roux GD, Roghani M, Palmer MA, Rifkin DB, Moscatelli DA, Lang RA. FGF suppresses apoptosis and induces differentiation of fiber cells in the mouse lens. Development (Camb) 1995;121:4383–4393. doi: 10.1242/dev.121.12.4383. [DOI] [PubMed] [Google Scholar]
- Cossarizza A, Kalashnikova G, Grassilli E, Chiappelli F, Salvioli S, Capri M, Barbieri D, Troiano L, Monti D, Franceschi C. Mitochondrial modifications during rat thymocyte apoptosis: a study at the single cell level. Exp Cell Res. 1994;214:323–330. doi: 10.1006/excr.1994.1264. [DOI] [PubMed] [Google Scholar]
- Counis MF, David JC, Chaudun E, Carre D. DNA polymerase, DNA ligase, and thymidine kinase activity in chicken lens, related to DNA X-ray lesion repair. Differentiation. 1981;20:188–195. doi: 10.1111/j.1432-0436.1981.tb01175.x. [DOI] [PubMed] [Google Scholar]
- Counis MF, Chaudun E, Allinquant B, Muel AS, Sanwal M, Skidmore C, Courtois Y. The lens: a model for chromatin degradation studies in terminally differentiating cells. Int J Biochem. 1989a;21:235–242. doi: 10.1016/0020-711x(89)90181-x. [DOI] [PubMed] [Google Scholar]
- Counis MF, Chaudun E, Courtois Y, Allinquant B. Lens fiber differentiation correlated with activation of two different DNAases in lens embryonic cells. Cell Differ Dev. 1989b;27:137–146. doi: 10.1016/0922-3371(89)90743-0. [DOI] [PubMed] [Google Scholar]
- Counis MF, Chaudun E, Courtois Y, Allinquant B. DNAase activities in embryonic chicken lens: in epithelial cells or in differentiating fibers where chromatin is progressively cleaved. Biol Cell. 1991;72:231–238. doi: 10.1111/j.1768-322x.1991.tb03020.x. [DOI] [PubMed] [Google Scholar]
- Fromm L, Shawlot W, Gunning K, Butel JS, Overbeek PA. The retinoblastoma protein-binding region of simian virus 40 large T antigen alters cell cycle regulation in lenses of transgenic mice. Mol Cell Biol. 1994;14:6743–6754. doi: 10.1128/mcb.14.10.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao CY, Bassnett S, Zelenka PS. Cyclin B, p34cdc2, and H1-kinase activity in terminally differentiating lens fiber cells. Dev Biol. 1995;169:185–194. doi: 10.1006/dbio.1995.1136. [DOI] [PubMed] [Google Scholar]
- Hess JF, Fitzgerald P. Lack of DNase I mRNA sequences in murine lenses. Mol Vis. 1996;2:8. [PubMed] [Google Scholar]
- Kerr JFR, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman NJ, Spector A. DNA single strand breaks in human lens epithelial cells from patients with cataract. Curr Eye Res. 1993;12:423–431. doi: 10.3109/02713689309024624. [DOI] [PubMed] [Google Scholar]
- Kuwabara T, Imaizumi M. Denucleation process of the lens. Invest Ophthalmol Vis Sci. 1974;13:973–981. [PubMed] [Google Scholar]
- Liao T-H. The subunit structure and active site sequence of porcine spleen deoxyribonuclease. J Biol Chem. 1985;260:10708–10713. [PubMed] [Google Scholar]
- Modak S, Bollum FJ. Terminal lens cell differentiation. III. Initiator activity of DNA during nuclear degeneration. Exp Cell Res. 1970;62:421–432. doi: 10.1016/0014-4827(70)90573-2. [DOI] [PubMed] [Google Scholar]
- Modak SP, Bollum FJ. In situ detection of single strand breaks in DNA of fixed cell nuclei. Experientia (Basel) 1971;27:439. [Google Scholar]
- Modak SP, Bollum FJ. Detection and measurement of singlestrand breaks in nuclear DNA in fixed lens sections. Exp Cell Res. 1972;75:307–313. doi: 10.1016/0014-4827(72)90434-x. [DOI] [PubMed] [Google Scholar]
- Modak SP, Perdue SW. Terminal lens cell differentiation I. Histological and microspectrophotometric analysis of nuclear degeneration. Exp Cell Res. 1970;59:43–56. doi: 10.1016/0014-4827(70)90622-1. [DOI] [PubMed] [Google Scholar]
- Montague JW, Gaido ML, Frye C, Cidlowski JA. A calcium-dependent nuclease from apoptotic rat thymocytes is homologous with cyclophilin. Recombinant cyclophilins A, B, and C have nuclease activity. J Biol Chem. 1994;269:18877–18880. [PubMed] [Google Scholar]
- Morgenbesser SD, Williams BO, Jacks T, DePinho RA. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature (Lond) 1994;371:72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- Muel AS, Chaudun E, Courtois Y, Modak SP, Counis MF. Nuclear endogenous Ca2+-dependent endodeoxyribonuclease in differentiating chick embryonic lens fibers. J Cell Physiol. 1986;127:167–174. doi: 10.1002/jcp.1041270120. [DOI] [PubMed] [Google Scholar]
- Muel AS, Laurent M, Chaudun E, Alterio J, Clayton R, Courtois Y, Counis MF. Increased sensitivity of various genes to endogenous DNase activity in terminal differentiating chick lens fibers. Mutat Res. 1989;219:157–164. doi: 10.1016/0921-8734(89)90010-6. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- Ostling O, Johanson KJ. Microelectrophoretic study of radiationinduced DNA damages in individual mammalian cells. Biochem Biophys Res Commun. 1984;123:291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- Pan H, Griep AE. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- Pan H, Griep AE. Temporally distinct patterns of p53-dependent and p53-independent apoptosis during mouse lens development. Genes Dev. 1995;9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. Lens differentiation in vertebrates. Differentiation. 1981;19:134–153. doi: 10.1111/j.1432-0436.1981.tb01141.x. [DOI] [PubMed] [Google Scholar]
- Peitsch MC, Polzar B, Stephan H, Crompton T, MacDonald HR, Mannherz HG, Tschopp J. Characterization of the endogenous deoxyribonuclease involved in nuclear degradation during apoptosis (programmed cell death) EMBO (Eur Mol Biol Organ) J. 1993;12:371–377. doi: 10.1002/j.1460-2075.1993.tb05666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C. Über den bau und die entwicklung der linse. III Teil: die linse der säugethiere. Ruckblick und schluss. Z Wiss Zool. 1899;67:1–138. [Google Scholar]
- Robinson ML, MacMillan-Crow LA, Thompson JA, Overbeek P. Expression of a truncated FGF receptor results in defective lens development in transgenic mice. Development (Camb) 1995;121:3959–3967. doi: 10.1242/dev.121.12.3959. [DOI] [PubMed] [Google Scholar]
- Sandilands A, Prescott AR, Carter JM, Hutcheson AM, Quinlan RA, Richards J, Fitzgerald PG. Vimentin and CP49/filensin form distinct networks in the lens which are independently modulated during lens fibre cell differentiation. J Cell Sci. 1995;108:1397–1406. doi: 10.1242/jcs.108.4.1397. [DOI] [PubMed] [Google Scholar]
- Sanwal M, Muel AS, Chaudun E, Courtois Y, Counis MF. Chromatin condensation and terminal differentiation process in embryonic chicken lens in vivo and in vitro. Exp Cell Res. 1986;167:429–439. doi: 10.1016/0014-4827(86)90183-7. [DOI] [PubMed] [Google Scholar]
- Schmid DS, Tite JP, Ruddle NH. DNA fragmentation: manifestation of target cell destruction mediated by cytotoxic T-cell lines, lymphotoxin-secreting helper T-cell clones, and cell-free lymphotoxin-containing supernatant. Proc Natl Acad Sci USA. 1986;83:1881–1885. doi: 10.1073/pnas.83.6.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S-R, Key ME, Kalra KL. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J Histochem Cytochem. 1991;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- Torriglia A, Chaudun E, Chany-Fornier F, Jeanny J-C, Courtois Y, Counis M-F. Involvement of Dnase II in nuclear degeneration during lens cell differentiation. J Biol Chem. 1995;270:28579–28585. doi: 10.1074/jbc.270.48.28579. [DOI] [PubMed] [Google Scholar]
- Vrensen GFJM, Graw J, DeWolf A. Nuclear breakdown during terminal differentiation of primary lens fibers in mice: a transmission electron microscopic study. Exp Eye Res. 1991;52:647–659. doi: 10.1016/0014-4835(91)90017-9. [DOI] [PubMed] [Google Scholar]
- Watt JA, Pike CJ, Walencewicz-Wasserman AJ, Cotman CW. Ultrastructural analysis of β-amyloid-induced apoptosis in cultured hippocampal neurons. Brain Res. 1994;661:147–156. doi: 10.1016/0006-8993(94)91191-6. [DOI] [PubMed] [Google Scholar]
- Weis M. Cellular events in Fas/APO-1-mediated apoptosis in JURKAT T lymphocytes. Exp Cell Res. 1995;219:699–708. doi: 10.1006/excr.1995.1281. [DOI] [PubMed] [Google Scholar]
- Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature (Lond) 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman LE, Font RL. Congenital malformations of the eye: some recent advances in knowledge of the pathogenesis and histopathological characteristics. J Am Med Assoc. 1966;196:684–692. [Google Scholar]