Abstract
Skin wound healing depends on cell migration and extracellular matrix remodeling. Both processes, which are necessary for reepithelization and restoration of the underlying connective tissue, are believed to involve the action of extracellular proteinases. We screened cDNA libraries and we found that six matrix metalloproteinase genes were highly expressed during rat skin wound healing. They were namely those of stromelysin 1, stromelysin 3, collagenase 3, gelatinase A (GelA), gelatinase B, and membrane type-1 matrix metalloproteinase (MT1-MMP). The expression kinetics of these MMP genes, the tissue distribution of their transcripts, the results of cotransfection experiments in COS-1 cells, and zymographic analyses performed using microdissected rat wound tissues support the possibility that during cutaneous wound healing pro-GelA and pro-gelatinase B are activated by MT1-MMP and stromelysin 1, respectively. Since MT1-MMP has been demonstrated to be a membrane-associated protein (Sato, H., T. Takino, Y. Okada, J. Cao, A. Shinagawa, E. Yamamoto, and M. Seiki. 1994. Nature (Lond.). 370: 61–65), our finding that GelA and MT1-MMP transcripts were expressed in stromal cells exhibiting a similar tissue distribution suggests that MT1-MMP activates pro-GelA at the stromal cell surface. This possibility is further supported by our observation that the processing of proGelA to its mature form correlated to the detection of MT1-MMP in cell membranes of rat fibroblasts expressing the MT1-MMP and GelA genes. These observations, together with the detection of high levels of the mature GelA form in the granulation tissue but not in the regenerating epidermis, suggest that MT1-MMP and GelA contribute to the restoration of connective tissue during rat skin wound healing.
Skin wound healing is a complex process characterized by reepithelization and restoration of the underlying connective tissue. During this process, keratinocytes, endothelial cells, fibroblasts, and inflammatory cells proliferate and/or migrate to the site of injury, interacting with each other and with extracellular matrices (ECMs)1 (Gailit and Clark, 1994).
The migration of cells and the remodeling of tissues during wound healing require the controlled degradation of the ECM and the activation or release of growth factors (Vassalli and Saurat, 1996). These processes are achieved by extracellular proteases, particularly those belonging to the serine protease and matrix metalloproteinase (MMP) families. It has been demonstrated that urokinase-deficient mice develop nonhealing skin ulcerations (Carmeliet and Collen, 1995) and that plasminogen-deficient mice have a marked defect in the closure of skin wounds (Rømer et al., 1996). However, the exact contribution of MMPs to wound healing has not been well established yet. The expression of gelatinase A (GelA) was localized in the connective tissue and that of gelatinase B (GelB) in the migrating epithelial sheet during human skin wound healing (Salo et al., 1994). In healing human burn wounds, interstitial collagenase (Col1) RNA has been detected in epithelial cells, hair follicles, and eccrine sweat structures (Stricklin et al., 1993). Furthermore, in vitro studies indicate that Col1 may play a role in the formation of new blood vessels (Fisher et al., 1994).
When the present study was initiated, ten human MMPs were known (Col1-3, stromelysin1-3 [ST1-3], GelA, GelB, matrilysin, and the macrophage metalloelastase; reviewed in Birkedal-Hansen, 1995), while only four rat MMPs (ST1, Matrisian et al., 1986a ; ST2, Breathnach et al., 1987; Col3 [According to Knäuper et al. (1996), the rat collagenase identified by Quinn et al. (1990) should be considered as the rat counterpart of collagenase 3], Quinn et al., 1990; GelA, Marti et al., 1993) had been identified. To gain insight into the contribution of MMPs to wound healing, we designed a two-step procedure for screening cDNA libraries, which allowed the identification of MMPs expressed highly during rat skin wound healing. We thus identified six rat MMP cDNAs, including ST1, ST3, Col3, GelA, GelB, and a novel member of the MMP family. The latter one was found to exhibit a high level of homology with a human MMP cloned by Sato et al. (1994), and now known as membrane-type-1 MMP (MT1-MMP; Sato and Seiki, 1996).
MT1-MMP is the first member of a new MMP subgroup characterized by its localization at the cell surface, where it is anchored through a transmembrane domain located at the COOH-terminal portion of the molecule. Three additional MT-MMPs (MT2-MMP, Will and Hinzmann, 1995; MT3-MMP [Although the MT-MMP described by Takino et al. (1995) was initially termed MT2-MMP, this MMP should be now referred as MT3-MMP and that found by Will and Hinzmann (1995) as MT2-MMP] Takino et al., 1995; MT4-MMP, Puente et al., 1996) have so far been described. While the function of MT4-MMP is still unknown, the three other MT-MMPs have been shown to be cell surface activators of pro-GelA. MT1-MMP was predominantly detected in malignant cells of lung and gastric carcinomas (Sato et al., 1994; Nomura et al., 1995; Tokuraku et al., 1995). This has led to the proposition that MT1-MMP activates pro-gelA at the cancer cell surface (Sato and Seiki, 1996). However, MT1-MMP transcripts were predominantly detected in fibroblastic cells of colon, breast, and head and neck carcinomas (Okada et al., 1995a ; Heppner et al., 1996). These observations suggest that pro-GelA activation by MT1-MMP also occurs at the stromal cell surface, which is supported by the specific expression of MT1-MMP in microglial cells of brain tissues (Yamada et al., 1995). Furthermore, MT1-MMP gene expression has been recently demonstrated in mesenchymal cells of mouse embryos (Kinoh et al., 1996), indicating that this MMP could contribute to pro-GelA activation in physiological conditions.
In the present study, we show that MT1-MMP and GelA RNAs are both expressed in stromal cells of rat skin wounds, and that high levels of the mature GelA form are detected in the wound stroma, but not in the regenerating epithelium. These findings suggest that during rat skin wound healing, GelA is activated at the stromal cell surface by MT1-MMP and contributes to the remodeling processes implicated in the restoration of the connective tissue.
Materials and Methods
Animals
Pathogen- and virus-free 10-wk-old female Wistar rats were obtained from IFFA-CREDO (L'Arbresle, France). Animals were maintained under the guidelines of the Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC). Rats were fed with “D 04” (Usine Alimentation Rationnelle, Epinay-sur-Orge, France) and tap water ad libitum. Rat dorsal skin was shaved and cleaned with isopropyl alcohol under anesthesia, and the skin was linearly incised (3-cm long) up to the level of the subcutaneous adipose tissue. Incisions were made on either side of the midline using a #10 sterile surgical scalpel. A sterile dressing was then placed circumferentially around the trunk in order to protect against infection. On days 1, 3, 5, 7, 10, and 14 after wounding, the entire dorsal skin encompassing healing wounds was excised. For RNA and protein extractions, wound tissues were immediately frozen in liquid nitrogen. For RNA in situ hybridization and histopathological examination, the samples were embedded in OCT compound (Miles, Elkhart, IN), and frozen in liquid nitrogen. Adequate sampling was ensured by obtaining wound specimen from at least three separate animals for each time point.
Cell Lines
COS-1 cells (Amer. Type Culture Collection, Rockville, MD), and RAT1 and FR3T3 rat fibroblasts (Matrisian et al., 1985) were grown and maintained in DMEM supplemented with 5% or 10% FCS, or 10% calf serum, respectively. Con A (Sigma Chem. Co., St. Louis, MO) treatment of rat fibroblasts in culture was carried out in serum-free DMEM supplemented with 50 μg/ml Con A for 24 h (Overall and Sodek, 1990). After treatment, conditioned media and cells were harvested for protein analysis and RNA extraction.
RNA Extraction
Tissues were homogenized in 4 M guanidinium thiocyanate (Merck, Darmstadt, Germany). Total RNA was purified by ultracentrifugation (SW60 rotor, Beckman Instrs., Fullerton, CA, 35,000 rpm) through a 2.5-ml cushion of 5.7 M cesium chloride. Cells in culture were washed with PBS, and RNA was extracted using the single-step method of Chomczynski and Sacchi (1987).
Construction of a Rat Skin Wound cDNA Library and Cloning of cDNAs Encoding MT1-MMP
Poly(A)+RNA from rat skin wounds was purified using the Pharmacia purification kit (Uppsala, Sweden). The RNA thus isolated was used to construct a cDNA library with the Uni-ZAP™XR Vector kit (Stratagene, La Jolla, CA). This library was screened with size-selected (∼350–550 bp) RTPCR products. For the RT-PCR experiments, poly(A)+RNA from rat skin wounds was reverse-transcribed using an oligo (dT) primer, and the resulting cDNA was amplified using a set of degenerate primers corresponding to the cysteine-switch and zinc-binding domains of rat MMPs (5'-C(CT)N(AC)GNTGTGGN(AG)(AT)NCCNGA, and 5′-(CT)C(AG) A(AG)(CT)TC (AG)TGNGC(AT)GC(AC)AC, respectively). Additional screening was conducted using cDNA probes (see Results) encoding rat GelA (nucleotides 482-2306; accession number No. U65656), GelB (nucleotides 517-2392), ST1 (nucleotides 418-1771), ST3 (nucleotides 644-1810), and Col3 (nucleotides 1387-2606). The clones which were only recognized by the RT-PCR probe, were plaque-purified and used for plasmid rescue. After cross-hybridization experiments, all independent clones were sequenced from their 5′-terminus. The full nucleotide sequence of rat MT1MMP cDNA was determined on both strands as previously described (Okada et al., 1995a ). Nucleotide and amino acid sequences were analyzed using the sequence analysis software of the Wisconsin program package.
Cloning of cDNAs Encoding Rat TIMP2 and TIMP3
Rat tissue inhibitor of metalloproteinase type-2 (TIMP2) (Santoro et al., 1994) and TIMP3 (Wu and Moses, 1996) cDNA fragments containing the complete open reading frame sequence were obtained by PCR amplification from a previously described skin wound library (Okada et al., 1994) and a 13.5-d placenta library in the λ ZAPII vector, respectively. The PCR products were cloned into the pBluescript vector and sequenced.
RNA Analyses
For Northern blot analysis, 10 μg of total RNA from rat tissues was electrophoresed on formaldehyde-agarose (1%) gels and transferred to Hybond-N membranes (Amersham, Buckinghamshire, UK), according to the supplier's instructions. Hybridizations were carried out with 32P-labeled cDNA probes in 50% formamide at 42°C. The cDNA probes used for these hybridizations were identical to those used for the differential screening of the skin wound cDNA library. An additional probe used for hybridization was rat MT1-MMP cDNA (nucleotides 911-2410).
For in situ hybridization, tissue sections from rat normal skin and skin wounds were collected as described above. Sections were cut perpendicularly to the cutaneous incision. 35S-labeled RNA sense and antisense probes were synthesized from DNA templates identical to those used for Northern blot analyses, using either the T3 or T7 polymerase. Probe length was reduced to an average size of 150 nucleotides by limited alkaline hydrolysis before hybridization as previously described (Okada et al., 1995a ). Frozen sections were fixed with 4% paraformaldehyde, dehydrated, and hybridized with 35S-labeled RNA probes. After washing, autoradiography was performed using NTB2 emulsion (Kodak, Rochester, NY). Slides were then developed and counterstained with toluidine blue.
Construction of Expression Plasmids for Rat MMPs and TIMPs
To express rat MT1-MMP in E. coli, a cDNA fragment encoding the putative catalytic domain and the hemopexin-like domain (amino acids 112508) was inserted into the pET15b vector containing an NH2-terminal 6-histidine tag (Novagen, Madison, WI). For expression in COS-1 cells, cDNA fragments containing the complete open reading frame sequence of rat MT1-MMP, GelA, GelB, ST1, ST3, TIMP1 (Okada et al., 1994), TIMP2, and TIMP3, were inserted into the pTL1 expression vector as previously described (Okada et al., 1995b ). The resulting plasmids, pET15bMT1-MMP (MT1-MMP, amino acids 112-508), pTL1MT1-MMP (pro-MT1-MMP), pTL1GelA (pro-GelA), pTL1GelB (pro-GelB), pTL1ST1 (pro-ST1), pTL1ST3 (pro-ST3), pTL1TIMP1 (TIMP1), pTL1TIMP2 (TIMP2), and pTL1TIMP3 (TIMP3) were verified by restriction enzyme digestion and DNA sequencing.
Transient Transfection into COS-1 Cells
pTL1 expression plasmids for rat MMPs and TIMPs,and the control pTL1 vector alone were transiently transfected into COS-1 cells using the calcium phosphate procedure. 5 × 105 COS-1 cells per 10-cm diameter culture dish were grown in DMEM supplemented with 5% FCS for 16 h, and transfected with 3 μg of each expression plasmid and 12 μg of carrier DNA (pBluescript vector) in fresh culture medium. After a 1-d incubation, cells were washed with DMEM and incubated in 3 ml of serum-free DMEM for another day.
Gelatin Zymography
Zymography was performed as previously described (Okada et al., 1995b ), using conditioned media or rat tissues. 20 μl of each conditioned medium was mixed with a same volume of Laemmli's 2× SDS sample buffer in the absence of a reducing agent, and incubated for 30 min at 22°C. The samples were electrophoresed on 8%-polyacrylamide gels containing 0.2% gelatin. After electrophoresis, gels were soaked in 50 mM Tris-HCl (pH 7.6) containing 2.5% Triton X-100 for 30 min, and incubated with 50 mM Tris-HCl (pH 7.6) containing 150 mM NaCl, 10 mM CaCl2, and 0.02% Brij-35, at 37°C for 16 h. Gels were subsequently stained with 0.5% Coomassie brilliant blue R-250. Protein molecular standards were purchased from Bio-Rad (Hercules, CA).
Normal rat skin epidermis and dermis, and proliferative epidermis or granulation tissue from rat skin wounds were carefully microdissected under a light microscope from 30-μm thick frozen sections. After microdissection, individual samples were suspended in Laemmli's 2× SDS sample buffer (40 μl/mg of fresh tissue). After a 30-min incubation at 22°C, the extracted proteins were analyzed by gelatin zymography as described above.
Western Blot Analysis
Anti-rat MT1-MMP mAb (1MMP-1C1) was obtained by immunizing Balb/c mice with recombinant rat MT1-MMP (amino acids 112-508) purified from E. coli inclusion bodies. The antigen was purified by Ni2+-NTA-Agarose (Qiagen Inc., Chatsworth, CA) chromatography according to the manufacturer's instruction. The specificity of mAb 1MMP-1C1 for MT1-MMP was demonstrated by immunocytochemical analysis of COS-1 cells transfected with cDNAs for MT1-MMP, MT2-MMP (Will and Hinzmann, 1995), or MT3-MMP (Takino et al., 1995).
Subconfluent transfected COS-1 cells, and RAT1 and FR3T3 rat fibroblasts incubated with serum-free DMEM in the absence or presence of 50 μg/ml Con A for 24 h, were scraped and lysed in PBS containing 1% NP40 and 5 mM EDTA (100 μl/ 10-cm diam culture plate). After centrifugation at 4,000 g for 15 min, supernatants corresponding to crude cell membrane extracts were collected and used for Western blot analysis (Okada et al., 1997). Proteins were separated by electrophoresis performed on 10%-polyacrylamide gels under reducing conditions. The proteins were then electrophoretically transferred to a nylon membrane (Immobilon-P, Millipore, Bedford, MA). After treatment with a blocking solution, the nylon membrane was incubated with anti-rat MT1-MMP mouse mAb 1MMP-1C1, diluted 1/1,000 in PBS containing 2% BSA. The nylon membrane was washed and incubated with peroxidase-conjugated goat anti– mouse IgG antibody (Jackson ImmunoResearch Laboratory, West Grove, PA). After washing in PBS containing 0.05% Tween-20, proteins were detected using a chemiluminescence procedure (Renaissance, DuPont, Boston, MA).
Results
Cloning of Rat MT1-MMP from a Skin Wound cDNA Library
To investigate the contribution of MMPs to skin wound healing, we devised a two-step procedure to identify MMP genes expressed during this process. As a first approach, screening of a 5-d rat skin wound library constructed in the λgt10 vector (Okada et al., 1994) with six human MMP cDNA probes (ST2, ST3, Col1, GelA, GelB, and matrilysin), led to the identification of four rat MMP cDNAs (ST1, ST3, GelA, and GelB) (Okada et al., 1995b , 1997, and Okada, A., unpublished results). However, ST2, matrilysin, and the rat homologue of human Col1 were not found in the library. A nearly full-length cDNA for rat Col3 (nucleotides 145-2583) was obtained by screening the library with a partial rat Col3 cDNA fragment (nucleotides 865-1262, Quinn et al., 1990).
The next step was to determine whether other MMPs were expressed during rat skin wound healing. We constructed a second cDNA library in the Uni-ZAP™XR vector (Stratagene, La Jolla, CA) using RNA prepared from healing wounds on days 3 and 5 after cutaneous incision, which corresponded to the period of concomitant expression of the MMP genes already identified (Fig. 1 A). For screening this second library, a probe was designed in such a way that any known or novel members of the MMP family would be identified. We amplified 3- and 5-d rat skin wound RNA by RT-PCR, using a set of degenerate primers derived from nucleotide sequences corresponding to the highly conserved cysteine-switch and zinc-binding domains of the six rat MMPs known at that time (ST3, GelA, GelB, ST1, ST2, and Col3) (Fig. 2).
Figure 1.
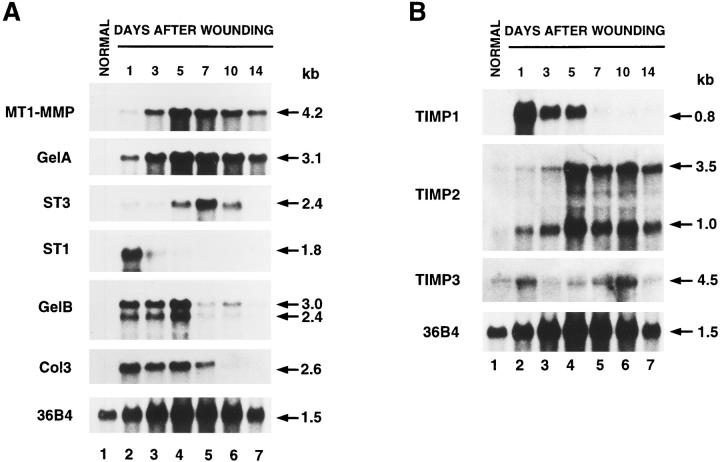
Northern blot analysis of MMP and TIMP RNAs during rat skin wound healing. Total RNA (10 μg) from normal skin (lanes 1) and skin wounds on days 1, 3, 5, 7, 10, and 14 after cutaneous incision (lanes 2–7), were electrophoresed, transferred to nylon membranes, and hybridized with 32P-labeled cDNA probes for rat MT1-MMP, GelA, ST3, ST1, GelB, Col3 (A); TIMP1, TIMP2, TIMP3 (B). Blots were reprobed with the 36B4 cDNA used as a loading control (Masiakowski et al., 1982).
Figure 2.

Amino acid sequence alignments of rat MMPs. Amino acids are represented using the one-letter code. The predicted amino acid sequences of MT1-MMP, ST3, GelA, GelB, and ST1 were derived from cDNAs cloned from two distinct rat skin wound libraries (Okada et al., 1994, and the present study). One amino acid residue of ST1 (isoleucine 189) differs from the corresponding residue (threonine) in the sequence reported by Matrisian et al. (1985). The sequences of ST2, Col3, and matrilysin were obtained from the PIR-protein data bank. These sequences were aligned using the CLUSTAL program of the Wisconsin program package. Identical amino acid residues in all MMPs are indicated by asterisks below the sequences. The cysteine-switch region (P-/LRCGV/NPD), and the zinc-binding domain (XVAXHXHEL/FGHXL/MGLXHS/T) are in the boxed regions. The two regions selected for designing the degenerated oligoprimers used for RT-PCR amplification are indicated by arrows above the sequences. The putative transmembrane segment (amino acid 539563) of rat MT1-MMP is overlined.
Sixty thousand plaques of the ZAP library were screened with the RT-PCR probe. After dehybridization, the same membranes were hybridized with the five rat MMP cDNA probes (ST3, GelA, GelB, ST1, and Col3) previously isolated. Among the clones that were detected selectively by the RT-PCR probe and not by the MMP cDNA probes, we identified 10 cDNAs which had been previously described that did not belong to the MMP family, and 11 novel cDNA sequences (data not shown). One of them was found to correspond to a putative novel member of the MMP family, and by sequence homology we proposed that it represented the rat homologue of human MT1MMP (Sato et al., 1994; Okada et al., 1995a ). Although three other human MMPs containing a transmembrane domain have since been described (Will and Hinzmann, 1995; Takino et al., 1995; Puente et al., 1996), the rat MMP that we have identified exhibits the highest sequence homology with human MT1-MMP until now (data not shown).
Expression of MMP RNAs during Skin Wound Healing
MT1-MMP RNA was found to be expressed in most normal adult rat tissues (data not shown). However, higher levels of MT1-MMP RNA were detected during rat skin wound healing. MT1-MMP RNA expression was maximal on day 5 after cutaneous incision (Fig. 1 A). This corresponded to a period of intense stromal activity, filling of granulation tissue in the incisional space, maximal neovascularization, and when collagen fibers were present at the incision margins where the scar tissue was being formed. The expression kinetics of GelA RNA was identical to that of MT1-MMP (Fig. 1 A). However, ST3 RNA was highly expressed only on day 5 and reached a peak on day 7. The maximal expression of ST1 RNA was detected on day 1 after cutaneous incision. GelB and Col3 RNAs were also highly expressed on day 1, and maintained high expression levels until day 5.
We then examined MMP transcript distribution by in situ hybridization on normal skin sections and skin wound sections from days 1–14 after cutaneous incision. Each wound was examined at its extreme and median portions. The results of analyses performed on days 3 and 5 are presented in Fig. 3. On day 3, various stages of granulation tissue and immature capillary lumens were observed, together with spurs of epithelial cells migrating from both edges of wounds beneath the surface scab (Fig. 3 A and data not shown). MT1-MMP transcripts were detected in cells of the granulation tissue and those of the superficial dermis juxtaposed to the proliferative epithelial cell layer (Fig. 3 B). The transcripts were also observed in stromal cells surrounding hair follicles and especially in the dermal papilla. Most cells expressing MT1-MMP transcripts were of the fibroblastic type, thus likely corresponding to fibroblasts or myofibroblasts. However, some cells were round or oval, suggesting that other types of stromal cells, including endothelial cells or macrophages, may also express the MT1-MMP gene. While it is clear that additional studies are required to better define the precise nature of these stromal cells, MT1-MMP transcripts could not be detected in any epithelial cell. The localization of GelA transcripts was similar to that observed for MT1-MMP, although the expression levels were higher for GelA than for MT1MMP (Fig. 3 C). ST3 transcripts were undetectable, except in a few stromal cells surrounding hair follicles (Fig. 3 D). ST1 transcripts were detected in some proliferative epithelial basal cells, and in a few stromal cells juxtaposed to the proliferative epithelial layer (Fig. 3 E). GelB and Col3 RNAs were highly expressed in the epithelial basal cell layer, but not in the dermis (Fig. 3, F and G). On day 5, though granulation tissue was still observed in the median portion of incision (data not shown), reepithelization had been completed at the wound extremities, where thickened epidermis, scar tissue and tissue contraction were observed (Fig. 3 H). MT1-MMP, GelA, and ST3 transcripts were detected in cells of the scar tissue, below the thickened epithelial cell layer (Fig. 3, I–K). The distribution of MT1-MMP transcripts (Fig. 3 I) was more narrow than that of GelA (Fig. 3 J), but broader than that of ST3 (Fig. 3 K). MT1-MMP, GelA, and ST3 transcripts were not detected in epithelial cells. ST1 transcripts were not detected (Fig. 3 L). GelB and Col3 transcripts were intensely expressed in some basal cells of the thickened epidermis, and in some cells of the hair follicle sheath (Fig. 3, M and N).
Figure 3.
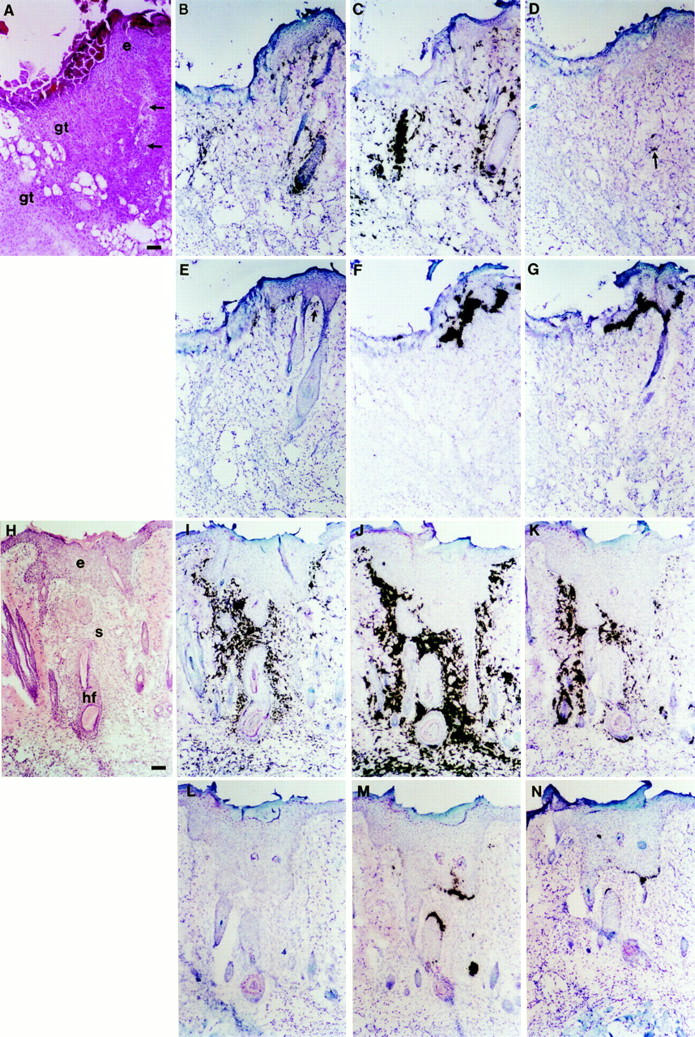
In situ hybridization of rat MMP RNAs in cutaneous wounds. Serial frozen sections of rat skin wounds on day 3 (A–G), or day 5 (H–N) after cutaneous incision, were hybridized with 35S-labeled antisense RNA probes derived from cDNA templates for rat MT1MMP (B and I), GelA (C and J), ST3 (D and K), ST1 (E and L), GelB (F and M), Col3 (G and N), or stained with hematoxylin and eosin (A and H). Note that the hair follicle which is observed on sections B and C, is only partially visible on A (arrows). In D and E, arrows indicate transcripts expressed in stromal cells surrounding a hair follicle and juxtaposed to the proliferative epithelial cell layer, respectively. No signal above background could be detected with the corresponding 35S-labeled sense RNA probes (data not shown). gt, granulation tissue; e, epithelial layer; hf, hair follicle; s, scar tissue. Bar, 100 μm.
MMP Interactions during Skin Wound Healing
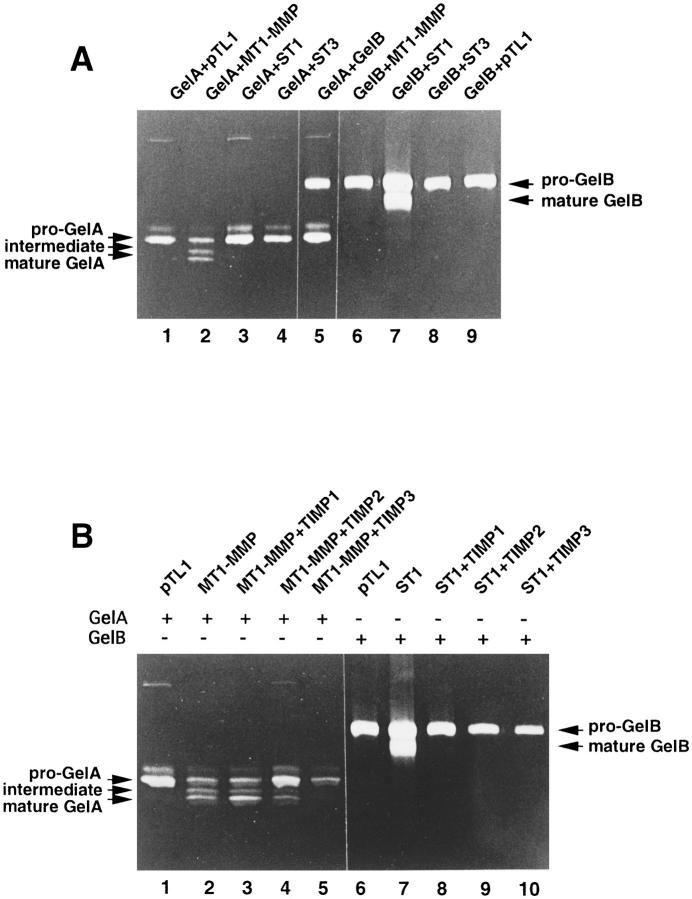
Expression vectors encoding rat MT1-MMP, GelA, GelB, ST1, and ST3, were transiently transfected into COS-1 cells, which do not produce any detectable levels of these MMPs endogenously (Okada et al., 1995b , 1997, and data not shown). The expression plasmids were transfected in pairs into COS-1 cells. Conditioned media were analyzed by gelatin zymography for GelA and GelB production. As shown in Fig. 4 A, pro-GelA was selectively activated by MT1-MMP, and pro-GelB by ST1 only. Other combinations which could not be analyzed by gelatin zymography, were investigated by Western blot analyses and immunoprecipitation, using specific antibodies against ST1 (Matrisian et al., 1986b ), ST3 (mAb 5ST-4C10, Santavicca et al., 1995), and MT1-MMP (mAb 1MMP-1C1). None of the other pairs led to activation of the MMP proform (data not shown). Furthermore, MT1-MMP was found exclusively in crude cell membrane extracts and could not be detected in conditioned media (data not shown).
Figure 4.
Pro-GelA and proGelB activation by MT1MMP and ST1, in the absence or presence of TIMPs. (A) COS-1 cells were transiently cotransfected with expression plasmids for MMPs whose transcripts were found to be expressed during rat skin wound healing (Figs. 1 and 3), according to the following combinations: pTLlGelA and pTL1 (lane 1) or pTL1MT1MMP (lane 2) or pTL1ST1 (lane 3) or pTL1ST3 (lane 4) or pTL1GelB (lane 5); pTL1GelB and pTL1MT1MMP (lane 6) or pTL1ST1 (lane 7) or pTL1ST3 (lane 8) or pTL1 (lane 9). (B) COS-1 cells were transiently cotransfected with pTL1GelA and pTL1 (lane 1), or pTL1GelA and pTL1MT1-MMP (lanes 2–5), or pTL1GelB and pTL1 (lane 6), or pTL1GelB and pTL1ST1 (lanes 7–10), in the absence (lanes 1, 2, 6, and 7) or presence of one of the TIMP expression plasmids: pTL1TIMP1 (lanes 3 and 8), pTL1TIMP2 (lanes 4 and 9), pTL1TIMP3 (lanes 5 and 10). In A and B, 20 μl of each conditioned medium was analyzed by gelatin zymography.
The possibility that MT1-MMP interacts with GelA, and ST1 with GelB, respectively, during rat skin wound healing, appears consistent both with the expression kinetics of their RNAs (Fig. 1 A) and their expression patterns in wound tissues (Fig. 3). Thus, MT1-MMP and GelA transcripts were detected in stromal cells exhibiting a similar tissue distribution (Fig. 3, B, C, I, and J). In the case of ST1 and GelB, the expression patterns of their transcripts were not superimposable on day 5 after cutaneous incision (Fig. 3, L and M). However, ST1 and GelB transcripts were co-expressed in the proliferative epithelial basal cell layer on day 1 (data not shown) and to some extent on day 3 (Fig. 3, E and F), when the mature form of GelB was detected in wound tissues (see below). We also note that the GelB and Col3 genes exhibited comparable expression kinetics (Fig. 1 A) and distribution of transcripts in tissues (Fig. 3, F, G, M, and N), suggesting that GelB and Col3 could also interact during rat skin wound healing. However, we could not further evaluate this possibility in the present study, since we could not obtain any full-length rat Col3 cDNA for protein expression.
Pro-GelA Activation by MT1-MMP Is Regulated by TIMP2 and TIMP3
Activity of mature MMP forms and activation of proforms are regulated by their physiological inhibitors, the TIMPs (Stetler-Stevenson et al., 1996). In particular, TIMP2 has been shown to be both a more efficient inhibitor than TIMP1 in pro-GelA activation (Atkinson et al., 1995) and an active agent in promoting pro-GelA binding to MT1MMP (Strongin et al., 1995). The effect of TIMP3 on the activation of pro-GelA has not been reported yet. As shown in Fig. 1 B, TIMP1 RNA was not expressed in normal skin, while TIMP2 and TIMP3 RNAs were weakly expressed. The induction kinetics of TIMP1 RNA after cutaneous incision was comparable to that of ST1, while that of TIMP2 was identical to that of MT1-MMP and GelA (compare Fig. 1, A and B). However, the induction kinetics of TIMP3 RNA was different from that of the other TIMPs and the six MMPs examined. TIMP3 kinetics exhibited a biphasic pattern, with the highest RNA levels on day 1 after cutaneous incision, which corresponds to the onset of epithelial and mesenchymal cell migrations, and on day 10, when cell migration terminates.
To test the influence of TIMPs on pro-GelA activation by MT1-MMP, we performed triple transfection experiments in COS-1 cells. Different TIMPs and pro-GelA/ MT1-MMP expression plasmids were used for these triple transfections. Pro-GelA activation by MT1-MMP was not affected by the presence of TIMP1 (Fig. 4 B, lane 3) and only partially prevented by TIMP2 (Fig. 4 B, lane 4). However, TIMP3 was found to completely inhibit pro-GelA activation by MT1-MMP (Fig. 4 B, lane 5). In contrast to the observations made for GelA and MT1-MMP, pro-GelB activation by ST1 was similarly prevented by all the three TIMPs (Fig. 4 B, lanes 7–10). These observations strongly suggest that TIMP3 may be a physiological inhibitor of proGelA activation by MT1-MMP.
Pro-GelA Is Activated by MT1-MMP-Expressing Rat Fibroblasts
During rat skin wound healing, MT1-MMP and GelA transcripts were specifically found in wound stromal cells (Fig. 3, B, C, I, and J). This finding prompted us to examine MT1-MMP and GelA gene expression in two different rat fibroblast cell lines, RAT1 and FR3T3. Both cell lines were found to express high levels of MT1-MMP and GelA transcripts (Fig. 5 A, lanes 1 and 3). However, the MT1-MMP protein could not be detected either in crude cell membrane extracts or in cytosolic fractions by Western blot analysis (Fig. 5 B, lanes 1 and 3, and data not shown). Consistently, media conditioned by these cells contained mainly pro-GelA, as shown by gelatin zymography (Fig. 5 C, lanes 3 and 5). Since pro-GelA activation has been demonstrated to be induced by Con A treatment in human fibroblasts (Overall and Sodek, 1990), we then examined whether treatment with con A of these rat fibroblasts was associated with pro-GelA activation. Although both cell lines stimulated by Con A showed only a weak increase in MT1-MMP and GelA transcript levels (Fig. 5 A, lanes 2 and 4), the MT1-MMP protein could now be easily detected by Western blot analysis of crude cell membrane fractions prepared from the Con A–treated cells (Fig. 5 B, lanes 2 and 4). Furthermore, as shown by gelatin zymography, the mature form of GelA was found to be the major GelA species represented in media conditioned by Con A–treated cells (Fig. 5 C, lanes 4 and 6).
Figure 5.
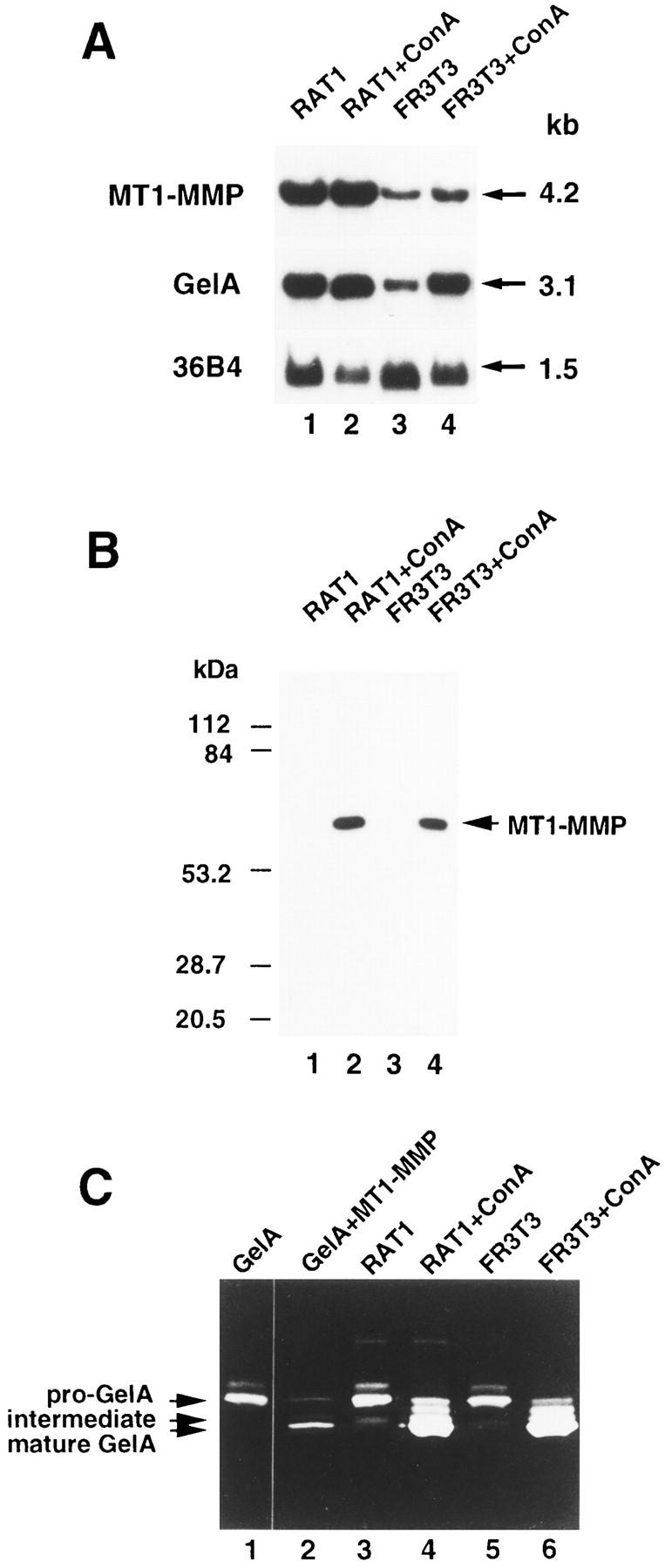
MT1-MMP gene expression in rat fibroblasts and proGelA activation in the presence of Con A. (A) Northern blot analysis. 10 μg of total RNA obtained from RAT1 (lanes 1 and 2) or FR3T3 (lanes 3 and 4) rat fibroblasts cultured in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of Con A (50 μg/ml) were used in each lane. Blots were hybridized with 32P-labeled rat MT1-MMP and GelA cDNA probes. Blots were reprobed with the 36B4 cDNA used as a loading control (Masiakowski et al., 1982). (B) Western blot analysis. Crude cell membrane extracts corresponding to half a culture plate of subconfluent RAT1 (lanes 1 and 2) or FR3T3 (lanes 3 and 4) fibroblasts cultured in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of Con A (50 μg/ml) were analyzed. Proteins were detected using anti-rat MT1-MMP mouse monoclonal antibody, 1MMP-1C1. (C) Gelatin zymography. 20 μl of medium conditioned by COS-1 cells transiently transfected with pTL1GelA (lane 1), or pTL1GelA and pTL1MT1-MMP (lane 2), and 20 μl of medium conditioned by RAT1 (lanes 3 and 4) and FR3T3 (lanes 5 and 6) rat fibroblasts cultured in the absence (lanes 3 and 5) or presence (lanes 4 and 6) of Con A (50 μg/ml), were used.
Detection of High Levels of GelA and GelB Activities in the Granulation Tissue
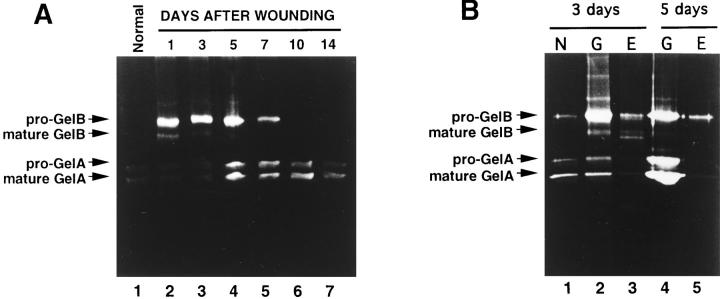
To directly evaluate pro-GelA activation during rat skin wound healing, we analyzed microdissected tissue samples from frozen sections by gelatin zymography. These samples were collected at different times after cutaneous incision. In wound tissues containing both the stroma and epithelium, protein species corresponding to pro-GelA/GelA were detected from days 1–14, and pro-GelB or pro-GelB/ GelB were detected from days 1–7 (Fig. 6 A). The level of pro-GelA was maximal on day 5, when the granulation tissue was most active, and pro-GelB expression was elevated all through from days 1–5. However, the mature form of GelB was only detected on days 1 and 3, while the mature form of GelA was observed, along with its corresponding pro-form, in all the tissue samples examined (Fig. 6). When the granulation tissue and the proliferative epithelial cell layer from healing wounds on days 3 and 5 were analyzed separately, pro-GelA/GelA was almost exclusively detected in the granulation tissue (Fig. 6 B). ProGelB/GelB was also predominantly detected in the granulation tissue, with only low levels associated with the proliferative epithelial cell layer. These observations indicate that the wound stroma is the predominant site of action for both GelA and GelB during rat skin wound healing.
Figure 6.
Gelatinolytic activities during rat skin wound healing. Tissues were microdissected from cryostat sections, incubated with SDS sample buffer, and subjected to gelatin zymography. (A) Normal skin containing both epidermis and dermis (lane 1), and healing skin wounds at days 1, 3, 5, 7, 10, and 14 after cutaneous incision (lanes 2–7). (B) Normal skin adjacent to the wound tissue and containing both epidermis and dermis (N) (lane 1), and granulation tissue (G) (lanes 2 and 4) or proliferative epithelial layer (E) (lanes 3 and 5) of healing skin wounds at days 3 (lanes 2 and 3) and 5 (lanes 4 and 5) after cutaneous incision. In A and B, proteins extracted from ∼1 mg of fresh tissue were analyzed in each lane.
Discussion
MMP Gene Expression during Rat Skin Wound Healing
In the present study, we found that six MMP (ST1, ST3, GelA, GelB, Col3, MT1-MMP) genes were expressed during rat skin wound healing. However, we could not detect rat ST2 and matrilysin cDNAs, nor the rat homologue of human Col1 cDNA in the skin wound cDNA libraries used for our screening. Furthermore, we could detect neither MT2-MMP nor MT3-MMP RNA in wound tissues by Northern blot analysis (Okada, A., unpublished results).
Among the six MMP genes expressed during rat skin wound healing, ST1, GelB, and Col3 were mainly expressed between days 1 and 5 after cutaneous incision, while the three others, MT1-MMP, GelA, and ST3, continued to be expressed at high levels at least until day 7. Interestingly, the transcripts corresponding to the early genes were found predominantly in migrating epithelial cells while those corresponding to the late genes were specifically detected in wound stromal cells. However, MT1-MMP and GelA transcripts were both found in granulation and scar tissues, while ST3 expression was not detected in granulation tissue.
Pro-GelB Activation by ST1
Ogata et al. (1992) have demonstrated that human proGelB was processed to its mature form by human ST1 when both enzymes were incubated together in vitro. In agreement with this observation, we found that rat proGelB was processed to its mature form by rat ST1 when the two cDNAs corresponding to GelB and ST1 were transiently transfected into COS-1 cells. The expression kinetics of GelB and ST1 genes, the tissue distribution of cells expressing their transcripts, and the detection of mature GelB protein in wound tissue from days 1 to 3 after cutaneous incision all together support the possibility that pro-GelB is activated by ST1 during rat skin wound healing. The most convincing argument is that while GelB transcripts and protein were detected at similar high levels from days 1 to 5 after cutaneous incision, the active form of GelB was only detected from days 1 to 3, consistent with maximal ST1 gene expression on day 1. Finally, we observed that pro-GelB/GelB were predominantly detected in wound stroma, although their transcripts were predominantly expressed in the epithelial basal cell layer. Taken together, these observations support the proposition of Salo et al. (1994) that GelB plays a role in epithelial cell migration, assuming that the substrate for GelB is found in the wound stroma immediately surrounding the migrating epithelial cells.
Pro-GelA Activation by MT1-MMP
Our finding that pro-GelA was processed to its corresponding mature form when rat MT1-MMP and GelA cDNAs were transiently expressed in COS-1 cells, is consistent with the previous observation of Sato et al. (1994) made with the human enzymes. However, GelB, ST1, or ST3 could not activate pro-GelA. The possibility that proGelA activation by MT1-MMP also occurs during rat skin wound healing is supported by some of our observations. First, MT1-MMP and GelA transcripts were found to be expressed by wound stromal cells exhibiting a similar tissue distribution. Second, the expression kinetics after cutaneous incision were identical for both genes. Third, high levels of the mature form of GelA were observed in the wound stroma, particularly on day 5 after cutaneous incision when both the MT1-MMP and GelA RNAs attained their highest expression levels. However, the ratios between the amounts of mature GelA and pro-GelA were found to be similar in all the wound tissue samples examined, and they did not significantly differ from that observed in normal skin. In this respect, pro-GelA activation by MT1-MMP during rat skin wound healing differs from that of pro-GelB by ST1, although both MMPs are predominantly found in the wound stroma. Another difference observed between GelA and GelB, is that the cells expressing GelA transcripts are not found specifically at the stromal-epithelial interface. The GelA transcripts are in fact distributed throughout the wound stroma, both in the granulation tissue and in the scar tissue. This observation suggests that GelA activated by MT1-MMP contributes to the remodeling processes implicated in the restoration of connective tissue.
MT1-MMP as a Stromal Cell Surface Activator of Pro-GelA
MT1-MMP is believed to correspond to a transmembrane proteinase whose active site is orientated extracellularly (Sato et al., 1994). Our finding that the MT1-MMP and GelA genes are coexpressed in stromal cells, suggests that MT1-MMP activates pro-GelA at the stromal cell surface during rat skin wound healing. Consistent with this suggestion, we observed that the induction of MT1-MMP by Con A treatment in two different rat fibroblast cell lines, RAT1 and FR3T3, resulted in pro-GelA activation. We note that similar observations have been made in human skin fibroblasts, where the processing of pro-GelA was coincident with MT1-MMP expression induced by Con A treatment (Atkinson et al., 1995). Furthermore, we have previously shown that MT1-MMP and GelA transcripts were specifically detected in fibroblast-like cells of breast, colon, and head and neck carcinomas (Okada et al., 1995a ). More recently, MT1-MMP transcripts and the MT1-MMP protein have been shown to be expressed by mesenchymal cells during embryonic development (Kinoh et al., 1996). It remains to be seen, however, whether GelA acts at the same cell surface where it has been activated by MT1-MMP, or on the surface of other cells, or maybe after binding to the ECM. The possibility that GelA binds to cell surface sites distinct from the extracellular domain of MT1-MMP is supported by a recent observation that active forms of GelA bind to integrin αvβ3on the surface of angiogenic blood vessels (Brooks et al., 1996).
Further studies are required to precisely define the sites where GelA is functionally active in remodeling tissues, and to determine whether MT1-MMP and molecules such as integrin αvβ3 cooperate in targeting active GelA to the cell surface. The expression of integrinαvβ3 and/or molecules with similar GelA-binding function at the stromal cell surface during wound healing would be consistent with our finding that the mature form of GelA is specifically found in the stroma. In this case, by activating proGelA, MT1-MMP would contribute to the migration of stromal cells and ECM remodeling implicated in the restoration of the connective tissue during wound healing.
Acknowledgments
We thank P. Chambon for his support, J.M. Garnier for helpful discussions, R. Kannan for critical reading, I. Stoll for protein purification, S. Vicaire for DNA sequencing, and G. Duval for animal care. We are grateful to Dr. Matrisian (Vanderbilt University, Nashville, TN) for anti-ST1 antibody, to Dr. Will (InViTek GmbH, Berlin-Buch, Germany) for human MT2-MMP cDNA, and to Dr. Seiki (Kanazawa University, Kanazawa, Japan) for human MT3-MMP cDNA.
This work was supported by funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Centre Hospitalier Universitaire Régional, the BristolMyers Squibb Pharmaceutical Research Institute, the Association pour la Recherche sur le Cancer, the Ligue Nationale Française contre le Cancer and the Comité du Haut-Rhin, the Fondation de France, the Programme Hospitalier de Recherche Clinique 1995, the BIOMED 2 (contract no BMH4CT96-0017) and BIOTECH 2 (contract no ERBBIO4CT96-0464) Programmes, and a grant to P. Chambon from the Fondation Jeantet. A.Okada was a recipient of fellowships from the Ministère des Affaires Etrangères and the Fondation pour la Recherche Médicale Française.
Abbreviations used in this paper
- Col1
interstitial collagenase
- Col2
neutrophil collagenase
- Col3
collagenase 3
- ECM
extracellular matrix
- GelA and GelB
gelatinase A and B
- MMP
matrix metalloproteinase
- MT1-MMP
MT2-MMP, MT3-MMP, and MT4-MMP, membrane-type-1, -2, -3, and -4 MMP
- ST1
ST2, and ST3, stromelysin 1, 2, and 3
- TIMP1
2, and 3, tissue inhibitor of metalloproteinase type-1, -2, and -3
Footnotes
Address all correspondence to Paul Basset, Institut de Génétique et de Biologie Moléculaire et Cellulaire, Centre National de la Recherche Scientifique/Institut National de la Santé et de la Recherche Médicale/Université Louis Pasteur, BP 163, 67404 Illkirch Cedex, France. Tel.: 33 3 88 65 34 25. Fax: 33 3 88 65 32 01. E-Mail: basset@igbmc.u-strasbg.fr
The current address of A. Okada is the Department of Cancer Cell Research, Institute of Medical Science, University of Tokyo, Tokyo 108, Japan.
References
- Atkinson SJ, Crabbe T, Cowell S, Ward RV, Butler MJ, Sato H, Seiki M, Reynolds JJ, Murphy G. Intermolecular autolytic cleavage can contribute to the activation of progelatinase A by cell membranes. J Biol Chem. 1995;270:30479–30485. doi: 10.1074/jbc.270.51.30479. [DOI] [PubMed] [Google Scholar]
- Birkedal-Hansen H. Proteolytic remodeling of extracellular matrix. Curr Opin Cell Biol. 1995;7:728–735. doi: 10.1016/0955-0674(95)80116-2. [DOI] [PubMed] [Google Scholar]
- Breathnach R, Matrisian LM, Gesnel MC, Staub A, Leroy P. Sequences coding for part of oncogene-induced transin are highly conserved in a related rat gene. Nucleic Acids Res. 1987;15:1139–1151. doi: 10.1093/nar/15.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks PC, Strömblad S, Sanders LC, von Schalscha TL, Aimes RT, Stetler-Stevenson WG, Quigley JP, Cheresh DA. Localization of matrix metalloproteinase MMP-2 to the surface of invasive cells by interaction with integrin αvβ3. Cell. 1996;85:683–693. doi: 10.1016/s0092-8674(00)81235-0. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Collen D. Gene targeting and gene transfer studies of the plasminogen/plasmin system: implications in thrombosis, hemostasis, neointima formation, and atherosclerosis. FASEB (Fed Am Soc Exp Biol) J. 1995;9:934–938. doi: 10.1096/fasebj.9.10.7615162. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Fisher C, Gilbertson-Beadling S, Powers EA, Petzold G, Poorman R, Mitchell MA. Interstitial collagenase is required for angiogenesis in vitro. Dev Biol. 1994;162:499–510. doi: 10.1006/dbio.1994.1104. [DOI] [PubMed] [Google Scholar]
- Gailit J, Clark RAF. Wound repair in the context of extracellular matrix. Curr Opin Cell Biol. 1994;6:717–725. doi: 10.1016/0955-0674(94)90099-x. [DOI] [PubMed] [Google Scholar]
- Heppner KJ, Matrisian LM, Jensen RA, Rodgers WH. Expression of most matrix metalloproteinase family members in breast cancer represents a tumor-induced host response. Am J Pathol. 1996;149:273–282. [PMC free article] [PubMed] [Google Scholar]
- Kinoh H, Sato H, Tsunezuka Y, Takino T, Kawashima A, Okada Y, Seiki M. MT-MMP, the cell surface activator of proMMP-2 (pro-gelatinase A), is expressed with its substrate in mouse tissue during embryogenesis. J Cell Sci. 1996;109:953–959. doi: 10.1242/jcs.109.5.953. [DOI] [PubMed] [Google Scholar]
- Knäuper V, López-Otin C, Smith B, Knight G, Murphy G. Biochemical characterization of human collagenase-3. J Biol Chem. 1996;271:1544–1550. doi: 10.1074/jbc.271.3.1544. [DOI] [PubMed] [Google Scholar]
- Marti HP, McNeil L, Davies M, Martin J, Lovett DH. Homology cloning of rat 72 kDa type IV collagenase: cytokine and second-messenger inducibility in glomerular mesangial cells. Biochem J. 1993;291:441–446. doi: 10.1042/bj2910441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiakowski P, Breathnach R, Bloch J, Gannon F, Krust A, Chambon P. Cloning of cDNA sequence of hormone-regulated genes from the MCF-7 human breast cancer cell line. Nucleic Acids Res. 1982;10:7895–7903. doi: 10.1093/nar/10.24.7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisian LM, Glaichenhaus N, Gesnel MC, Breathnach R. Epidermal growth factor and oncogenes induce transcription of the same cellular mRNA in rat fibroblasts. EMBO (Eur Mol Biol Organ) J. 1985;4:1435–1440. doi: 10.1002/j.1460-2075.1985.tb03799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisian LM, Leroy P, Ruhlmann C, Gesnel MC, Breathnach R. Isolation of the oncogene and epidermal growth factor-induced transin gene: complex control in rat fibroblasts. Mol Cell Biol. 1986a;6:1679–1686. doi: 10.1128/mcb.6.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisian LM, Bowden GT, Krieg P, Fürstenberger G, Briand JP, Leroy P, Breathnach R. The mRNA coding for the secreted protease transin is expressed more abundantly in malignant than in benign tumors. Proc Natl Acad Sci USA. 1986b;83:9413–9417. doi: 10.1073/pnas.83.24.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura H, Sato H, Seiki M, Mai M, Okada Y. Expression of membrane-type matrix metalloproteinase in human gastric carcinomas. Cancer Res. 1995;55:3263–3266. [PubMed] [Google Scholar]
- Ogata Y, Enghild JJ, Nagase H. Matrix metalloproteinase 3 (stromelysin) activates the precursor for the human matrix metalloproteinase 9. J Biol Chem. 1992;267:3581–3584. [PubMed] [Google Scholar]
- Okada A, Garnier JM, Vicaire S, Basset P. Cloning of the cDNA encoding rat tissue inhibitor of metalloproteinase 1 (TIMP-1), amino acid comparison with other TIMPs, and gene expression in rat tissues. Gene (Amst) 1994;147:301–302. doi: 10.1016/0378-1119(94)90088-4. [DOI] [PubMed] [Google Scholar]
- Okada A, Bellocq JP, Rouyer N, Chenard MP, Rio MC, Chambon P, Basset P. Membrane-type matrix metalloproteinase (MT-MMP) gene is expressed in stromal cells of human colon, breast, and head and neck carcinomas. Proc Natl Acad Sci USA. 1995a;92:2730–2734. doi: 10.1073/pnas.92.7.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada A, Santavicca M, Basset P. The cDNA cloning and expression of the gene encoding rat gelatinase B. Gene (Amst) 1995b;164:317–321. doi: 10.1016/0378-1119(95)00447-e. [DOI] [PubMed] [Google Scholar]
- Okada A, Saez S, Misumi Y, Basset P. Rat stromelysin3: cDNA cloning from healing skin wound, activation by furin and expression in rat tissues. Gene (Amst) 1997;185:187–193. doi: 10.1016/s0378-1119(96)00615-4. [DOI] [PubMed] [Google Scholar]
- Overall CM, Sodek J. Concanavalin A produces a matrix-degradative phenotype in human fibroblasts. Induction and endogenous activation of collagenase, 72-kDa gelatinase, and Pump-1 is accompanied by the suppression of the tissue inhibitor of matrix metalloproteinases. J Biol Chem. 1990;265:21141–21151. [PubMed] [Google Scholar]
- Puente XS, Pendás AM, Llano E, Velasco G, López-Otín C. Molecular cloning of a novel membrane-type matrix metalloproteinase from a human breast carcinoma. Cancer Res. 1996;56:944–949. [PubMed] [Google Scholar]
- Quinn CO, Scott DK, Brinckerhoff CE, Matrisian LM, Jeffrey JJ, Partridge NC. Rat collagenase. Cloning, amino acid sequence comparison, and parathyroid hormone regulation in osteoblastic cells. J Biol Chem. 1990;265:22342–22347. [PubMed] [Google Scholar]
- Rømer J, Bugge TH, Pyke C, Lund LR, Flick MJ, Degen JL, Danø K. Impaired wound healing in mice with a disrupted plasminogen gene. Nature Med. 1996;2:287–292. doi: 10.1038/nm0396-287. [DOI] [PubMed] [Google Scholar]
- Salo T, Mäkelä M, Kylmäniemi M, Autio-Harmainen H, Larjava H. Expression of matrix metalloproteinase-2 and -9 during early human wound healing. Lab Invest. 1994;70:176–182. [PubMed] [Google Scholar]
- Santavicca M, Noël A, Chenard MP, Lutz Y, Stoll I, Segain JP, Rouyer N, Rio MC, Wolf C, Bellocq JP, Basset P. Characterization of monoclonal antibodies against stromelysin-3 and their use to evaluate stromelysin-3 levels in breast carcinoma by semi-quantitative immunohistochemistry. Int J Cancer. 1995;64:336–341. doi: 10.1002/ijc.2910640510. [DOI] [PubMed] [Google Scholar]
- Santoro M, Battaglia C, Zhang L, Carlomagno F, Martelli ML, Salvatore D, Fusco A. Cloning of the rat tissue inhibitor of metalloproteinases type 2 (TIMP-2) gene: analysis of its expression in normal and transformed thyroid cells. Exp Cell Res. 1994;213:398–403. doi: 10.1006/excr.1994.1215. [DOI] [PubMed] [Google Scholar]
- Sato H, Seiki M. Membrane-type matrix metalloproteinases (MTMMPs) in tumor metastasis. J Biochem. 1996;119:209–215. doi: 10.1093/oxfordjournals.jbchem.a021223. [DOI] [PubMed] [Google Scholar]
- Sato H, Takino T, Okada Y, Cao J, Shinagawa A, Yamamoto E, Seiki M. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature (Lond) 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- Stetler-Stevenson WG, Hewitt R, Corcoran M. Matrix metalloproteinases and tumor invasion: from correlation and causality to the clinic. Semin Cancer Biol. 1996;7:147–154. doi: 10.1006/scbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Stricklin GP, Li L, Jancic V, Wenczak BA, Nanney LB. Localization of mRNAs representing collagenase and TIMP in sections of healing human burn wounds. Am J Pathol. 1993;143:1657–1666. [PMC free article] [PubMed] [Google Scholar]
- Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J Biol Chem. 1995;270:5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- Takino T, Sato H, Shinagawa A, Seiki M. Identification of the second membrane-type matrix metalloproteinase (MT-MMP-2) gene from a human placenta cDNA library. MT-MMPs form a unique membrane-type subclass in the MMP family. J Biol Chem. 1995;270:23013–23020. doi: 10.1074/jbc.270.39.23013. [DOI] [PubMed] [Google Scholar]
- Tokuraku M, Sato H, Murakami S, Okada Y, Watanabe Y, Seiki M. Activation of the precursor of gelatinase A/72 kDa type IV collagenase/MMP-2 in lung carcinomas correlates with the expression of membrane-type matrix metalloproteinase (MT-MMP) and with lymph node metastasis. Int J Cancer. 1995;64:355–359. doi: 10.1002/ijc.2910640513. [DOI] [PubMed] [Google Scholar]
- Vassalli JD, Saurat JH. Cuts and scrapes? Plasmin heals! . Nature Med. 1996;2:284–285. doi: 10.1038/nm0396-284. [DOI] [PubMed] [Google Scholar]
- Will H, Hinzmann B. cDNA sequence and mRNA tissue distribution of a novel human matrix metalloproteinase with a potential transmembrane segment. Eur J Biochem. 1995;231:602–608. doi: 10.1111/j.1432-1033.1995.tb20738.x. [DOI] [PubMed] [Google Scholar]
- Wu IM, Moses MA. Cloning and expression of the cDNA encoding rat tissue inhibitor of metalloproteinase 3 (TIMP-3) Gene (Amst) 1996;168:243–246. doi: 10.1016/0378-1119(95)00783-0. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yoshiyama U, Sato H, Seiki M, Shinagawa A, Takahashi M. White matter microglia produce membrane-type matrix metalloprotease, an activator of gelatinase A, in human brain tissues. Acta Neuropathol. 1995;90:421–424. doi: 10.1007/BF00294800. [DOI] [PubMed] [Google Scholar]