Abstract
The Fas/APO-1/CD95 ligand (CD95L) and the recently cloned TRAIL ligand belong to the TNFfamily and share the ability to induce apoptosis in sensitive target cells. Little information is available on the degree of functional redundancy between these two ligands in terms of target selectivity and intracellular signalling pathway(s). To address these issues, we have expressed and characterized recombinant mouse TRAIL. Specific detection with newly developed rabbit anti-TRAIL antibodies showed that the functional TRAIL molecule released into the supernatant of recombinant baculovirus-infected Sf9 cells is very similar to that associated with the membrane fraction of Sf9 cells. CD95L resistant myeloma cells were found to be sensitive to TRAIL, displaying apoptotic features similar to those of the CD95L- and TRAIL-sensitive T leukemia cells Jurkat. To assess if IL-1β-converting enzyme (ICE) and/or ICE-related proteases (IRPs) (caspases) are involved in TRAIL-induced apoptosis of both cell types, peptide inhibition experiments were performed. The irreversible IRP/caspase-inhibitor AcYVAD-cmk and the reversible IRP/caspase-inhibitor Ac-DEVD-CHO blocked the morphological changes, disorganization of plasma membrane phospholipids, DNA fragmentation, and loss of cell viability associated with TRAIL-induced apoptosis. In addition, cells undergoing TRAIL-mediated apoptosis displayed cleavage of poly(ADP)-ribose polymerase (PARP) that was completely blocked by Ac-DEVD-CHO.
These results indicate that TRAIL seems to complement the activity of the CD95 system as it allows cells, otherwise resistant, to undergo apoptosis triggered by specific extracellular ligands. Conversely, however, induction of apoptosis in sensitive cells by TRAIL involves IRPs/caspases in a fashion similar to CD95L. Thus, differential sensitivity to CD95L and TRAIL seems to map to the proximal signaling events associated with receptor triggering.
Recently, a new member of the TNF family, the TRAIL/APO-2 ligand has been cloned and shown to induce apoptosis in sensitive target cells (Wiley et al., 1995; Pitti et al., 1996). Within the TNF family, human TRAIL shares the highest similarity (28% homology at the amino acid level) with CD95L.
The FAS/APO-1/CD95 ligand (CD95L)1 (Suda et al., 1993; Suda and Nagata, 1994) is a member of the TNF family, that induces apoptosis in sensitive target cells (for review see Krammer et al., 1994; Nagata and Golstein, 1995). Highly expressed by activated T cells, CD95L has been shown to mediate T cell cytotoxicity (Kägi et al., 1994; Lowin et al., 1994; Hanabuchi et al., 1994; Stalder et al., 1994), activation-induced T cell death (Dhein et al., 1995; Ju et al., 1995; Brunner et al., 1995), regulation of activated B cells by Th1 CD4+ T cells (Rothstein et al., 1995) and liver damage (Ogasawara et al., 1993; Rensing-Ehl et al., 1995; Galle et al., 1996).
The CD95 receptor (CD95) is expressed on a wide variety of normal and transformed cells (for review see Krammer et al., 1994). Induction of apoptosis requires oligomerization of the receptor on the cell surface either by CD95L or agonistic monoclonal antibodies (mAb). Within seconds after receptor oligomerization, an adaptor molecule, FADD/MORT1, is found associated with the functional receptor (Boldin et al., 1995; Chinnaiyan et al., 1995; Kischkel et al., 1995). The death effector domain of FADD, in turn, has been recently shown to interact with an ICE- related protease (IRP) called FLICE/MACH1α (Boldin et al., 1996; Muzio et al., 1996) or caspase-8, according to the new nomenclature proposed by Alnemri et al. (1996). Recruitment of FLICE/MACH1α to the signaling complex is believed to lead to proteolytic activation of FLICE itself and of other apoptosis-mediating IRPs /caspases, thereafter (Muzio et al., 1996). Sequential activation of ICE-like and CPP32-like proteases was found to occur in CD95- mediated apoptosis (Enari et al., 1995; Chinnaiyan et al., 1996; Duan et al., 1996).
The finding that among the TNF family members, TRAIL and CD95L share the highest homology and show a similar potency in inducing apoptosis (Wiley et al., 1995), raises the question of the extent of redundancy existing between these two systems. To address this issue, we have expressed and characterized recombinant mouse TRAIL using the baculovirus expression system, as previously reported for CD95L (Mariani et al., 1996). In the present study we compare the target specificity and the intracellular pathway(s) activated by TRAIL and CD95L. We show that mouse myeloma cells, that are resistant to CD95L, are sensitive to TRAIL and that inhibition of IRPs/caspases by synthetic peptides prevents all TRAIL-induced apoptotic events analyzed: i.e., morphological changes, disorganization of plasma membrane phospholipids, poly(ADP)- ribose polymerase (PARP) cleavage, DNA fragmentation, and cell death.
Materials and Methods
Materials
The tetrapeptide chloromethylketone Acetyl-Tyr-Val-Ala-Asp-cmk (AcYVAD-cmk) (an irreversible inhibitor of IRPs/caspases) and the tetrapeptide aldehyde Acetyl-Asp-Glu-Val-Asp-CHO (Ac-DEVD-CHO) (a reversible inhibitor of IRPs/caspases) were obtained from Bachem (Switzerland). Stock solutions of peptide inhibitors (40 mM) were prepared in DMSO and stored at −80°C. Working solutions were made in culture medium immediately before use. The recombinant mouse wild-type CD95L, the mouse gld CD95L (Lynch et al., 1994; Takahashi et al., 1994) and the human CD95L (Suda et al., 1993; Suda and Nagata, 1994) expressed in Sf9 cells (Mariani et al., 1996) as well as the anti-CD95L antibody have been described previously (Mariani et al., 1995). The anti-PARP Ab was kindly provided by A. Bürkle.
Cells
The CD95L-resistant mouse myeloma cells Ag8, the CD95L-sensitive human T leukemia cells Jurkat, the human B lymphoma cells, Bjab and REH, and the mouse T hybridoma cells 2H11 were cultured in RPMI1640 medium supplemented with 10% heat-inactivated FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, 10 mM Hepes (pH 7.2) and 2 mM glutamine at 37°C with 5% CO2 in a humidified atmosphere. The cell viability was greater than 95% in each cell preparation used. Sf9 insect cells were purchased from GIBCO BRL (Gaithersburg, MD) and cultured at 27°C in Grace's insect medium (GIBCO BRL) supplemented with 10% heat-inactivated FCS.
Preparation of Rabbit Anti-TRAIL Antibodies and Control Mouse mAb
Rabbit anti-TRAIL antibodies were developed as previously described (Mariani et al., 1994, 1995) in accordance with institutional guidelines. Briefly, the 21-mer peptide NEHLIDMDHEASFFGAFLVGC was conjugated to keyhole lympet hemocyanin (KLH) by m-maleimidobenzoyl- N-hydroxysuccinimide ester (MBS) (Pierce, Rockford, IL) and mixed with complete Freund's adjuvant (CFA). Rabbits were immunized with 700 μg of the conjugated peptide. The animals were boosted at biweekly intervals with half the dose of the same immunogen in incomplete Freund's adjuvant (IFA) and bled 7–8 d after each boost. Rabbit IgG was purified by affinity chromatography on protein A–Sepharose (Pharmacia, Uppsala, Sweden) according to the manufacturer's instructions. The hybridoma secreting the mouse mAb SM 22 (IgG,k) was established from the spleen of a mouse immunized with baculovirus-infected Sf9 cells. In immunoblots, it specifically recognizes baculovirus-derived proteins with an apparent molecular mass of 27–32 kD. It does not react with non-infected Sf9 cells or a variety of eukaryotic cells.
Cloning of Mouse TRAIL cDNA
Mouse TRAIL cDNA was synthesized by RT-PCR using the following primers: 5′ sense: 5′ ATA CCC GGG GCC ACC ATG CCT TCC TCA GGG GCC CTG AAG 3′ and 3′ antisense: CCT CGG AAA AAT TAA TTG ATT CTT AAG ATA 5′. Sequences corresponding to TRAIL cDNA are underlined. The cDNA was cloned into the baculovirus transfer vector pVL1393 (Invitrogen, San Diego, CA) downstream from the polyhedrin promoter according to standard protocols (O'Reilly et al., 1994). The sequence of the cloned cDNA was confirmed by double strand cDNA sequencing and found to be identical to that published (Wiley et al., 1995).
Expression of Recombinant Mouse TRAIL in Sf9 Insect Cells
TRAIL was expressed as previously described for recombinant mouse and human CD95L (Mariani et al., 1996). Sf9 insect cells were cotransfected with TRAIL cDNA-containing transfer vector and linearized Autographa californica nuclear polyhedrosis virus (AcNPV) DNA (BaculoGold DNA, Pharmingen, San Diego, CA) according to the manufacturer's instructions. 4 d after transfection, supernatants containing the recombinant virus were harvested from transfected cells and used to infect Sf9 cells that had been plated in 60-mm petri dishes at 2 × 106 cells/dish. After infection, the cell monolayer was overlaid with 1% Seaplaque agarose (FMC BioProducts, Rockland, ME). Individual viral plaques were isolated and tested for expression of TRAIL (O'Reilly et al., 1994). The titer of the recombinant virus was determined by end-point dilution. The recombinant virus was further expanded to generate a stock virus by infecting Sf9 cells at a multiplicity of infection (MOI) of 0.1. Infection of Sf9 cells for protein production was performed with an MOI of 10 (Summers and Smith, 1987). Supernatants were collected 4 d after infection from culture flasks with a confluent monolayer of Sf9 cells, spun at 1,000 g for 10 min and stored at 4°C. Cells were further washed with PBS and stored as frozen pellets at −80°C.
SDS-PAGE and Immunoblotting of Recombinant TRAIL
SDS-PAGE and immunoblotting were performed as previously described (Mariani et al., 1994). Briefly, following SDS-PAGE, proteins were transferred to nitrocellulose membranes by semi-dry electroblotting. Membranes were blocked with 5% nonfat dry milk in PBS and then incubated overnight with the primary antibody. Membranes were washed five times with PBS/0.5% Tween 20 and incubated with HRPO-conjugated affinitypurified mouse anti–rabbit IgG Ab or HRPO-conjugated affinity-purified goat anti–mouse IgG Ab (Dianova, Hamburg, FRG) for 2 h. Membranes were then washed five times and the reaction developed using the ECL detection system (Amersham, Frankfurt, FRG).
Determination of Cell Death
Cell death was assessed as described previously (Darzynkiewicz et al., 1992; Chrest et al., 1993; Mariani et al., 1996). Briefly, serial dilutions of supernatants of Sf9 cells expressing mouse TRAIL or mouse wild-type and gld CD95L were incubated with target cells overnight in 96-well microtiter plates. Cell death was determined by forward/side scatter analysis and PI staining using a FACScan (Becton Dickinson, Heidelberg, FRG). The target cells were the mouse myeloma cells Ag8, the human CD95Lsensitive T lymphoblastoid cells Jurkat, and the mouse T hybridoma cells 2H11. Blocking and control reagents were added to the plates immediately before addition of the target cells. Results are expressed as percent specific cell death calculated according to the formula: (Experimental − Background)/(100 − Background) × 100. Background apoptosis was measured in target cells incubated with serial dilutions of Sf9 culture medium.
MC540 Staining and Flow Cytometry
MC540 staining was performed as previously described (Fadok et al., 1992). Briefly, cells incubated with recombinant TRAIL or CD95L were stained with the dye Merocyanine 540 (MC540), freshly diluted in PBS (5 μg/ml), a few minutes before analysis. Uptake of MC540 was determined by flow cytometric analysis. Cells undergoing loss of physiological asymmetry in plasma membrane phospholipids expose phosphatidylserine residues with a high uptake of MC540. Results are expressed as percent cells with high MC540 uptake. The morphology of cells undergoing TRAIL- and CD95L-mediated apoptosis was not affected by MC540 staining.
DNA Fragmentation
DNA fragmentation was analyzed as previously described (Wyllie and Morris 1982; Mariani et al., 1996). Briefly, target cells incubated with recombinant TRAIL or CD95L were washed and lysed for 30 min at 4°C in hypotonic buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 0.2% Triton X-100). Lysates were centrifuged at 13,000 g for 10 min. Soluble fragmented DNA was precipitated overnight from the supernatant by adding 0.5 M NaCl and an equal volume of isopropanol. The precipitated DNA was recovered by centrifugation at 13,000 g for 15 min, dissolved in TE (10 mM Tris, pH 7.4, 1 mM EDTA) and electrophoresed on a 1% agarose gel at 10 V/ cm. DNA bands were visualized by ethidium bromide staining and digitized by high resolution optical scanning; volumetric integration of signal intensities was performed using the Scan Analysis software. Results are expressed as relative density units.
Results
Expression of Recombinant Mouse TRAIL and Generation of Rabbit Anti-TRAIL Antibodies
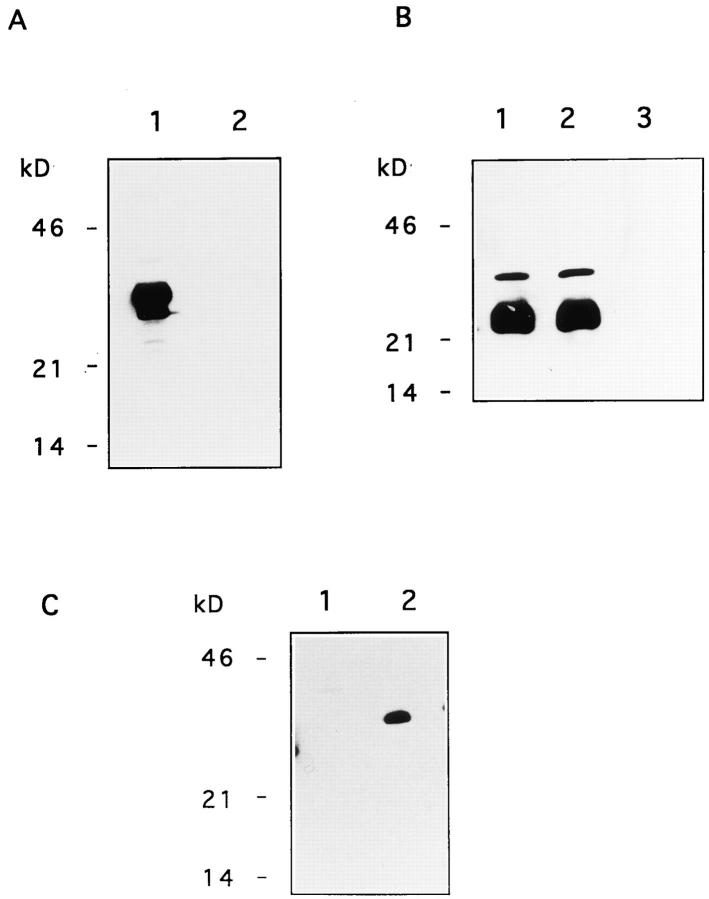
To produce functional recombinant TRAIL, full-length mouse TRAIL cDNA was subcloned into the baculovirus expression vector pVL1393 under the strong polyhedrin promoter. Sf9 cells were infected with the recombinant virus and tested for expression of recombinant TRAIL. To specifically detect TRAIL, a rabbit polyclonal antibody was developed in rabbits immunized with a peptide corresponding to the carboxy-terminal region of TRAIL. As shown in Fig. 1 A, the Ab specifically detects a protein of ∼32–33 kD in lysates of Sf9 cells expressing TRAIL (lane 1) but not of Sf9 cells expressing CD95L (lane 2). No reactivity was found in both cell lysates with purified rabbit IgG. Incubation of the same cell lysates with the mouse mAb SM22 to a baculovirus-derived product showed an equal loading of cell extract (Fig. 1 B).
Figure 1.
Immunoblot analysis of recombinant mouse TRAIL. (A) Whole cell lysates of Sf9 cells expressing recombinant mouse TRAIL (lane 1) or mouse CD95L (lane 2) were tested with purified rabbit anti-TRAIL Ab. Molecular weight markers are indicated. HRPO-labeled mouse anti–rabbit IgG antibodies were used as the detecting reagent. No reactivity was present with purified rabbit IgG. (B) Whole cell lysates of Sf9 cells expressing recombinant mouse TRAIL (lane 1) or mouse CD95L (lane 2) and cell lysates of non-infected Sf9 cells (lane 3) were tested with the mouse mAb SM22 to a baculovirus-related product. Molecular weight markers are indicated. HRPO-labeled rabbit anti–mouse IgG antibodies were used as the detecting reagent. No reactivity was present with an isotype-matched control mAb. (C) SN of Sf9 cells expressing mouse TRAIL (lane 2) or mouse CD95L (lane 1) were tested with purified rabbit anti-TRAIL antibody. Molecular weight markers are indicated. HRPO-labeled mouse anti–rabbit IgG antibodies were used as the detecting reagent. No reactivity was found with purified rabbit IgG.
Immunoblot analysis of the supernatant (SN) of infected Sf9 cells showed that during the lytic infection, recombinant TRAIL is released into the culture medium (Fig. 1 C, lane 2). The apparent molecular weight, determined by SDS-PAGE under reducing conditions, suggests that TRAIL present in the SN is similar to the membranebound form present in Sf9 cells (Fig. 1 A) and in Sf9 membrane preparations (not shown). A similar phenomenon was observed for recombinant mouse and human CD95L produced in the same expression system (Mariani et al., 1996).
Differential Sensitivity of the Mouse Myeloma Cells Ag8 to TRAIL and CD95L
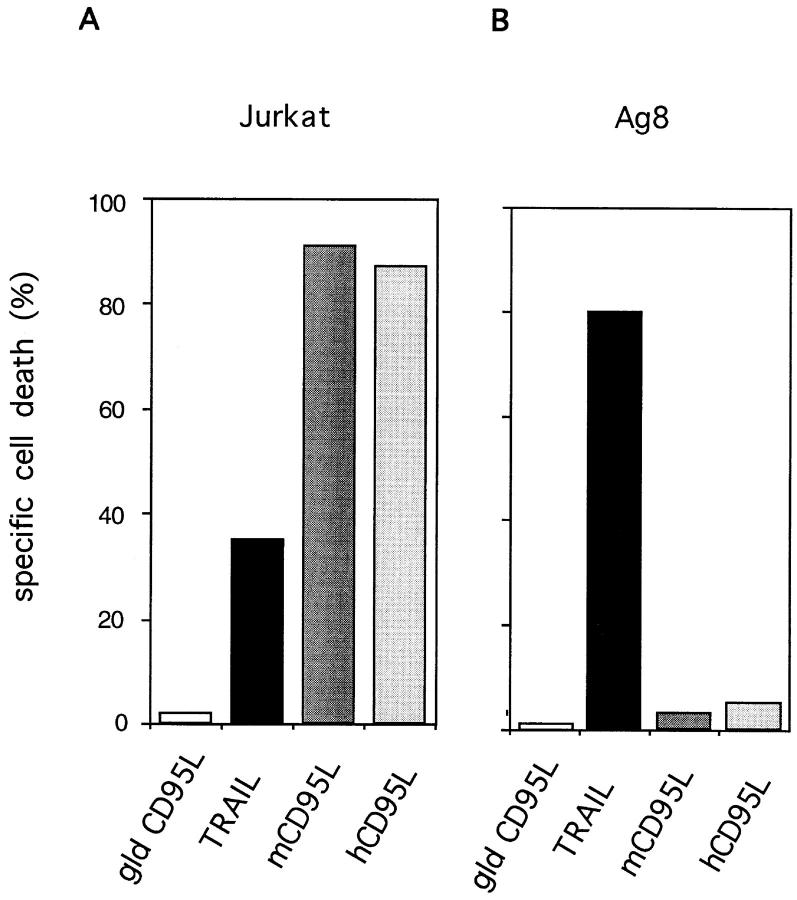
Recombinant TRAIL was tested for functional activity by incubating SN from TRAIL-expressing Sf9 cells with the human leukemic cells Jurkat. TRAIL killed the target cells but no cell death was observed in Jurkat cells incubated with control medium or SN of Sf9 cells expressing mouse gld CD95L. (Fig. 2 A). To assess the ability of recombinant TRAIL to affect mouse cells, a similar assay was performed with the mouse myeloma cells Ag8 as target. As shown in Fig. 2 B, Ag8 cells were killed very efficiently by TRAIL but not by the control mouse gld CD95L. It is of note that the same target cells were almost completely resistant to recombinant mouse and human CD95L. Specificity was further indicated by the lack of cell death in T hybridoma cells incubated with mouse TRAIL and CD95L (Fig. 3 A).
Figure 2.
Target cells resistant to mouse CD95L are sensitive to mouse TRAIL. CD95L-sensitive human leukemia Jurkat cells (Jurkat) (A) and CD95L-resistant mouse myeloma cells Ag8 (Ag8) (B) were incubated overnight with SN from Sf9 cells expressing mouse gld CD95L (gld CD95L), mouse TRAIL (TRAIL), mouse wild-type CD95L (mCD95L), and human CD95L (hCD95L). Results are expressed as percent specific cell death. Background apoptosis of the target cells incubated with SN of mock-infected Sf9 cells was <15%.
Figure 3.
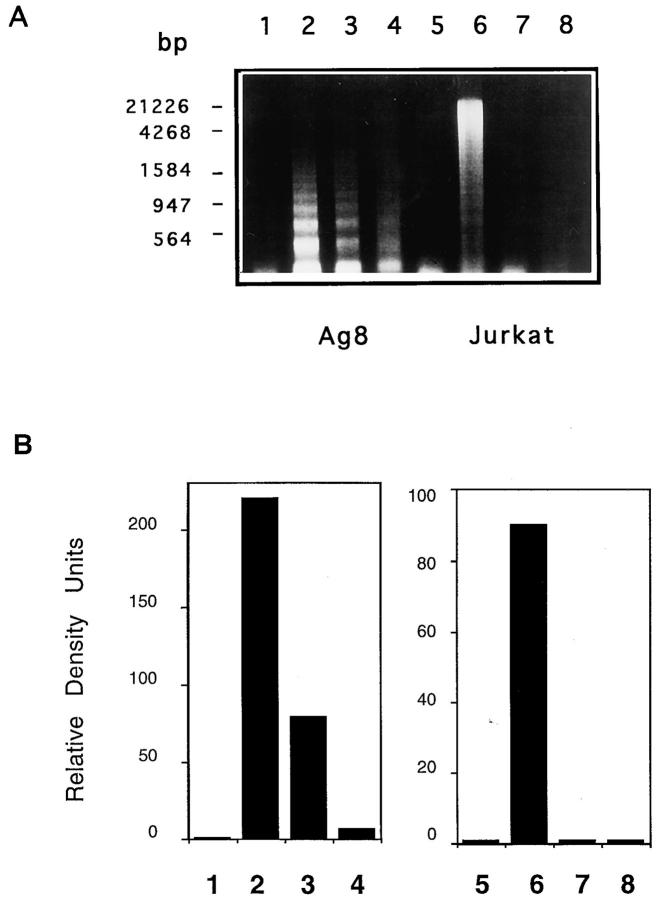
Induction of apoptosis and DNA fragmentation in Ag8 cells by recombinant mouse TRAIL. (A) The human T leukemia cells Jurkat (Jurkat) (left panels), the mouse myeloma cells Ag8 (Ag8) (central panels), and the mouse T hybridoma cells 2H11 (TcHy) (right panels) were incubated with SN from Sf9 cells expressing mouse TRAIL (TRAIL) (upper panels), or mouse wildtype CD95L (CD95L) (lower panels). Results are presented as forward/side scatter analysis of cellular morphology. Apoptotic cells are shown in gate R1. (B) Soluble DNA was extracted from Ag8 (Ag8) (lanes 1–3) and Jurkat cells (Jurkat) (lanes 4–6) (0.5 × 106 cells/lane) incubated with SN from mock-infected Sf9 (lanes 1 and 4) or from Sf9 cells expressing mouse CD95L (lanes 2 and 5) or mouse TRAIL (lanes 3 and 6). Extracted DNA was separated on an agarose gel and visualized by ethidium bromide staining. Molecular mass markers (MM) are shown. No soluble DNA was extracted from TRAIL-resistant T hybridoma cells incubated with either ligand.
Features of Apoptosis Induced by TRAIL in Myeloma and Leukemia Cells
To characterize the cell death induced by TRAIL, morphology, DNA integrity, and membrane lipid organization of target cells were examined. Ag8 cells exposed to TRAIL showed membrane blebbing, reduction in cell size, increase in surface rugosity (increase in side scatter) (Fig. 3 A), nuclear condensation, and release of apoptotic bodies (not shown). Similar events were seen also in Jurkat target cells, as previously described (Wiley et al., 1995). Analysis of soluble DNA from the target cells showed that a significant quantity of fragmented DNA was present in the cytoplasm of target Ag8 (lane 3) and Jurkat cells (lane 6) incubated for 6 h with TRAIL (Fig. 3 B). Specificity is indicated by the lack of oligonucleosomal DNA fragments in extracts of Ag8 cells incubated with CD95L (lane 2) and of T hybridoma cells incubated with both ligands (not shown). It is of note that the pattern of TRAIL-induced DNA fragmentation is different between Ag8 and Jurkat cells. As shown in lanes 5 and 6, the same pattern of DNA fragmentation is present in Jurkat cells undergoing TRAIL- or CD95L-mediated apoptosis. Thus, the phenotypic difference in the average size of the DNA fragments between Ag8 and Jurkat cells, seems to be independent of the effector molecule and the extent of apoptosis. It may reflect the activation of different nucleases and/or a different susceptibility of cellular DNA to enzymatic fragmentation.
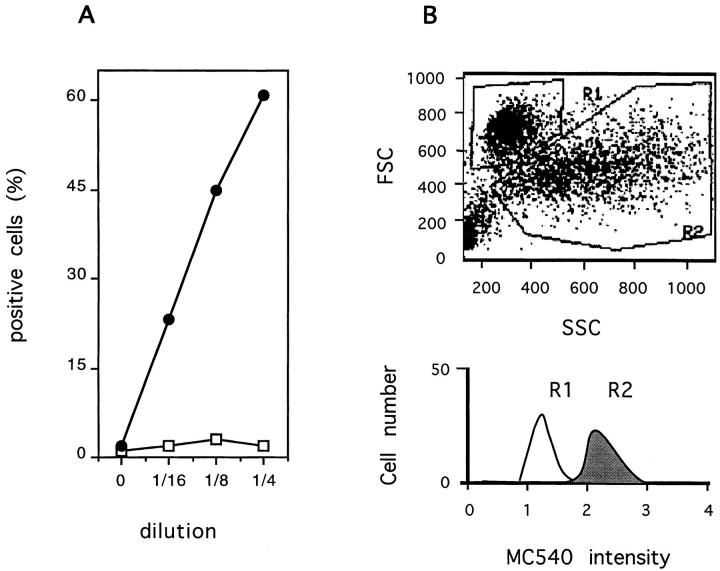
Distribution of phospholipids in the cellular plasma membranes was determined by MC540 staining (Fadok et al., 1992). As shown in Fig. 4 A, incubation of Jurkat cells with increasing concentrations of TRAIL, led to an increasingly higher uptake of MC540, suggesting an alteration of the membrane lipids and increased exposure of phosphatidylserine, as previously described for other apoptosis-inducing agents (Martin et al., 1995). Similar results were obtained with Ag8 cells (not shown). Specificity is indicated by the lack of increased MC540 uptake by Jurkat cells incubated with mouse gld CD95L (Fig. 4 A). Higher uptake of MC540 was correlated with the apoptotic phenotype of the target cells. As shown in Fig. 4 B, only the apoptotic cells undergoing shrinkage and increase in surface rugosity (gate R2) showed increased uptake of MC540. A kinetic analysis indicated that high uptake of MC540 by target cells preceded loss of cell viability as determined by propidium iodide permeability (not shown). These results indicate that TRAIL-induced apoptosis is associated with loss of membrane phospholipid asymmetry not only in normal T cells (Marsters et al., 1996) but also in T cells with a transformed phenotype.
Figure 4.
Induction of plasma membrane phospholipid disorganization by recombinant mouse TRAIL. The T leukemia cells Jurkat were incubated overnight with serial dilutions of SN from Sf9 cells expressing mouse TRAIL (filled circles) or mouse gld CD95L (empty squares). Results are expressed as percent cells with high uptake of the lipophilic dye MC540 (A). In B, results are presented as forward/side scatter analysis of cells incubated with recombinant TRAIL. MC540 uptake is shown for cells with nonapoptotic morphology (gate R1) (white histogram) and for cells with apoptotic morphology (gate R2) (gray histogram). No difference in cell morphology was observed in Jurkat cells exposed to TRAIL in the presence or absence of MC540.
The IRP/Caspase-Inhibitors Ac-YVAD-cmk, and Ac-DEVD-CHO Block the Morphological and Structural Changes Associated with TRAIL-induced Apoptosis in Myeloma and Leukemia Cells
To assess if IRPs/caspases are involved in the intracellular signaling events triggered by TRAIL receptor engagement, peptide inhibition experiments were performed. The mouse myeloma cells Ag8 exposed to TRAIL in the presence of the synthetic peptide Ac-YVAD-cmk did not undergo the morphological changes characteristic of TRAIL- mediated apoptosis, i.e., membrane blebbing and nuclear condensation, as determined by optical microscopy. The lack of cell shrinkage and of increased cellular density are presented in the forward/side scatter analysis of Fig. 5 A. Equal concentrations of the vehicle (dmso) did not prevent TRAIL-induced cell death. Very similar results were obtained with the peptide Ac-DEVD-CHO (not shown). Analysis of the dose-effect relationship (Fig. 5 B) showed that maximal inhibition was achieved at a concentration of 200 μM of Ac-YVAD-cmk, with an IC50 (concentration yielding 50% inhibition) of ∼50 μM. Very similar results were obtained when the human leukemic cells Jurkat were used as target. In these cells, the Ac-YVAD-cmk peptide blocked TRAIL-induced apoptosis with an IC50 of ∼75 μM (Fig. 6 A). To determine whether Ac-YVAD-cmk could prevent loss of asymmetry in the plasma membrane phospholipids, exposure of phosphatidylserine residues was determined in target cells by measuring uptake of the lipophilic dye MC540 (Fadok et al., 1992). As shown in Fig. 6 B Jurkat cells exposed to TRAIL in the presence of increasing concentrations of Ac-YVAD-cmk displayed a dose-dependent inhibition of the TRAIL-induced alteration in plasma membrane phospholipids. Similar results were obtained with Ag8 cells as target or with Ac-DEVDCHO as inhibitor (not shown).
Figure 5.
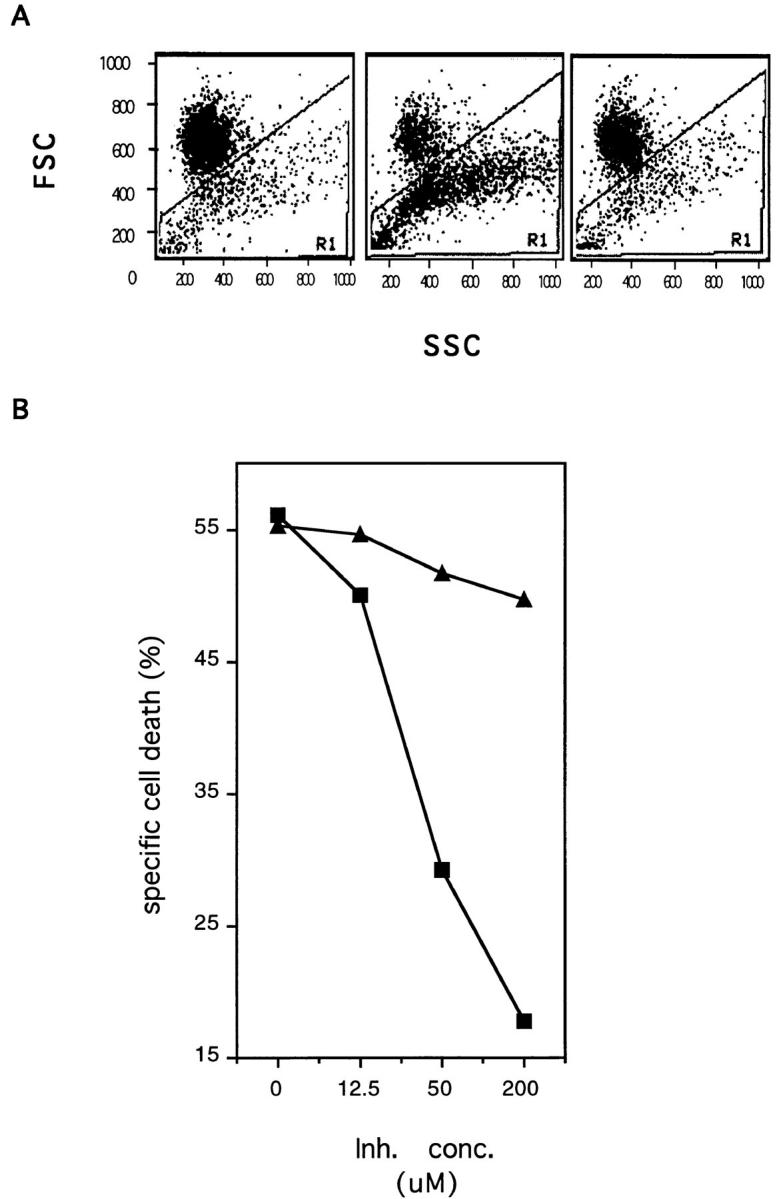
The IRP/caspase-inhibitor Ac-YVAD-cmk blocks TRAILinduced apoptosis in mouse myeloma cells. (A) The mouse myeloma cells Ag8 were incubated overnight with SN of Sf9 cells expressing mouse TRAIL (central and right panel) in the presence of the peptide inhibitor Ac-YVAD-cmk (200 μM) (right panel) or an equal concentration of vehicle (dmso) (central panel). As a specificity control Ag8 cells were incubated with SN of mockinfected Sf9 cells (left panel). Results are presented as forward/ side scatter analysis of cellular morphology. Apoptotic cells are shown in gate R1. (B) Ag8 cells were incubated overnight with SN of Sf9 cells expressing mouse TRAIL in the presence of increasing concentrations of the peptide inhibitor Ac-YVAD-cmk (squares). Equal concentrations of vehicle (triangles) were used as a specificity control. Results are expressed as percent specific cell death. Background apoptosis of target cells was <15%.
Figure 6.
The IRP/caspase-inhibitor Ac-YVAD-cmk blocks TRAILinduced apoptosis in human leukemia cells. (A) The human leukemia cells Jurkat were incubated overnight with SN of Sf9 cells expressing mouse TRAIL in the presence of increasing concentrations of the peptide inhibitor Ac-YVAD-cmk (circles). Equal concentrations of vehicle (squares) were used as a specificity control. Results are expressed as percent specific cell death. Background apoptosis of target cells was less than 15%. (B) Jurkat cells were incubated for 8 h with SN of Sf9 cells expressing mouse TRAIL in the presence of increasing concentrations of the peptide inhibitor Ac-YVAD-cmk (circles). Equal concentrations of vehicle (squares) were used as a specificity control. Results are expressed as percent of cells with high uptake of the MC540 dye.
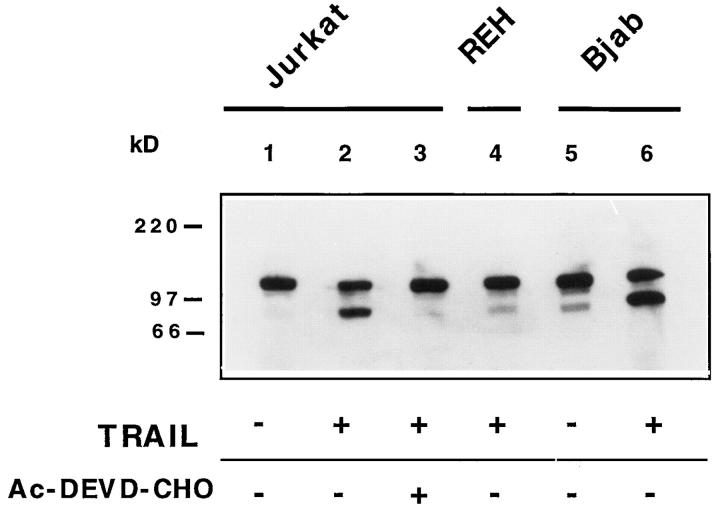
The nuclear repair enzyme PARP is cleaved by IRPs/ caspases during apoptosis induced by CD95 receptor triggering (Tewari et al., 1995) and by a variety of other agents (Henkart, 1996). To determine whether PARP is cleaved in cells undergoing TRAIL-mediated apoptosis, immunoblotting experiments were performed. As shown in Fig. 7, PARP (∼116 kD) was cleaved in sensitive Jurkat (lane 2) and Bjab cells (lane 6) with generation of a shorter fragment of ∼85 kD following incubation with TRAIL. Specificity is indicated by the lack of PARP cleavage in Jurkat (lane 1) and Bjab cells (lane 5) incubated with SN of mock-infected Sf9 cells and in TRAIL-resistant REH cells (lane 4). Triggering of TRAIL receptor in the presence of the peptide inhibitor Ac-DEVD-CHO completely blocked generation of the shorter fragment in Jurkat (lane 3) and Bjab cells (not shown). No inhibition was present with the vehicle alone. Thus, Ac-DEVD-CHO-inhibitable proteases are involved in TRAIL-induced cleavage of PARP in sensitive cells.
Figure 7.
The IRP/caspase inhibitor Ac-DEVD-CHO blocks TRAIL-induced PARP cleavage. Whole cell lysates of Jurkat cells incubated with SN of Sf9 cells expressing mouse TRAIL (lanes 2–3) or of mock-infected Sf9 cells (lane 1) in the presence (lane 3) or absence of Ac-DEVD-CHO (lanes 1–2) were tested with the anti-PARP Ab. The TRAIL-resistant REH (lane 4) and the TRAIL-sensitive BJAB cells (lanes 5–6) incubated with (lanes 4 and 6) or without TRAIL (lane 5) were used as specificity controls. Molecular markers are indicated. HRPO-labeled goat anti–mouse IgG Ab were used as the detecting reagent.
As shown previously, TRAIL-induced apoptosis is associated with extensive fragmentation of cellular DNA. To define the functional involvement of IRPs/caspases in this process, inhibition experiments were performed with the peptide inhibitors. As shown in Fig. 8, TRAIL-induced fragmentation of DNA from Ag8 and Jurkat cells was completely prevented in the presence of the IRP/caspaseinhibitor Ac-DEVD-CHO (lanes 4 and 8). No difference in the DNA fragmentation pattern or intensity was found in the presence or absence of the vehicle (not shown). Also the IRP/caspase inhibitor Ac-YVAD-cmk prevented TRAIL-induced DNA fragmentation in both cell types, but to a lower extent than the Ac-DEVD-CHO inhibitor in Ag8 cells. These results indicate that Ac-YVAD-cmk- and Ac-DEVD-CHO-sensitive IRPs/caspases are involved in TRAIL-induced DNA fragmentation and apoptosis of sensitive human and mouse cells.
Figure 8.
The Ac-YVAD-cmk and Ac-DEVD-CHO peptide inhibitors block TRAIL-induced DNA fragmentation in mouse and human cells. (A) Soluble DNA was extracted from mouse myeloma cells Ag8 (lanes 1–4) (Ag8) or from the human leukemia cells Jurkat (lanes 5–8) (Jurkat) (0.5 × 106 cells/lane) incubated with SN from mock-infected Sf9 (lanes 1 and 5) or with SN from TRAIL-expressing Sf9 cells (lanes 2–4 and 6–8) in the presence of the peptide inhibitor Ac-YVAD-cmk (lanes 3 and 7) or Ac-DEVD-CHO (lanes 4 and 8) (200 μM). An equal concentration of vehicle (dmso) was used as a specificity control (lanes 2 and 6). Extracted DNA was separated on an agarose gel and visualized by ethidium bromide staining. Molecular mass markers (MM) are shown. In B, results are shown as relative density of fragmented DNA in each lane (relative density units) as determined by densitometric analysis.
Discussion
In the present study, we have compared the apoptotic responses induced by TRAIL and CD95L, which share the highest degree of homology among the members of the TNF family so far characterized (Wiley et al., 1995). Mouse myeloma cells that are quite resistant to mouse CD95L show a high sensitivity to recombinant mouse TRAIL. The morphological and structural changes observed are very similar to those displayed by CD95L-sensitive Jurkat cells exposed to TRAIL or CD95L. The differential sensitivity to CD95L and TRAIL, however, is not uniquely found in mouse Ag8 cells, since also the mouse fibrosarcoma cells L929 and the human T lymphoma cells HPB were found to be sensitive to TRAIL- but not CD95L-mediated apoptosis. TRAIL function(s), therefore, seem to complement the activity of the CD95 system allowing a wider variety of cells to be sensitive to apoptosis triggered by specific extracellular ligands. The extent of the functional diversification of the two systems will be more defined after detailed analysis of a larger panel of normal and transformed cell types.
The different effector potential of TRAIL and CD95L may reflect differences in the proximal signaling complexes (receptor and receptor-associated molecules) or in the more downstream signaling pathway(s). IRPs/caspases were shown to be the major mediators of intracellular proteolysis which leads to the apoptotic demise of target cells exposed to a variety of apoptosis-inducing agents (for review see Martin and Green, 1995; Fraser and Evan, 1996; Henkart, 1996; Takahashi and Earnshaw, 1996). Different intracellular proteins were found to be proteolytically cleaved during CD95-mediated apoptosis. It is still not clear, however, which are the critical targets that determine the irreversibility of the apoptotic response (Ashkenas and Werb, 1996). The proteolytic cascade activated by CD95- and TNF-RI- triggering can be blocked by crmA, a serpin-like molecule (Tewari and Dixit, 1995) and by synthetic peptides that represent specific target sequences of ICE and/or other IRPs/caspases, Acetyl-Tyr-Val-Ala-Asp-cmk (Ac-YVADcmk), and Acetyl-Asp-Glu-Val-Asp-aldehyde (Ac-DEVDCHO) (Enari et al., 1995; Los et al., 1995). ICE (caspase-1), FLICE/MACH1α (caspase-8), Mch3/ICE-LAP3/CMH-1 (caspase-7), and CPP32/YAMA (caspase-3) were shown, so far, to be the major intracellular effectors of CD95- and TNF-mediated apoptotic signals (Enari et al., 1995; Kuida et al., 1995; Los et al., 1995; Boldin et al., 1996; Chinnaiyan et al., 1996; Duan et al., 1996; Enari et al., 1996; Muzio et al., 1996).
Here, we provide evidence that similarly to CD95L, intracellular signaling following TRAIL receptor engagement, involves an IRP/caspase pathway. Dissociation of TRAIL- from CD95L-sensitivity seems therefore to map quite near to the receptor complexes. Inhibition of IRPs/ caspases was found to prevent all apoptotic events analyzed: (1) membrane blebbing, (2) cell shrinking, (3) disorganization of plasma membrane phospholipids, (4) PARP cleavage, (5) DNA fragmentation, and (6) cell death. These findings are consistent with the observation that transfection with CrmA of TRAIL-sensitive B lymphoma cells (Mariani, S., unpublished observations) or Hela cells (Marsters et al., 1996) confers almost complete protection from TRAIL-induced apoptosis. The concentrations of peptide inhibitors required to block TRAIL-mediated apoptosis are very similar to those described in CD95L-induced apoptosis for the Ac-YVAD-cmk inhibitor (Enari et al., 1995) and in camptothecin-induced apoptosis for the AcDEVD-CHO inhibitor (Nicholson et al., 1995). It is of note that both Ac-YVAD-cmk- and Ac-DEVD-CHO-inhibitable IRPs/caspases are involved in the apoptotic response triggered by TRAIL in mouse myeloma and human leukemia cells. The presence of PARP cleavage in cells undergoing TRAIL-induced apoptosis suggests an active role for CPP32- like proteases (Lazebnick et al., 1994; Fernandes-Alnemri et al., 1995; Nicholson et al., 1995; Duan et al., 1996; Lippke et al., 1996) as previously shown for CD95-mediated apoptosis (Tewari et al., 1995). It remains to be defined which are the critical IRPs/caspases involved and whether there is a cellular and/or a receptor specificity for any of them.
We have shown here that mouse myeloma cells are quite resistant to CD95L. Similarly to other cell types, Ag8 cells express CD95 on their surface (Mariani et al., 1994), and develop sensitivity after sensitization with protein synthesis inhibitors (Mariani, S., unpublished results). It is still not clear which are the cellular mechanisms that underly this phenotypic conversion. They seem, however, to reside in the signaling cascade proximal to the receptor, as we have shown that Ag8 cells display a high sensitivity to TRAIL and therefore possess an apparently intact downstream proteolytic cascade. It cannot, however, be excluded that distal events, i.e., negative regulators of the proteolytic cascade, are unique to each receptor system and are specifically released by cycloheximide in the presence of CD95L. Similarly to Ag8 cells, also the mouse myeloma cells J558L were shown to be resistant to CD95- mediated apoptosis (Onel et al., 1995). In the human system, two out of five myelomas (U266 and Hs) were found to be resistant even though they expressed CD95 on their surface. Similar results were reported for tumor cells freshly isolated from myeloma patients (Shima et al., 1995). The mechanisms underlying resistance to CD95triggering and potential sensitization to CD95-mediated apoptosis by cycloheximide were not addressed in these studies. From a practical point of view, the finding that myeloma cells are very sensitive to TRAIL suggests that development of agonistic mAbs to the mouse TRAIL receptor, still to be characterized, may be a tricky endeavor. Availability of a TRAIL-resistant myeloma variant for cell fusion may help to overcome this problem.
While much attention is focused on the biological functions of the CD95 system, no information is available, at present, on the physiological relevance of the TRAIL system. Availability of recombinant TRAIL described here will help in elucidating its function(s). In this study we have employed a membrane-like form of recombinant mouse TRAIL. We have shown previously that Sf9 cells lytically infected with recombinant virus release functional mouse and human CD95L in the culture medium, respectively (Mariani et al., 1996). These ligands, with a relative molecular weight similar to that of the membrane-bound form, were found to be associated with small membrane vescicles, thus mimicking the physiological expression of membrane-bound CD95L. A very similar phenomenon is observed here for recombinant mouse TRAIL. So far, recombinant TRAIL has been stably expressed only as a soluble form. Wiley et al. (1995) showed that purified soluble TRAIL has a low effector activity. Cross-linking with an anti-FLAG mAb led to an ∼30-fold increase in cytotoxic activity. Marster et al. (1996), on the other hand, prepared a His-tagged soluble human TRAIL that was used at very high concentrations (1 μg/ml), considering the specific activity of 10 and 6.5 × 106 U/mg estimated for TNF and CD95L, respectively (Suda et al., 1994). These data would suggest that oligomerization of TRAIL is required for effective triggering of its receptors, as previously found for the CD95 system (Krammer et al., 1994; Peitsch and Tschopp, 1995). Since recombinant mouse TRAIL, produced by Sf9 cells seems to have the characteristics of a membrane-associated protein, it may represent a useful source of functional TRAIL with a more physiological interaction at the receptor level. It remains, in fact, to be established whether TRAIL is released physiologically as a soluble mediator from mouse and human cells, as previously described for human CD95L (Mariani et al., 1995; Tanaka et al., 1995).
Our finding that IRPs/caspases are involved in TRAILinduced apoptosis underlines the critical role played by this family of proteolytic enzymes, as they appear to mediate almost every known form of apoptosis (Henkart, 1996). For therapy (Enari et al., 1995; Nicholson et al., 1995; Thornberry et al., 1995; Rouquet et al., 1996) this could imply an excessive nonspecificity with the risk of a generalized block of apoptosis. More selective strategies will have to be devised, if a specific block of only one ligand/receptor system is desired: i.e., inhibitors targeted at the ligand/receptor interaction, at the proteins associated with the proximal signaling complexes and/or at specific IRPs/caspases with potentially unique selectivity.
Acknowledgments
We thank A. Hekele and J.P. Medema for critical reading of the manuscript and M. Pach for skillful technical assistance.
Abbreviations used in this paper
- Ab
antibody
- CD95
CD95/APO-1/ Fas receptor
- CD95L
CD95/APO-1/Fas ligand
- CHO
aldehyde
- cmk
chloromethylketone
- ICE
IL-1β converting enzyme
- IRP
ICE-related protease
- PARP
poly(ADP)-ribose polymerase
- SN
supernatant
Footnotes
Please address all correspondence to P.H. Krammer, Tumor Immunology Program, Division of Immunogenetics, German Cancer Research Center, Im Neuenheimer Feld 280, D-69120 Heidelberg, Germany. Tel.: 49 6221 423717. Fax: 49 6221 411715. E-Mail: P.Krammer@DKFZ-Heidelberg.de
References
- Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- Ashkenaz J, Werb Z. Proteolysis and the biochemistry of life-ordeath decisions. J Exp Med. 1996;183:1947–1951. doi: 10.1084/jem.183.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldin MP, Varfolomeev EE, Pancer Z, Mett IL, Camonis JH, Wallach D. A novel protein that interacts with the death domain of FAS/ APO-1 contains a sequence motif related to the death domain. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- Brunner T, Mogil RJ, LaFace D, Yoo NJ, Mahboubi A, Echeverry F, Martin SJ, Force WR, Lynch DH, Ware CF, et al. Cell-autonomous Fas (CD95)/Fas-ligand interaction mediates activation-induced apoptosis in T-cell hybridoma. Nature (Lond) 1995;373:441–444. doi: 10.1038/373441a0. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan AM, O'Rourke KO, Tewari M, Dixit VM. FADD, a novel death-domain containing protein interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan AM, Orth K, O'Rourke KO, Duan H, Poirier GG, Dixit VM. Molecular ordering of the cell death pathway. J Biol Chem. 1996;271:4573–4577. doi: 10.1074/jbc.271.9.4573. [DOI] [PubMed] [Google Scholar]
- Chrest FJ, Buchholz MA, Kim YH, Kwon TK, Nordin AA. Identification and quantitation of apoptotic cells following anti-CD3 activation of murine G0cells. Cytometry. 1993;14:883–890. doi: 10.1002/cyto.990140806. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Bruno S, Del Bino G, Gorzyca W, Hotz MA, Lassota P, Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- Dhein J, Walczak H, Bäumler C, Debatin KM, Krammer PH. Autocrine T-cell suicide mediated by APO-1/(Fas/CD95) Nature (Lond) 1995;373:438–441. doi: 10.1038/373438a0. [DOI] [PubMed] [Google Scholar]
- Duan H, Chinnaiyan AM, Hudson PL, Wing JP, He WW, Dixit VM. ICE-LAP3, a novel mammalian homologue of the C. eleganscell death protein ced-3 is activated during Fas and TNF-mediated apoptosis. J Biol Chem. 1996;271:1621–1625. doi: 10.1074/jbc.271.3.1621. [DOI] [PubMed] [Google Scholar]
- Enari M, Hug H, Nagata S. Involvement of an ICE-like protease in Fas-mediated apoptosis. Nature (Lond) 1995;375:78–81. doi: 10.1038/375078a0. [DOI] [PubMed] [Google Scholar]
- Enari M, Talanian RV, Wong WW, Nagata S. Sequential activation of ICE-like and CPP32-like proteases during Fas-mediated apoptosis. Nature (Lond) 1996;380:723–726. doi: 10.1038/380723a0. [DOI] [PubMed] [Google Scholar]
- Fadok VA, Volker DR, Campbell PA, Cohen JJ, Bratton DL, Herron PM. Exposure of phospatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Takahashi A, Armstrong R, Kress J, Fritz L, Tomaselli KJ, Wang L, Wu Z, Croce CM, Salveson G, et al. Mch3, a novel human apoptotic cysteine protease highly related to CPP32. Cancer Res. 1995;55:6045–6052. [PubMed] [Google Scholar]
- Fraser A, Evan G. A license to kill. Cell. 1996;85:781–784. doi: 10.1016/s0092-8674(00)81005-3. [DOI] [PubMed] [Google Scholar]
- Galle PR, Hofmann WJ, Walczak H, Schaller H, Otto G, Stremmel W, Krammer PH, Runkel L. Involvement of the CD95 (APO-1/Fas) receptor and ligand in liver damage. J Exp Med. 1995;182:1223–1230. doi: 10.1084/jem.182.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanabuchi S, Koyonagi M, Kawasaki A, Shinohara N, Matsuzawa A, Nishimura Y, Kabayashi Y, Yonehara S, Yagita H, Okumura K. Fas and its ligand in a general mechanism of T-cell mediated cytotoxicity. Proc Natl Acad Sci USA. 1994;91:4930–4934. doi: 10.1073/pnas.91.11.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkart PA. ICE family proteases: mediators of all apoptotic cell death? . Immunity. 1996;4:195–196. doi: 10.1016/s1074-7613(00)80428-8. [DOI] [PubMed] [Google Scholar]
- Ju ST, Panka DJ, Cui H, Ettinger R, El-Khatib M, Sherr DH, Stanger BZ, Marshak-Rothstein A. Fas(CD95)/FasL interactions required for programmed cell death after T-cell activation. Nature (Lond) 1995;373:444–448. doi: 10.1038/373444a0. [DOI] [PubMed] [Google Scholar]
- Kägi D, Vignaux F, Lederman B, Burkin K, Depretere V, Nagata S, Hengartner H, Golstein P. Fas and perforin pathway as major mechanisms of T-cell mediated cytotoxicity. Science (Wash DC) 1994;265:528–530. doi: 10.1126/science.7518614. [DOI] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME. Cytotoxicity-dependent APO-1 (Fas/CD95) associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO (Eur Mol Biol Organ) J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer PH, Dhein J, Walczak H, Behrman I, Mariani S, Matiba B, Fath M, Daniel PT, Knipping E, Westendorp MO, et al. The role of APO-1-mediated apoptosis in the immune system. Immunol Rev. 1994;142:175–191. doi: 10.1111/j.1600-065x.1994.tb00889.x. [DOI] [PubMed] [Google Scholar]
- Kuida K, Lippke J, Ku G, Harding MW, Livingston DJ, Su M, Flavell RA. Altered cytokine export and apoptosis in mice deficient in interleukin-1β converting enzyme. Science (Wash DC) 1995;267:2000–2002. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP)-ribose polymerase by a proteinase with properties like ICE. Nature (Lond) 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- Lippke JA, Gu Y, Sarnecki C, Caron PR, Su MS. Identification and characterization of CPP32/Mch2 homolog 1, a novel cysteine protease similar to CPP32. J Biol Chem. 1996;271:1825–1828. doi: 10.1074/jbc.271.4.1825. [DOI] [PubMed] [Google Scholar]
- Los M, Van de Craen M, Penning LC, Schenk H, Westendorp M, Bauerle PA, Droge W, Krammer PH, Fiers W, Schulze K, Osthoff Requirement of an ICE/CED3 protease for Fas/APO-1 mediated apoptosis. Nature (Lond) 1995;375:81–83. doi: 10.1038/375081a0. [DOI] [PubMed] [Google Scholar]
- Lowin B, Hahne M, Mattmann C, Tschopp J. Cytolytic T-cell cytotoxicity is mediated through perforin and Fas ligand pathways. Nature (Lond) 1994;370:650–653. doi: 10.1038/370650a0. [DOI] [PubMed] [Google Scholar]
- Lynch DH, Watson ML, Alderson MR, Baum PR, Miller RE, Tough T, Gibson M, Davis-Smith T, Smith CA, Hunter K, et al. The mouse Fas-ligand gene is mutated in gldmice and is part of a TNF family gene cluster. Immunity. 1994;1:131–136. doi: 10.1016/1074-7613(94)90106-6. [DOI] [PubMed] [Google Scholar]
- Mariani SM, Matiba B, Armandola EA, Krammer PH. The APO-1/Fas (CD95) receptor is expressed in homozygous MRL/lprmice. Eur J Immunol. 1994;24:3119–3123. doi: 10.1002/eji.1830241231. [DOI] [PubMed] [Google Scholar]
- Mariani SM, Matiba B, Bäumler C, Krammer PH. Regulation of cell surface APO-1/Fas (CD95) ligand expression by metalloproteases. Eur J Immunol. 1995;25:2303–2307. doi: 10.1002/eji.1830250828. [DOI] [PubMed] [Google Scholar]
- Mariani SM, Matiba B, Sparna T, Krammer PH. Expression of biologically active mouse and human CD95/Fas ligand in the baculovirus system. J Immunol Methods. 1996;193:63–70. doi: 10.1016/0022-1759(96)00051-8. [DOI] [PubMed] [Google Scholar]
- Marsters SA, Pitti RM, Donahue CJ, Ruppert S, Bauer KD, Ashkenazi A. Activation of apoptosis by APO-2 ligand is independent of FADD but blocked by crmA. Curr Biol. 1996;6:750–752. doi: 10.1016/s0960-9822(09)00456-4. [DOI] [PubMed] [Google Scholar]
- Martin S, Green D. Protease activation during apoptosis: death by a thousand cuts? . Cell. 1995;82:349–352. doi: 10.1016/0092-8674(95)90422-0. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Reutelingsperger CPM, Mc AJ, Gahon, Rader JA, van Schrie RC, LaFace DM, Green DR. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of bcl2 and abl. J Exp Med. 1995;182:1545–1553. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M, Chinnaiyan AM, Kischkel FC, O'Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, et al. FLICE, a novel FADD-Homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- Nagata S, Golstein P. The Fas death factor. Science (Wash DC) 1995;267:1449–1455. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- Nicholson DW, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Garron Y, Griffin PR, Labelle M, Lazebnik YA, Munday NA, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature (Lond) 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- Ogasawara J, Watanabe-Fukunaga R, Adachi M, Matsuzawa A, Kasugai T, Kitamura Y, Itoh N, Suda T, Nagata S. Lethal effect of the antiFas antibody in mice. Nature (Lond) 1993;364:806–809. doi: 10.1038/364806a0. [DOI] [PubMed] [Google Scholar]
- Onel KB, Tucek-Szabo CL, Ashany D, Lacy E, Nikolic-Zugic J, Elkon KB. Expression and function of the murine CD95/FasR/APO-1 receptor in B cell ontogeny. Eur J Immunol. 1995;25:2940–2947. doi: 10.1002/eji.1830251034. [DOI] [PubMed] [Google Scholar]
- O'Reilly, D.R., L.K. Miller, and V.A. Luckow. 1994. Baculovirus expression vectors. New York Oxford University Press, New York, NY. pp. 109–204.
- Peitsch M, Tschopp J. Comparative molecular modeling of the Fasligand and other members of the TNF family. Mol Immunol. 1995;32:761–768. doi: 10.1016/0161-5890(95)00016-8. [DOI] [PubMed] [Google Scholar]
- Pitti RM, Marsters SA, Rupper S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by APO-2 ligand, a new member of the TNF cytokine family. J Biol Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- Rensing-Ehl A, Fre K, Flury R, Matiba B, Mariani SM, Weller M, Aebischer P, Krammer PH, Fontana A. Local Fas/APO-1 (CD95) ligand-mediated tumor cell killing in vivo. . Eur J Immunol. 1995;25:2253–2258. doi: 10.1002/eji.1830250821. [DOI] [PubMed] [Google Scholar]
- Rothstein TL, Wang JKM, Panka DJ, Foote LC, Wang Z, Stanger B, Cui H, Ju S, Marshak-Rothstein A. Protection against Fas-dependent Th1-mediated apoptosis by antigen receptor engagement in B cells. Nature (Lond) 1995;374:163–165. doi: 10.1038/374163a0. [DOI] [PubMed] [Google Scholar]
- Rouquet N, Pages JC, Molina T, Briand P, Joulin V. ICE inhibitor YVAD-cmk is a potent therapeutic agent against in vivoliver apoptosis. Curr Biol. 1996;6:1192–1195. doi: 10.1016/s0960-9822(02)70688-x. [DOI] [PubMed] [Google Scholar]
- Shima Y, Nishimoto N, Ogata A, Fujii Y, Yoshizaki K, Kishimoto T. Myeloma cells express FAS/APO-1 (CD95) but only some are sensitive to anti-Fas antibody resulting in apoptosis. Blood. 1995;85:757–764. [PubMed] [Google Scholar]
- Stalder T, Hahn S, Erb P. Fas antigen is the major target molecule of CD4+T cell-mediated cytotoxicity. J Immunol. 1994;152:1127–1132. [PubMed] [Google Scholar]
- Suda T, Nagata S. Purification and characterization of the Fas ligand that induces apoptosis. J Exp Med. 1994;179:873–879. doi: 10.1084/jem.179.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Takahashi T, Golstein P, Nagata S. Molecular cloning and expression of the Fas ligand: a novel member of the tumor necrosis factor family. Cell. 1993;75:1169–1178. doi: 10.1016/0092-8674(93)90326-l. [DOI] [PubMed] [Google Scholar]
- Summers, M.D., and G.E. Smith. 1987. A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agric. Exp. Stat. Bull. No. 1555.
- Takahashi A, Earnshaw WC. ICE-related proteases in apoptosis. Curr Biol. 1996;6:50–55. doi: 10.1016/s0959-437x(96)90010-6. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Tanaka M, Brannan CI, Jenkins NA, Copeland NG, Suda T, Nagata S. Generalized lymphoproliferative disease in gldmice, caused by a point mutation in the Fas ligand. Cell. 1994;76:969–976. doi: 10.1016/0092-8674(94)90375-1. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Suda T, Takahashi T, Nagata S. Expression of the functional soluble form of human Fas ligand in activated lymphocytes. EMBO (Eur Mol Biol Organ) J. 1995;14:1129–1135. doi: 10.1002/j.1460-2075.1995.tb07096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari M, Dixit VM. Fas- and TNF-induced apoptosis is inhibited by the poxvirus crmA gene product. J Biol Chem. 1995;270:3255–3260. doi: 10.1074/jbc.270.7.3255. [DOI] [PubMed] [Google Scholar]
- Tewari M, Quan LT, O'Rourke K, Desnoyers S, Zeng Z, Beidler DR, Poirier GG, Salveson GS, Dixit VM. YAMA/CPP32β, a mammalian homolog of CDE-3 is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP)-ribose polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- Thornberry NC, Miller D, Nicholson D. Interleukin-1β converting enzyme and related proteases as potential targets in inflammatory apoptosis. Perspectives in Drug Discovery and Design. 1995;2:389–399. [Google Scholar]
- Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Davis T, Smith, Rauch C, Smith CA, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- Wyllie AH, Morris RG. Hormone-induced cell death: purification and properties of thymocytes undergoing apoptosis after glucocorticoid treatment. Am J Path. 1982;109:78–87. [PMC free article] [PubMed] [Google Scholar]