Abstract
We have analyzed the progressive changes in the spatial distribution of telomeres during meiosis using three-dimensional, high resolution fluorescence microscopy. Fixed meiotic cells of maize (Zea mays L.) were subjected to in situ hybridization under conditions that preserved chromosome structure, allowing identification of stage-dependent changes in telomere arrangements. We found that nuclei at the last somatic prophase before meiosis exhibit a nonrandom, polarized chromosome organization resulting in a loose grouping of telomeres. Quantitative measurements on the spatial arrangements of telomeres revealed that, as cells passed through premeiotic interphase and into leptotene, there was an increase in the frequency of large telomere-to-telomere distances and a decrease in the bias toward peripheral localization of telomeres. By leptotene, there was no obvious evidence of telomere grouping, and the large, singular nucleolus was internally located, nearly concentric with the nucleus. At the end of leptotene, telomeres clustered de novo at the nuclear periphery, coincident with a displacement of the nucleolus to one side. The telomere cluster persisted throughout zygotene and into early pachytene. The nucleolus was adjacent to the cluster at zygotene. At the pachytene stage, telomeres rearranged again by dispersing throughout the nuclear periphery. The stagedependent changes in telomere arrangements are suggestive of specific, active telomere-associated motility processes with meiotic functions. Thus, the formation of the cluster itself is an early event in the nuclear reorganizations associated with meiosis and may reflect a control point in the initiation of synapsis or crossing over.
Anearly universal element in the choreography of genetic material during meiosis is the pairing of homologous chromosomes. Exactly how the homology search and chromosome pairing processes are accomplished remains a mystery, despite decades of relevant cytological studies (34, 40, 41). The complete pairing of homologs for species with large genomes is a formidable task likely to require the ability to move chromosomes and to reorganize the nucleus. Although some cases of premeiotic pairing have been observed (42, 64), the absence of premeiotic pairing is generally well documented (5, 12, 33, 54). Thus, some force or active organizing mechanism must be operative to ensure timely chromosome alignment and synapsis. Indications of such forces are found, for example, in the dramatic changes in chromosome morphology associated with the initiation of synapsis in maize (12). In that study, Dawe et al. described a transition stage called prezygotene, during which sister chromatids were slightly separated and the normally spherical blocks of heterochromatin called knobs were axially elongated. Additionally, prezygotene marks the first stage at which pairing of homologous knobs could be observed. The telomeric knobs, which relocated to the nuclear envelope at prezygotene, were the first to pair followed by pairing of interstitial knobs later in zygotene. We wanted to determine whether the behavior of the telomeric knobs observed by Dawe et al. (12) reflected the general behavior of all telomeres, and, if so, could we correlate their behavior with the homology search and chromosome synapsis.
Many studies have drawn attention to a particular organization of meiotic chromosomes in which the ends of the chromosomes are pointed towards the same side of the nucleus, usually touching the nuclear envelope. This polarized organization of chromosome ends, historically called the bouquet stage, occurs during meiotic prophase (16). The bouquet stage is widely conserved in nature, having been observed in yeast, plants, and animals (13). The bouquet stage always coincides with chromosome pairing, leading to the conclusion that the bouquet, as a nuclear structure, is needed for the chromosome pairing process (11, 21, 54).
The exact role of the bouquet during meiotic prophase is difficult to discern because of the inherent complexities of chromosome pairing itself. Cytogenetic and molecular genetic studies have shown that meiotic chromosome pairing is composed of two separable events, chromosome alignment and chromosome synapsis. Chromosome alignment, occasionally referred to as “pairing,” is generally thought to result from homology searching interactions, the timing and nature of which are poorly understood (38, 64). Synapsis, on the other hand, can be clearly defined as the intimate juxtaposition of chromosomes when joined by the synaptonemal complex, a conserved tripartite structure connecting meiotic chromosomes along their length (62). While alignment of homologues is generally thought to precede and contribute to the synapsis, the two processes can be uncoupled. For instance, homologous chromosomes can align, recombine, and segregate in the absence of synapsis (1, 34, 40). In addition, a synaptonemal complex can form between nonhomologous chromosomes or chromosome regions, illustrating that synapsis is not dependent on homologue alignment (34). The extent to which synapsis has occurred delineates the first three stages of meiotic prophase: leptotene, zygotene, and pachytene; these stages are characterized by synapsis of none, part, or all of their chromosomes, respectively. The correct identification of these stages is critical for our goal of defining the relationship between the behavior of meiotic chromosomes and the structure of the meiotic nucleus during the bouquet stage.
Towards this end, it was necessary to establish the exact timing and duration of the bouquet stage. To begin, we wanted to know whether the bouquet forms de novo or simply reveals a preexisting nuclear organization that becomes evident upon chromosome condensation. This question has not been answered by conventional cytology because the identification of telomeres requires the resolution of individual chromosomes. Therefore, the location of telomeres at the onset of chromosome condensation in meiotic prophase is ambiguous. To overcome this problem, one can use fluorescence in situ hybridization (FISH)1 with probes that are specific for telomeres. This approach avoids the limitations described above because it does not rely on the ability to resolve the axial paths of individual chromosomes. In a recent study using telomere FISH probes on sectioned testes of mouse and human, Scherthan et al. found that telomeres undergo dramatic changes in distribution during early meiosis (51, 54). Telomeres in mouse spermatagonia (premeiotic) nuclei were not clustered, whereas telomeres at zygotene were clustered in the classical bouquet configuration. A progression of telomere arrangements was observed within these nuclei; scattered telomeres at the spermatagonial stage became peripheral at preleptotene, and eventually peripheral and clustered at zygotene. The inability to identify nuclei at the leptotene stage resulted in an incomplete description of sequential developmental stages in which preleptotene was followed by zygotene (54). Therefore, the timing of the bouquet stage relative to the onset of meiotic prophase could not be directly determined.
To precisely ascertain the organization of the telomeres in nuclei known to be at premeiotic interphase, leptotene, or zygotene, we have applied telomere FISH to intact maize meiotic cells. The inherent cytological clarity and developmental synchrony of these cells along with the excellent genetics of maize have provided workers with a model system to investigate structure–function relationships operating during meiosis (6, 19, 24, 25, 49, 56). The chromosomes of maize meiocytes undergo normal chiasmate meiosis (6, 49). In the early literature of McClintock and others describing the meiotic chromosomes of maize, little mention is made of the bouquet stage (49). However, the presence of a bouquet structure in maize is clearly documented in ultrastructural analyses using serial section reconstructions of entire nuclei (22, 23, 46). In these studies, the ends of the chromosomes in zygotene and pachytene staged nuclei were shown to be polarized, clustering on or near a small region of the nuclear envelope.
To take full advantage of the maize meiocyte system, we have adapted a FISH protocol for use with higher plant material, using methodologies established to study the threedimensional organization of the nucleus (2, 12, 14, 15, 31, 43, 60). Telomere kinetics were monitored in optically sectioned nuclei spanning a wide developmental range, including the last mitosis before meiosis. A quantitative spatial analysis of telomere distributions was carried out to document the extent of the changes that were observed. This work represents the first detailed three-dimensional analysis of telomere dynamics at stages before and during early meiotic prophase, a period that has been only briefly examined in previous studies. As a consequence of our analysis, we have found that the bouquet structure forms de novo, at the end of leptotene, just before chromosome synapsis. Thus, the duration of the bouquet stage was observed to be limited to meiotic prophase, suggestive of a meiosis-specific telomere movement system. Finally, we discuss the significance of this timing in relation to other nuclear changes associated with the initiation of synapsis.
Materials and Methods
Plant Materials and Fixation of Reproductive Organs
Inbred lines of maize (Zea mays L., 2n = 20) were grown and harvested throughout the year at the Oxford greenhouses (Berkeley, CA). Three inbred lines, KYS+, W23+, and A344+, were used to obtain the majority of the nuclei characterized in this study. A total of 57 nuclei was completely modeled and the models were analyzed mathematically (see below). In addition to the standard inbreds, two nuclei (one pachytene and one diakinesis) were derived from a BMS line (without B chromosomes) that was ditelocentric and tetrasomic for chromosome 3 (2n = 22, gift from R. Staub).
Anthers from preemerged tassels were fixed as previously described (12) with the following modifications. Whole anthers (20–40) were fixed in 5 ml 4% freshly prepared formaldehyde in meiocyte buffer A (15 mM Pipes-NaOH, pH 6.8, 80 mM KCl, 20 mM NaCl, 0.5 mM EGTA, 2 mM EDTA, 0.15 mM spermine tetra HCL, 0.05 mM spermidine, 1 mM DTT, 0.32 M sorbitol) for 2 h at room temperature. After fixation, anthers were washed (four times, 15 min each) with meiocyte buffer A and used immediately or stored in air-tight containers at 4°C in sterile-filtered meiocyte buffer A. The majority of data was collected on anthers used immediately or within several days of fixation. For some of the zygotene and pachytene stages, fixed anthers were stored for up to 8 wk without any noticeable deterioration in the cells or nuclei.
Acrylamide Embedding of Meiocytes
We used a novel polyacrylamide embedding technique originally developed for Drosophila polytene nuclei (60). A 15-mm-diam ring of nail polish (Sally Hanson's Hard as Nails) was painted onto ethanol-washed glass slides and allowed to dry for at least 4 h. Fixed anthers, submerged in 60 μl of meiocyte buffer A, were cut open at their tips and the meiocytes were gently extruded out of the open end. Meiocytes suspended in meiocyte buffer A were transferred (12 μl) by micropipetting onto the slide followed by immediate addition of 6 μl of activated acrylamide stock. The activated acrylamide stock was made by addition of 25 μl of 20% ammonium persulfate and 25 μl of 20% sodium sulfite to 500 μl of gel stock (15 mM Pipes-NaOH, pH 6.8, 80 mM KCl, 20 mM NaCl, 0.5 mM EGTA, 2 mM EDTA, 0.15 mM spermine tetra HCL, 0.05 mM spermidine, 1 mM DTT, 0.3 M sorbitol, 15% polyacrylamide [from a 30% 29:1 acrylamide/bis acrylamide stock]). The slides were rocked and rotated for 15–20 s until mixed, and a lens paper-cleaned coverslip (22 × 22 × 1.5) was placed on top for 20 min, and then removed, leaving a thin pad of acrylamide with embedded meiocytes attached to the slide. All washing steps below consisted of placing a drop (100–200 μl) on top of the face-up acrylamide pad followed by rotary shaking at the maximum speed possible for 10 min at room temperature unless specified otherwise. Solutions were removed by aspiration. Although the 5% acrylamide sheet can be rather delicate, typically the acrylamide pad made it through the procedure for 70% of the slides.
Oligonucleotide Probe and In Situ Hybridization
We used a modification of the three-dimensional FISH methods established by Dernburg et al. to be compatible with chromosome preservation (14, 15). This protocol uses a high temperature codenaturation of target and probe sequences by placement of glass slides on a PCR block. Newly polymerized acrylamide pads were washed (four times, as described above) with buffer (15 mM Pipes-NaOH, pH 6.8, 80 mM KCl, 20 mM NaCl, 0.5 mM EGTA, 2 mM EDTA) to remove unpolymerized acrylamide, followed by four equilibration washes with prehybridization buffer (50% deionized formamide, 2× SSC). Hybridization buffer (50% deionized formamide, 2× SSC, 1–3 μg/ml telomere oligonucleotide) was added, removed, and added a second time. The telomere oligonucleotide probe had the sequence 5′-CCCTAAACCCCTAAACCCTAAACCCTAAA-3′. The oligonucleotide was labeled its 5′ end by direct incorporation of “Fluorescein-ON phosphoramidite” (catalogue 5235; Clontech Laboratories, Inc., Palo Alto, CA) as the last step in the synthesis (HHMI DNA Synthesis Facility, University of California, San Francisco). The initial oligonucleotide powder was dissolved in 10 mM Tris-HCL (pH 8.0), and its concentration was determined spectrophotometrically using the formula 1.0 A260 = 33 μg/ml. A final 20–30 μl drop of hybridization buffer was added, sealed under a coverslip using rubber cement, and then placed on a 40°C warming plate for 30 min. The chromosomes were denatured on a PCR block at 94°C for 6 min, followed by overnight incubation at 37°C. The use of various competitors and blocking reagents including yeast tRNA, calf thymus DNA, unlabeled excess oligonucleotides, and Denhardt's solution did not show consistent improvement of signal to noise ratios and was therefore excluded from future experiments (data not shown). Because we found the optimal denaturation temperature varied between anthers and experiments, slides were usually prepared in triplicate and denatured for 6 min each at 92°, 94°, or 96°C. Too much heat caused noticeable disruption of chromosomes and cytoplasm, and such samples were not used (see below) despite their typically high signal to noise ratios.
After overnight incubation, the samples were washed sequentially with 2× PBS containing 0.1% Tween-20 (four times, as described above) and 1× PBS (three times), and then were stained with 10 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) in 1× PBS for 30 min at room temperature. Excess DAPI was removed by washing with 2× PBS (three times), followed by 1× PBS (three times). Slides were equilibrated in an FITC-stabilizing medium (95% glycerin, 50 mM Tris base, 2% N-propyl gallate) three times for 20 min each and mounted in the same medium. Data sets were collected immediately thereafter and up until 48 h. Within a single slide, we occasionally found isolated nuclei (no cytoplasm) with relatively bright FISH signals compared with those from intact cells. We reasoned that the nuclei from intact cells would be less likely to have rearranged as a result of handling or manipulation, despite the potential cost of reduced signal to noise ratios. For these reasons, data collection was predominantly limited to those nuclei (>90% in this paper) within intact cells or tissues (not shown). Since meiocytes have a distinctly dense cytoplasm lacking large vacuoles, assessment of cellular preservation could be readily made.
Three-dimensional Microscopy and Image Processing
All images were recorded using an IMT-2 wide-field microscope (Olympus Corp., Lake Success, NY), making use of one of two oil immersion lenses, ×60 NA 1.4 PlanApo (Olympus Corp.) or ×100 NA 1.4 PlanApo (Nikon Inc., Garden City, NY) (30). In both cases, the data were oversampled in the X, Y, and Z dimensions with typical XYZ voxel dimensions of 0.07 × 0.07 × 0.2 μm3. The computerized light microscope workstation has been described elsewhere (14, 15). Original data collection was made on an area extending at least 2 μm beyond the nuclear border in Z, and extending at least 5 μm in X and Y from the widest edges of the nuclei. After three-dimensional iterative deconvolution (7) of the original fullsized data sets, individual nuclei were computationally cropped in all dimensions. The resulting data subsets were used for all subsequent model building and image display, resulting in the exclusion of the surrounding cytoplasm and neighboring cells from the figures presented. The images presented were adjusted for brightness and contrast using linear scaling of the minimum and maximum intensities, but no additional image processing such as edge or local contrast enhancements were used, except for the volume-rendered projections in Fig. 3, C and E.
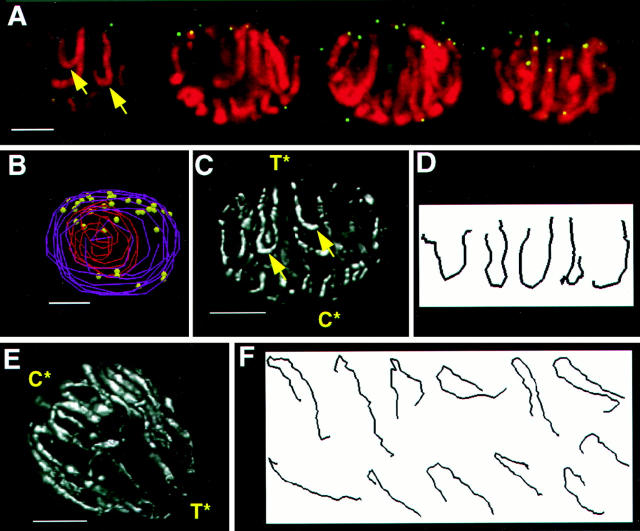
Figure 3.
Polarized chromosomes and telomere grouping at the last prophase before meiosis. 3-dimensional telomere FISH was carried out on acrylamide-embedded cells (see Materials and Methods) from 0.2-mm anthers. Two nuclei at the last prophase before meiosis are shown (A–D, and E–F). To convey the 3-dimensional data in 2-dimensional images, the original data sets (60–80 optical sections each) were converted to a series of four sequential projections each with an effective focal planes of 1/4 the nuclear depth (∼5 microns). The four 1/4 nucleus projections are displayed in sequence to give visual access to the full complement of data in the original data set (A). Pseudocolor overlays showing the DNA (DAPI image, red) and the telomeres (bright dots in the FITC images, green/yellow) for each projection show a loose grouping telomere signals (green dots). (B) A 3-dimensional model of the nucleus in A is shown. The edges of the nucleus (purple wire) and the nucleolus (red wire) are indicated along with the positions of the telomeres (yellow spheres). (C) Volume-rendered projections of the entire nucleus is also shown to convey the overall paths of the chromosomes. The telomere hemisphere (T*) and the presumed centromere hemisphere (C*) are indicated. (D) The axial paths of continuous chromosome were determined and they are individually displayed side by side. (E) A volume-rendered projection of a different nucleus is shown along with the paths of the 12 longest chromosome tracings (F). The tracings in F are rotated clockwise about 10° relative to the projection in E. Each nucleus has a large, singular internal nucleolus (not shown for nucleus in E). Bar, 5 μm.
Modeling Building and Spatial Analysis of Telomere Positions
Individual nuclei were modeled using the Priism software program 3DModel (8). Each model file contains a list of points with the real-space coordinates of three objects: the edge of the nucleus (Object-1), the edge of the nucleolus (Object-2), and the telomeres (all the points in Object-3). Negative control experiments were carried out to confirm that none of the bright spots identified as telomeres could be explained as nonspecific background artifacts. An FITC-labeled oligonucleotide probe (produced as described above) homologous to the noncoding strand of a single copy maize gene was used under standard conditions (see above). The resulting FITC images lacked bright dots, but exhibited nucleolar staining similar to the nonspecific background staining seen with the telomere probe (see Fig. 2 B). In this study, we have decided to define telomeres as the discrete spots (three to six pixels across in the X and Y dimensions) in the FISHtreated nuclei. We also required that these spots be brighter (by a factor of > 1.5-fold) than the average intensity of pixels in the nucleolus, and within or touching the DAPI-revealed chromatin of the nucleus. These criteria allowed us to identify telomeres in decondensed chromatin where the DAPI images provide little to no assistance in recognizing the ends of chromosomes. Typically, 30–80% of the expected telomeres were observed.
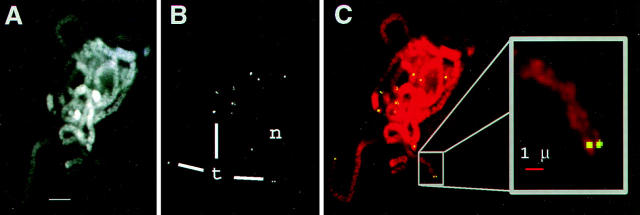
Figure 2.
Specificity of oligonucleotide telomere FISH. A partially ruptured and flattened pachytene nucleus following FISH with telomere oligonucleotide probe (see Materials and Methods). Images show the relationship of chromosomes (A, DAPI image) and telomeres signals (B, FITC image), by two-colored overlay (C) with inset enlargement. The nucleolus (n) and some telomeres (t) are indicated. Bars: (A) 5 μm; (inset) 1 μm.
We analyzed the spatial distribution of telomeres in two different ways. The first method compares the distribution of distances (Euclidean pointto-point distances) between pairs of telomeres to the distribution generated from randomized points in nuclei but outside the nucleolus. To accomplish this task, we used a surface harmonic expansion to fit surfaces to the boundaries of the nucleus and nucleolus, as defined by the the coordinates specified in the 3DModel files (43). To generate a random curve for comparison, we averaged the pairwise distance values (normalized to the maximum diameter) for 16 nuclei (eight at premeiotic interphase, and eight at leptotene). Monte Carlo simulations were used to generate a list of 5,000 randomly placed points within a given nucleus but outside the nucleolus. The diameter-normalized pairwise distances between all possible pairs of the 5,000 points (12.5 million distances) were determined. The distributions of random distances were remarkably similar for different nuclei at premeiotic interphase and leptotene (not shown). The diameternormalized pairwise distances were determined for the positions of the telomere signals (yellow spheres in 3DModels), and the values were averaged according to stage. A nucleus with 30 telomere signals yields 435 nonreciprocal pairwise distances. The largest pairwise distances of Object-1 and Object-2 of the 3DModel files are presented in Table I as diameter of the nucleus and nucleolus, respectively.
Table I.
Measurements of Telomere Distributions and Sizes of Nucleus and Nucleolus*
| Measurement | Premeiotic prophase‡ | Premeiotic interphase | Leptotene | Bouquet | Pachytene | Diakinesis‡ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of cells | 2 | 28 | 11 | 11 | 4 | 1 | ||||||
| Mean pairwise distance§ | 46, 48 | 49.8 (±3.9) | 52.2 (±3.7) | 25.2 (±7.5) | 45.2 (±6.1) | 57 | ||||||
| Median pairwise distance§ | 48, 51 | 51.3 (±4.8) | 53.5 (±5.1) | 22.4 (±8.6) | 44.9 (±7.6) | 58 | ||||||
| Nucleus diameter (mm)‖ | 19.9, 20.0 | 18.1 (±1.3) | 17.4 (±2.1) | 22.9 (±1.9) | 25.1 (±4.4) | 24.1 | ||||||
| Nucleolus diameter (mm)‖ | 11.6, 11.6 | 10.9 (±0.98) | 10.2 (±0.84) | 12.0 (±0.62) | 11.3 (±0.73) | 11.0 | ||||||
| Number of FISH Signals | 33, 21 | 18.0 (±4.5) | 16.8 (±4.3) | 21.6 (±8.1) | 35.2 (±18) | 41 | ||||||
| % signals inner 50% ¶ | 17 (54) | 24 (188) | 27 (165) | 5.6 (195) | 13 (141) | nd |
Based on average values for the same nuclei analyzed for Figs. 7 and 8, standard deviations are shown in parentheses.
Single nucleus measurements, not averages, are shown.
Nonreciprocal pairwise distances normalized the nuclear diameter.
Based on the largest single point-to-point distance derived from the 3D model objects (see Materials and Methods).
See Materials and Methods, number in parentheses is the total number of telomeres.
For the determination of telomere positions relative to the inner and outer 50% of the nuclear volume, we used the method described above to partition the space in the nucleus into an inner and outer half, excluding the space occupied by the nucleolus (43). For the 5,000 randomly placed points in each nucleus, the distance to the nuclear envelope was determined, and the median value of these distances was used as the threshold to classify telomeres as being in the inner or outer half of the nuclear volume.
Results
Preservation of Nuclear Morphology Is Accomplished with a Three-Dimensional Telomere FISH Protocol
We have used molecular cytology and computerized light microscopy to monitor the dynamic behavior of telomeres during meiosis. We wished to avoid the limitations and possible artifacts associated with conventional cytological methods in which samples are routinely squashed, dehydrated, or physically sectioned. To specifically preserve the morphology of the meiotic nuclei, we have adapted for use with higher plant cells a novel polyacrylamide-embedding technique originally developed for analysis of Drosophila polytene chromosomes (60). The acrylamide-embedded, formaldehyde-fixed cells were subjected to FISH with a fluorescently labeled telomere probe (see below). The FISH protocol we used was derived from other three- dimensional FISH protocols known to be compatible with the preservation of chromatin and chromosome structure (14, 15). The final protocol developed for the maize meiocytes is detailed in the Materials and Methods. Throughout the initial stages of developing these procedures, we required that the nucleus remain intact and that the chromosome morphology should resemble that of cells that had been fixed and stained with the DNA-specific dye, DAPI.
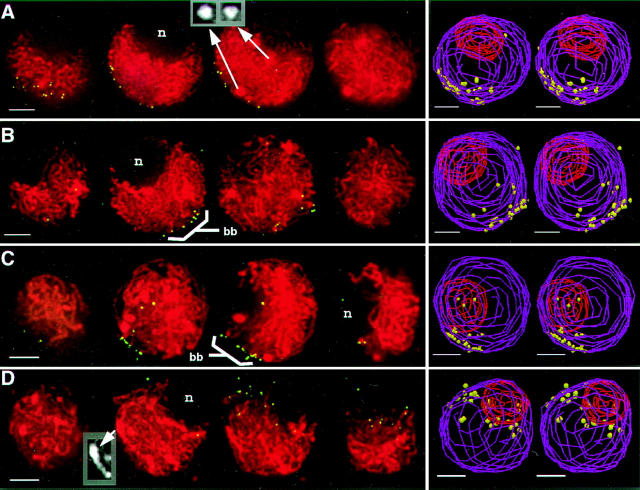
Fig. 1 shows representative DAPI images of staged nuclei following successful in situ hybridization. These images illustrate that nuclei and the chromosomes within them were well preserved. The morphological detail seen in the DAPI images is used in conjunction with several other criteria to stage the meiocytes. Although meiocytes from a single anther are synchronous, we typically pooled meiocytes from 3–6 similarly sized anthers on each slide. Consequently, stages were identified on a nucleus by nucleus basis using the criteria detailed in Fig. 1 (see legend). The multiple criteria for staging are detailed here because proper staging is essential for accurately sorting out the temporal relationships among multiple meiotic processes.
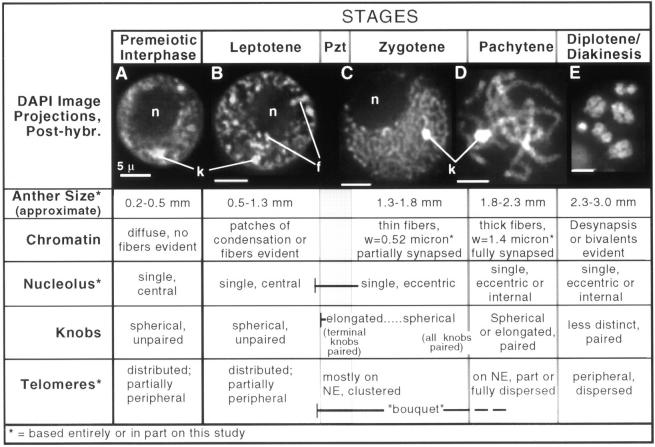
Figure 1.
Criteria for staging meiocytes after FISH. A summary of the multiple criteria used to determine meiotic stages in maize meiotic is shown, along with DAPI images of representative nuclei (FITC telomere signals not shown). DAPI images are shown as single optical sections (A and B) or projections of optical sections spanning 2–3 microns in the Z dimension (C, D, and E). Each image is from a data stack subvolume originally containing an entire nucleus (see Materials and Methods). Chromatin and chromosome fiber appearances, the most definitive characteristics of meiotic stages, are considered together with anther size, nucleolus position, and knob morphology in classifying individual nuclei. Asterisks (*) indicate those criteria established in part or in full from this study; additional staging criteria are taken in part or in full from the literature (12, 22, 23, 46). Heterochromatic knobs (k), condensed fibers (f), and the position of the nucleolus (n) are indicated. The five conventional stages of the prophase of meiosis I (leptotene, zygotene, pachytene, diplotene, and diakinesis) are indicated at the top. Chromosomes of leptotene, zygotene, and pachytene nuclei are not yet synapsed, partially synapsed, or completely synapsed, respectively. Chromosomes of diplotene and diakinesis have desynapsed and are further condensed. Additionally, premeiotic interphase and prezygotene (Pzt, and see text) are indicated.
Equally critical to the proper interpretation of molecular cytological data is the assurance that discrete FISH signals are reliably reporting the position of the intended target sequences. For the detection of telomeres, we designed an oligonucleotide probe cosynthetically labeled at the 5′ end with FITC. The sequence chosen was based on the Arabidopsis telomere-repeat sequence, (TTTAGGG)n, which had been successfully used with other grass species (50). Fig. 2 shows the telomere FISH results from a pachytene nucleus that was purposely ruptured and flattened in order to reveal isolated chromosome arms. The chromosomes (DAPI image, Fig. 2 A) and the telomeres (t, FITC image, Fig. 2 B) from a single optical section capturing most of the flattened nucleus are overlaid (Fig. 2 C) to show that the FISH signals were found at the ends of the chromosomes (see enlarged inset from Fig 2 C). The restriction of telomere FISH signals to the ends of chromosomes was consistently observed in numerous meiotic nuclei at pachytene or later, as well as in somatic nuclei undergoing mitosis (Bass, H.W., unpublished observations). Occasionally, spots were observed to be outside the nucleus. Subsequent analyses with multiple wavelength imaging revealed that the cytoplasmic spots (from zero to several per cell) typically resulted from nonspecific autofluorescence (not shown). The double dots at the ends of the pachytene chromosomes (e.g., inset Fig. 2 C) are interpreted to represent the ends of the homologous chromosomes. The sister chromatids were not resolved in our images (for zygotene or pachytene, and by inference leptotene). From these analyses, we found no evidence for telomeric sequences at internal chromosomal loci.
Chromosomes Are Polarized at the Last Prophase before Meiosis
Having established the conditions for direct labeling of telomeres on pachytene chromosomes, our analysis was extended to include the earlier stages where little is known about telomere arrangements. It was possible to directly test whether the bouquet arrangement of the nucleus existed before meiotic prophase. We first asked whether we could find any evidence for a nonrandom orientation of chromosome ends in nuclei developmentally committed to the meiotic pathway. The persistence of the anaphase configuration of chromosomes throughout an entire cell cycle results in a polarized nucleus with telomeres and centromeres occupying opposite hemispheres of the nucleus. Such polarized nuclei are said to be in a Rabl configuration, as first described by Rabl in 1885 (47) and subsequently documented for a number of different species (10, 20, 26, 32, 43, 47, 55, 61). The Rabl orientation is most easily identified in nuclei at mitotic prophase when condensation reveals the chromosome arrangements.
We therefore started our analysis at the last mitosis preceding meiosis where we found evidence for a Rabl-like configuration as shown in Fig. 3. The telomeric hemisphere was identified by the congregation of telomere FISH signals (A, green spots; B, yellow spheres) near the top of this nucleus. To identify the centromeric hemisphere, we took advantage of previous observations that anaphase movements of metacentric or submetacentric chromosomes create “U” or “J” shaped chromosome paths in which the turn around point reveals the centromere position (10, 13, 20). In our data, these “U-turns” in the chromosome (arrows, A and C, Fig. 3) were interpreted as the approximate centromere positions, best seen in volume-rendered projections of whole nuclei (Fig. 3, C and E). We traced the paths of the chromosomes in the original datasets of the nuclei used to make the projections shown in C and E (Fig. 3). The nucleus in A yielded the traces shown in D, and the nucleus used to make the projection in E yielded the traces shown in F (see legend). Within each nucleus, the chromosome paths showed a consistent orientation. Measuring the size ratios of the chromosome arms on each side of a U-turn, we obtained ratio values (data not shown) that were comparable to the actual arm ratio values known for maize (42). Although we were unable to specifically identify each chromosome, we believe that the chromosome paths and the arm ratios together provide a reasonable estimate of where the centromeres are located in the nucleus (C*, Fig. 3). Thus, the chromosomes in these somatic prophase nuclei were polarized in a Rabl-like configuration, associated with a definite, but loose grouping of many of the telomeres in one half of the nucleus.
Telomere Grouping Is Reduced during Premeiotic Interphase and Leptotene
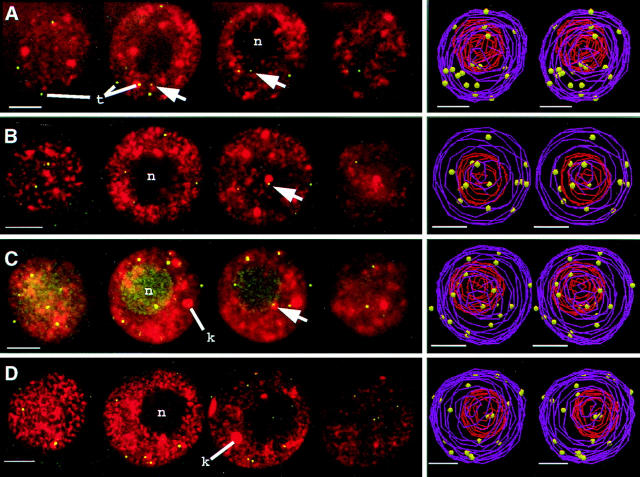
Having observed a telomere grouping associated with a polarized nucleus at the last prophase before meiosis, we asked to what extent did this grouping persist into the meiosis. To our surprise, we found that premeiotic interphase and even leptotene telomeres were not persistently grouped, nor did they tend to be located exclusively near the nuclear envelope. Examples of these findings are shown in Fig. 4. The first two panels (Fig. 4, A and B) show nuclei at premeiotic interphase. For all nuclei at premeiotic interphase, some telomeres could be observed to be more than one micron from the nuclear envelope (arrows). Spherical knobs (k) and an internalized nucleolus (n) are evident. We modeled the positions of the telomeres and the boundaries of the nucleus and nucleolus for 28 premeiotic nuclei and 11 leptotene nuclei. The stereo-pair projections of these models show that some of the premeiotic interphase nuclei have a slight grouping of telomeres (Fig. 4 A) whereas others did not (Fig. 4 B). Two leptotene nuclei (Fig. 4, C and D) show telomeres distributed throughout the nuclear volume with little resemblance to the grouping seen at the last prophase before meiosis (compare models of Fig. 4, A and B with that from Fig. 2 B).
Figure 4.
Telomere distribution at premeiotic interphase and leptotene. Sequential color overlay projections of four nuclei subjected to telomere FISH are shown for the DAPI (DNA, red) and FITC (telomeres, green) images as described in Fig. 3. (A and B) Two premeiotic interphase nuclei show an internal nucleolus (n) and distributed telomeres (t). Arrows indicate examples of individual telomeres more than one micron from the nuclear periphery. 3-dimensional models (see Materials and Methods) are presented as cross-eyed stereo-pair projections at right showing the edges of the nucleus (purple wire) the nucleolus (red wire), and the telomeres (yellow spheres). (C and D) Two leptotene nuclei also show telomeres distributed throughout the nucleus and an internalized nucleolus. Bar, 5 μm.
A Telomere Cluster Forms de novo; Direct Detection of the Bouquet
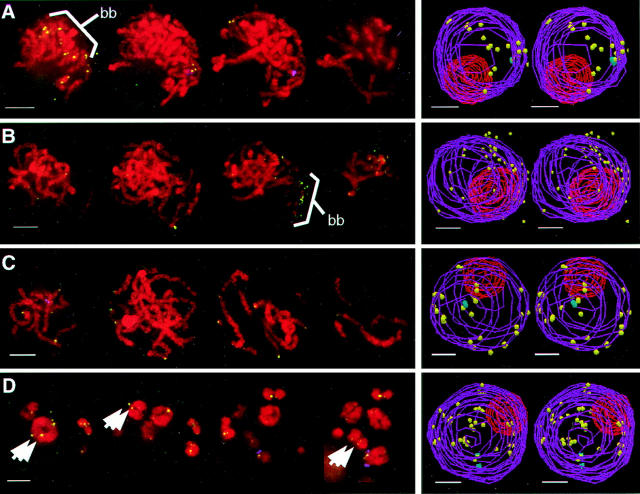
In striking contrast to the telomere distribution patterns in premeiotic and interphase nuclei, the telomeres of the bouquet stage were clustered at the nuclear periphery. The four bouquet nuclei shown in Fig. 5 illustrate that the nucleus has undergone a substantial reorganization. We found that the earliest bouquet stage nuclei occurred in nuclei identified as late leptotene or prezygotene stage. For example, the nucleus in Fig. 5 A does not yet show signs of prezygotene-specific morphology (elongated knobs), whereas the nucleus in Fig. 5 B, from a similarly sized anther shows elongated knobs and thin chromosome fibers with no signs of synapsis. These early bouquet nuclei were relatively rare and in them the telomere cluster site (bb in Fig. 5 b) was on the opposite side of the nucleus from the nucleolus (n, Fig. 5). At zygotene, the telomere cluster was consistently colocalized with the nucleolus (Fig. 5, C and D) such that the nuclei could be bisected into two distinct hemispheres, one containing the full complement of telomere signals and the nucleolus, and the other devoid of either (Fig. 5, C and D). In all cases, whether early bouquet or zygotene bouquet, the nucleolus took up an eccentric position in the nucleus. Although the bouquet stage can be identified in many species by the parallel arrangement of chromosome ends, this arrangement is not obvious in maize (6, 12, 36). Therefore our results show that telomere FISH signals or nucleolus position can be used to identify the bouquet stage in maize.
Figure 5.
Telomere distribution at the bouquet stage. Sequential color overlay projections of four nuclei subjected to telomere FISH are shown for the DAPI (DNA, red) and FITC (telomeres, green) images as described in Fig. 3. (A and B) Nuclei identified to be at the “early bouquet” stage (see Results and Discussion) show clustering of telomeres at the nuclear periphery. The nucleolus (n) is remote from the base of the bouquet (bb) in these early bouquet nuclei. (C and D) Two representative zygotene nuclei show colocalization of the base of the bouquet with the nucleolus (n). Gray scale images of knobs are included (insets with arrows) to illustrate the spherical (A) and elongated (D) shapes of these heterochromatic blocks, features used to stage the nuclei (see text and Fig. 1).
Telomeres Disperse, but Remain Peripheral during Middle-Late Pachytene
The telomere cluster was found to disperse throughout pachytene as shown in Fig. 6. We used the progressive decrease in chromosome crowding to indicate the early, middle, and late substages of pachytene. Nuclei identified as early or middle pachytene (Fig. 6, A and B) were found to have a telomere distribution that was intermediate between the zygotene bouquet cluster and the dispersed, peripheral distribution found at late pachytene (Fig. 6 C). Pachytene nuclei, like those in zygotene, typically gave good hybridization signals resulting in detection of more than 80% of the expected telomeres. Most of these signals were found on or near the nuclear periphery as seen by stereo viewing of the models (Fig. 6 C). By the end of pachytene, the telomeres were fully dispersed (Fig. 6 C). By diakinesis the bivalent organization of the chromosomes was evident (Fig. 6 D). At this stage, the synaptonemal complex has presumably dissolved and the telomere pairs at the end of homologous chromosomes (double arrows Fig. 6 D) were connected only via the nearest chiasmata; thereby increasing the space between homologous telomeres.
Figure 6.
Telomere distribution at pachytene and diakinesis. Sequential color overlay projections of four nuclei subjected to telomere FISH are shown for the DAPI (DNA, red) and FITC (telomeres, green) images as described in Fig. 3. (A–C) Examples of pachytene nuclei identified as early pachytene (A), middle pachytene (B), and late pachytene (C) are shown. (D) The bivalents of a diakinesis have widely separated homologous telomeres (double arrows), reflecting the desynapsis of homologues. In all cases the nucleolus is seen to be peripherally located, but less common examples of an internal nucleolus at late pachytene and diakinesis have been observed (not shown).
Spatial Analysis of Telomere Positions Confirm that the Bouquet Structure Forms de novo
Mathematical analyses, based on the spatial X, Y, and Z coordinates of each telomere, were used to objectively quantify the changes in telomere positions that were revealed by inspection of the images and models (Figs. 3–6). The analyses of overall telomere distributions are shown in Figs. 7 and 8 and summarized in Table I. We first analyzed the distribution of distances between the telomeres relative to the diameter of the nucleus. The Euclidean distances between all possible pairs of points in a given volume has a characteristic distribution (27). For comparison with the telomere data, the distribution of distances that characterizes the available space in nuclei with large internal nucleoli were determined as follows. We used a Monte Carlo simulation to generate 5,000 randomly placed points within individual nuclei, but outside the nucleolus (see Materials and Methods). The distribution of pairwise distances was determined for the randomly placed points in order to derive a “random” curve that took into account the presence of the large nucleolus. This allowed us to compare the distribution of telomeres to the distribution of available space as shown in Fig. 7. The distributions of pairwise distances for telomeres in nuclei preceding the bouquet stage are similar to each other, and when compared to the random curve, show a tendency to have greater than expected numbers of large distances (Fig. 7 A). In striking contrast, the clustering of telomeres at the bouquet stage is clearly evident in the dramatic shift towards smaller values in the pairwise distances curve. The dispersal of telomeres after the bouquet stage is revealed in Fig. 7 (B) by the shift in the pairwise distances curve back towards the larger pairwise distance categories. The proximity of homologous telomeres in paired chromosomes (as seen for example in Fig. 2 C), is reflected in the greater than expected frequency of short distances at the left-most region of the curve (compare the values at different stages for the smallest pairwise distance category, 10%). These data indicate that the telomere cluster is not formed by a gradual coming together of telomeres during premeiotic interphase and leptotene, but rather by a relatively abrupt mechanism.
Figure 7.
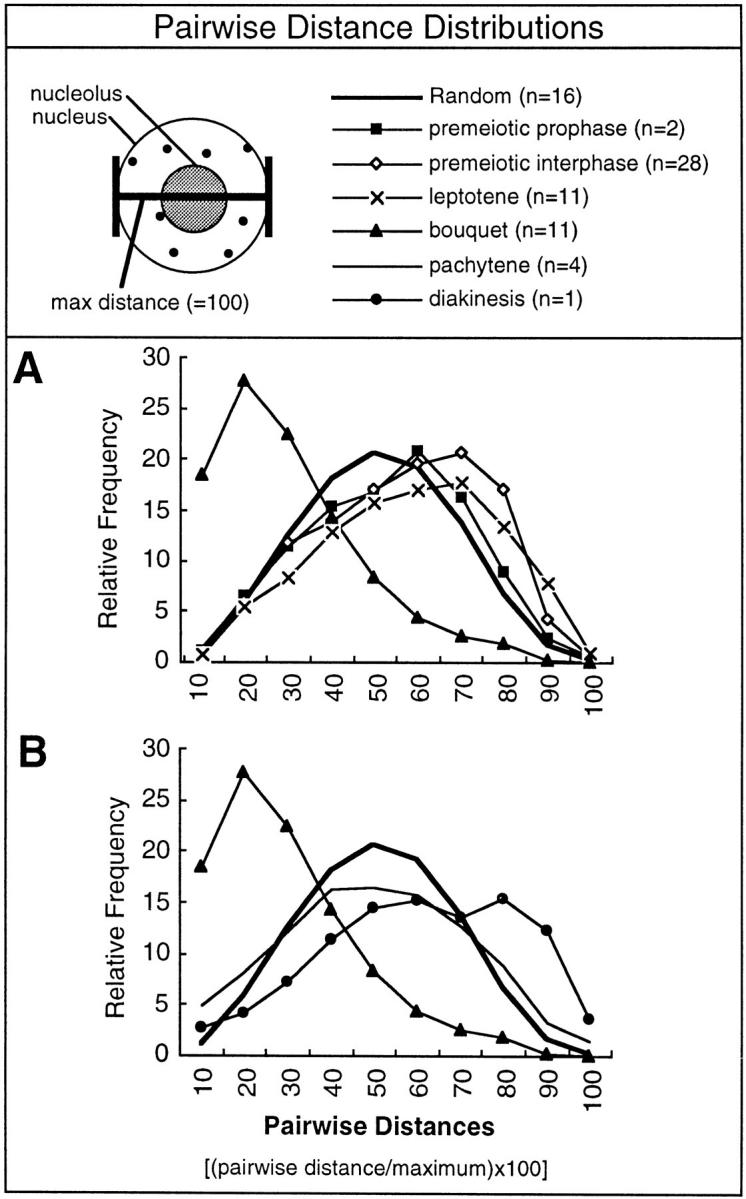
Analysis of the distribution of distances between telomeres. The Euclidean straight-line distances between all the pairs of telomeres were determined for each nucleus and normalized to the maximum distance, generating a distance distribution curve that can be directly compared for nuclei that differ in volume and telomere signal count (see Materials and Methods). The relative frequencies of these distances were binned by increments of 10% of the maximum (see Materials and Methods). These binned normalized pairwise distance values were averaged to yield a single curve for each stage (stage is indicated in the legend at top, n, number of nuclei averaged). The “Random” curve was made from modeled nuclei as described in Materials and Methods, and is plotted for comparison in both graphs (thick line). (A) The distribution of distances for nuclei before and during the bouquet stage. (B) The distribution of distances during and after the bouquet stage.
Figure 8.
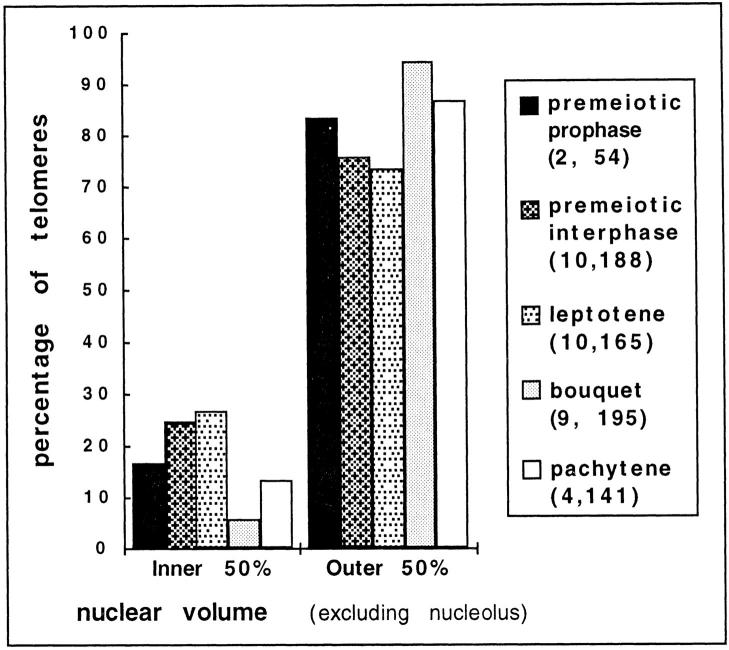
Analysis of telomere locations within the nuclear volume. Telomeres were scored as being located in the inner or outer half of the nuclear volume (see Materials and Methods). For each stage, all the telomeres were pooled and plotted. The two numbers in parentheses refer to the total number of nuclei pooled (1st number) and the total number of telomeres from those pooled nuclei (2nd number).
In addition to the spatial relationship between individual telomeres (above), we were interested in the spatial relationship between the individual telomeres and the edge of the nucleus. We defined the edges of the nucleus and nucleolus using a surface harmonic expansion as described by Marshall et al. (43). This allowed us to use Monte Carlo simulations to partition the non-nucleolar space within each nucleus into two equal, but exclusive sub-volumes, the inner and the outer half of each nucleus. Each telomere was scored as being in the inner or outer half of the available nuclear space and the results are shown in Fig. 8. We found that the 83% of the telomeres from the last premeiotic prophase nuclei were located in the outer half of the nucleus, followed by a gradual redistribution throughout the available nuclear volume at premeiotic interphase and leptotene. At the bouquet stage, the inner telomeres drop significantly in abundance from 27% at leptotene to 6% at zygotene (Fig. 8). The data unexpectedly revealed that as cells progress from premeiotic interphase through leptotene, the telomeres were not approaching the nuclear envelope, but were increasingly partitioned into the interior spaces in the nucleus. These observations were similar to those derived from the distributions of pairwise distances in that they indicated a tendency for telomeres to become increasingly randomized as cells progressed towards the beginning of the bouquet stage (Figs. 7 and 8, and Table I). Thus, we could conclude that the meiotic telomere cluster of the widely conserved bouquet stage forms de novo at the end of leptotene, with no apparent contribution from the telomere arrangements of preceding stages.
Discussion
One of the great advantages of analyzing meiosis in maize is the ability to clearly stage nuclei by multiple criteria (Fig. 1). Accurate staging relies on the ability to detect changes in the structure of chromosomes and the morphology of the nucleus. Using a FISH protocol that preserves the nucleus, it was possible to monitor the distribution of telomeres as a function of progression through the cell cycle. This work provides the first detailed account of telomere kinetics using 3-dimensional optical reconstructions of entire nuclei spanning a full cell cycle. In particular, we have documented and mathematically analyzed the dynamics of telomere behavior at premeiotic interphase, leptotene, and zygotene, stages that are typically difficult to discern using FISH (52, 54, 64). This allowed us to identify the end of leptotene as the exact time point for the formation of the meiotic telomere cluster, the defining feature of the bouquet stage. Thus we could rule out a mechanistic linkage between the anaphase-based telomere grouping of the Rabl configuration of chromosomes and the nuclear envelope-associated telomere clustering of the bouquet structure. The de novo formation of the bouquet just before zygotene implies the presence of a meiosis-specific telomere movement system that is employed by meiotic cells to affect chromosome synapsis.
The Timing of Initial Telomere Clustering
We have identified in maize a relatively abrupt starting point for the initiation of the telomere cluster or bouquet arrangement. This clustering is coincident with, or immediately precedes prezygotene, the transition stage previously shown to be associated with transient changes in chromatin organization and initiation of pairing (12). We have been able to detect the telomere grouping resembling a Rabl configuration (Fig. 3) at the last premeiotic mitosis. This grouping is gradually lost as cells pass through premeiotic interphase and leptotene (Figs. 4, 7, and 8 Table I). Given that premeiotic S can be 8–10 times longer than mitotic S, there is ample time for Rabl-grouped telomeres to scatter (41). Our data are consistent with the possibility that the localization of telomeres at premeiotic interphase is not strictly regulated and given enough time, they would eventually be randomly dispersed. The availability of telomere FISH, employed by Scherthan et al. (54) and ourselves to examine premeiotic nuclei in very different organisms, has allowed us to demonstrate that the bouquet structure is formed de novo during meiotic prophase. In both studies, premeiotic telomere FISH signals were found to be distributed throughout the nuclear volume. Our data on the mode and timing of bouquet formation in maize are in general agreement with the recent data of Scherthan et al. (54), with one notable exception (54). In the mouse, telomeres in late-preleptotene nuclei are distributed around the nuclear envelope before their colocalization into the bouquet arrangement. However in maize, the distribution of telomeres was either scattered or clustered at the nuclear periphery. We have considered two possible explanations for this discrepancy: One is that the two systems are in fact different; in the mouse, telomeres attach to a broad area of the nuclear envelope before congregating into the bouquet arrangement, but in maize the telomeres may attach to the nuclear envelope and congregate at the same time. The other possibility is that we did not detect an attachment of maize telomeres to a broad area of the nuclear envelope because of the brevity of this stage. We repeatedly made attempts to identify an intermediate between scattered and clustered telomere arrangements. Invariably, meiocytes from anthers expected to yield late leptotene to early zygotene meiocytes showed either scattered or clustered telomere arrangements. Interestingly, these very same samples yielded the relatively rare cells in which the nucleolus and the telomere cluster sites were spatially separated. Such nuclei (Fig. 4, A and B) were referred to as “early bouquet” nuclei in which all but the presumptive NOR-linked telomeres were at the bouquet site. Our early bouquet stage nuclei could be interpreted as reflecting a transition stage toward bouquet formation in which all telomeres have clustered except for those which are slower to move because of linkage to the nucleolus.
For every nucleus subjected to data collection, deconvolution, and model building, at least five additional nuclei were imaged or observed while scanning the slides. In all cases where telomere clustering could be detected, the nucleolus was at the edge of the nucleus and almost always adjacent to the bouquet. This observation suggests that in maize, the eccentric position of the nucleolus may be used as an indicator that the bouquet stage has commenced. A critical piece of evidence in concluding that telomere clustering initiates at the end of leptotene, as opposed to prezygotene or early zygotene, is the observation that the relocation of the nucleolus to the nuclear periphery occurs at the end of leptotene (Dawe, R.K., personal communication, see also Fig. 2 C of Dawe et al., 1994). The end of leptotene is distinct from prezygotene because the knobs are still spherical (Fig. 1). In fact, the first signs of knob elongation are diagnostic for the end of leptotene and the beginning of prezygotene (12). Therefore, the nucleus shown in Fig. 5 A provides an example of telomere clustering before prezygotene. Additional evidence in support of this claim comes from a partial reconstruction of a leptotene nucleus by serial sectioning electron microscopy (23). In this study on maize, the ends of the chromosomes were attached to a small region of the nuclear envelope. Similarly, the bouquet stage has been observed as early as leptotene in human and rye, and at the leptotene-zygotene transition in locust, respectively (44, 48, 59).
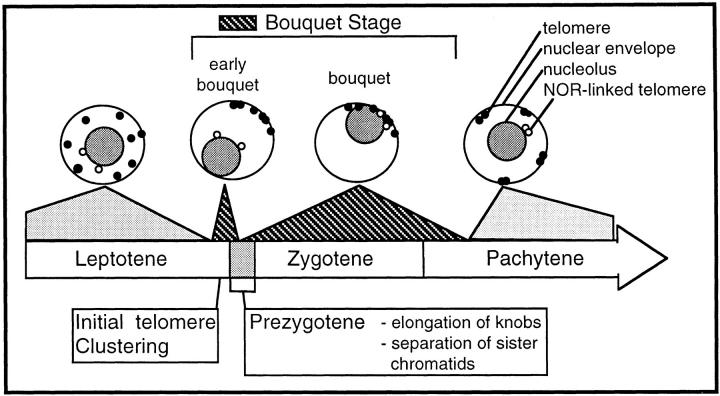
We have illustrated our results in Fig. 9 to summarize the temporal relationship between the bouquet stage, defined by the bouquet structure, and the canonical early stages of meiotic prophase, leptotene, zygotene, and pachytene. The initial telomere clustering is shown to occur at the end of leptotene just preceding prezygotene. The “early bouquet” nuclei are the first to exhibit telomere clustering where all but the NOR-linked telomeres have reached the cluster site. The NOR in maize is located near the end of the short arm of chromosome 6. The spatial separation of the NOR from the telomere cluster site may reflect a slower rate at which the NOR-linked telomeres can move to the cluster site. Alternatively, the nucleolus may be positioned in these early bouquet nuclei by a telomere-independent mechanism that overrides the telomere clustering mechanism (28). Prezygotene is then followed by zygotene, during which the most commonly encountered bouquet arrangement was observed (indicated by “bouquet,” Fig. 9, above zygotene). The dispersal of telomeres during pachytene is indicated by the predominantly peripheral localization of telomeres near the nuclear envelope (nucleus diagrammed above “pachytene,” Fig. 9). This summary illustrates that the de novo formation of the telomere cluster is among the very first steps in nuclear changes associated with synapsis.
Figure 9.
Diagram of the timing of meiotic telomere clustering. A summary of the fine scale timing of events associated with the onset of synapsis is presented based on the observations from this work and from Dawe et al. (1994). Leptotene telomeres (legend at right) are shown as either NOR-associated (open circles) or non-NOR (closed circles, representing the other 38 of 40 maize telomeres) are scattered throughout the chromatin, and the nucleolus is fully internal and approximately concentric with the nucleus. The first signs of telomere rearrangements give rise to the “early bouquet” (see text). The early bouquet nucleus (as in Fig. 5, A and B) is first seen at the end of leptotene, just preceding prezygotene. Next, prezygotene occurs, comprising the major transition between leptotene and zygotene as described (12). By zygotene, all of the telomeres are at the cluster site resulting in the eccentric nucleolus showing an obligatory colocalization with the telomere cluster (as in Fig. 5, C and D). At pachytene the telomere bouquet disperses and the paired homologous telomeres remain at the nuclear periphery.
We have interpreted our results to indicate that the changes in telomere arrangements are continuous and essentially irreversible. However, the harvesting, fixation, or hybridization of cells could result in alterations of the nuclear organization that normally occur in vivo. To directly address such a possibility we will need to compare these results with those of living cells. Although our study was carried out on fixed material, we have described the pooled results from three different inbred lines of maize, harvested throughout the year. Thus we have attempted to describe observations of a general nature, avoiding description of observations that may be peculiar to a particular genetic line or growth condition.
Possible Mechanisms of Meiotic Telomere Movements
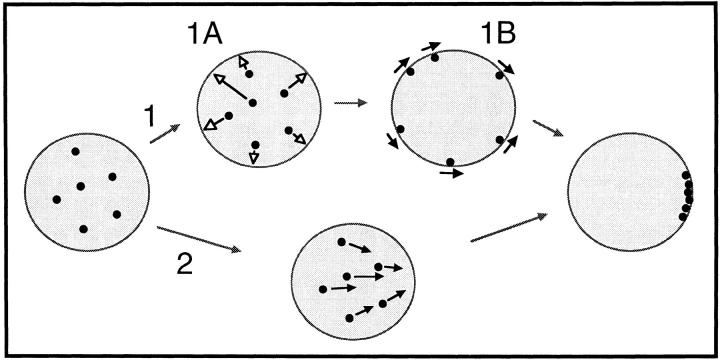
The present study describes the sudden formation and the gradual dissolution of the meiotic telomere cluster. In light of these findings, we now consider the possible mechanisms underlying these dynamic changes in telomere localization. The abruptness of the initial cluster formation allows us to rule out one class of mechanisms relying on passive diffusion. Specifically, our data do not support a scenario in which a region of the nuclear envelope suddenly acquires an affinity for telomeres, and collects the telomeres on the basis of chance interaction. What then is the mechanistic basis for the formation of the telomere cluster? To begin to address this question, two possible mechanisms of bouquet formation are proposed in Fig. 10. The first mechanism involves a two-step process (movements indicated by arrows of nuclei 1A and 1B of Fig. 10) in which the telomeres first attach to any region of the nuclear envelope (arrows of 1A), followed by the movement of telomeres in the plane of the nuclear envelope (arrows of 1B) to end up at the final cluster site. The second mechanism would involve a one-step process (movements indicated by arrows of nucleus 2, Fig. 10) in which telomeres move directly to the region of the nuclear envelope that will be the final cluster site, the bouqet base (referred to as bb in Figs. 5 and 6). The two-step clustering mechanism is consistent with the observations on mouse spermatocytes (54). Our data on meiosis in maize does not allow us to distinguish between the two paths shown in Fig. 10. Although this model only addresses the mechanisms responsible for telomere movements, we recognize that telomeres are but one of many cellular components that show altered distributions at the bouquet stage (13).
Figure 10.
Model of possible mechanisms of telomere cluster formation. The de novo clustering of randomly distributed telomeres is proposed to occur by one of two different mechanisms (1 and 2). Mechanism 1 proposes a two-step model; 1A, telomeres move from their positions in the nucleus to the nuclear envelope where they become attached, and 1B nuclear envelope-attached telomeres move over the nuclear surface to the final cluster site. Mechanism 2 proposes a one-step model in which telomeres move from their positions in the nucleus directly to the region of the nuclear envelope that will be the final cluster site.
Evidence that clustering of telomeres is associated with active forces on chromosomes or nuclei come from a series of studies on the bouquet stage conducted by Hiraoka (29). In his research on meiotic nuclei in living cells of Equisetum and other plants, he observed that when the bouquet configuration is formed, nuclei take up an eccentric position in the cell with the telomeric cluster regions predicting the direction of nuclear migration. Chikashige et al. (9) showed that a similar phenomenon, i.e., meiotic nuclear migration, explains the dramatic distortions in meiotic prophase nuclei (the horsetail stage) of the fission yeast, S. pombe (9). In this case, the entire nucleus, led by the telomeres and the spindle pole body, is translocated through the cytoplasm. Thus, some of the morphological changes in chromatin and nuclei are consistent with the possibility that telomeres serve as connection points for a cytoplasmic motility system, whose function during meiosis remains to be discerned. It is noteworthy that there may be a difference between the forces that form the bouquet, as drawn in Fig. 10, and the forces that result in movement of the whole nucleus.
Analysis of nuclear motility along with ultrastructural studies on meiotic chromosomes has revealed that the microtubule cytoskeleton may be involved in these telomere- based actions. These data are based primarily on observations in many species that the base of the bouquet is juxtaposed to the microtubule organizing center, be it a centriole or spindle pole body (13). It remains to be determined whether a microtubule organizing center is near the telomere cluster in angiosperms such as maize, which lack cytologically distinct microtubule organizing centers. Another indirect line of evidence linking microtubules to bouquet formation is based on disruption of meiotic chromosome pairing by treatment with the microtubule depolymerizing drug, colchicine (40). In addition, Driscoll and Darvey provide compelling evidence that chromosome pairing is sensitive to colchicine treatments (18). They applied colchicine to a genetic stock of wheat carrying an isochromosome, a compound chromosome in which the right and left arms are identical, but with mirror image orientation. The isochromosome alone was resistant to the chiasmata-reducing effect of colchicine leading the authors to conclude that microtubules participated in the first step of chromosome pairing, bringing homologous chromosomes into close spatial proximity (18). This concept is consistent with the possibility that telomere movements on the nuclear envelope (as shown for 1B of Fig. 10) may involve interactions with the microtubule cytoskeleton.
Even if the cytoskeleton participates in the nuclear motility seen at meiotic prophase or the movement of telomeres along the nuclear envelope, it remains to be determined how the telomeres arrive at and attach to the nuclear envelope. Although we refer to telomeres in the cytological sense in this paper, we do not know what role, if any, the telomere repeat sequences actually play in movement or attachment of the ends of chromosomes to the nuclear envelope. It is also recognized that we did not directly localize the nuclear envelope in this study, but inferred its location from the edge of the DAPI-stained chromatin.
Functional Significance of the Bouquet Stage
Having considered the when and how of telomere clustering, we are still left with the basic question of why, i.e., the functional significance of the bouquet. The inherently complex nature of the processes taking place during meiotic prophase preclude any simple assessment of bouquet function on the basis of temporal coincidence alone. Despite its invariant association with zygotene, the bouquet arrangement could have a role in chromosome alignment, homology search, resolution of interlocks, synapsis, synaptonemal complex formation, or crossover control.
Scherthan et al. (54) have argued that the bouquet may function to facilitate alignment of homologues as extended chromosome fibers (54). Once aligned, synapsis could commence in a relatively efficient manner while minimizing entanglements and search time (54). Dawe et al. (12) showed that heterochromatic, homologous knob loci were on average more than 7 microns apart at leptotene. Similar observations were made using a FISH probe for the euchromatic 5S rDNA locus (Bass, H.W., unpublished observations). Therefore, homologous loci might have a considerable distance to travel to encounter their homologues in maize, which has a normal diploid genome, similar in size to that of human (3). However, it is not clear that reduction of the homology search volume is sufficient to explain conservation of the bouquet because even fission and budding yeast, with small genomes and small nuclei, have a bouquet stage (9, 17, 53). Furthermore, budding yeast may have already accomplished a substantial degree of homologous chromosome alignment before the bouquet stage (38, 64).
A very different type of role for the bouquet might involve crossover control. The existence of crossover control is inferred from the observations that in most species, crossovers (chiasmata) between homologous chromosomes occur at very low, but non-zero rates, and the distribution of crossovers is not random (i.e., crossover interference). In a recently published model, Kleckner has suggested that crossover control occurs “via the imposition and relief of stress” (37). It is suggested that such stress derives from differential compaction of axis-associated chromatin. In this scenario, a role for bouquet-related pulling forces on the ends of the chromosomes could be easily envisioned. The idea that the bouquet, and in particular the forces that giving rise to nuclear movement, may be involved in crossover control is consistent with the observation that the initiation of synapsis, and the formation of chiasmata predominantly occur near the ends of chromosomes in many species (4, 35, 39, 45, 57, 58, 63).
The two possible functions of the bouquet discussed above might be predicted to manifest quite different phenotypes in the event of a failure of the bouquet to form or function. In the case of a role in chromosome pairing, a bouquet failure would be predicted to result in the accumulation of unpaired chromosomes at metaphase I, causing a reduction or loss of fertility. Whereas in the case of a role in crossover control, a bouquet failure might result in a change of the position or frequency of chiasmata. If these changes involved a loss of chiasmata, the result would be infertility associated with the production of univalents. A full understanding of the role of the bouquet structure will require that we can test its function by experimental or genetic disruption, in conjunction with cytological anlysis. While this work reports on the discovery that the bouquet forms de novo at the end of leptotene, it also presents a powerful experimental approach that can be used to elucidate the basic structure-function relationships of meiotic nuclei.
Acknowledgments
We thank H. Scherthan for discussion of unpublished results during the early stages of this work; A.F. Dernburg and S.J. Parmelee for their excellent technical advice on 3-D FISH methodologies; D.D. Hughes and H. Chen for their generous assistance with the distance analyses; and A.E. Franklin, J.L. Paluh, L.C. Harper, and J.C. Fung for critical reading of the manuscript. The first author (H.W. Bass) is especially grateful to W.Z. Cande and J.W. Sedat for their guidance, support, and enthusiasm throughout this project.
Footnotes
H.W. Bass was supported in the early years of this work by a National Institutes of Health (NIH) grant to J.W. Sedat (R01-GM-25101-16). H.W. Bass is currently supported as a D.O.E. postdoctoral fellow of the Life Sciences Research Foundation. This work was supported by the (NIH) grants to W.Z. Cande (R01-GM-48547) to J.W. Sedat (R01-GM-2510116), and to D.A. Agard (R01-GM-31627). D.A. Agard is an investigator of the Howard Hughes Medical Institute.
1. Abbreviations used in this paper: DAPI 4′,6-diamidino-2-phenylindole, dihydrochloride; FISH, fluorescence in situ hybridization; FITC, fluorescein isothiocyanate.
Please address all correspondence to W.Z. Cande, 341 LSA, Department of Molecular and Cell Biology, University of California, Berkeley, CA. Tel.: (510) 642-1669. Fax: (510) 643-6791.
References
- 1.Bahler J, Wyler T, Loidl J, Kohli J. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J Cell Biol. 1993;121:241–256. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belmont AS, Sedat JW, Agard DA. A three-approach to mitotic chromosome structure: evidence for a complex hierarchical organization. J Cell Biol. 1987;105:77–92. doi: 10.1083/jcb.105.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett MD, Smith JB, Heslop-Harrison JS. Nuclear DNA amounts in angiosperms. Proc R Soc Lond Ser B. 1983;216:179–199. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- 4.Burnham CR, Stout JT, Weinheimer WH, Knowles RV, Phillips RL. Chromosome pairing in maize. Genetics. 1972;71:111–125. doi: 10.1093/genetics/71.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns JA. Preleptotene chromosome contraction in Nicotianaspecies. J Hered. 1972;63:175–178. [Google Scholar]
- 6.Chang MT, Neuffer MG. Maize microsporogenesis. Genome. 1989;32:232–244. [Google Scholar]
- 7.Chen, H., J.R. Swedlow, M.A. Grote, J.W. Sedat, and D.A. Agard. 1995. The collection, processing, and display of digital three-dimensional images of biological specimens. In Handbook of Biological Confocal Microscopy. J.B. Pawley, editor. Plenum Press, New York. 197–210.
- 8.Chen H, Hughes DD, Chan T-A, Sedat JW, Agard DA. IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J Struct Biol. 1996;116:56–60. doi: 10.1006/jsbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 9.Chikashige Y, Ding D-Q, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science (Wash DC) 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- 10.Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schrock E, Speicher MR, Mathiew U, Jauch A, Emmerich P, et al. Role of chromosomal territories in the functional compartmentalization of the cell nucleus. Cold Spring Harbor Symp Quant Biol. 1993;43:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- 11.Darlington, C.D., editor. 1937. Recent Advances in Cytology. Second edition. Churchill, Ltd., London. 650 pp.
- 12.Dawe RK, Sedat JW, Agard DA, Cande WZ. Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell. 1994;76:901–912. doi: 10.1016/0092-8674(94)90364-6. [DOI] [PubMed] [Google Scholar]
- 13.Dernburg, A.F., J.W. Sedat, W.Z. Cande, and H.W. Bass. 1995. Cytology of telomeres. In Telomeres. E.H. Blackburn and C.W. Grieder, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 295–338.
- 14.Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 15.Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- 16.Digby L. On the archesporial and meiotic mitoses of . Osmunda Ann Bot (Lond) 1919;33:135–172. [Google Scholar]
- 17.Dresser ME, Giroux CN. Meiotic chromosome behavior in spread preparations of yeast. J Cell Biol. 1988;106:567–574. doi: 10.1083/jcb.106.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Driscoll CJ, Darvey NL. Chromosome pairing: effect of colchicine on an isochromosome. Science (Wash DC) 1970;162:290–291. doi: 10.1126/science.169.3942.290. [DOI] [PubMed] [Google Scholar]
- 19.Freeling, M., and V. Walbot, editors. 1994. The Maize Handbook. Springer-Verlag, Inc, New York. 759 pp.
- 20.Fussell CP. Interphase chromosome order: a proposal. Genetica. 1984;62:193–201. [Google Scholar]
- 21.Gelei J. Weitere Studien über die Oogenese des Dendrocoelum lacteum. II. Die Längskonjugation der Chromosomen. Arch Zellforsch. 1921;16:88–169. [Google Scholar]
- 22.Gillies CB. Ultrastructural analysis of maize pachytene karyotypes by three-dimensional reconstruction of the synaptonemal complexes. Chromosoma (Berl) 1973;43:145–176. [Google Scholar]
- 23.Gillies CB. An ultrastructural analysis of chromosomal pairing in maize. Carlsberg Res Commun. 1975;40:135–162. [Google Scholar]
- 24.Golubovskaya IN. Meiosis in maize: meigenes and conception of genetic control of meiosis. Adv Genet. 1989;26:149–192. [Google Scholar]
- 25.Golubovskaya I, Avalkina NA, Sheridan WF. Effects of several meiotic mutations on female meiosis in maize. Dev Genet. 1992;13:411–424. [Google Scholar]
- 26.Gruenbaum Y, Hochstrasser M, Mathog D, Saumwever H, Agard DA, Sedat JW. Spatial organization of the Drosophila nucleus: a three-dimensional cytogenetic study. J Cell Sci Suppl. 1984;1:223–234. doi: 10.1242/jcs.1984.supplement_1.14. [DOI] [PubMed] [Google Scholar]
- 27.Hammersley J M. The distribution of distances in a hypersphere. Ann Math Stat. 1950;21:447–452. [Google Scholar]
- 28.Hiraoka T. Observational and experimental studies of meiosis with special reference to the bouquet stage. XI. Locomotory movement of the nucleolus in the bouquet stage. Cytologia (Tokyo) 1952;17:201–209. [Google Scholar]
- 29.Hiraoka T. Observational and experimental studies of meiosis with special reference to the bouquet stage. XIV. Some considerations on a probable mechanism of the bouquet formation. Cytologia (Tokyo) 1952;17:292–299. [Google Scholar]
- 30.Hiraoka Y, Swedlow JR, Paddy MR, Agard DA, Sedat JW. Three-dimensional multiple wavelength fluorescence microscopy for the structural analysis of biological phenomena. Semin Cell Biol. 1991;2:153–165. [PubMed] [Google Scholar]
- 31.Hiraoka Y, Dernburg AF, Parmelee SJ, Rykowski MC, Agard DA, Sedat JW. The onset of homologous chromosome pairing during Drosophila melanogasterembryogenesis. J Cell Biol. 1993;120:591–600. doi: 10.1083/jcb.120.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hochstrasser M, Mathog D, Gruenbaum Y, Saumweber H, Sedat JW. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. . J Cell Biol. 1986;102:112–123. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.John B. Myths and mechanisms of meiosis. Chromosoma (Berl) 1976;54:295–325. doi: 10.1007/BF00292812. [DOI] [PubMed] [Google Scholar]
- 34.John, B., editor. 1990. Meiosis. In Developmental and Cell Biology Series. Cambridge University Press, New York. 396 pp.
- 35.Kasha KJ, Burnham CR. The location of interchange breakpoints in barley. II. Chromosome pairing and the intercross method. Can J Genet Cytol. 1965;7:620–632. [Google Scholar]
- 36.Kezer J, Sessions SK, Leon P. The meiotic structure and behavior of the strongly heteromorphic X/Y sex chromosomes of neotropical plethodontid salamanders of the genus Oedipina. . Chromosoma (Berl) 1989;98:433–442. [Google Scholar]
- 37.Kleckner N. Meiosis, how could it work? . Proc Natl Acad Sci USA. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleckner N, Weiner BM. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harbor Symp Quant Biol. 1993;43:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- 39.Laurie DA, Hulten MA. Further studies on chiasma distribution and interference in the human male. Ann Hum Genet. 1985;49:203–214. doi: 10.1111/j.1469-1809.1985.tb01694.x. [DOI] [PubMed] [Google Scholar]
- 40.Loidl J. The initiation of meiotic pairing: the cytological view. Genome. 1990;33:759–778. doi: 10.1139/g90-115. [DOI] [PubMed] [Google Scholar]
- 41.Lu, B.C.K. 1996. Chromosomes, mitosis, and meiosis. In Fungal Genetics. C.J. Bos, editor. Marcel Dekker, Inc., New York. 119–176.
- 42.Maguire M. Evidence for homologous pairing of chromosomes prior to meiotic prophase in maize. Chromosoma (Berl) 1967;21:221–231. [Google Scholar]
- 43.Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. . Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moens PB. The fine structure of meiotic chromosome polarization and pairing in Locusta migratoriaspermatocytes. Chromosoma (Berl) 1969;28:1–25. doi: 10.1007/BF00325986. [DOI] [PubMed] [Google Scholar]
- 45.Moens PB, Bernelot-Moens C, Spyropoulos B. Chromosome core attachment to meiotic nuclear envelope regulates synapsis in Chloealtis(Orthoptera) Genome. 1989;32:601–610. [Google Scholar]
- 46.Mogensen HL. Ultrastructural analysis of female pachynema and the relationship between synaptonemal complex length and crossingover in Zea mays. Carlsberg Res Commun. 1977;42:475–497. [Google Scholar]
- 47.Rabl C. Über Zelltheilung. Morpholog Jahrbuch. 1885;10:214–330. [Google Scholar]
- 48.Rasmussen SW, Holm PB. Human meiosis II. Chromosome pairing and recombination nodules in human spermatocytes. Carlsberg Res Commun. 1978;43:275–327. [Google Scholar]
- 49.Rhoades MM. Meiosis in maize. J Hered. 1950;41:59–67. doi: 10.1093/oxfordjournals.jhered.a106089. [DOI] [PubMed] [Google Scholar]
- 50.Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. . Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- 51.Scherthan, H. 1996. Chromosome behavior in earliest meiotic prophase. In Chromosomes Today. J.S. Parker and M. Puertas, editors. Capman and Hall, London.
- 52.Scherthan H, Cremer T. Nonisotopic in situ hybridization in paraffin-embedded tissue sections. Methods Mol Genet. 1994;5:223–238. [Google Scholar]
- 53.Scherthan H, Bahler J, Kohli J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J Cell Biol. 1994;127:273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scherthan H, Weich S, Schwegler H, Heyting C, Härle M, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS. In situ localization of parental genomes in a wide hybrid. Ann Bot (Lond) 1989;64:315–324. [Google Scholar]
- 56.Sprague, G.F., and J.W. Dudley. 1988. Corn and corn improvement. In Agronomy. Vol. 18. ASA, CSSA, SSSA, New York.
- 57.Tabata M. Chromosome pairing in intercrosses between stocks of interchanges involving the same two chromosomes in maize. Diakinesis configurations. Cytologia (Tokyo) 1962;27:410–417. [Google Scholar]
- 58.Tabata M. Chromosome pairing in intercrosses between stocks of interchanges involving the same two chromosomes in maize. II. Pachytene configurations. Cytologia (Tokyo) 1963;28:278–292. [Google Scholar]
- 59.Thomas JB, Kaltsikes PJ. A bouquet-like attachment plate for telomeres in leptotene of rye revealed by heterochromatin staining. Heredity. 1976;36:155–162. [Google Scholar]
- 60.Urata Y, Parmelee SJ, Agard DA, Sedat JW. A three-dimensional structural dissection of Drosophilapolytene chromosomes. J Cell Biol. 1995;131:279–295. doi: 10.1083/jcb.131.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanderlyn L. Somatic mitosis in the root tip of Allium cepa—a review and orientation. Bot Rev. 1948;14:270–318. [Google Scholar]
- 62.von Wettstein D, Rasmussen SW, Holm PB. The synaptonemal complex in genetic segregation. Ann Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- 63.Wallace BMN, Hulten MA. Meiotic chromosome pairing in the normal human female. Ann Hum Genet. 1985;49:215–226. doi: 10.1111/j.1469-1809.1985.tb01695.x. [DOI] [PubMed] [Google Scholar]
- 64.Weiner BA, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]