Abstract
Basonuclin is a zinc finger protein that was thought to be restricted to keratinocytes of stratified squamous epithelia. In epidermis, basonuclin is associated with the nuclei of mitotically active basal cells but not in terminally differentiating keratinocytes. We report here the isolation of a novel form of basonuclin, which we show is also expressed in stratified epithelia. Most unexpectedly, we find both forms in testis, where a surprising localization pattern was uncovered. While basonuclin RNA expression occurs in mitotically active germ cells, protein was not detected until the meiotic stage, where basonuclin localized to the appendage of the distal centriole of spermatocytes and spermatids. Near the end of spermiogenesis, basonuclin also accumulated in the acrosome and mitochondrial sheath surrounding the flagellum. Intriguingly, a perfect six– amino acid residue mitochondrial targeting sequence (Komiya, T., N. Hachiya, M. Sakaguchi, T. Omura, and K. Mihara. 1994. J. Biol. Chem. 269:30893–30897; Shore, G.C., H.M. McBride, D.G. Millar, N.A. Steenaart, and M. Nguyen. 1995. Eur. J. Biochem. 227: 9–18; McBride, H.M., I.S. Goping, and G.C. Shore. 1996. J. Cell. Biol. 134:307–313) is present in basonuclin 1a but not in the 1b form. Moreover, three distinct affinity-purified peptide antibodies gave this unusual pattern of basonuclin antibody staining, which was confirmed by cell fractionation studies. Our findings suggest a unique role for basonuclin in centrosomes within the developing spermatid, and a role for one of the protein forms in germ cell mitochondrial function. Its localization with the acrosome suggests that it may also perform a special function during or shortly after fertilization.
Mammalian testes consist of numerous seminiferous tubules, which converge toward common ducts, i.e., the epididymis, through which mature sperm travel to exit the male animal (Fig. 1 A). Within each seminiferous tubule, the epithelial cells, referred to as Sertoli cells, anchor and provide nourishment for the developing spermatozoa (Fig. 1 A; Browder et al., 1991). The germ stem cells within each tubule reside at the tubule periphery and give rise to proliferating spermatogonial cells. Only the most primitive spermatogonia, i.e., those that contribute to the stem cell population, complete their cytoplasmic divisions. All other daughter cells are linked through their cytoplasms and undergo a series of synchronous differentiation steps that culminate in the production of mature sperm.
Figure 1.
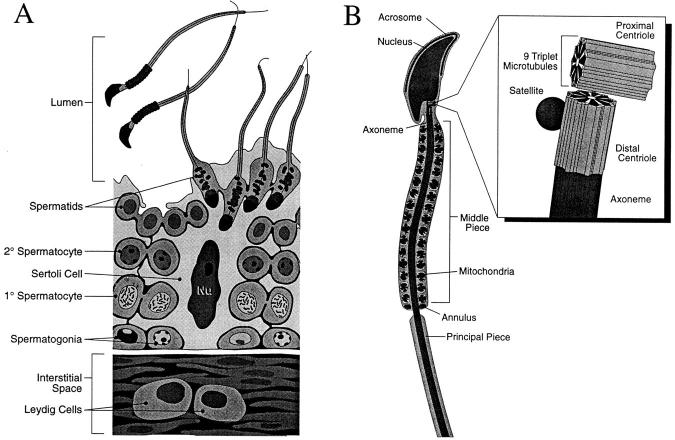
Schematic of male germ cell development. (A) In the seminiferous tubules, stem cells and mitotically active spermatogonia are located at the periphery. As the germ cells differentiate, they move inward toward the lumen of the tubule. Spermatocytes undergo two rounds of meiotic divisions before they embark upon spermiogenesis, a process culminating in the production of mature spermatids. Spermatids are released into the lumen as sperm, leaving their cytoplasm behind. The large Sertoli cells provide nutrients and support for the developing germ cells; seminiferous tubules are surrounded by interstitial space containing blood vessels and also Leydig cells, the hormone-producing cells of the testis. (B) A diagram of a mature sperm, revealing in more detail its unique structures. The tail flagellum is a 9 + 2 axoneme of microtubules, which at its upper end is surrounded by a sheath of specialized mitochondria (Middle Piece). The head of the sperm is composed of an elongated nucleus, which in its upper hemisphere is surrounded by a membrane-encased acrosome. The acrosome is thought to be a storage vessel used during fertilization. At the base of the crescent-shaped nucleus is the pair of sperm centrioles. The proximal centriole attaches to the nuclear envelope but does not appear to have microtubule organizing activity. It is surrounded at its base by pericentriolar material but is otherwise naked. The distal centriole is decorated by an appendage or satellite structure and is the only centriole capable of assembling the 9 + 2 axoneme of the flagellum. We have only visualized one appendage at any one time on ultrathin sections of mouse spermatid centrioles (artwork by Christa Wellman; adapted from Larsen, 1993).
As spermatogonia differentiate, they leave the basement membrane and transit towards the lumen of the tubule. During the early stages, the cells continue mitoses, but when they differentiate first into primary and then secondary spermatocytes, they undergo two sequential meiotic divisions resulting in the production of haploid spermatids (Fig. 1 A). In the later stages of differentiation, referred to as spermiogenesis, spermatids mature into fully motile sperm. An acrosomal cap forms at the anterior tip of the nucleus and continues to spread over an entire half (Fig. 1 B). The acrosome appears to be a storage vessel for factors needed in the fertilization process. As this cap develops, the spermatid nucleus becomes elongated and flattened, and nuclear chromatin condenses and moves towards the periphery of the caudal hemisphere. A cylinder of microtubules, referred to as the manchette, assembles downward from the posterior margin of the acrosomal cap. The manchette has been implicated in sperm head elongation and in organizing condensed chromatin on the opposing side of the nuclear envelope. The pair of centrioles migrates and attaches to the base of the nucleus, where the distal centriole, distinguished at the ultrastructural level by its association with pericentriolar “satellite” appendages, serves as the organizing center for the 9 + 2 axoneme of the sperm flagellum (Fig. 1 B; for review see Browder et al., 1991; Lange and Gull, 1995). As the sperm tail matures, the upper portion (middle piece) becomes ensheathed by mitochondria, the manchette disappears, and the sperms become motile and are released into the lumen, leaving the cytoplasm of the spermatids behind (Fig. 1 B).
The process of spermatogenesis and spermiogenesis involves three distinct and unusual cytoskeletal networks of microtubules, which are likely to assemble from specialized organizing centers. During meiosis 2, the spindle must form in the absence of a preceding round of DNA synthesis. While the mechanism underlying this process in spermatogenesis is still not well understood, genetic differences between these stages have been identified, and morphological differences between meiotic and mitotic centrosomes have been reported (Gonzalez et al., 1988, 1990; Staiger and Cande, 1990; Messinger and Albertini, 1991; Kubiak et al., 1992; Wickramasinghe and Albertini, 1992; Fuge, 1994; Matthies et al., 1996; de Vant'ery et al., 1996). Another unusual feature of the microtubule architecture in sperm is the development of a tail flagellum. The ability of spermatid centrioles to assemble these 9 + 2 axonemes implies that the distal centriole acquires some component(s) that enables it to orchestrate this unique microtubule assembly process. Finally, the formation of the manchette is perhaps the least understood of the microtubule assembly processes that occur during spermatogenesis. The manchette does not seem to emanate from a centrosome, but rather assembles from a membranous ring that circumvents the equator of the spermatid nucleus.
Very few of the genes involved in sperm development have been characterized at a molecular level. We report here the cloning and characterization of murine and human cDNAs that encode a novel form of a previously identified zinc finger protein called basonuclin. We demonstrate that basonuclin mRNA is expressed in the differentiating germ cells of seminiferous tubules, and the protein is made later during spermatogenesis and spermiogenesis. Using three affinity-purified, monospecific antibodies that we have made to different peptide sequences within the basonuclin protein, we show that the protein localizes to several interesting places during sperm morphogenesis. We first detect basonuclin in the centrosomes of meiotic spermatocytes. As differentiation proceeds, it maintains its centriolar location, but in addition, it accumulates in the acrosome. Basonuclin antibody also labels the mitochondrial sheath encompassing the midpiece of the flagellum, and intriguingly one of the two basonuclin forms has a perfect mitochondrial localizing signal. Our findings, supported by cell fractionation studies, are entirely unexpected and have important implications for our understanding of the specialized centrosomes, microtubule arrays, and mitochondria of late stage spermatogenesis and spermiogenesis.
Materials and Methods
Preparation of a Keratinocyte cDNA Library
Human epidermal keratinocytes were cultured (Rheinwald and Green, 1977), and poly(A)+ mRNAs were isolated using the procedure of Chomzcynski and Sacchi (1987). The RNA preparation was translated in a reticulocyte lysate system in the presence of [35S]methionine, and proteins were resolved by SDS-PAGE and autoradiography. Proteins >200 kD were translated from the mRNAs, verifying the quality of the preparation. These mRNAs were then used to engineer a λ-zap phage library (Stratagene, La Jolla, CA), made from a mixture of cDNAs that were synthesized using oligo dT and random hexamer oligonucleotide primers.
Isolation of Genomic Clones
A 688-bp EcoRI/EcoRD fragment encoding a 5′ segment of the human basonuclin 1b mRNA was used to screen a 129/sv mouse genomic library (Stratagene). Three hybridizing clones were identified and subsequently purified. Two clones, mBSN-1 and mBSN-2, were subcloned as ∼17 kb NotI restriction fragments into Bluescript KS+. These clones were then subjected to restriction map analyses and partial sequencing.
Preparation and Characterization of Basonuclin Antibodies
Three peptides corresponding to segments of the published basonuclin protein sequence (Tseng and Green, 1992) were synthesized, coupled to keyhole limpet hemacyanin, and used for generating polyclonal antisera in rabbits (Zymed Labs, Inc., S. San Francisco, CA). The three basonuclin peptides are: UC56, CRPPPSYPGSGEDSK (human sequence, corresponding to the published amino acid residues 449–463; Tseng and Green, 1992); Ab176, ESCGHRSASLPTPVD (mouse sequence, equivalent to human residues 208–222); and Ab372, ASPNPRLHAMNRNNR (mouse sequence, equivalent to human residues 404–419). All antisera were purified by affinity chromatography using the appropriate peptide-conjugated Sepharose columns. Antibodies were tested by immunoblot analyses on proteins extracted from the skin and testes of adult mice.
Immunoblot, Northern and In Situ Hybridizations
Immunoblot analyses were performed as described (Yang et al., 1996). RNA blots used for Northern analysis were purchased from Clontech (Palo Alto, CA), and hybridizations were performed as described by the manufacturer. A 576-bp radiolabeled human cDNA corresponding to sequences within exons 2–4 of basonuclin was used for hybridization. In situ hybridizations of frozen sections of mouse testes were performed using digoxygenin-labeled antisense and sense riboprobes as described (Chiang and Flanagan, 1996).
Immunofluorescence Microscopy on Frozen Tissue Sections
Frozen tissue sections (10 μm) were cut onto Superfrost plus slides. Sections were briefly fixed with methanol (−20°C) for 10 min and then washed 2× with PBS. Sections were preblocked with a solution containing 1% BSA, 0.1% Triton X-100, and 1% gelatin in PBS. Primary antibodies were then added to fresh solution and incubated with sections at room temperature for 1 h. Antibody concentrations used were: anti-BSN antibodies (1:20 to 1:500); anti–γ tubulin antibodies (1:200 dilution; gift of Dr. Harish C. Joshi, Emory University, Atlanta, GA; gift of Dr. Bruce Alberts, University of California at San Fransisco, San Fransisco, CA); H1 human autoantisera against centrosomes (1:250 dilution; gift of Dr. Thomas Medsgar, University of Pittsburgh, Pittsburgh, PA); and anti–PLCβ1 (purchased from Santa Cruz Biotechnology, Santa Cruz, CA). After washing the slides 3× with PBS for 10 min each, sections were incubated with fresh solution containing secondary Texas red or FITC-conjugated antibodies (1:100 dilution) for 30 min before washing as before and mounting. Nuclei were stained either with propidium iodide or with 4,6-diamidino-2-phenylindole (DAPI)1. Sections were examined using either a confocal microscope (model LSM 410; Carl Zeiss, Inc., Thornwood, NY) or an immunofluorescence microscope (model Axiophot; Carl Zeiss, Inc.).
Ultrastructural Analysis
For regular EM, tissues were fixed at room temperature for at least 1 h with 2.5% glutaraldehyde and 4% paraformaldehyde in 0.2 M sodium cacodylate buffer, pH 7.4. Samples were trimmed and washed three times with the same buffer and then postfixed with 1% aqueous osmium tetroxide for 1 h at room temperature. These samples were further washed with sodium cacodylate buffer, followed by maleate buffer, pH 5.1, and stained en bloc with 1% uranyl acetate for 1 h at room temperature. The samples were then washed several times with maleate buffer, dehydrated with cold ascending grades of ethanol and propylene oxide, embedded with LX-112 medium, and polymerized at 70°C for 48 h before sectioning.
Semithin sections (0.75 μm) were stained with toluidine blue, and these were visualized by light microscopy. Ultrathin sections (∼80 nm) were cut with a regular diamond knife, collected on 200-mesh, uncoated copper grids, and double stained with 50% saturated uranyl acetate and 0.2% lead citrate. These sections were then examined with a transmission electron microscope (model CX100; JEOL U.S.A., Inc., Peabody, MA) operated at 60 kV.
For postembedding immunolabeling, samples were placed in warm fixative containing 2% fresh paraformaldehyde, 0.05% glutaraldehyde in 0.1 M PBS, pH 7.4, for 10 min. The samples were trimmed, washed several times with the same PBS at 4°C, then dehydrated at −25°C with ascending grades of ethanol, infiltrated with different concentrations of Lowicryl K4M medium, embedded in gelatin capsules with fresh 100% Lowicryl K4M medium, and polymerized at −25°C with UV light for 5–7 d.
Ultrathin sections (∼90 nm) were cut with a diamond knife, collected on Nickel grids coated with Formvar and thin carbon films, and labeled with specific antibodies according to the tested dilution, followed by incubation with 10-nm colloid gold-conjugated secondary antibodies. The sections were briefly stained with uranyl acetate and lead citrate and examined with an electron microscope (model CX100; JEOL U.S.A., Inc.) operated at 60 kV.
Results
Isolation of a Novel cDNA Encoding a Known Zinc Finger Protein, and Genomic Mapping of Its Encoded Exons
In the course of our studies on epidermal differentiation, we hybridized a human keratinocyte cDNA library with a 517-bp PCR fragment corresponding to the zinc finger domain of human basonuclin (nucleotides 2657–3174 of hBSN1a), a protein known to be expressed in mitotically active basal epidermal cells (Tseng and Green, 1992). One clone contained a 4,606-bp insert, which upon sequencing was shown to harbor a complete open reading frame, followed by 1,547 bp of 3′ sequence containing a polyadenylation signal at nucleotide 4583. This cDNA encoded a protein that was nearly identical to the published sequence from amino acid residues 33–993 (Tseng and Green, 1992). However, it differed in its 5′ segment by the absence of a 32–amino acid residue sequence and the replacement of a novel 166-nucleotide residue sequence.
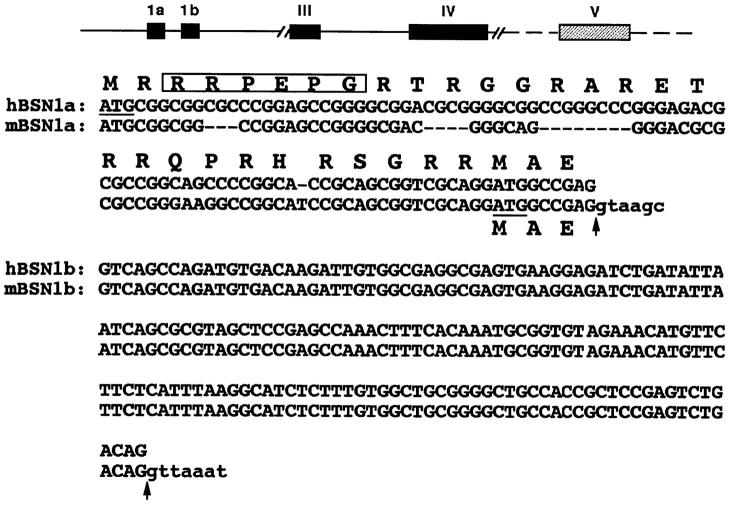
To assess whether both 5′ basonuclin sequences were bona fide, we screened a genomic library and mapped the positions of the two 5′ upstream sequences relative to the remainder of the basonuclin gene. Each sequence was contained within individual exons that were found within a single genomic clone (data not shown). The new sequence was located in an exon 3′ to the one present in the previously published sequence. These data confirmed the existence of the two sequences within the basonuclin gene. We refer to the two predicted forms as BSN1a (Tseng and Green, 1992) and BSN1b (this report), based upon the positioning of their respective exons within the basonuclin gene. The two basonuclin sequences were characterized from both mouse and human and are provided in Fig. 2. The BSN1b exon is highly conserved and is nearly identical between mouse and human. The BSN1a exon found in the originally reported sequence (Tseng and Green, 1992) is less conserved between the two species, and different translation start sites are predicted.
Figure 2.
Sequence relation between the two forms of basonuclin. Shown is a schematic of the mouse gene structure (top) and the two basonuclin forms (bottom). Hatched bars denote regions not fully sequenced; intron sizes are unknown. Human BSN1a sequence shown is from Tseng and Green (1992); the mBSN1a RNA is likely to use a downstream ATG for translation (underlined). The hBSN1b sequence, determined from a full-length cDNA, differs only in its 5′ sequence from hBSN1a. It is possible that an ATG shared by 1a and 1b is used for translation, since in vitro transcription/translation yields a >110-kD protein. The sequences encoding the unique segments of BSN1a and BSN1b are located on individual exons. Arrows denote splice sites; small case nucleotides represent intron sequences; putative mitochondrial localization sequence is boxed.
Interestingly, both mouse and human forms of BSN1a but not BSN1b contain the sequence RRPEPG, which has been shown to be a mitochondrial targeting sequence for proteins (Komiya et al., 1994; Shore et al., 1995; McBride et al., 1996). While not previously noted, a computer survey of all protein sequences known indicates a 60% chance of the BSN1a form localizing to the mitochondria, a prediction that we address experimentally later in the text.
Basonuclin mRNAs Are Abundantly Expressed in the Testis
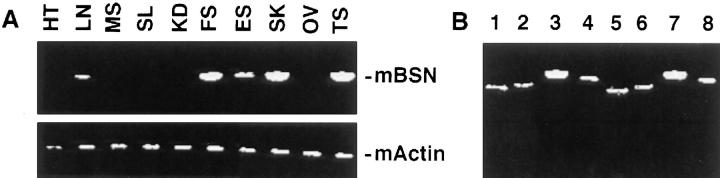
Previously, basonuclin was thought to be restricted in its expression to mitotically active cells of stratified squamous epithelia (Tseng and Green, 1994). In the course of examining basonuclin RNA expression in different mouse tissues, we were surprised to discover that PCR primers corresponding to the shared sequences of BSN1a and BSN1b detected a band in testis mRNA in addition to RNAs isolated from tissues known to contain keratinocytes (Fig. 3 A).
Figure 3.
Expression of both basonuclin RNA forms in testis as well as in stratified squamous epithelia. (A) Reversetranscriptase–PCR (RT-PCR) Analysis I. RNAs were isolated from the various mouse tissues indicated and subjected to RT-PCR as described above. Primer sets to a shared portion of BSN1a and BSN1b were used, along with an actin control. HT, heart; LN, lung; MS, muscle; SL, spleen; KD, kidney; FS, forestomach; ES, esophagus; SK, skin; OV, ovary; TS, testis. (B) RT-PCR Analysis II. RNAs were isolated from mouse skin and testis. These RNAs were subjected to RT-PCR analysis using primer sets specific for each of the two unique exons and for a shared segment of mouse BSN1a and BSN1b RNAs, respectively. Appropriate primer sets for β-actin were used as controls. DNA fragments generated were resolved by electrophoresis through 1% agarose gels. Lanes 1–4, skin RNAs; lanes 5–8, testes RNAs. Fragments shown were generated using primers specific for: lanes 1 and 5, BSN1a; lanes 2 and 6, BSN1b; lanes 3 and 7, BSN1a/1b; lanes 4 and 8, actin.
To assess whether our new form of basonuclin was expressed in both stratified epithelia and testis, we used reverse transcriptase and PCR on mouse skin and testis mRNAs in the presence of a 5′ oligonucleotide primer to one or the other of the 1a and 1b sequences. In each case, the 3′ primer corresponded to sequence shared by both cDNAs. As shown in Fig. 3 B, both primer sets produced a band of the expected size, indicating that both sequences are expressed by skin and testis. In our initial study, we have focused on using cRNA probes and antibodies that are shared by the two forms, and we refer generally to the properties of basonuclin (BSN).
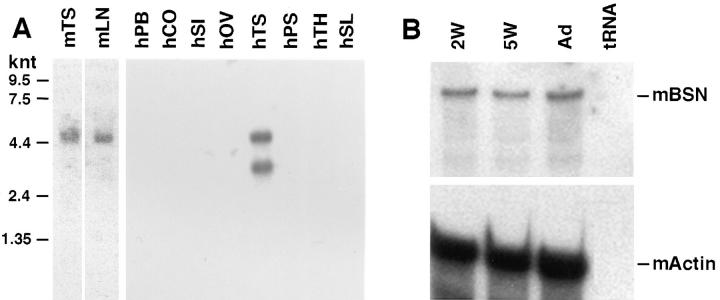
To examine BSN expression in testis in more detail, we first conducted Northern blot analysis. As shown in Fig. 4 A, a single RNA band of ∼4,600 nucleotides was obtained from mouse testis. This band was comparable in size to that seen in human keratinocyte mRNA preparations (Tseng and Green, 1994), and it corresponded to the size expected for BSN1a and BSN1b mRNAs, which have a long 3′ untranslated sequence. For human testis, two bands were detected in approximately comparable levels: one was an ∼4,600-nucleotide band, as expected, and the other was an ∼3,200-nucleotide band. This smaller band is large enough to encode full-length basonuclin, although our focus for the remainder of the study was on mouse, and we have not pursued the identity of this smaller band in human testis.
Figure 4.
Northern blot and RNase protection analyses. Northern blots of mouse and human RNAs were purchased from Clontech. Blots contained 2 μg polyA+ mRNAs, which were hybridized with a radiolabeled probe corresponding to a 576-bp fragment shared by BSN1a and BSN1b. As control, a radiolabeled probe corresponding to a 1-kb fragment of glyceraldehyde dehydrogenase was used. Note the presence of a 4,600-nucleotide band, predicted for the bona fide BSN1a and BSN1b RNAs, in testes samples (TS) from both mouse (m) and human (h); note the presence of an additional 3,200-nucleotide band in human testis. PB, peripheral blood; CO, colon; SI, small intestine; OV, ovary; TS, testis; PS, prostate; TH, thymus; SL, spleen. (B) RNAs were isolated from the testes of mice at 2 wk, 5 wk, and adult postnatally. RNAs were hybridized with radiolabeled mouse cRNas generated to either (a) a 576-nucleotide sequence common to BSN1a and BSN1b or (b) a 359-nucleotide actin sequence. After hybridization, RNase treatment was performed as described by Faus et al. (1994). Duplexes were resolved by PAGE, and the gel was exposed to x-ray film overnight. Note the presence of 487- and 250-nucleotide bands expected for BSN1a/BSN1b and actin RNAs, respectively, after hybridization with the probes and RNase digestion.
To verify that the hybridizing band(s) detected in the testis corresponded to bona fide BSN mRNA and to examine the testis-specific expression of BSN mRNA during sexual maturation, we conducted RNase protection assays. In mouse germ cell development, meiosis in the seminiferous tubules begins at about 2 wk postnatally, and production of mature sperm occurs by 5 wk of age. As shown in Fig. 4 B, a single band of the expected size was protected when mRNAs were used from mouse testes taken at 2 and 5 wk after birth and adult. The overall levels of BSN RNAs appeared to be comparable. These data demonstrated unequivocally that BSN mRNAs are expressed in testis and that their expression exists before sexual maturity.
Basonuclin mRNA Is Detected Early in Spermatogenesis
To determine where BSN mRNAs are expressed within the testis, we conducted in situ hybridizations on frozen sections of mouse testes isolated at various stages of postnatal development. A digoxygenin-labeled antisense BSN cRNA hybridized strongly in the seminiferous tubules of all testis samples examined (Fig. 5). Hybridization was detected at the periphery of the tubules and appeared to be present even at birth, before spermatogenesis (Fig. 5, A). Hybridization remained high throughout most of spermatogenesis. At 2 wk of postnatal development, hybridization was strongest in the centers of the tubules, where the primary spermatocytes are located, and weaker at the periphery, where the spermatogonia reside (Fig. 5, B). By 4–5 wk of age, spermatid formation, i.e., spermiogenesis, had begun (Rugh, 1990), and BSN RNAs were still detected throughout the tubules (Fig. 5, C–E). The persistence of BSN mRNA in late-stage spermiogenesis suggests that BSN RNAs are stable, as is the case with many mRNAs that are translated at this time (for review see Browder et al., 1991). Basonuclin cRNA hybridization was largely specific for derivatives of the germ cell population within the testis and was not detected in the interstitial Leydig cells. If present at all in Sertoli cells, the signal was reduced over that seen in germ cells. No hybridization was seen with the sense control cRNA (Fig. 5 F).
Figure 5.
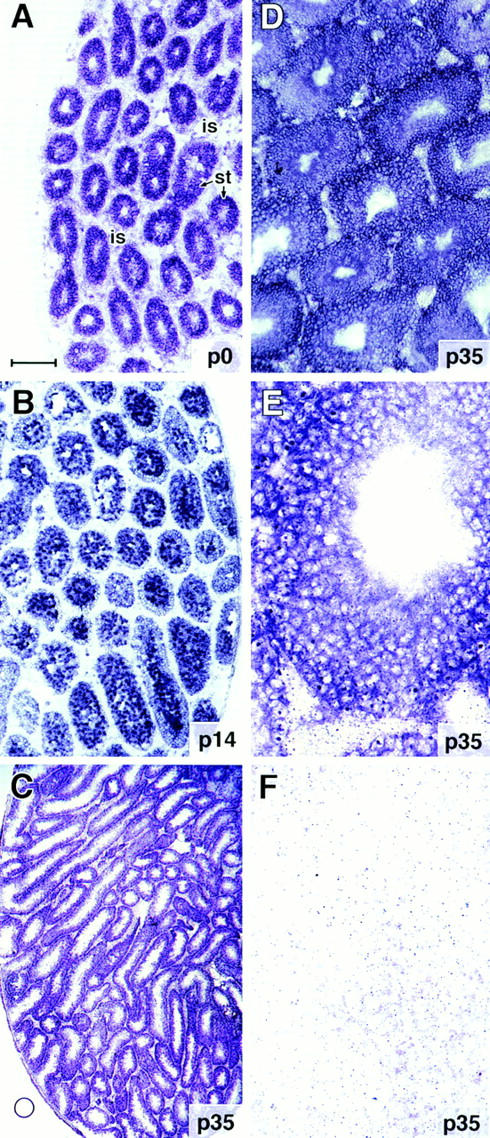
In situ hybridization of BSN RNAs in mouse testis. Testes were isolated from mice at various times after birth, and frozen sections (10 μm) were hybridized with a 576-nucleotide digoxygeninlabeled cRNA (A–E) or sense (F) control corresponding to the shared region of BSN1a and BSN1b RNAs. After hybridization, sections were washed extensively and developed for equal times (Yang et al., 1996). Shown are samples from: A, postnatal day zero (p0); B, p14; C–F, p35. Bar: (A and B) ∼175 μm; (C) 420 μm; (D) 100 μm; (E and F) 40 μm.
Basonuclin mRNAs Are Translated in the Testis, Where They Produce a 120-kD Protein
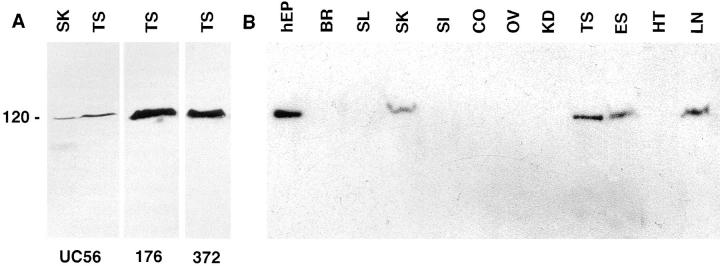
Both BSN1a and BSN1b cDNAs are predicted to encode 120-kD polypeptides; however, this has never been confirmed by immunoblot analysis. We therefore raised monospecific rabbit antibodies to three different peptide sequences present in the shared regions of these proteins (see Materials and Methods). As judged by immunoblot analysis, each of the three different affinity-purified peptide antibodies (UC56, 372, and 176) detected a single crossreacting band of 120 kD in protein extracts from mouse testis (Fig. 6 A). A band of this size was also detected in protein extracts from mouse skin and from other epithelial tissues known to contain keratinocytes (Fig. 6, A and B). Collectively, these findings: (a) establish the size of basonuclin in testis, skin, and other stratified epithelia; (b) verify the specificity of our antisera; and (c) suggest that, if other major forms of basonuclins exist, they either must be 120 kD in size or alternatively must have a most peculiar splice pattern, missing three different domains of the basonuclin protein.
Figure 6.
Immunoblot analysis with affinity-purified antiBSN UC56, 372, and 176 antibodies. (A) Total proteins were isolated from mouse testis and skin, and samples were resolved by electrophoresis through 8.5% SDS– polyacrylamide gels. Proteins were transferred by electroblotting to Immobilon-P membranes (Millipore Corp., Bedford, MA), and blots were subjected to immunoblot analysis as described by the manufacturer. Each of the three anti-BSN peptide antibodies were affinity column purified before use. In both testis and skin samples, a single band of 120 kD was detected; this band was not detected by secondary antibody alone or by preimmune sera. (B) Total proteins were isolated from a variety of adult mouse tissues and subjected to immunoblot analysis as outlined in A. Abbreviations are as indicated in the legend to Fig. 3, except: hEP, human epidermal keratinocytes; BR, brain.
Basonuclin Localizes to the Centrosomes of Spermatocytes and Developing Spermatids
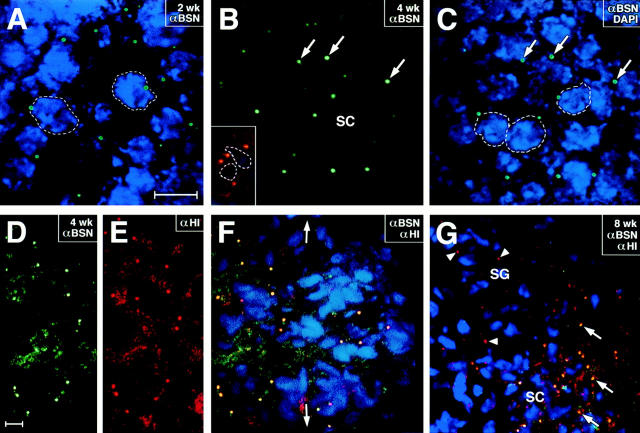
To assess the location of basonuclin within differentiating male germ cells, we conducted indirect immunofluorescence on frozen sections of developing mouse testes. Basonuclin protein was first detected in testis at 2 wk postnatally (Fig. 7 A). In contrast to BSN RNAs, which are expressed in mitotic spermatogonia, protein was not detected until the cells had differentiated into primary spermatocytes located at the midregion of the 2-wk seminiferous tubules. These cells, in the first meiotic phase of differentiating male germ cells, displayed a dotlike pattern of staining with the UC56 anti-BSN (αBSN) antibody. Double immunofluorescence with DAPI to stain chromatin indicated that the labeling was located near the nucleus (Fig. 7 A). Staining was more prevalent by 4 wk (Fig. 7, B and C), when primary and secondary spermatocytes exist (Rugh, 1990). While the majority of these spermatocytes contained single dots, a few seemingly contained double dots positioned at opposing sides of the nucleus (Fig. 7 B, inset). Similar staining patterns were observed with all three affinitypurified BSN antibodies, although antibodies UC56 (shown) and 372 gave the strongest staining. The pattern was not seen with secondary antibody alone.
Figure 7.
Detection of antibasonuclin immunofluorescence in the centrosomes of meiotic spermatocytes. Frozen sections (∼10 μm) of mouse testis (2, 4, or 8 wk old, as indicated in the panels) were subjected to double or triple immunofluorescence, and sections were viewed with a confocal microscope (Carl Zeiss, Inc.). Sections stained with anti-BSN antibodies (shown here are UC56 αBSN profiles) were visualized by costaining with an FITC-conjugated anti–rabbit IgG secondary antibody (green). Sections stained with human H1 autoantisera (αH1) were visualized by costaining with a Texas red–conjugated anti–human IgG secondary antibody (red). Nuclei were labeled with DAPI (blue). (A) Seminiferous tubule at 2 wk, containing a mixture of mitotically active spermatogonia at the periphery and primary spermatocytes in the midsection (Rugh, 1990); αBSN stained only midsection cells; (B and C) Seminiferous tubule at 4 wk, containing spermatogonia, primary spermatocytes, and secondary spermatocytes, located from the periphery to the centers of the tubules, respectively; view is of midsection, double stained with: αBSN (B and C) and DAPI (C). Arrows denote typical single dot pattern. Inset in B shows what is probably a secondary spermatocyte in late G2 or prophase, after duplication of the centrosomes; double labeling of dots is with αBSN and αH1 antiserum. (D–F) Region of spermatocytes, triple labeled with αBSN (D and F), αH1 (E and F), and DAPI (F). Yellow reveals costaining with αBSN and αH1. Portion of F to the left of the double arrows is the region shown in D and E. (G) 8-wk sexually mature seminiferous tubule costained with αBSN, αH1, and DAPI. View is of spermatogonia, primary spermatocytes, and secondary spermatocytes. Note that spermatogonial centrosomes at the periphery only labeled with αH1 (arrowheads), while spermatocytes colabeled with αBSN and αH1 (arrows). Additional note: αBSN antibodies 372 and 176 gave similar immunofluorescence staining patterns to those shown here. Dotted lines denote nuclear borders. Bars, 10 μm.
The dotlike staining pattern suggested that basonuclin might be localizing to centrosomes. To explore this possibility in greater detail, we used double immunofluorescence labeling with the H1 human autoimmune serum (αH1), known to cross-react with centrosomal proteins (Shu and Joshi, 1995). As shown in Fig. 7, D–F, the two antibodies displayed staining that was superimposable at the confocal microscopy level. This was further verified by staining serial cross-sections with same-species antibodies against γ-tubulin (not shown). Interestingly, only the centrosomes near the midregion of the seminiferous tubules costained with αBSN and αH1; centrosomes at the periphery stained with αH1, but not the UC56 sera (Fig. 7 G). Based upon these data, basonuclin appeared to be a specific component of the centrosomes of postmitotic, differentiating male germ cells.
Basonuclin Also Localizes to Acrosomes and to the Middle Piece of Developing Spermatids
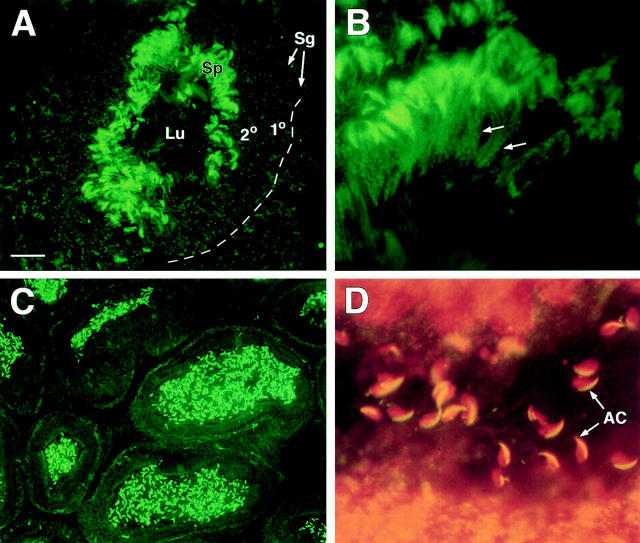
In sexually mature adult testes, anti-BSN antibodies strongly stained the spermatid heads (Fig. 8). Costaining with propidium iodide, which labels chromatin, indicated that this labeling was not nuclear. The crescent-shaped staining pattern, coupled with the appearance of this strong staining in the spermatid region of the seminiferous tubules, was reflective of that seen for acrosomal proteins in spermatids (Lepage and Roberts, 1995; Walensky and Snyder, 1995; Yoshiki et al., 1995). Interestingly, despite the fact that spermiogenesis in mouse is initiated by 4 wk, and that acrosomal caps are seen throughout the centers of the 4-wk seminiferous tubules, these caps did not stain with αBSN (not shown). The relatively late acquisition of anti-BSN staining in the acrosome suggested that basonuclin is a component of late-stage sperm acrosomes.
Figure 8.
Anti-BSN antibodies label the acrosomes and middle piece of developing spermatids. Frozen sections of adult mouse testis were processed as described in the legend to Fig. 7, except that propidium iodide (red) was used instead of DAPI to label chromatin. (A) Low magnification to show strong staining of spermatids (Sp) near the lumen (Lu) of all seminiferous tubules. Sg, spermatogonia; 1° and 2°, spermatocytes at the first and second meiotic stage, respectively. (B) Higher magnification of spermatids showing αBSN staining of both the head region and also the middle piece of the tail (arrows). (C) Lower magnification to show that spermatids in all seminiferous tubules of the adult testis labeled strongly with anti-BSN antibodies; note dotlike staining of spermatocyte centrosomes as well. (D) Double label of spermatids with propidium iodide and αBSN to show that the majority of αBSN labeling is in the acrosome and not the nucleus. Bar: (A) 20 μm; (B) 9 μm; (C) 30 μm; (D) 7 μm.
Finally, we observed αBSN staining within the middle piece of the tail of maturing spermatids (Fig. 8 B, arrows). This structure contains the mitochondrial sheath at the upper portion of the 9 + 2 axoneme (Fig. 1 B). This observation was surprising, given that mBSN1a was not predicted to contain mitochondrial localization signal seen at the amino end of hBSN1a.
Again, as was the case for the centrosomal staining, all three affinity-purified antibodies against basonuclin labeled the acrosomes and the middle piece. This said, the UC56 and 176 antibodies showed significantly stronger staining in the acrosome than did the 372 antibody. Since the three antibodies detected a single major 120-kD band by immunoblot analysis, we posit that these differences reflect variation in masking of the basonuclin epitopes in centrosomes and acrosomes.
Cell Fractionation Supports the Complex Localization Pattern of Basonuclin
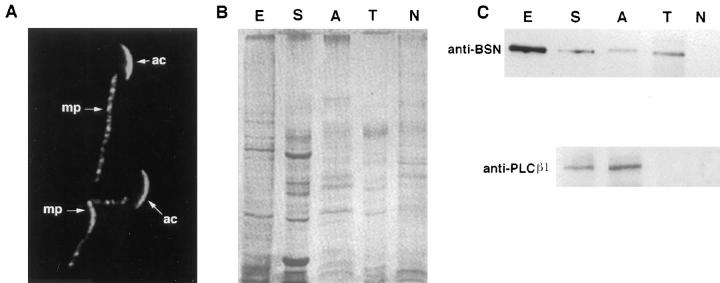
The pattern of BSN antibody staining was unexpected and diverse. To verify that the staining patterns reflected multiple locations for basonuclin protein, we conducted cell fractionation studies. Although procedures for isolation of centrosomes from testis tissue have not yet been developed, it is possible to dissociate isolated sperm into tail, acrosome, and headpiece by sonication and to subsequently resolve these fractions by sucrose gradient ultracentrifugation (Walensky and Snyder, 1995). We applied this procedure to mature sperm that we removed from the epididymis of adult mice. First, we verified that mature sperm, similar to spermatids, display αBSN UC56 immunofluorescence staining in the acrosome, middle piece of the tail, and centrosome. (Fig. 9 A; sperm centrosomal staining was more readily visible with the 372 antibody, which did not stain acrosomes so brightly.)
Figure 9.
Fractionation of sperm proteins confirms presence of the majority of basonuclin in the acrosome and sperm tail. Sperm were isolated from the epididymis of adult mice. They were first stained with αBSN UC56 antibodies to confirm the presence of acrosomal (ac) and middle piece (mp) staining in mature sperm (two sperm shown in A; bottom middle piece has a 90° kink). Sperm were then fractionated by sonication and sucrose gradient centrifugation as described by Walensky and Snyder (1995). Proteins were solubilized in 10 mM DTT, 2% SDS, and samples were resolved by electrophoresis through 8.5% SDS–polyacrylamide gels. Gels were analyzed by either staining with Coomassie blue (B) to visualize total proteins or immunoblot analysis (C) with antibodies against basonuclin or PLCβ1. E, total protein extract from epidermal keratinocytes; S, total sperm proteins; A, acrosomal fraction; T, tail fraction; N, nuclear fraction. Note: Sperm centrioles were lost in the fractionation procedure, as judged by immunofluorescent staining of each fraction.
Sperm fractions were examined by phase contrast and immunofluorescence microscopy to verify that the separation procedure was successful (not shown). Proteins from each fraction were then resolved by SDS-PAGE, and the gel was stained with Coomassie blue to visualize the proteins (Fig. 9 B). All three fractions contained different sets of proteins. As judged by immunoblot analysis, basonuclin was present in the sperm tail and acrosome fractions, but it was not present in appreciable amounts in the nuclear fraction (Fig. 9 C). The purity of fractions was confirmed by immunoblot analysis using an antibody against the established acrosomal marker PLCβ1 (Walensky and Snyder, 1995; Fig. 9 C) and a protamine nuclear antibody (not shown). These data were consistent with our immunofluorescence analysis. The absence of BSN immunoblot reaction in the nuclear fraction indicated that our failure to observe αBSN staining in sperm nuclei was due to the absence of protein, rather than the masking of BSN epitopes. We do not yet know whether basonuclin is present in nuclei of germ cells at earlier stages of spermatogenesis.
Immunoelectron Microscopy of Spermatids Reveals Basonuclin Protein in the Centriolar Appendages, in the Acrosomal Membrane, and in the Mitochondria of the Tail Middle Piece
To further examine basonuclin expression during spermiogenesis, we conducted electron and immunoelectron microscopy (Fig. 10). Fig. 10 A provides an example of the typical pair of centrioles that associates with the nuclear envelope during the acrosomal cap phase of spermiogenesis (also see diagram in Fig. 1 B). At this stage of differentiation, centrioles migrate to the nuclear pole opposing the acrosomal cap. The 9 + 2 axoneme assembly of the flagellum always initiates from the end of the distal centriole (Fig. 10, A and C, dis), which contains a prominent electron-dense satellite appendage often in close proximity to the nuclear envelope (Fig. 10 A and B, arrowheads). The proximal centriole (Fig. 10, A and C, px), laterally aligned with the nuclear envelope, is not directly involved in flagellar assembly, and its fate is unknown. Concomitant with the attachment of centrioles to the nuclear envelope, the cylinder of manchette microtubules forms around the lower half of the nucleus as it elongates (Fig. 10 B, ma).
Figure 10.
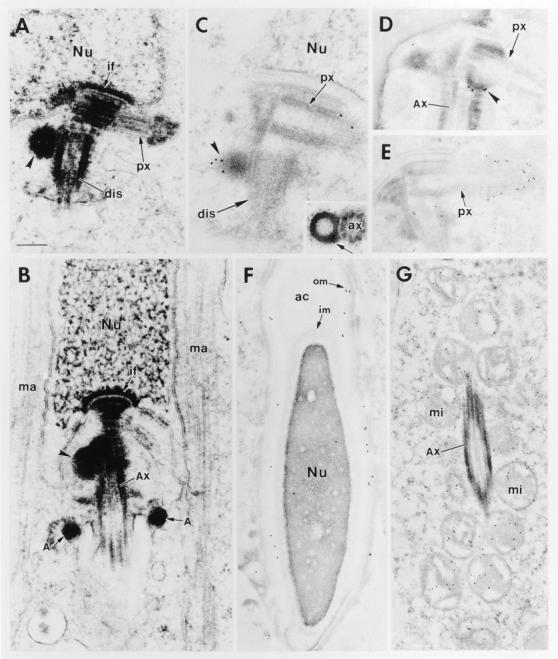
Electron and immunoelectron microscopy of mouse spermatids. Mouse testes from adult animals were fixed and processed for either regular or immunoelectron microscopy as described in the Materials and Methods. For immunoelectron microscopy, we used either αBSN (UC56) or anti–γ-tubulin (Shu and Joshi, 1995) antibodies. (A and B) Negatively stained sections of spermatid centrioles. The tip of the proximal centriole (px) is always associated with the nuclear envelope through implantation foci (if). Its free tip is always encased by a cloud of pericentriolar material; the distal centriole (dis) is always the site of 9 + 2 axoneme assembly and most sections revealed an attached appendage(s) or satellite structure(s) (arrowhead) located near the junction of the two centrioles and often in close proximity to the nuclear envelope. The manchette (ma) of microtubules that surrounds the lower hemisphere of the nucleus is formed at the acrosomal cap phase and disappears shortly after nuclear elongation and flagellar assembly (B). (C and D) Immunogold labeling of centrioles with αBSN. Note that the appendages of the distal centrioles (arrowheads) labeled specifically with the antibody. Inset shows a cross section of an appendage, revealing a hollow center to the structure. (E) Immunogold labeling of centrioles with anti–γ-tubulin. Note that this antibody heavily labeled the pericentriolar cloud at the tip of the proximal centriole and, to a lesser extent, showed some labeling near the distal appendages. (F) αBSN immunogold labeling of the acrosomal cap (ac) of a late-stage spermatid. Note that most of the labeling is concentrated near the outer membrane (om) of the cap rather than the inner membrane (im) or acrosomal space. Note the elongated nucleus, characteristic of nearly mature sperm. (G) Immunogold labeling of the sheath of mitochondria (mi) in the middle piece surrounding the axoneme (Ax). Additional abbreviations: Nu, nucleus; A, annulus. Bar: (A–D) 0.2 μm; (E and F) 0.3 μm; (G) 0.4 μm.
Antibodies against basonuclin specifically labeled the satellite appendages of distal centrioles (Fig. 10, C and D). Perfect cross-sections of these appendages revealed a hollow ringlike structure (Fig. 10 C, inset). The labeling of these structures with αBSN was largely distinct from anti–γ- tubulin, which specifically labeled the pericentriolar material surrounding the end of the proximal centriole (Fig. 10 E). γ-Tubulin labeling was not detected at the end of the distal centriole, i.e., at the site of assembly of the 9 + 2 axoneme. Given that the fate of the proximal tubule seems to be variable dependent upon species, this might explain why in Xenopus sperm, γ-tubulin has not been found associated with the pericentriolar material of flagellar centrioles (Stearns et al., 1991; Felix et al., 1994; Stearns and Kirschner, 1994), whereas in mouse sperm, it has (Palacios et al., 1993).
αBSN also labeled acrosomes of late-stage spermatids that had undergone nuclear elongation (Fig. 10 F). By immunoelectron microscopy, the labeling was most dense at the inner surface of the outer acrosomal membrane. Finally, as predicted from our immunofluorescence data, the mitochondria within the middle piece of the sperm tail were specifically and uniformly labeled with anti-BSN antibodies (Fig. 10 G). Based upon the human sequence, we would have presumed that this labeling represented BSN1a rather than BSN1b. Further studies will be necessary to determine whether there are multiple forms of basonuclin that are differentially localized in germ cells.
Discussion
For years, it has been known that zinc plays an important role in testis development, and a number of zinc finger proteins are expressed in male germ cells (Burke and Wolgemuth, 1992; Noce et al., 1992, 1993; Hosseini et al., 1994; Zambrowicz et al., 1994; Kundu and Rao, 1995; Passananti et al., 1995; Stassen et al., 1995; Mello et al., 1996; Supp et al., 1996). Where tested, these proteins have been found to be strictly nuclear. Our discovery that basonuclin is a testis protein adds another zinc finger protein to this growing list, but its location sets it apart from the others.
Given the prior studies of Tseng and Green (1992, 1994), we were surprised to find basonuclin expressed in testis at all, since it had been thought to be restricted to stratified squamous epithelia. However, BSN RNA expression was as high or higher in testis than in any other organ examined. BSN RNAs were detected early in the differentiative pathway of mouse germ cells, i.e., long before the animals reached sexual maturity. Despite basonuclin RNA expression in mitotically active spermatogonia, basonuclin protein was not detected until later, where antibody labeling was first seen in meiotic spermatocytes. While antibody masking is always a formal possibility, three different affinity-purified peptide antibodies failed to reveal labeling in spermatogonia. Thus, we conclude that if basonuclin protein is expressed earlier in development, it is present at reduced levels or in a very different complex than its location in spermiogenesis.
In meiotic spermatocytes, basonuclin appeared to be concentrated in centrosomes, a location that it then maintained throughout spermiogenesis. At least at later stages of spermiogenesis, basonuclin seemed to be localized to hollow ringlike appendages that were largely if not fully confined to the centriole forming the sperm flagellum. This was reminiscent of mitotic cells, where satellite structures are generally unique to the mature centriole and are not found on newly synthesized (immature) centrioles (for review see Lange and Gull, 1996). The restriction of appendages to the axonemal centriole of male germ cells has been described before (Browder et al., 1991; Lange and Gull, 1996).
Little is known about the functions or molecular complexity of satellite structures associated with centrioles. In fact, the first molecular marker for centriole maturation, cenexin (96 kD), was only recently discovered in mitotically active cells (Lange and Gull, 1995), and as yet cenexin has not been cloned. In contrast to cenexin, which is regulated with the cell cycle of mitotic cells, basonuclin may represent the first example of a centriolar appendage marker that, in male germ cells, is largely specific for spermatocytes and spermatids. The primarily distal centriole location in spermatids is particularly intriguing because: (a) This is the only centriole that nucleates microtubule assembly in the spermatid, and pericentriolar material surrounding centrioles has been implicated in orchestrating microtubule organizing activity (Gould and Borisy, 1977; Telzer and Rosenbaum, 1979; Calarco-Gilliam et al., 1983; Doxsey et al., 1994; Lange and Gull, 1995); and (b) so little is known about how microtubules organize into their unique and diverse arrays during spermiogenesis. Through its association with the distal centrioles of developing spermatids, basonuclin becomes a candidate for a protein that could be involved in tailoring the organization of the microtubules during male germ cell meiosis and spermatid differentiation. Moreover, by identifying one protein involved in these centrioles, basonuclin becomes a powerful tool for identifying additional proteins involved in centriole maturation during postmeiotic spermatogenesis. This should allow us to probe deeper into the differences that exist between the centrosomes of meiotic versus mitotic germ cells.
It is puzzling that BSN antibodies and cell fractionation studies also detect this protein in the acrosome and mitochondrial sheath of the mature sperm. Since the acrosome is a storage vessel for proteins used in fertilization, we surmise that basonuclin might perform a specialized role in this process. This notion is particularly interesting in light of the facts that: (a) Basonuclin appears to associate with the acrosome late in spermiogenesis; and (b) centrioles are absent in the oocytes of many species, including mouse (Schatten, 1994; Lange and Gull, 1996). In the future, it will be important to examine basonuclin expression during oogenesis and to track the fate of sperm basonuclin during fertilization.
While the localization of basonuclin in sperm acrosomes is consistent with the hypothesis that basonuclin performs a function in centrosomes and/or microtubule organization, we are at a loss to explain why basonuclin was also detected in the mitochondria of the flagellar midpiece. This said, BSN1a has a perfect mitochondrial targeting sequence at its amino terminus, and this sequence is evolutionarily conserved. Although mBSN1a seems to utilize a downstream ATG, the two putative forms may perform unique functions in separate compartments of the differentiating male germ cell.
To make matters more intriguing, basonuclin has what appears to be a reasonably bona fide nuclear localization signal. While computer analysis of known proteins indicates that basonuclin has only a 40% chance of being localized to the nucleus, the protein does associate with the nucleus in epidermal keratinocytes (Tseng and Green, 1994), and we have confirmed this with our antibodies (unpublished results). We did not detect basonuclin in isolated sperm nuclei, nor did we detect αBSN labeling in germ cell nuclei. Thus, it seems unlikely that basonuclin is nuclear in germ cells, although we cannot rule out the possibility that in the early stages of spermatogenesis, its antigenic determinants are masked by association with other nuclear proteins. Additionally, while evidence that basonuclin is a DNA-binding protein is lacking, its structural features predict that it has this potential.
One possibility is that basonuclin might be transiently associated with chromatin after nuclear envelope breakdown of meiotic germ cells. Since the cell cycle of meiotic mouse germ cells is so long, meiotic germ cells in the act of nuclear envelope breakdown and spindle formation are rare, making such analysis difficult. However, in this regard, it may be relevant that CP190, a recently described zinc finger protein in mitotic cells of Drosophila, is associated with centrosomes during mitosis and with chromatin during interphase (Oegema et al., 1995; Whitfield et al., 1995). Despite the lack of sequence similarity between CP190 and basonuclin, these findings suggest collectively that: (a) Zinc finger proteins may play important roles in the formation, structure, positioning, or function of centrosomes; and (b) the requirements for these proteins may differ in meiosis and mitosis. As future studies are conducted, the mysteries underlying the dynamic roles of basonuclin during spermiogenesis should become increasingly apparent.
Acknowledgments
We thank Christa Wellman for her artistic talents exerted in the design of Fig. 1 and Chuck Welleck for his artwork in preparation of the color figures. We thank Dr. Thomas Medsgar for his autoimmune sera against centriolar proteins, Dr. Bruce Alberts, and Dr. Harish Joshi for antibodies against γ-tubulin. We thank Dr. Gary Smith for his valuable discussions on spermiogenesis, Dr. Xiaoming Wang for her advice on RNase protection assays, and Dr. Satrajit Sinha for his computer analysis and identification of the mitochondrial localization signal.
Footnotes
1. Abbreviations used in this paper: BSN, basonuclin; DAPI, 4,6-diamidino-2-phenylindole.
Z.-H. Yang was supported by a grant from the National Institutes of Health (AR31737), and Dr. I. Gallicano was a postdoctoral fellow supported by a training grant awarded to the University of Chicago from the National Cancer Institute. Dr. E. Fuchs is an Investigator of the Howard Hughes Medical Institute.
Address all correspondence to Elaine Fuchs, Howard Hughes Medical Institute, Department of Molecular Genetics and Cell Biology, The University of Chicago, 5841 S. Maryland Avenue, Room N314, Chicago, IL 60637. Tel.: (773) 702–1347. Fax: (773) 702-0141.
References
- Browder, L.W., C. Erickson, and W. Jeffery. 1991. Spermatogenesis. In Developmental Biology, 3rd ed. Saunders College Publishing, New York. 22–53.
- Burke PS, Wolgemuth DJ. Zpf-37, a new murine zinc finger encoding gene, is expressed in a developmentally regulated pattern in the male germ line. Nucleic Acids Res. 1992;20:2827–2834. doi: 10.1093/nar/20.11.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco-Gillam PD, Siebert MC, Hubble R, Mitchison T, Kirschner M. Centrosome development in early mouse embryos as defined by an autoantibody against pericentriolar material. Cell. 1983;35:621–629. doi: 10.1016/0092-8674(83)90094-6. [DOI] [PubMed] [Google Scholar]
- Chiang MK, Flanagan JG. PTP-NP, a new member of the receptor protein tyrosine phosphatase family, implicated in development of nervous system and pancreatic endocrine cells. Development (Camb) 1996;122:2239–2250. doi: 10.1242/dev.122.7.2239. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- de Vant'ery C, Gavin AC, Vassalli JD. An accumulation of p34cdc2 at the end of mouse oocyte growth correlates with the acquisition of meiotic competence. Dev Biol. 1996;174:335–344. doi: 10.1006/dbio.1996.0078. [DOI] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Faus I, Hsu HJ, Fuchs E. Oct 6: a regulator of keratinocyte gene expression in stratified squamous epithelia. Mol Cell Biol. 1994;14:3263–3275. doi: 10.1128/mcb.14.5.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA, Antony C, Wright M, Maro B. Centrosome assembly in vitro: role of γ-tubulin recruitment in Xenopussperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuge H. Unorthodox male meiosis in Trichosia pubescens (Sciaridae)chromosome elimination involves polar organelle degeneration and monocentric spindles in first and second division. J Cell Sci. 1994;107:299–312. doi: 10.1242/jcs.107.1.299. [DOI] [PubMed] [Google Scholar]
- Gonzalez C, Casal J, Ripoll P. Functional monopolar spindles caused by mutation in mgr, a cell division gene of Drosophila melanogaster. . J Cell Sci. 1988;89:39–47. doi: 10.1242/jcs.89.1.39. [DOI] [PubMed] [Google Scholar]
- Gonzalez C, Saunders RD, Casal J, Molina I, Carmena M, Ripoll P, Glover DM. Mutations at the asp locus of Drosophila lead to multiple free centrosomes in syncytial embryos, but restrict centrosome duplication in larval neuroblasts. J Cell Sci. 1990;96:605–616. doi: 10.1242/jcs.96.4.605. [DOI] [PubMed] [Google Scholar]
- Gould RR, Borisy GG. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J Cell Biol. 1977;73:601–615. doi: 10.1083/jcb.73.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini R, Marsh P, Pizzey J, Leonard L, Ruddy S, Bains S, Dudley K. Restricted expression of a zinc finger protein in male germ cells. J Mol Endocrinol. 1994;13:157–165. doi: 10.1677/jme.0.0130157. [DOI] [PubMed] [Google Scholar]
- Komiya T, Hachiya N, Sakaguchi M, Omura T, Mihara K. Recognition of mitochondria-targeting signals by a cytosolic import stimulation factor, MSF. J Biol Chem. 1994;269:30893–30897. [PubMed] [Google Scholar]
- Kubiak JZ, Weber M, Geraud G, Maro B. Cell cycle modification during the transitions between meiotic M-phases in mouse oocytes. J Cell Sci. 1992;102:457–467. doi: 10.1242/jcs.102.3.457. [DOI] [PubMed] [Google Scholar]
- Kundu TK, Rao MR. DNA condensation by the rat spermatidal protein TP2 shows GC-rich sequence preference and is zinc dependent. Biochemistry. 1995;34:5143–5150. doi: 10.1021/bi00015a027. [DOI] [PubMed] [Google Scholar]
- Lange BM, Gull K. A molecular marker for centriole maturation in the mammalian cell cycle. J Cell Biol. 1995;130:919–927. doi: 10.1083/jcb.130.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BMH, Gull K. Structure and function of the centriole in animal cells: progress and questions. Trends Cell Biol. 1996;6:348–352. doi: 10.1016/0962-8924(96)10033-7. [DOI] [PubMed] [Google Scholar]
- Larsen, W.J. 1993. Human Embryology. Churchill Livingstone, Inc., Singapore. 1–479.
- Lepage N, Roberts KD. Purification of lysophospholipase of human spermatozoa and its implication in the acrosome reaction. Biol Reprod. 1995;52:616–624. doi: 10.1095/biolreprod52.3.616. [DOI] [PubMed] [Google Scholar]
- Matthies HJ, McDonald HB, Goldstein LS, Theurkauf WE. Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J Cell Biol. 1996;134:455–464. doi: 10.1083/jcb.134.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride HM, Goping IS, Shore GC. The human mitochondrial import receptor, hTom20p, prevents a cryptic matrix targeting sequence from gaining access to the protein translocation machinery. J Cell Biol. 1996;134:307–313. doi: 10.1083/jcb.134.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello GC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegansembryos. Nature (Lond) 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- Messinger SM, Albertini DF. Centrosome and microtubule dynamics during meiotic progression in the mouse oocyte. J Cell Sci. 1991;100:289–298. doi: 10.1242/jcs.100.2.289. [DOI] [PubMed] [Google Scholar]
- Noce T, Fujiwara Y, Sezaki M, Fujimoto H. Expression of a mouse zinc finger protein gene in both spermatocytes and oocytes during meiosis. Dev Biol. 1992;153:356–367. doi: 10.1016/0012-1606(92)90120-6. [DOI] [PubMed] [Google Scholar]
- Noce T, Fujiwara Y, Ito M, Takeuchi T, Hashimoto N, Yamanouchi M, Higashinakagawa T, Fujimoto H. A novel murine zinc finger gene mapped within the tw18 deletion region expresses in germ cells and embryonic nervous system. Dev Biol. 1993;155:409–422. doi: 10.1006/dbio.1993.1039. [DOI] [PubMed] [Google Scholar]
- Oegema K, Whitfield WGF, Alberts B. The cell cycle–dependent localization of the CP190 centrosomal protein is determined by the coordinate action of two separable domains. J Cell Biol. 1995;131:1261–1273. doi: 10.1083/jcb.131.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios MJ, Joshi HC, Simerly C, Schatten G. γ-Tubulin reorganization during mouse fertilization and early development. J Cell Sci. 1993;104:383–389. doi: 10.1242/jcs.104.2.383. [DOI] [PubMed] [Google Scholar]
- Passananti C, Corbi N, Paggi MG, Russo MA, Perez M, Cotelli F, Stefanini M, Amati P. The product of Zfp59 (Mfg2), a mouse gene expressed at the spermatid stage of spermatogenesis, accumulates in spermatozoa nuclei. Cell Growth Diff. 1995;6:1037–1044. [PubMed] [Google Scholar]
- Rheinwald JG, Green H. Epidermal growth factor and the multiplication of cultured human epidermal keratinocytes. Nature (Lond) 1977;265:421–424. doi: 10.1038/265421a0. [DOI] [PubMed] [Google Scholar]
- Rugh, R. 1990. The Mouse: Its Reproduction & Development. Oxford University Press, Oxford.
- Schatten G. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol. 1994;165:299–335. doi: 10.1006/dbio.1994.1256. [DOI] [PubMed] [Google Scholar]
- Shore GC, McBride HM, Millar DG, Steenaart NA, Nguyen M. Import and insertion of proteins into the mitochondrial outer membrane. Eur J Biochem. 1995;227:9–18. doi: 10.1111/j.1432-1033.1995.tb20354.x. [DOI] [PubMed] [Google Scholar]
- Shu HB, Joshi HC. γ-Tubulin can both nucleate microtubule assembly and self-assemble into novel tubular structures in mammalian cells. J Cell Biol. 1995;130:1137–1147. doi: 10.1083/jcb.130.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, Cande WZ. Microtubule distribution in dv, a maize meiotic mutant defective in the prophase to metaphase transition. Dev Biol. 1990;138:231–242. doi: 10.1016/0012-1606(90)90193-m. [DOI] [PubMed] [Google Scholar]
- Stassen MJ, Bailey D, Nelson S, Chinwalla V, Harte PJ. The Drosophilatrithorax proteins contain a novel variant of the nuclear receptor type DNA binding domain and an ancient conserved motif found in other chromosomal proteins. Mech Dev. 1995;52:209–223. doi: 10.1016/0925-4773(95)00402-m. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of γ-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. γ-Tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Supp DM, Witte DP, Branford WW, Smith EP, Potter SS. Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability, and male fertility. Dev Biol. 1996;176:284–299. doi: 10.1006/dbio.1996.0134. [DOI] [PubMed] [Google Scholar]
- Telzer BR, Rosenbaum JL. Cell cycle–dependent, in vitro assembly of microtubules onto the pericentriolar material of HeLa cells. J Cell Biol. 1979;81:484–497. doi: 10.1083/jcb.81.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H, Green H. Basonuclin: a keratinocyte protein with multiple paired zinc fingers. Proc Natl Acad Sci USA. 1992;89:10311–10315. doi: 10.1073/pnas.89.21.10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H, Green H. Association of basonuclin with ability of keratinocytes to multiply and with the absence of terminal differentiation. J Cell Biol. 1994;126:495–506. doi: 10.1083/jcb.126.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Snyder SH. Inositol 1,4,5-triphosphate receptors selectively localized to the acrosomes of mammalian sperm. J Cell Biol. 1995;130:857–869. doi: 10.1083/jcb.130.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield WGF, Chaplin MA, Oegema K, Parry H, Glover DM. The 190 kDa centrosome-associated protein of Drosophila melanogastercontains four zinc finger motifs and binds to specific sites on polytene chromosomes. J Cell Sci. 1995;108:3377–3387. doi: 10.1242/jcs.108.11.3377. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe D, Albertini DF. Centrosome phosphorylation and the developmental expression of meiotic competence in mouse oocytes. Dev Biol. 1992;152:62–74. doi: 10.1016/0012-1606(92)90156-b. [DOI] [PubMed] [Google Scholar]
- Yang Y, Dowling J, Yu Q-C, Kouklis P, Cleveland DW, Fuchs E. An essential cytoskeletal linker protein connecting actin microfilaments to intermediate filaments. Cell. 1996;86:655–665. doi: 10.1016/s0092-8674(00)80138-5. [DOI] [PubMed] [Google Scholar]
- Yoshiki T, Herr JC, Lee CYG. Purification and characterization of a sperm antigen recognized by HSA-5 monoclonal antibody. J Reprod Immunol. 1995;29:209–222. doi: 10.1016/0165-0378(95)00946-i. [DOI] [PubMed] [Google Scholar]
- Zambrowicz BP, Zimmermann JW, Harendza CJ, Simpson EM, Page DC, Brinster RL, Palmiter RD. Expression of a mouse Zfy-1/ lacZ transgene in the somatic cells of the embryonic gonad and germ cells of the adult testis. Development (Camb) 1994;120:1549–1559. doi: 10.1242/dev.120.6.1549. [DOI] [PubMed] [Google Scholar]