Abstract
Aminopeptidase I (API) is transported into the yeast vacuole by the cytoplasm to vacuole targeting (Cvt) pathway. Genetic evidence suggests that autophagy, a major degradative pathway in eukaryotes, and the Cvt pathway share largely the same cellular machinery. To understand the mechanism of the Cvt import process, we examined the native state of API. Dodecameric assembly of precursor API in the cytoplasm and membrane binding were rapid events, whereas subsequent vacuolar import appeared to be rate limiting. A unique temperature-sensitive API-targeting mutant allowed us to kinetically monitor its oligomeric state during translocation. Our findings indicate that API is maintained as a dodecamer throughout its import and will be useful to study the posttranslational movement of folded proteins across biological membranes.
The faithful transport of resident organellar proteins is a hallmark in maintaining the functional characteristics and integrity of eukaryotic cells. All nuclearencoded proteins delivered to their respective organelles must cross at least one membrane barrier. Posttranslational delivery of proteins into membrane compartments occurs by three known mechanisms: a protein-conducting channel, as observed for example in the import of proteins into mitochondria and ER (Pfanner and Neupert, 1990; Hannavy et al., 1993; Egner et al., 1995; Rapoport et al., 1996); a large nuclear pore–like complex (Davis, 1995; Görlich and Mattaj, 1996); and by vesicle-mediated transport and fusion events (Rothman and Wieland, 1996). The conformational state of proteins during this translocation step limits and defines the possible mechanisms by which proteins can be delivered to their final destination.
Proteins arrive at their respective organelles in either a partially unfolded state or a fully folded conformation. Most proteins enter the ER cotranslationally, while those that cross the membrane after synthesis assume an extended conformation to pass through the translocation complex in the ER membrane (Rapoport et al., 1996). Most mitochondrial proteins enter the organelle posttranslationally and must assume a partially unfolded state before translocation across the membrane through a proteinaceous channel (Hannavy et al., 1993; Hachiya et al., 1995; Ryan and Jensen, 1995). Small monomeric proteins or peptides may also be translocated across membranes through ATP binding cassette transporters (Egner et al., 1995). No proteins are known to enter the vacuole cotranslationally, and a protein-conducting channel has not been identified in the vacuole membrane.
A vacuolar version of a channel equivalent to the nuclear pore complex would allow the passage of folded proteins into the vacuole lumen. However, there is no morphological evidence for such pore complexes on the vacuole membrane. Furthermore, the vacuole maintains a membrane potential through the action of a vacuolar ATPase (Klionsky et al., 1990). The corresponding requirement for a sealed membrane would preclude such pore complexes from existing on the vacuole.
In contrast to the extended conformation used by proteins imported into mitochondria and ER, conformational studies of peroxisomal proteins revealed that substrates destined for this organelle can enter the peroxisomal matrix in a fully folded state. In fact, large, preassembled oligomeric complexes can be imported into the peroxisomal lumen (Subramani, 1993; Rachubinski and Subramani, 1995). The dimeric thiolase, CAT trimers, and alcohol oxidase octamers all serve as substrates for import into the peroxisome (Walton et al., 1992; Glover et al., 1994; McNew and Goodman, 1994). The lack of ideal marker proteins that undergo proteolytic maturation steps, however, has complicated the analysis of this pathway. Accordingly, whether peroxisomal substrates enter the organelle by a translocation pore or through a vesicular mechanism remains to be resolved.
The majority of resident vacuolar proteases are transported posttranslationally via part of the secretory pathway. Upon vacuolar delivery, most of these hydrolases are processed by cleavage of a propeptide region to produce the active mature form of the enzymes. In contrast, aminopeptidase I (API)1 is delivered to the vacuole by the nonclassical cytoplasm to vacuole targeting (Cvt) pathway (Klionsky et al., 1992; Harding et al., 1995). After synthesis as a cytosolic 61-kD precursor, API becomes proteolytically processed in the vacuole by the removal of its NH2-terminal propeptide, resulting in the 50-kD mature protease (Klionsky et al., 1992). Thus, the molecular mass shift of API upon vacuolar delivery serves as a useful marker for correct import. Using this criterion, a set of mutants (cvt) defective in API processing was recently isolated (Harding et al., 1995, 1996). Surprisingly, the cvt mutants show extensive genetic and phenotypic overlap with two sets of mutants defective in autophagy, apg and aut (Tsukada and Ohsumi, 1993; Thumm et al., 1994), which suggests that the Cvt pathway and the autophagy pathway use much of the same cellular machinery (Harding et al., 1996; Scott et al., 1996).
The study of the mechanism of API import promises to contribute to our understanding of the basic processes of not only the Cvt pathway but also autophagy. Autophagy plays a central role in protein and organelle turnover in all eukaryotic cells by delivering cytoplasmic components to the lysosome/vacuole. In addition, autophagy has been implicated in cellular remodeling during development and differentiation and removal of damaged cellular components, and is critical for survival during stress conditions such as nutrient deprivation (Glaumann et al., 1981; Marzella and Glaumann, 1987; Mortimore et al., 1989; Hilt and Wolf, 1992; Egner et al., 1993; Dunn, 1994). Despite the genetic overlap between the autophagy and cvt mutants, some distinctions between the pathways are still apparent. Whereas bulk autophagy is a nonselective, degradative pathway, the delivery of API is a selective, biosynthetic process. Similarly, vacuolar protein uptake by bulk autophagy is relatively slow and cannot account for the rapid kinetics of API import (Egner et al., 1993; Knop et al., 1993; Scott et al., 1996). Finally, import of API is constitutive, occurring under vegetative growth as well as starvation conditions.
The molecular components and mechanisms of both autophagy and the Cvt pathway remain to be elucidated. In this study, we examined the oligomerization state of API during its import process to understand how it enters the vacuole. Previous studies have indicated that API exists in the vacuole as a dodecamer of identical subunits (Metz et al., 1977; Löffler and Röhm, 1979). We hypothesized that examining the kinetics of API assembly and transit through the Cvt pathway would offer insight into the question of whether large oligomeric complexes are imported into the vacuole as well as elucidating possible intermediate steps of the biosynthetic Cvt pathway. Further, we used a unique API-targeting mutant that allowed us to kinetically examine the assembly state of API during the membrane crossing event. Our findings indicate that precursor API is rapidly oligomerized into a dodecameric complex that is subsequently transported into the vacuole without disassembly.
Materials and Methods
Strains and Media
The Saccharomyces cerevisiae strains used in this study were SEY6210, MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL and SEY6211, MAT a leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 ade2-101 suc2- Δ9 GAL (Robinson et al., 1988); DYY101 MATα leu2-3,112 ura3-52 his3- Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL ape1Δ::LEU2 (Klionsky et al., 1992); THY101 MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL ape1Δ::LEU2 (Oda et al., 1996).
cvt strains 1 to 17 were derived from SEY6210 and SEY6211 (Harding et al., 1995, 1996). The plasmid construction of the API propeptide deletions (Δ3–5, Δ6–8, Δ9–11, Δ12–14, Δ15–17, Δ18–20, Δ25–27, Δ28–30, Δ31– 33, Δ34–36, Δ37–39, Δ40–42, Δ2–45) and the K12R API mutation were described previously (Oda et al., 1996). The propeptide deletion mutation plasmids were introduced into strain DYY101, while the K12R mutant was transformed into strain THY101.
Reagents
The Vistra enhanced chemifluorescence (ECF) Western Blotting System was obtained from Amersham Corp. (Arlington Heights, IL); oxalyticase was from Enzogenetics (Corvallis, OR); Express35S35S-label was from Dupont/NEN Research Products (Boston, MA); proteinase K, Pefabloc, and the molecular mass standards used for the native gel and glycerol gradient analyses were obtained from Boehringer Mannheim Corp. (Indianapolis, IN); Sulfo-NHS-Biotin and avidin agarose were from Pierce (Rockford, IL); all other reagents were from Sigma Chemical Co. (St. Louis, MO). Antisera against the following proteins were prepared as described previously: API (Klionsky et al., 1992), carboxypeptidase Y (CPY) and proteinase A (PrA) (Klionsky et al., 1988), and alkaline phosphatase (ALP) (Klionsky and Emr, 1989); antiserum against phosphoglycerate kinase (PGK) was provided by Dr. Jeremy Thorner (University of California, Berkeley, CA) (Baum et al., 1978).
Preparation of Crude Cell Extracts, Spheroplasting, Cell Fractionation, and Labeling
Labeling of whole cells has been described previously. In brief, cells were grown to an OD600 of 1 and resuspended in synthetic minimal medium (SMD; 0.067% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as needed) at 20–30 OD600/ml. The resuspended cells were labeled with 10–20 μCi of 35S Express label/OD600 for the indicated times and temperatures, followed by a chase reaction in which the labeled cells were diluted to 1 OD/ml in SMD supplemented with 0.2% yeast extract, 4 mM methionine, and 2 mM cysteine.
For preparation of crude cell extracts used for glycerol gradient analysis, 5–15 OD600 units of labeled or unlabeled cells were resuspended in 300 μl of 20 mM K-Pipes, pH 6.8, containing a cocktail of protease inhibitors (10 mM benzamidine, 4 mM Pefabloc or 2 mM PMSF, 2 μg/ml pepstatin, and 10 mM NaN3). Acid-washed glass beads were then added to a level 50% of the resuspended cell volume. The cell suspension was lysed by vortexing for 1 min, and the cell extract was collected after a 3-min centrifugation at 12,000 g.
Spheroplasts were prepared using a modified procedure of one previously described (Klionsky et al., 1992). Cells were grown in SMD to an OD600 of 1 and incubated for 15 min at 30°C in a buffer of 0.1 M Tris-SO4, pH 9.4, 10 mM DTT. After a 5,000-g spin for 5 min, the cells were resuspended in an osmotically supportive spheroplast labeling medium (1 M sorbitol, 1% glucose, 1% proline, Wickerham's salts, pH 7.5 [Guthrie and Fink, 1991]), containing 1 μg/OD600 of oxalyticase and incubated for 30 min at 30°C. After a 5-min spin at 3,000 g, the spheroplasts were resuspended in fresh spheroplast labeling medium, pH 5.0, at 20–50 OD600/ml and labeled with 10–20 μCi of 35S Express label/OD600 for 5–10 min at the indicated temperatures. The labeling reaction was chased by diluting the spheroplasts to 1–3 OD600/ml in SMD supplemented with 0.2% yeast extract, 1 M sorbitol, 4 mM methionine, and 2 mM cysteine for the times and temperatures indicated.
Subcellular fractionations of spheroplasts by differential osmotic lysis has been described previously (Scott and Klionsky, 1995). The pellet fraction was resuspended in 20 mM K-Pipes, pH 6.8, with protease inhibitor cocktail to release the vacuolar lumenal contents as well as to release API bound to the pellet.
Protease Studies
Spheroplasts of the THY101 (ape1Δ) strain harboring the K12R API single copy plasmid were labeled for 5 min, followed by a chase reaction for 30 min at 38°C. The labeled spheroplasts were subjected to differential osmotic lysis in import buffer (20 mM K-Pipes, pH 6.8, 100 mM sorbitol, 100 mM KCl, 50 mM KOAc, 5 mM Mg[OAc]2). Permeabilized cells were then fractionated by centrifugation at 5,000 g for 3 min, resulting in a supernatant fraction and a pellet fraction that was then resuspended in import buffer. The pellet fraction contains intact vacuoles (Scott and Klionsky, 1995). The fractions were treated with 100 μg/ml proteinase K alone, or with the addition of 0.2% Triton X-100 for 15 min, on ice. Nonradioactive pellet and supernatant fractions were added to the labeled supernatant and pellet samples, respectively, to keep the protein concentration constant during the protease digestion procedure and subsequent protein recovery steps.
Biotinylation Studies
Spheroplasts were subjected to pulse/chase analysis and subcellular fractionation exactly as in the protease protection experiment above. The pellet fraction (from 5 OD600 of radiolabeled spheroplasts) was resuspended in 500 μl of import buffer (pH adjusted to 7.4) containing 0.5 mg/ml SulfoNHS-Biotin in the presence or absence of 0.2% Triton X-100. The samples were incubated for 20 min at room temperature. The reaction was quenched by the addition of 100 mM Tris-Cl, pH 7.5, followed by precipitation with 10% trichloroacetic acid. Proteins were recovered by immunoprecipitation with antibodies against API and CPY followed by recovery on avidin agarose beads (Scott and Klionsky, 1995). Nonradioactive pellet and supernatant fractions were added to the labeled supernatant and pellet samples, respectively, to keep the protein concentration constant during the cross-linking and protein recovery procedures.
Glycerol Gradients, Immunoprecipitations, and Western Blotting
For gradient analysis, 200 μl of cell extracts or fractionated spheroplast samples were loaded on a 1.8-ml, 20–50% glycerol step-gradient prepared in 20 mM K-Pipes, pH 6.8, with a cocktail of protease inhibitors. The gradients were spun in a centrifuge (model TL-100; Beckman Instrs., Fullerton, CA) for 4 h at 270,000 g, and at 15°C using a TLS-55 rotor (Beckman Instrs.). Fractions were collected from the top of the gradients and immunoprecipitated as previously described (Klionsky et al., 1992). The Western blotting procedure described previously (Oda et al., 1996) was modified and immunodetection was executed using a Vistra ECF chemifluorescent substrate (Amersham Corp.). Identical gradients were run with a mixture of molecular mass standards consisting of ovalbumin (45 kD), aldolase (158 kD), catalase (240 kD), apoferritin (450 kD), and thyroglobulin (669 kD); the collected fractions were precipitated as described above and subjected to SDS-PAGE and Coomassie brilliant blue staining.
Quantitation of immunoprecipitations and Western blots was performed using a phosphorimager (model Storm; Molecular Dynamics, Sunnyvale, CA) equipped with both phosphorescent and chemifluorescent scanning capabilities.
Native Gel Sample Preparation and Electrophoresis
Sample preparation, native gel electrophoresis, and Hedrick-Smith analysis procedures have been previously described in detail (Hedrick and Smith, 1968; Tomashek et al., 1996).
Results
Precursor and Mature API Are Both Dodecamers in Steady-State Conditions
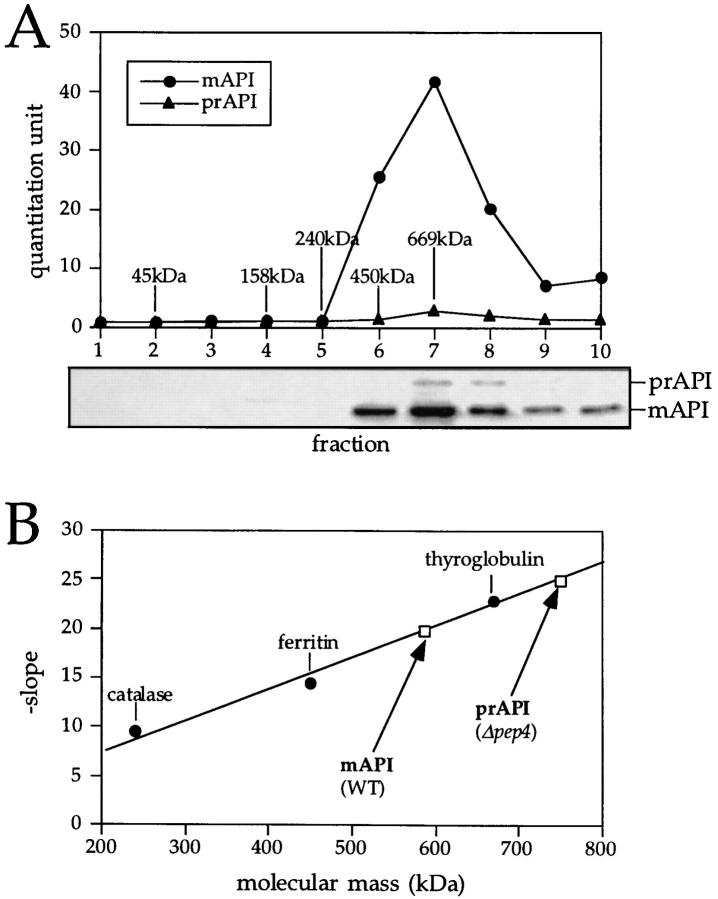
The native state of API was examined by glycerol density gradients and native gel electrophoresis. Cell extracts were prepared from wild-type cells and separated on 20–50% glycerol density gradients as described in Materials and Methods. After centrifugation, the proteins in the collected fractions were precipitated and subjected to Western blot analysis. Under steady-state conditions, the majority of API in wild-type cells exists as the processed, mature form of the molecule. However, when cells were grown to midlog phase, a small population of precursor API could also be detected (Fig. 1 A). The peak concentrations of both precursor and mature API appeared in fraction 7 of the gradient (Fig. 1 A), cofractionating with the 669-kD molecular mass standard, thyroglobulin. This finding is consistent with the dodecameric stoichiometry of mature API subunits (12 × 50 kD = 600 kD) that was proposed previously (Metz et al., 1977; Löffler and Röhm, 1979). In addition, the appearance of precursor API in fraction 7 suggests that it also forms a dodecameric complex (12 × 61 kD = 732 kD). To confirm that these large complexes were in fact API homooligomers, cross-linking studies were performed using radiolabeled cell extracts. As before, precursor API-containing cross-linked products were recovered from fraction 7 of glycerol gradients (data not shown). On reduction of the cross-link, only API was detected, suggesting that the API oligomer recovered from fraction 7 is a homooligomer. These data do not rule out the possibility, however, that other proteins transiently associate with the API oligomer.
Figure 1.
Molecular mass determination of native precursor and mature API under steady-state conditions. (A) Glycerol gradient analysis of precursor and mature API under steady-state conditions. Wild-type (SEY6210) cells were grown to midlog phase, lysed with glass beads, and the resulting cell extracts were separated on 20–50% glycerol gradients. Collected fractions were subjected to Western blotting with antiserum to API. The reaction of a chemifluorescent substrate (ECF) with an alkaline phosphatase– conjugated secondary antibody allowed for the quantitation of the Western blots by a chemifluorescence scanner (Molecular Dynamics) as shown in the graph corresponding to the Western blot signals. Molecular mass standards indicated are hen egg albumin (45 kD), aldolase (158 kD), catalase (240 kD), ferritin (450 kD), and thyroglobulin (669 kD). Both mature (mAPI) and precursor API (prAPI) peaks appeared in fraction 7, cofractionating with the thyroglobulin molecular mass standard. (B) Hedrick-Smith calculation of the molecular masses of precursor and mature API. Relative mobilities were measured for protein standards resolved on native gels of 4.0 to 5.5% acrylamide and the negative slopes were determined by plotting the relative mobilities as a function of gel percentage. A standard curve was then generated by plotting the negative slopes of the protein standards as a function of molecular mass. Extracts from wild-type and pep4Δ strains were run on the same gels, and their molecular masses were calculated with reference to the standard curve. Molecular masses of mAPI and precursor API were determined to be 592 and 752 kD, respectively.
To obtain a separate estimation of the molecular mass of the API oligomer, native gels were run and analyzed by the Hedrick-Smith method (Hedrick and Smith, 1968; Tomashek et al., 1996). Native gels ranging from 4 to 5.5% acrylamide were run on extracts from wild-type and pep4Δ strains, and the relative mobilities of both mature and precursor API were measured. Molecular mass standards were run on the same gels, blotted, and stained with Amido black as cited in Materials and Methods. The relative mobilities were plotted as a function of gel percent and the negative slopes of these graphs were then plotted as a function of molecular mass (Fig. 1 B). The molecular masses of mature API from a wild-type strain and precursor API from a pep4Δ strain were determined to be 592 and 752 kD, respectively, consistent with the predicted molecular mass of 600 kD for the mature API dodecamer and 732 kD for the dodecameric precursor hydrolase (Fig. 1 B). Because the native gel electrophoresis analyses were in close agreement with the glycerol gradient results, subsequent kinetic studies were performed using the gradient method.
Oligomerization of Precursor API Is an Early Step in Its Import
The half-time of API import and processing is 30–45 min (Klionsky et al., 1992). We wanted to determine the point during biosynthesis at which API oligomerization occurs. To observe the oligomerization process in vivo, whole cells were labeled for 2 min, followed by chase reactions of 0, 3, 6, and 10 min with excess cold methionine and cysteine. After each chase point, the labeled cells were immediately lysed with glass beads and centrifuged for 1 min to remove cellular debris, and the extracts were separated on 20–50% glycerol density gradients. Fractions were collected from the gradient and analyzed by immunoprecipitation. Oligomerization did not appreciably occur after cell lysis (data not shown), presumably because of the resulting substantial dilution.
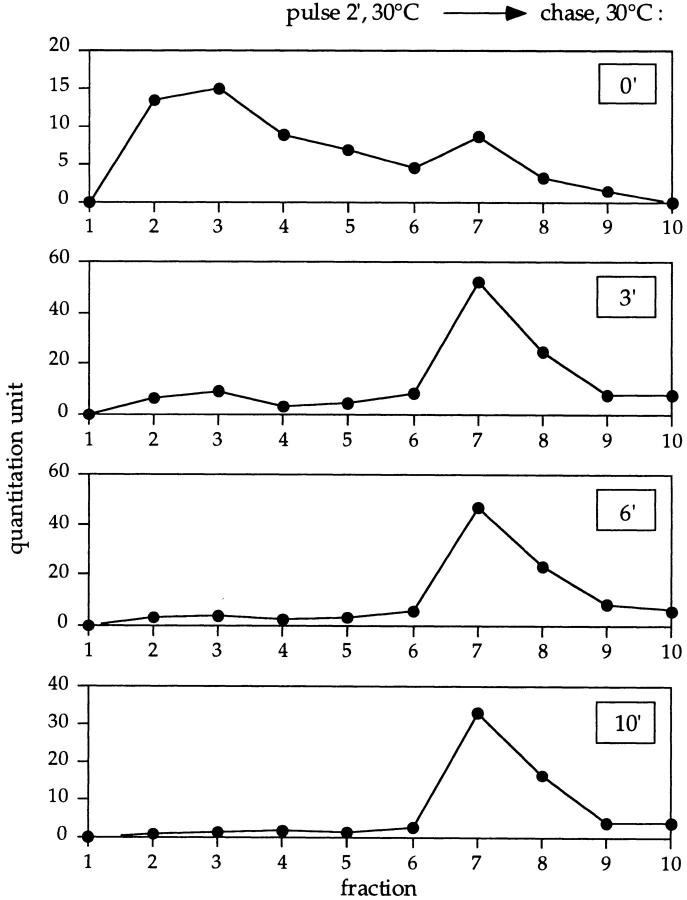
After a 2-min pulse without a chase, the majority of labeled precursor API was recovered in fractions 2 and 3, consistent with the size of the precursor API monomer (61 kD; Fig. 2, top panel). Interestingly, even at this early time point, a small peak at fraction 7 corresponding to oligomeric precursor could be detected. After a 3-min chase, the relative distribution of precursor API monomer and oligomer was reversed, with the majority of the labeled precursor API assembling into the oligomeric form and the concomitant depletion of the labeled monomer (Fig. 2, second panel). Oligomerization of labeled precursor API was nearly complete after 6 min of the chase reaction, and no monomer was detected after 10 min of chase (Fig. 2, third and fourth panels, respectively). These data indicate a half-time of oligomerization of ∼2 min. Therefore, the in vivo kinetics of precursor API oligomerization are far more rapid than the half-time of API maturation, suggesting that oligomer assembly is an early step in the import of API into the vacuole.
Figure 2.
Oligomerization kinetics of precursor API. Wild-type cells were pulse labeled for 2 min at 30°C followed by nonradioactive chase reactions. Aliquots were removed at the indicated chase times and lysed with glass beads, and the resulting cell extracts were separated on 20–50% glycerol gradients. Fractions were collected, immunoprecipitated with antiserum to API, and resolved by SDS-PAGE. Molecular mass standards corresponding to the fractions: 45 kD, fraction 2; 158 kD, fraction 4; 240 kD, fraction 5; 450 kD, fraction 6; and 669 kD, fraction 7. Quantitation of the radioactive signals was performed using a phosphorimager (model Storm; Molecular Dynamics).
Precursor API Oligomerizes in the Cytoplasm and Then Binds to a Membrane
To determine the subcellular location of precursor API oligomerization, subcellular fractionation experiments were performed. Cells were converted into spheroplasts, labeled for 5 min, chased for 3 min, and subjected to a differential osmotic lysis procedure that disrupts the plasma membrane while preserving the integrity of the vacuole (Scott and Klionsky, 1995). Subsequent centrifugation resulted in a cytosolic supernatant fraction and an organellar membrane fraction that included the unlysed vacuoles. The faithful segregation of vacuolar markers (ALP and PrA, vacuolar membrane and lumenal hydrolases, respectively) to the pellet fraction and a cytosolic marker (PGK) to the supernatant fraction indicated the efficiency of this subcellular fractionation procedure (Fig. 3 A).
Figure 3.

Oligomeric precursor API assembly occurs in the cytoplasm before membrane binding. Spheroplasts were labeled for 5 min at 30°C followed by a 3-min nonradioactive chase reaction. The samples were subjected to differential osmotic lysis and separated into a supernatant fraction and a pellet fraction containing intact vacuoles. An aliquot of the supernatant and pellet fractions was removed and immunoprecipitated with the indicated antisera, and the remainder was separated on 20–50% glycerol gradients. (A) Quantitation of the pellet fraction immunoprecipitated with vacuolar markers PrA and ALP, and the cytosolic marker PGK. The recovery of marker protein in the pellet was quantified using a Storm phosphorimager. The percent recovery was calculated as the ratio of the protein in the pellet fraction to the protein in the pellet and supernatant fractions. The supernatant (sup) fraction (B) and the pellet fraction (C) were separated on a glycerol gradient and immunoprecipitated with antiserum to API. Quantitation of the radioactive signals, represented by the graphs, indicates that the supernatant fraction contains both the precursor monomer and oligomer, while only the precursor API oligomer is bound to the pellet fraction.
Under these identical labeling and chase conditions, both the precursor API monomer and dodecamer were detected (Fig. 3 B). After subcellular fractionation, the labeled supernatant and resuspended pellet fractions were separated by 20–50% glycerol gradients and examined by immunoprecipitation. Monomeric precursor appears only in the supernatant fraction along with the fully assembled oligomeric precursor, suggesting that the API assembly process occurs in the supernatant fraction containing the released cytoplasm (Fig. 3 B). In contrast, primarily the oligomeric precursor was detected in the pellet fraction (Fig. 3 C). Furthermore, oligomeric precursor binding was very rapid, as 51% of the precursor API dodecamer was already bound to the membrane fraction after the 3-min chase reaction. Previous experiments have shown that the membrane-bound API is on the import pathway as it chases into the vacuole in vitro (Scott and Klionsky, 1995). These findings indicate that precursor assembly in the cytoplasm and subsequent membrane binding mark the early steps of API import to the vacuole.
cvt and Propeptide Deletion Mutants Are Not Defective in API Oligomerization
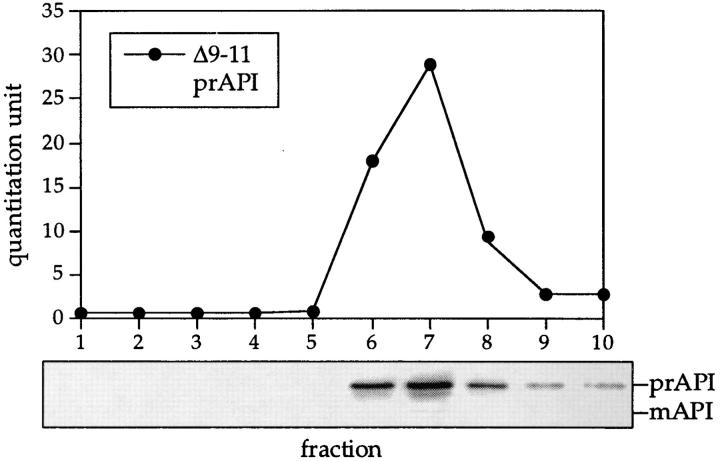
The first amphipathic helix of the API propeptide is critical for proper targeting of the enzyme (Oda et al., 1996). Specifically, deletions in this region inhibit API from binding to the membrane fraction, thus preventing subsequent import and processing to the mature form (Oda et al., 1996). We examined whether the targeting defect in these propeptide deletion mutants was caused by an oligomerization defect. Cell extracts were prepared from the DYY101 strain deleted for the gene encoding API (ape1Δ) but harboring a single copy plasmid encoding API with the propeptide deletions. Western blot analysis of glycerol gradient fractions indicated that precursor API in all of the propeptide deletion mutants peaked in fraction 7 (Fig. 4), indicating that it was properly oligomerized. In addition, an API molecule with a deletion of the entire propeptide also formed API oligomers (data not shown). These results indicate that, while the propeptide is necessary for binding to the membrane fraction and subsequent import of API into the vacuole, it does not encode any sequence determinants necessary for API oligomerization. In addition, these results demonstrate that dodecameric precursor API assembly can occur in the absence of membrane binding or transport to the vacuole, in agreement with the kinetic and subcellular fractionation data (Figs. 2 and 3).
Figure 4.
API-targeting mutants are not defective in API oligomerization. Strain DYY101 (ape1Δ) harboring a single copy plasmid encoding the propeptide deletions was grown to midlog phase, lysed with glass beads, and separated on a 20–50% glycerol gradient. The Δ9–11 API deletion mutant shown here is a representative example of the analysis of the cvt (1 to 17) and propeptide deletion mutants (Δ3–5, Δ6–8, Δ9–11, Δ12–14, Δ15– 17, Δ18–20, Δ25–27, Δ28–30, Δ31–33, Δ34–36, Δ37–39, Δ40–42, and Δ2–45). Collected fractions were subjected to Western blotting with antiserum to API and the quantitation of the Western blots (shown in the graph) was performed as in Fig. 1.
In addition to API propeptide mutants, we identified a series of chromosomal mutants (cvt) that are defective for API localization. The cvt mutants were isolated based on their accumulation of precursor API (Harding et al., 1995, 1996). To examine if the localization defect in any of these mutants was due to a failure in API oligomerization, cell extracts of cvt 1 to 17 were examined by glycerol gradients and Western blotting. Native precursor API from all of the cvt mutants appeared in fraction 7 in glycerol density gradients, consistent with correct precursor API assembly (data not shown). These results suggest that cvt mutants do not have defects in genes whose products are necessary for API oligomerization.
A Unique Temperature-sensitive API Propeptide Mutant (K12R) Accumulates in the Membrane Fraction
The oligomerization and membrane-binding steps of API targeting appear early in the overall import process. We next examined the oligomeric nature of API during the remainder of the targeting steps. The membrane binding of precursor API oligomer is followed by import into the vacuole and cleavage of the propeptide by proteinase B (PrB), which yields mature API (Klionsky et al., 1992). To study the oligomeric state of precursor API as it enters the vacuolar lumen from the membrane-bound state, we exploited the unique characteristics of an API-targeting mutant in which the twelfth lysine residue in the predicted amphipathic helix of the propeptide was changed to an arginine. The K12R point mutation rendered API defective for import at nonpermissive temperature, 38°C (Fig. 5 D, middle; Oda et al., 1996), while wild-type kinetics were observed at permissive temperature, 30°C (Fig. 5 D, bottom).
Figure 5.
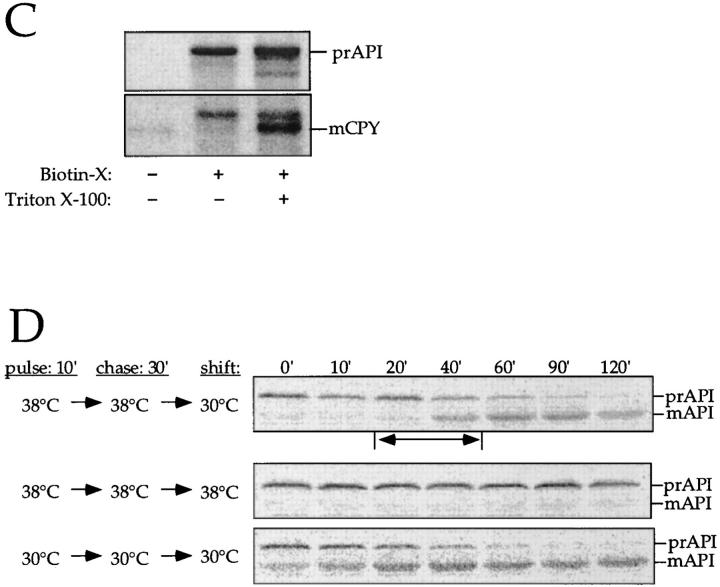
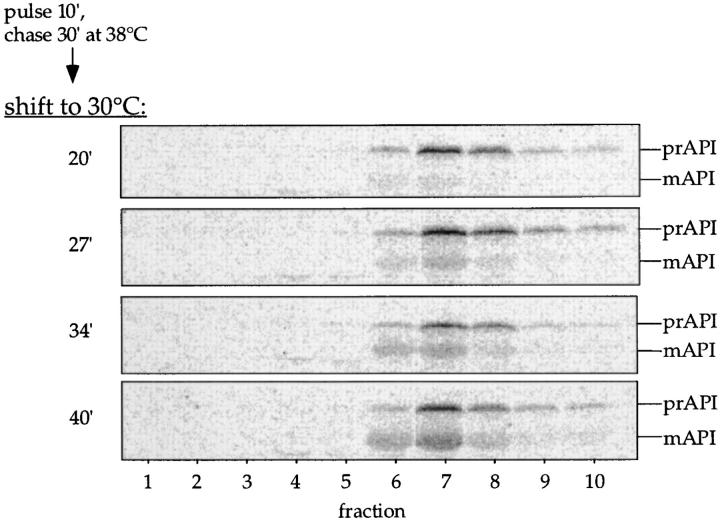
The membrane accumulation phenotype of the K12R API ts mutant is thermally reversible. (A) K12R API accumulates in the pellet fraction at nonpermissive temperature. Spheroplasts were labeled for 10 min at 38°C followed by nonradioactive chase. Aliquots were removed at the indicated chase times and separated into supernatant (sup) and pellet fractions. Samples were immunoprecipitated with antiserum to API, and the radiolabeled signals were quantitated. The percent radiolabeled precursor at a given chase point represents the ratio of precursor from each supernatant or pellet fraction to the total API combined in both fractions. (B) Protease accessibility of K12R API in the supernatant and pellet fractions. Spheroplasts were labeled for 5 min, chased for 30 min at 38°C, and separated into a supernatant and pellet fraction after differential osmotic lysis. The supernatant and pellet fractions were subjected to proteinase K and Triton X-100 as indicated and immunoprecipitated with antiserum to API (left). An aliquot of the recovered pellet fraction was also immunoprecipitated with antisera to the vacuolar marker CPY and the cytosolic marker PGK before protease treatment (right). The percent of marker proteins recovered in the pellet fraction was calculated as described for Fig. 3. (C) Accessibility of K12R API in the pellet fraction to cross-linking with Sulfo-NHS-biotin. Labeled spheroplasts were fractionated exactly as in B. The pellet fraction was cross-linked with Sulfo-NHS-biotin (Biotin-X) in the presence or absence of Triton X-100. API and CPY were recovered by immunoprecipitation followed by precipitation with avidin agarose beads. (D) Thermal reversibility of the K12R membrane-accumulation phenotype. The K12R API mutant was pulse-labeled for 10 min, chased for 30 min at 38°C, and then shifted to 30°C (top). Samples were removed at the indicated times during the shift period at 30°C and lysed with glass beads. The resulting cell extracts were immunoprecipitated with antiserum to API and resolved by SDS-PAGE. The double-headed arrow in the top panel marks the 20–40-min window of time when mature API increases from 10 to 56% during the 30°C shift. The K12R API strain was also pulse labeled, chased, and incubated all at 38°C (middle) or 30°C (bottom) in this experiment.
To examine the localization of precursor API in the K12R mutant at nonpermissive temperature, spheroplasts were labeled for 10 min at 38°C and chased from 0 to 2 h at 38°C. At each chase time point, samples were separated into supernatant and pellet fractions after differential osmotic lysis and analyzed by immunoprecipitation. Over the course of the chase period, the labeled precursor API was depleted from the supernatant fraction and accumulated in the pellet fraction (Fig. 5 A). The kinetics of membrane binding were essentially identical for K12R API at 38°C and the permissive temperature of 30°C (data not shown). Therefore, the unique targeting defect of the K12R API mutation did not affect the membrane-binding step, but rather a subsequent step in the import process.
The topology of K12R API accumulated at the membrane-bound stage was examined by protease digestion experiments. As above, labeled K12R API was allowed to accumulate in the membrane fraction of spheroplasts by a 10-min labeling and a 30-min chase reaction at 38°C. After differential osmotic lysis, the supernatant and pellet fractions were treated with proteinase K and examined by immunoprecipitation (Fig. 5 B). The vacuoles were shown to be intact as the majority of the lumenal vacuolar hydrolase CPY was localized in the pellet fraction, while the cytosolic marker, PGK, was concentrated in the supernatant fraction (Fig. 5 B, right). The majority of K12R API that accumulated in the membrane fraction was accessible to protease digestion, suggesting that the bound precursor API was exposed to the cytoplasmic environment and not protected within a lumenal compartment (Fig. 5 B, left). The small amount of precursor API remaining in the pellet fraction after protease treatment is consistent with unlysed spheroplasts that segregated to the pellet fraction (Fig. 5 B, right); this precursor API population was digested upon addition of detergent (Fig. 5 B, left).
The results of the protease-protection experiment were confirmed by assessing the accessibility of K12R API and CPY recovered in the pellet fraction to modification by Sulfo-NHS-biotin, a water-soluble cross-linker conjugated to biotin. Proteins modified by this cross-linker can be specifically recovered by precipitation with avidin agarose beads (Scott and Klionsky, 1995). When the pellet fraction was treated with Sulfo-NHS-biotin, precursor K12R API was recovered by avidin agarose regardless of whether detergent was present during the cross-linking reaction (Fig. 5 C). In the same samples, biotinylation of mature CPY and the small amount of mature API that escaped the temperature block required the vacuoles to be solubilized by detergent before the addition of cross-linker. Together with Fig. 5 B, these results indicate that the K12R precursor API that accumulates at 38°C is not in the vacuole, where it would be protected from both protease digestion and biotinylation. The fact that the mutant precursor is accessible to biotinylated cross-linker, while mature vacuolar hydrolases are not, suggests that it accumulates in a membrane-bound state that is exposed to the cytosolic environment before vacuolar delivery.
To determine if the K12R import block was thermally reversible, temperature shift experiments were performed. Cells were labeled for 10 min and chased for 30 min at 38°C to accumulate K12R precursor API on the membrane. These cells were then shifted to 30°C for various lengths of time before immunoprecipitation (Fig. 5 D, top). After 20 min at the permissive temperature, essentially all of the API was still present as the precursor form. However, between 20–40 min after the 30°C shift (Fig. 5 D, top, two-headed arrow), maturation of API increased from 10 to 56%. Thus, the thermal reversal of the targeting defect allowed nearly half of the labeled precursor API to enter the vacuole and become processed to the mature hydrolase between these two time points. Complete maturation of the precursor API was observed after 90 min at 30°C, whereas labeled cells maintained at 38°C during the shift phase of the experiment remained defective for import (Fig. 5 D, middle). The thermal reversibility of the K12R API indicated that it was on the authentic import pathway.
The K12R ts Mutant Traces the Import of API Oligomer during the Last, Rate-limiting Stages of the Cvt Pathway
A key question that we wanted to address was the oligomeric nature of precursor API during the transport step. The K12R API bound to the membrane at the nonpermissive temperature represented a synchronized population of labeled precursor, making it possible to follow its import into the vacuole upon reversal of the accumulation phenotype. Cells were labeled for 10 min and chased for 30 min at 38°C to accumulate precursor API oligomers on the membrane (Fig. 5 A). The labeled cells were then shifted to 30°C to reverse the block and allow precursor API to chase into the vacuole. The oligomeric state of API was examined during the 20–40 min window of the shift phase, when nearly half of the accumulated precursor API was chased into the vacuole and processed. Samples were removed at 20, 27, 34, and 40 min of the 30°C shift period and analyzed by glycerol gradients and immunoprecipitation. As expected, at the 20-min shift point all of the API is present as the precursor form, and by 40 min we see approximately half of the protein as the mature species (Fig. 6). At all time points, however, we detected only the oligomeric form of API, either as the precursor or mature dodecamer recovered in fraction 7 of the glycerol gradients. The kinetic examination of import indicated that oligomeric precursor API maintained its dodecameric state during the period of transport from the membrane-bound step to entry and processing in the vacuole (Fig. 6).
Figure 6.
Membrane-associated API is imported into the vacuole as an oligomer. The K12R API mutant was pulse labeled for 10 min and chased for 30 min at 38°C and then shifted to 30°C. Samples were removed at the indicated times during the 20–40-min window of the 30°C shift period and lysed with glass beads, and the resulting extract was separated by 20–50% glycerol gradients. Fractions were collected, immunoprecipitated with antiserum to API, and resolved by SDS-PAGE. Both mature (mAPI) and precursor API (prAPI) peaks appeared in fraction 7, cofractionating with the thyroglobulin molecular mass standard (669 kD).
While we could not detect monomeric API during the import process, it is possible that the signal for a monomeric translocation intermediate was below our detection level. However, if such disassembly occurred, then the API monomers would be exposed to the hydrolases of the vacuole lumen before reassembly into oligomers. In addition, a protein that goes through a translocation pore would probably assume a partially unfolded conformation (Schatz and Dobberstein, 1996). To mimic this disassembled state, we examined the protease susceptibility of newly synthesized monomeric precursor API. Cells were labeled for 2 min to yield predominantly monomeric API, lysed with glass beads, and subjected to proteolysis. These experiments indicated that monomeric API was completely degraded even after a brief treatment with proteinase K (data not shown), suggesting that newly synthesized API monomers would not survive the hydrolytic environment of the vacuolar lumen. In contrast, mature API is resistant to proteinase K digestion (Oda et al., 1996). This result provides additional support for our model that API is maintained as a dodecamer throughout its transport to the vacuole.
Discussion
Our study demonstrates that a large oligomeric protein is capable of being imported into the vacuole in a biosynthetic manner and suggests a possible mechanism of API transport by the Cvt pathway. Examination of the oligomeric state of API during transport allowed us to dissect this process into a number of discrete steps. Kinetic analyses indicated that precursor API oligomerizes rapidly into a homododecamer (Fig. 2) as the majority of precursor monomers assembled into oligomers within the first 5 min of the pulse-label/chase reaction. Subcellular fractionation localized the precursor API assembly process to the cytosol (Fig. 3 B). The subsequent membrane binding of the oligomeric precursor API was also rapid, with over half of the labeled precursor associating with the membrane within this brief labeling and chase period. These results demonstrate that the early steps of API import, oligomerization and membrane association, are not rate limiting, and the time required for these early steps constitutes only a fraction of the time needed for API import and maturation in the vacuole.
Steady-state analyses of the cvt and API propeptide deletion mutants indicated that neither encodes gene products or contains API sequence determinants necessary for API oligomerization, respectively (Fig. 4). However, a K12R point mutation in the propeptide resulted in a unique targeting defect that was useful for our examination of the later stages of API import.
The majority of the time required for API import was invested in the transport of the enzyme into the vacuole lumen from the membrane-bound state, indicating that this stage of the import process is rate limiting. To study this event, we exploited the novel targeting defect of K12R API. This temperature-sensitive mutant accumulated the oligomeric precursor at the membrane-associated step (Fig. 5 A), with the hydrolase exposed to the cytoplasmic milieu as determined by its accessibility to protease and biotinylated cross-linker (Fig. 5, B and C). Furthermore, the thermal reversibility of the targeting defect allowed us to kinetically examine the oligomerization state of API during the membrane transport event (Figs. 5 D and 6). Between 20–40 min of shift to the permissive temperature, nearly half of the membrane-accumulated oligomeric precursor was chased into the vacuole to the mature form (Fig. 5 D, top). When the oligomeric state of API was examined during this process, no disassembly of oligomeric precursor API was observed (Fig. 6). These results strongly suggest that the dodecameric precursor API maintains its oligomeric conformation throughout its import into the vacuole.
Oligomerization and membrane targeting do not appear to be rate-limiting steps in API import. The efficient assembly of the API dodecamer may require cytosolic chaperones for proper oligomerization as well as to prevent protein aggregation (Hartl, 1996; Jaenicke, 1996). We are currently pursuing detailed cross-linking studies to isolate possible transient API-associated cytosolic factors. Although oligomerization appears to stabilize API from proteolytic degradation (data not shown), a question that remains is whether oligomerization itself is necessary for correct targeting of the protein or for its enzymatic activity. Preliminary results suggest that the COOH terminus of API may contain an oligomerization domain; truncations in this region render API incompetent for import and subsequent processing in the vacuole (Oda et al., 1996). Although these deletion mutants are also defective for oligomerization (data not shown), the possibility that they are simply misfolded cannot be ruled out.
The target of the newly synthesized oligomeric precursor is a membrane fraction containing intact vacuoles. However, whether the initial membrane binding of precursor API oligomer occurs at the vacuole surface or perhaps at a prevacuolar compartment remains to be resolved. Overproduction of precursor API causes the accumulation of the cytosolic precursor, suggesting that there is a saturable receptor for API import (Klionsky et al., 1992). Assembly of a multimeric receptor may be necessary to accommodate multimeric propeptide binding. Recruitment of additional cytosolic factors may also contribute to the long half-time of API import.
Others have suggested that API uses a translocation mechanism to enter the vacuole (Seguí-Real et al., 1995). In this model, precursor API binds to the membrane as a monomer and inserts its propeptide extension into the vacuolar membrane before membrane translocation. Our current findings indicate that precursor API binds to the membrane as a fully assembled dodecamer rather than as a monomer. In order for membrane-bound API dodecamer to enter the vacuole through a protein channel, a disassembly of the oligomeric complex into partially unfolded monomers would be necessary before translocation into the vacuole. However, examination of the oligomeric state of API during membrane transport indicates that the dodecameric conformation is maintained throughout the import process. The precursor is directly processed to the mature hydrolase without the appearance of a kinetic intermediate API form as proposed by others (Seguí-Real et al., 1995). The sheer size of the membrane-bound oligomeric precursor and the instability of monomeric API make unlikely its passage into the vacuole by known posttranslational, translocation mechanisms.
The transport of oligomeric API across the vacuolar membrane is consistent with a vesicle-mediated mechanism. Vesicle-mediated transport events, including vacuolar vesicle fusion, are dependent on GTP-binding proteins (Zerial and Stenmark, 1993; Nuoffer and Balch, 1994; Stack et al., 1995). The finding that in vitro import of API is inhibited by nonhydrolyzable GTP analogues suggests a role for GTPases and the potential participation of vesicle intermediates in the Cvt pathway (Scott and Klionsky, 1995). We are currently characterizing vesicular intermediates that mediate API transport (Scott, S.V., and D.J. Klionsky, manuscript in preparation). From our results, two models of the API import process can be envisioned.
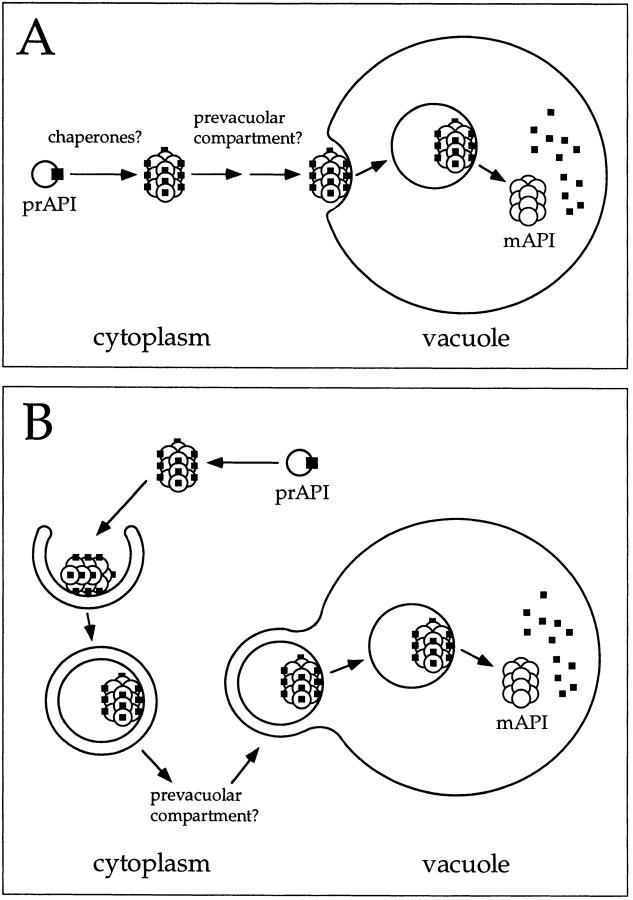
In the first model, the precursor oligomer binds directly to the vacuole and enters in an endocytic-like manner (Fig. 7 A). This is followed by the breakdown of the API-containing vesicles and processing of precursor API to the mature hydrolase. Alternatively, oligomeric precursor API may initially bind to a prevacuolar compartment before delivery to the vacuole via transport vesicles. The second model (Fig. 7 B) is based on the significant genetic overlap of the Cvt pathway with autophagy, which suggests that these two routes to the vacuole share largely the same cellular machinery (Harding et al., 1996; Scott et al., 1996). Specifically, the double-membrane autophagic vesicles (autophagosomes) that nonselectively capture and deliver cytoplasmic contents (Baba et al., 1994) to the vacuole may also be used by precursor API for its transport to this organelle. However, unlike autophagy, the delivery of API is constitutive. Thus, a continuous, basal-level supply of autophagic vesicles would be required for the selective delivery of API to the vacuole. In this model (Fig. 7 B), API initially binds to putative cusp-shaped progenitors of the double-membrane autophagosomes (Baba et al., 1994). Upon enclosure and autophagosome formation, API is transported to the vacuole, where the outer vesicle membrane fuses with the vacuole, releasing a single-membrane vesicle into the vacuolar lumen. These vesicles (autophagic bodies) are then degraded in a PrB-dependent manner (Takeshige et al., 1992), exposing the oligomeric precursor API to the vacuolar lumen for processing.
Figure 7.
Models for API import via the Cvt pathway. Precursor API is synthesized and assembled into dodecamers in the cytoplasm, followed by membrane binding. During the rate-limiting step of the pathway, both models A and B propose a vesicle-mediated mechanism of API entry into the vacuole followed by the breakdown of the API-containing vesicles and cleavage of the API propeptide in a PrB-dependent manner to yield the mature hydrolase. The role of molecular chaperones and the location of the initial binding of precursor API remain to be resolved. (A) Oligomeric precursor API directly binds to the vacuolar or prevacuolar membrane before the vesicle-mediated entry into the vacuole. (B) Genetic analyses have revealed a large overlap between the autophagy and Cvt pathways. In this model, API is delivered to the vacuole via double-membrane autophagic vesicles. Upon reaching the vacuole, these vesicles fuse to the vacuolar membrane, releasing a single-membrane vesicle (autophagic body) containing precursor API. This is followed by the breakdown of the vesicles and subsequent processing of the hydrolase to the mature enzyme.
The molecular details of the autophagy and Cvt pathways are not well understood. This study places the events of API oligomerization and import into discrete steps along the Cvt pathway. A detailed analysis of API import will help elucidate the molecular basis of both the Cvt and autophagy pathways.
Acknowledgments
We thank K.A. Morano for his advice and assistance and M.U. Hutchins for critical reading of the manuscript.
This work was supported by a National Institutes of Health Molecular and Cellular Biology Training Grant to J. Kim and by a Public Health Service Grant GM53396 from the National Institutes of Health to D.J. Klionsky.
Footnotes
1. Abbreviations used in this paper: ALP, alkaline phosphatase; API, aminopeptidase I; CPY, carboxypeptidase Y; Cvt, cytoplasm to vacuole targeting; ECF, enhanced chemifluorescence; PGK, phosphoglycerate kinase; PrA and PrB, proteinase A and B; SMD, synthetic minimal medium containing 2% glucose, essential amino acids and ammonium sulfate.
Address all correspondence to Daniel J. Klionsky, Section of Microbiology, University of California, Davis, CA 95616. Tel.: (916) 752-0277. Fax: (916) 752-9014.
References
- Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P, Thorner J, Honig L. Identification of tubulin from the yeast Saccharomyces cerevisiae. . Proc Natl Acad Sci USA. 1978;75:4962–4966. doi: 10.1073/pnas.75.10.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- Dunn WA., Jr Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Egner R, Thumm M, Straub M, Simeon A, Schuller HJ, Wolf DH. Tracing intracellular proteolytic pathways. Proteolysis of fatty acid synthase and other cytoplasmic proteins in the yeast Saccharomyces cerevisiae. . J Biol Chem. 1993;268:27269–27276. [PubMed] [Google Scholar]
- Egner, R., Y. Mahé, R. Pandjaitan, V. Huter, A. Lamprecht, and K. Kuchler. 1995. ATP binding cassette transporters in yeast: from mating to multidrug resistance. In Membrane Protein Transport. Vol. 2. S.S. Rothman, editor. JAI Press, Greenwich, CT. 57–96.
- Glaumann H, Ericsson JL, Marzella L. Mechanisms of intralysosomal degradation with special reference to autophagocytosis and heterophagocytosis of cell organelles. Int Rev Cytol. 1981;73:149–182. doi: 10.1016/s0074-7696(08)61288-7. [DOI] [PubMed] [Google Scholar]
- Glover JR, Andrews DW, Rachubinski RA. Saccharomyces cerevisiaeperoxisomal thiolase is imported as a dimer. Proc Natl Acad Sci USA. 1994;91:10541–10545. doi: 10.1073/pnas.91.22.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Mattaj IW. Nucleocytoplasmic transport. Science (Wash DC) 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–438. [PubMed] [Google Scholar]
- Hachiya N, Mihara K, Suda K, Horst M, Schatz G, Lithgow T. Reconstitution of the initial steps of mitochondrial protein import. Nature (Lond) 1995;376:705–709. doi: 10.1038/376705a0. [DOI] [PubMed] [Google Scholar]
- Hannavy K, Rospert S, Schatz G. Protein import into mitochondria: a paradigm for the translocation of polypeptides across membranes. Curr Opin Cell Biol. 1993;5:694–700. doi: 10.1016/0955-0674(93)90142-d. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature (Lond) 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hedrick JL, Smith AJ. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch Biochem Biophys. 1968;126:155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- Hilt W, Wolf DH. Stress-induced proteolysis in yeast. Mol Microbiol. 1992;6:2437–2442. doi: 10.1111/j.1365-2958.1992.tb01419.x. [DOI] [PubMed] [Google Scholar]
- Jaenicke R. Protein folding and association: in vitro studies for self-organization and targeting in the cell. Curr Top Cell Regul. 1996;34:209–314. doi: 10.1016/s0070-2137(96)80008-2. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO (Eur Mol Biol Organ) J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Banta LM, Emr SD. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol Cell Biol. 1988;8:2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiaeis localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiffer HH, Rupp S, Wolf DH. Vacuolar/lysosomal proteolysis: proteases, substrates, mechanisms. Curr Opin Cell Biol. 1993;5:990–996. doi: 10.1016/0955-0674(93)90082-2. [DOI] [PubMed] [Google Scholar]
- Löffler HG, Röhm KH. Comparative studies on the dodecameric and hexameric forms of yeast aminopeptidase I. Z Naturforsch Sect C J Biosci. 1979;34C:381–386. doi: 10.1515/znc-1979-5-610. [DOI] [PubMed] [Google Scholar]
- Marzella, L., and H. Glaumann. 1987. Autophagy, microphagy, and crinophagy as mechanisms for protein degradation. In Lysosomes: Their Role in Protein Breakdown. H. Glaumann and F.J. Ballard, editors. Academic Press, New York. 319–367.
- McNew JA, Goodman JM. An oligomeric protein is imported into peroxisomes in vivo. J Cell Biol. 1994;127:1245–1257. doi: 10.1083/jcb.127.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz G, Marx R, Röhm KH. The quaternary structure of yeast aminopeptidase I. 1. Molecular forms and subunit size. Z Naturforsch Sect C J Biosci. 1977;32C:929–937. doi: 10.1515/znc-1977-11-1209. [DOI] [PubMed] [Google Scholar]
- Mortimore GE, Poso AR, Lardeux BR. Mechanism and regulation of protein degradation in liver. Diabetes Metab Rev. 1989;5:49–70. doi: 10.1002/dmr.5610050105. [DOI] [PubMed] [Google Scholar]
- Nuoffer C, Balch WE. GTPases: multifunctional molecular switches regulating vesicular traffic. Annu Rev Biochem. 1994;63:949–990. doi: 10.1146/annurev.bi.63.070194.004505. [DOI] [PubMed] [Google Scholar]
- Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD, Klionsky DJ. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J Cell Biol. 1996;132:999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Neupert W. The mitochondrial protein import apparatus. Annu Rev Biochem. 1990;59:331–353. doi: 10.1146/annurev.bi.59.070190.001555. [DOI] [PubMed] [Google Scholar]
- Rachubinski RA, Subramani S. How proteins penetrate peroxisomes. Cell. 1995;83:525–528. doi: 10.1016/0092-8674(95)90091-8. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membrane. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science (Wash DC) 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Ryan KR, Jensen RE. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science (Wash DC) 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Scott SV, Klionsky DJ. In vitro reconstitution of cytoplasm to vacuole protein targeting in yeast. J Cell Biol. 1995;131:1727–1735. doi: 10.1083/jcb.131.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguí-Real B, Martinez M, Sandoval IV. Yeast aminopeptidase I is post-translationally sorted from the cytosol to the vacuole by a mechanism mediated by its bipartite N-terminal extension. EMBO (Eur Mol Biol Organ) J. 1995;14:5476–5484. doi: 10.1002/j.1460-2075.1995.tb00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Horazdovsky B, Emr SD. Receptor-mediated protein sorting to the vacuole in yeast: roles for a protein kinase, a lipid kinase and GTP-binding proteins. Annu Rev Cell Dev Biol. 1995;11:1–33. doi: 10.1146/annurev.cb.11.110195.000245. [DOI] [PubMed] [Google Scholar]
- Subramani S. Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. . FEBS (Fed Exp Biol Soc) Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Tomashek JJ, Sonnenburg JL, Artimovich JM, Klionsky DJ. Resolution of subunit interactions and cytoplasmic subcomplexes of the yeast vacuolar proton-translocating ATPase. J Biol Chem. 1996;271:10397–10404. doi: 10.1074/jbc.271.17.10397. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. . FEBS (Fed Exp Biol Soc) Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Walton PA, Gould SJ, Rachubinski RA, Subramani S, Feramisco JR. Transport of microinjected alcohol oxidase from Pichia pastorisinto vesicles in mammalian cells: involvement of the peroxisomal targeting signal. J Cell Biol. 1992;118:499–508. doi: 10.1083/jcb.118.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, Stenmark H. Rab GTPases in vesicular transport. Curr Opin Cell Biol. 1993;5:613–620. doi: 10.1016/0955-0674(93)90130-i. [DOI] [PubMed] [Google Scholar]