Abstract
Laminin trimers composed of α, β, and γ chains are major components of basal laminae (BLs) throughout the body. To date, three α chains (α1–3) have been shown to assemble into at least seven heterotrimers (called laminins 1–7). Genes encoding two additional α chains (α4 and α5) have been cloned, but little is known about their expression, and their protein products have not been identified. Here we generated antisera to recombinant α4 and α5 and used them to identify authentic proteins in tissue extracts. Immunoprecipitation and immunoblotting showed that α4 and α5 assemble into four novel laminin heterotrimers (laminins 8–11: α4β1γ1, α4β2γ1, α5β1γ1, and α5β2γ1, respectively). Using a panel of nucleotide and antibody probes, we surveyed the expression of α1-5 in murine tissues. All five chains were expressed in both embryos and adults, but each was distributed in a distinct pattern at both RNA and protein levels. Overall, α4 and α5 exhibited the broadest patterns of expression, while expression of α1 was the most restricted. Immunohistochemical analysis of kidney, lung, and heart showed that the α chains were confined to extracellular matrix and, with few exceptions, to BLs. All developing and adult BLs examined contained at least one α chain, all α chains were present in multiple BLs, and some BLs contained two or three α chains. Detailed analysis of developing kidney revealed that some individual BLs, including those of the tubule and glomerulus, changed in laminin chain composition as they matured, expressing up to three different α chains and two different β chains in an elaborate and dynamic progression. Interspecific backcross mapping of the five α chain genes revealed that they are distributed on four mouse chromosomes. Finally, we identified a novel full-length α3 isoform encoded by the Lama3 gene, which was previously believed to encode only truncated chains. Together, these results reveal remarkable diversity in BL composition and complexity in BL development.
Laminins are components of all basal laminae (BLs)1 throughout the bodies of vertebrates and invertebrates. In mammals they play at least three essential roles. First, they are major structural elements of BLs, forming one of two self-assembling networks (the other is composed of the collagens IV) to which other glycoproteins and proteoglycans of the BL attach (for review see Yurchenco and O'Rear, 1994; Timpl, 1996). Second, they interact with cell surface components such as dystroglycan to attach cells to the extracellular matrix (for review see Henry and Campbell, 1996). Third, they are signaling molecules that interact with cellular receptors such as the integrins to convey morphogenetically important information to the cell's interior (for review see Clark and Brugge, 1995; Mercurio, 1995; Yamada and Miyamoto, 1995). For example, laminin promotes myogenesis in skeletal muscle, outgrowth of neurites from central and peripheral neurons, and mesenchymal to epithelial transitions in kidney (Foster et al., 1987; Klein et al., 1988; Reichardt and Tomaselli, 1991; Vachon et al., 1996).
Laminin was initially isolated from tumor cells as a heterotrimer of A, B1, and B2 subunits (Chung et al., 1979; Timpl et al., 1979), later renamed α1, β1, and γ1 (Burgeson et al., 1994). Molecular cloning revealed that the three subunits were encoded by distinct but homologous genes (Martin and Timpl, 1987). Subsequently, homologues of the α1 chain (merosin, or α2; Ehrig et al., 1990) and the β1 chain (s-laminin, or β2; Hunter et al., 1989b ) were isolated, revealing a previously unsuspected heterogeneity of laminins. Five additional laminin chains have now been identified, all of which clearly belong to the α, β, or γ subfamilies (α3–5, β3, and γ2; Kallunki et al., 1992; Ryan et al., 1994; Aberdam et al., 1994b ; Richards et al., 1994; Gerecke et al., 1994; Miner et al., 1995). All native laminins isolated to date are composed of one α, one β, and one γ chain, and seven distinct heterotrimers have been identified (for review see Engvall and Wewer, 1996). The existence of multiple chains that oligomerize with a defined stoichiometry provides a means to generate functional diversity within a common structural framework (Sanes et al., 1990).
Here we focus on the α subfamily of laminin chains. This is the largest subfamily, with five members identified to date in mammals. Interactions of cells with laminin α chains are critical for cell–matrix interactions. For example, at least six distinct integrin heterodimers (α1β1, α2β1, α3β1, α6β1, α7β1, and αvβ3), as well as dystroglycan, heparin, and the adhesion molecule–associated glycoconjugate HNK-1/L2, bind to sites on laminin α chains (Rao and Kefalides, 1990; Gee et al., 1993; Hall et al., 1993; Sung et al., 1993; Mercurio, 1995; Mecham and Hinek, 1996; Colognato, H., and P.D. Yurchenco. 1996. Mol. Biol. Cell. 7(Suppl.):67a). Moreover, both dystroglycan and some integrins can distinguish amongst different α chains, suggesting that α chain diversity is functionally significant (Mercurio, 1995; Pall et al., 1996). In direct support of this notion, mutations in two α chains, α2 and α3, lead to congenital muscular dystrophy and junctional epidermolysis bullosa, respectively (Sunada et al., 1994; Xu et al., 1994; Helbling-Leclerc et al., 1995; McGrath et al., 1995). In Drosophila, mutation of the only known laminin α chain is embryonically lethal, leading to defects in numerous tissues (Henchcliffe et al., 1993; Yarnitzky and Volk, 1995; Garcia-Alonso et al., 1996).
Despite their importance, limited information is available about the distribution of the α chains (see, e.g., Engvall et al., 1990; Sanes et al., 1990; Vuolteenaho et al., 1994; Virtanen et al., 1995, 1996). Moreover, the α1 chain has been studied most intensively in human tissues with a single mAb (4C7; Engvall et al., 1986, 1990) whose specificity for α1 has been questioned (Ekblom, 1996). For the recently discovered α4 and α5 chains, no direct evidence has been presented to show that they form heterotrimers, and no data on cellular localization have been reported. Accordingly, we have generated and characterized antibodies to the α4 and α5 chains and used them to identify four novel laminin heterotrimers: laminin-8 (α4β1γ1), laminin-9 (α4β2γ1), laminin-10 (α5β1γ1), and laminin-11 (α5β2γ1). Using a panel of antibodies and cDNA probes, we analyzed the distribution of all five laminin α chains in embryonic and adult mice. We show that the α chains are expressed in overlapping but distinct patterns, with each BL containing at least one of the known α chains. Moreover, we demonstrate that some individual BLs contain different complements of α chains at distinct stages of their development. Finally, we identify a novel isoform of α3 and report the chromosomal locations of all five α chains in mice.
Materials and Methods
Isolation of cDNAs
Laminin α3B.
We performed the reverse transcription–coupled PCR (RT-PCR) using embryonic day (E) 17.5 mouse lung RNA and primers designed to amplify sequences encoding the NH2-terminal portion of domain VI of laminin α5. Surprisingly, the reaction generated a novel 209-bp fragment that was similar but not identical to known α chains. We hypothesized that this fragment could be part of a laminin α3B cDNA extending 5′ of that reported by Galliano et al. (1995). To test this hypothesis, two primers were used in RT-PCR from adult lung RNA to attempt to amplify the potentially intervening cDNA: sense, 5′AGCGGGACCCAGAGGTC3′ (from the novel product); antisense, 5′TGCCTCACAGACAATCTCACC3′ (from near the 5′ end of the sequence in Galliano et al. [1995]). RT-PCR conditions were as described (Miner and Sanes, 1994), with the addition of Taq Extender PCR Additive (Stratagene Cloning Systems, La Jolla, CA). A 2.2-kb fragment was sequenced with a Taq DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems, Inc., Foster City, CA), and sequence was analyzed on the BLAST server at the National Center for Biotechnology Information (Altschul et al., 1990). Sequence was obtained from multiple clones to resolve errors introduced by Taq polymerase amplification.
Laminin α4.
A mouse laminin α4 cDNA fragment was amplified by RT-PCR from E17.5 placenta using degenerate primers based on the human amino acid sequence (Richards et al., 1994). The primers, designed to amplify the mouse segment homologous to nucleotides 1,800–2,607, were: sense, 5′TCNATGATGTTYGAYGGNCARTC3′; antisense, 5′CGNCCRCTRCTRAANCCRAARTC3′. The fragment was isolated on a low melting point gel and ligated into the pCRII vector (Invitrogen, San Diego, CA). The DNA and deduced amino acid sequences were determined and have been deposited into GenBank under accession number U88352.
RNA Analyses
RNA was prepared from mouse tissues by acid guanidinium phenol/chloroform extraction (Chomczynski and Sacchi, 1987). RNase protections were performed as described (Miner and Wold, 1991) using [32P]UTP-labeled probes and 5 μg (E17.5) or 7.5 μg (adult) of total RNA per hybridization. A probe for elongation factor 1α was included to control for the quality and amount of input RNA. For Northern analysis, a filter containing poly(A)-selected RNA from several adult mouse tissues (Clontech, Palo Alto, CA) was hybridized according to the manufacturer's instructions. In situ hybridizations were performed with 35S-UTP–labeled probes as described (Lentz et al., 1997). The laminin α chain probes used for RNase protection assays and in situ hybridizations were as described (Lentz et al., 1997).
Antibodies
Rat mAbs to mouse laminin α1 (clones 198 and 200; Sorokin et al., 1992) were gifts from Lydia Sorokin (Institute for Experimental Medicine, Erlangen, Germany). A rabbit antiserum to human laminin α2 cross-reactive with the mouse protein (Vachon et al., 1996) was kindly provided by Peter Yurchenco (Robert Wood Johnson Medical School, Piscataway, NJ). A rabbit antiserum to mouse laminin α3 (Aberdam et al., 1994a ,b) was a gift from Daniel Aberdam (INSERM U385, Nice, France). Mouse mAbs to rat laminin chains β1 (C21, C22), β2 (D7, D19, D27), and γ1 (D18) were produced and characterized in our laboratory and have been described previously (Sanes and Chiu, 1983; Hunter et al., 1989b ; Sanes et al., 1990; Green et al., 1992). A guinea pig antiserum against a recombinant COOHterminal fragment of laminin β2 was produced as described (Sanes et al., 1990). A rat mAb to laminin γ1 was purchased from Chemicon (Temecula, CA). Second antibodies were purchased as follows: fluorescein- and HRP-conjugated goat anti–rabbit antibodies from Boehringer Mannheim Biochemicals (Indianapolis, IN); fluorescein-conjugated goat anti–rat antibodies from Cappel/Organon Teknika (Durham, NC); Cy3-conjugated goat anti–rabbit antibodies from Jackson ImmunoResearch Laboratories (West Grove, PA); biotinylated goat anti–guinea pig antibodies from Sigma Chemical Co. (St. Louis, MO).
To generate antibodies to the laminin α4 and α5 chains, the laminin α4 cDNA described above, another containing nucleotides 3,670–4,391 (described in Lentz et al., 1997), and a laminin α5 fragment comprising nucleotides 4,243–4,926 (SacI to EcoRV) were each cloned in frame into the pET 23 vector (Novagen, Madison, WI). Proteins were produced in BL21(DE3) bacteria, and inclusion bodies were isolated according to a protocol supplied by Novagen. Fusion proteins were gel isolated as described (Miner and Sanes, 1994) and used to immunize rabbits (Caltag, Healdsburg, CA). For both α4 and α5, two separately immunized rabbits generated antisera that displayed qualitatively similar patterns of reactivity on both sections and immunoblots. The higher titer antiserum to each immunogen was used for the studies reported here.
Immunohistochemistry
Mouse tissues were frozen fresh and sectioned at 4–8 μm on a cryostat. Antibodies were diluted in 1% (wt/vol) BSA in PBS and incubated on sections for 1–2 h. After rinsing off unbound primary antibody with PBS, secondary antibodies were applied for 1–2 h. Sections were rinsed again, and then mounted in glycerol-para-phenylenediamine and observed with epifluorescent illumination. Since the laminin α4 antisera only recognized denatured antigen, the following protocol was used when staining with these antibodies: sections were fixed in 2% paraformaldehyde in PBS for 20 min, rinsed in PBS, incubated with 100 mM glycine in PBS for 10 min, incubated in 0.05% SDS in PBS for 30 min at 50°C, and then rinsed in PBS before the antibody was applied. Anti-α5 stained untreated and SDS- denatured sections in qualitatively similar patterns.
Western Blots and Immunoprecipitations
For immunoblotting, tissues from rat were used because a greater range of mAbs was available against rat than against mouse laminin β and γ chains (see above). Lungs and kidneys from saline-perfused adult rats were homogenized in ice-cold 40 mM Tris, pH 7.5, 15 mM NaCl, and 2 mM CaCl2 (H buffer) containing protease inhibitors (0.1 mM PMSF, 1 mM benzamidine, and 1 μg/ml soybean trypsin inhibitor) using a Polytron. Crude membrane fractions were recovered at 20,000 g, washed once with H buffer containing PMSF, resuspended in H buffer containing 10 mM EDTA, 2 mM EGTA, PMSF, and soybean trypsin inhibitor (H+E buffer), and stored at −10°C. To improve immunoblotting sensitivity for BL components, the crude pellet was extracted by brief sonication in H+E buffer with 0.1 M NaCl and 1% Triton X-100, pelleted at 50,000 g, and resuspended in H+E buffer. However, data qualitatively similar to those presented in Results were obtained with the crude pellet. Protein content was assayed with bicinchoninic acid reagents (Pierce Chemical Co., Rockford, IL) using BSA as a standard. Purified Engelbreth-Holm-Swarm (EHS) laminin-1 was obtained from Gibco BRL (Gaithersburg, MD).
Samples were solubilized by boiling in SDS gel loading buffer with or without DTT. The proteins were then separated by SDS-PAGE and transferred to nitrocellulose using standard methods. Fusion proteins, reduced native laminins, and nonreduced native laminins were separated on 12%, 7%, and 3.5% polyacrylamide gels, respectively. After blotting, filters were blocked with nonfat dry milk/0.3% Tween-20 in PBS, and then incubated with antibodies overnight. For detecting the fusion proteins, antisera were preadsorbed to an unrelated fusion protein containing the common pET 23 leader and His tag sequences. Bound antibodies were detected with either HRP-conjugated second antibody (for rabbit) or biotinylated second antibody with HRP-conjugated Z-avidin (Zymed Laboratories, South San Francisco, CA) (for guinea pig), and Renaissance chemiluminescent substrate (DuPont/New England Nuclear, Boston, MA).
For immunoprecipitations, laminins were first partially purified from adult rat lung by the protocol of Lindblom et al. (1994), modified as follows: crude membranes were prepared as described above, and then extracted repeatedly over 12 h with 50 mM Tris, pH 7.5, 0.15 M NaCl, 10 mM EDTA, and 10 mM EGTA (TBS/EDTA). The pooled extracts were diluted to 90 mM NaCl, adjusted to pH 8.3, and loaded onto a DEAE– Sepharose CL-4B column (Pharmacia, Uppsala, Sweden). The column was eluted with 1.0 M NaCl, and fractions containing laminin β2 (detected by immunoblotting) were pooled and brought to 50% saturation with ammonium sulfate. Precipitated material was resuspended in TBS/EDTA, brought to 10% glycerol, 0.6 M KCl, and 0.05% Tween-20, and then fractionated on a Sepharose CL-4B column. Fractions containing laminin β2 were pooled and passed through CM–Sepharose CL-6B; the flow-through was resubjected to DEAE–Sepharose chromatography. Protein eluted with 0.5 M NaCl was stored at −70°C with 0.1 mM PMSF. Protein was measured by Bradford assay (Bio Rad Laboratories, Hercules, CA). SDSPAGE of this fraction under reducing conditions displayed a heterogeneous population of proteins >180 kD, along with the laminin-binding protein entactin (150 kD) as a major constituent.
Laminins were immunoprecipitated essentially by the method of Green et al. (1992), modified as follows: laminin samples were incubated with antiβ1 mAbs (a mixture of C21 and C22) or anti-β2 antibodies (a mixture of D7, D19, and D27) in 50 mM Tris, pH 8.0, 100 mM NaCl, 2 mM EDTA, 1% NP-40, and 0.05% sodium deoxycholate (IP buffer). Immune complexes were isolated using protein A–Sepharose 4B (Pharmacia) that was preblocked with 4 mg/ml IgG-free BSA (Sigma Chemical Co.) and washed in IP buffer. Rabbit anti–mouse IgG (Fc) antibodies (Jackson ImmunoResearch Laboratories) were included to bridge mAbs to protein A. Precipitated laminins were dissolved by boiling in SDS-PAGE sample buffer, separated by SDS-PAGE, and then detected by Western blotting as above.
Interspecific Mouse Backcross Mapping
Interspecific backcross progeny were generated by mating (C57BL/6J × Mus spretus) F1 females and C57BL/6J males as described (Copeland and Jenkins, 1991). DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, and Southern blot transfer and hybridization were performed essentially as described (Jenkins et al., 1982). All blots were prepared with Zetabind nylon membrane (Cuno, Inc., Meriden, CT). Probes, which were specific for each locus, were labeled with [32P]dCTP using a random primed labeling kit (Amersham Corp., Arlington Heights, IL) or a nick translation labeling kit (Boehringer Mannheim Biochemicals); washing was done to a final stringency of 0.8–1.0 × SSC and 0.1% SDS at 65°C. The probe and restriction fragment length polymorphisms (RFLPs) for Lama1 have been described previously (Okazaki et al., 1993). The Lama2 probe, a ∼360-bp fragment of mouse cDNA from domain G, detected fragments of 10.5 kb in C57BL/6J (B) DNA and 5.4 and 4.2 kb in M. spretus (S) DNA after digestion with PstI. The Lama3 probe, a ∼500-bp fragment of mouse cDNA from domains I/II, detected EcoRV fragments of 10.5 kb (B) and ∼23.0 kb (S). A second probe, a genomic clone containing a portion of domain VI of laminin α3B, gave results identical to those obtained with the domain I/II probe. The Lama4 probe, a ∼800-bp fragment of mouse cDNA from domain G, detected BglII fragments of 4.4 and 1.7 kb (B) and 6.0 kb (S). The Lama5 probe, a ∼7.5-kb fragment of mouse genomic DNA from domains V and IVb, detected BamHI fragments of 4.2, 3.7, and 3.3 kb (B) and 4.2, 3.3, 2.3, and 1.8 kb (S). The presence or absence of M. spretus–specific fragments was followed in backcross mice. A total of 205 N2 mice were used to map each Lama locus.
Descriptions of most of the probes and RFLPs for the loci used to position the Lama loci in the interspecific backcross have been reported. These include: Gnas, chromosome 2 (Wilkie et al., 1992); Myb, Fyn, and Ros1, chromosome 10 (Justice et al., 1990); Fert and Tik, chromosome 17 (Fishel et al., 1993; Okazaki et al., 1993); and Tpl2, Cdh2, and Ttr, chromosome 18 (Justice et al., 1992, 1994). One locus has not been reported previously for this interspecific backcross: the Mc3r probe, a 2.0-kb BamHI/ XhoI fragment of mouse cDNA, was kindly provided by Roger Cone (Vollum Institute, Portland, OR) and detected SphI fragments of 3.5 kb (B) and 5.9 kb (S). Recombination distances were calculated as described (Green, 1981) using the computer program SPRETUS MADNESS. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
Results and Discussion
The Laminin α Chain Family: Identification of Full-Length α3
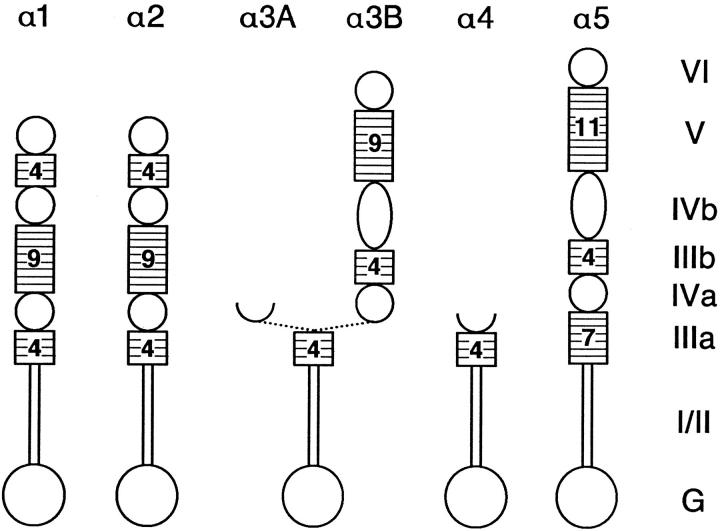
The α subfamily of laminin chains currently contains five members. Fig. 1 shows the domain structure of these chains based on the nomenclature of Sasaki et al. (1988). All α chains contain a carboxyl-terminal globular G domain and α-helical domains I and II. The previously described fulllength α1 and α2 chains contain six additional domains (IIIa–VI) that alternate between cysteine-rich stretches containing EGF-like repeats (IIIa, IIIb, and V) and globular regions (IVa, IVb, and VI) (Engvall and Wewer, 1996). The newest member of the family, α5, is also a full-length chain, but it is larger than α1 or α2, owing to the greater number of EGF-like repeats in domain V and a larger domain IVb (Miner et al., 1995). In contrast, laminins α3A and α4 are severely truncated chains that contain only a single cysteine-rich domain (IIIa) downstream of a short aminoterminal domain (Ryan et al., 1994; Galliano et al., 1995; Iivanainen et al., 1995; Richards et al., 1996). Of the vertebrate α chains, α5 may be most like the ancestral α chain, because of its similarity in both domain structure and sequence to the only known invertebrate α chain, Drosophila A (Miner et al., 1995).
Figure 1.
The laminin α chain subfamily. Numbering of domains is based on accepted nomenclature (Sasaki et al., 1988; Engvall and Wewer, 1996). Numbers of laminin-type cysteine-rich (EGFlike) repeats, rounded to the nearest integer, are indicated within each domain IIIa, IIIb, or V. Note that laminins α3A and α3B share domains G, I/II, and IIIa but have distinct NH2 termini.
The only laminin gene so far shown to encode more than a single polypeptide is the laminin α3 gene. The two known products, α3A and α3B, differ at their amino termini, resulting from alternative splicing and/or alternative promoter usage. The shorter α3A chain was the first to be identified by both immunological methods (Rousselle et al., 1991) and by cDNA cloning (Aberdam et al., 1994b ). Subsequently, however, multiple α3 cDNAs were identified in human (Ryan et al., 1994) and mouse (Galliano et al., 1995), suggesting the existence of a second, longer isoform called α3B. Sequence analysis (Galliano et al., 1995) indicated that α3B contains two cysteine-rich (IIIa and IIIb) and two globular (IVa and IV′′) domains in addition to the G and I/II domains, but no domains V or VI. This would make it the sole “mid-sized” α chain. However, we have obtained mouse cDNAs (see Materials and Methods) that extend the previously reported sequence 5′ by ∼2.2 kb (Fig. 2). This novel sequence encodes the NH2-terminal portion of domain IVb, a complete domain V, and what is likely all but a few amino acids of a domain VI. These domains are most similar in sequence and predicted tertiary structure to the analogous domains of laminin α5. For example, the amino acid sequence of domain VI is 74% identical to that of α5, but only 54 and 51% identical to those of α1 and α2, respectively. A probe from the 5′ end of this sequence recognized ∼10-kb bands on Northern blots of mouse RNA from adult brain, lung, and kidney (data not shown). The proposed α3B structure is shown in Fig. 1, indicating its unique NH2 terminus and the COOH terminus it shares with α3A.
Figure 2.

Nucleotide and deduced amino acid sequences of the laminin α3B chain, 5′ and NH2-terminal to those reported by Galliano et al. (1995). Overlapping nucleotides are in lowercase. The Galliano sequence contains a single deoxyguanosine (parentheses) not found in our sequence, which shifts the reading frame and leads to an encoded methionine (bottom line, italics). Domains are marked as in Fig. 1. This was hypothesized to be the initiator for translation. An adhesive tripeptide sequence, LRE (Hunter et al., 1989a ), is indicated by bullets; another LRE is located in the G domain (Galliano et al., 1995). These sequence data are available from GenBank under accession number U88353.
There is a discrepancy between our sequence and that of Galliano et al. (1995): a single base near the 5′ end of their reported sequence is absent from our cDNAs (see last line of Fig. 2). As a result of this insertion, the methionine residue that Galliano et al. (1995) suggested to initiate translation of α3B and the following amino acid are encoded in a different reading frame than is the extended sequence reported here (Fig. 2). Possible explanations for this discrepancy include a polymorphism between mouse strains, alternative splicing of exons that differ by only one base, RNA editing, sequencing error, or cDNA cloning artifact. We cannot exclude any of these possibilities, although we consider the first three unlikely and note that our sequence, derived from multiple cDNAs, generates a single uninterrupted open reading frame (∼9.8 kb) extending through the region of discrepancy. Therefore, we favor the interpretation that there is no mid-sized form of laminin α3, but only the severely truncated α3A and the full-length α3B form reported here. Full-length α3B would thus contain ∼3,300 amino acids and have a molecular mass of ∼360 kD. Thus, the α subfamily of laminin chains currently consists of four long (α1, α2, α3B, and α5) and two short (α3A and α4) proteins.
Differential Expression of Laminin α Chain Genes in Embryos and Adults
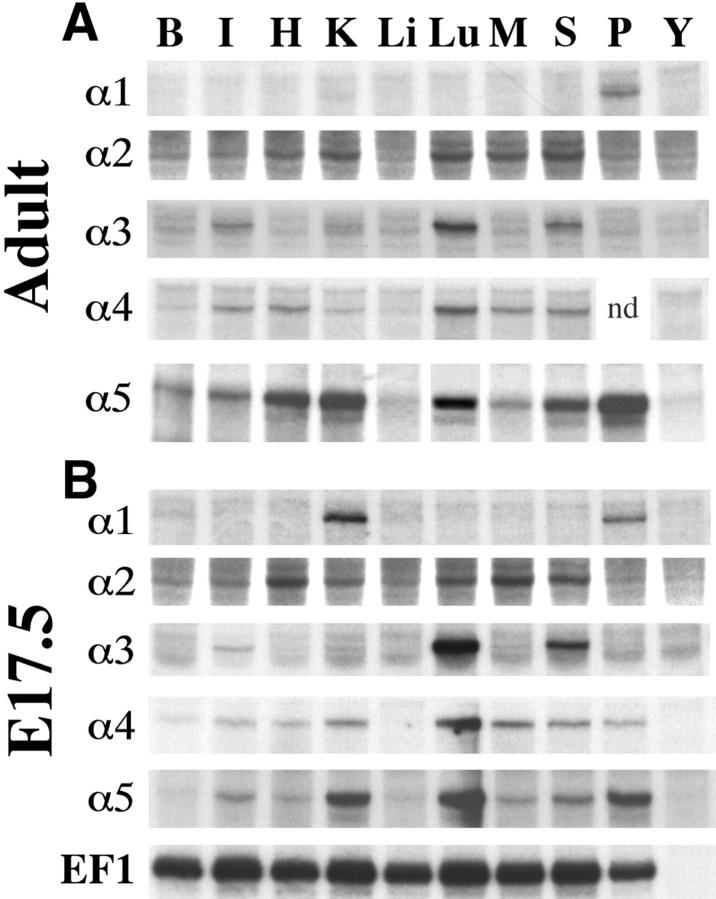
As a first step in assessing the distribution of the laminin α chains, we performed RNase protection analyses on RNA isolated from a set of eight adult tissues plus a late-term (E17.5) placenta (Fig. 3 A). Laminin α1 was readily detectable only in placenta, but long autoradiographic exposures revealed low levels in kidney. Laminin α2 RNA was present at levels above background (yeast RNA lane) in heart, kidney, lung, muscle, and skin. Laminin α3A/B was expressed primarily in lung, skin, and intestine, although very low levels of this RNA were also detectable in kidney. In contrast, laminin α4 RNA was present in all tissues at low (liver) to moderate (lung) levels. Laminin α5 transcripts were also easily detectable in all tissues, although levels were very low in liver. Thus, each laminin α chain is expressed in a distinct pattern. Interestingly, the two most recently discovered laminins, α4 and α5, are the most widely expressed. The α2 and α3 chains show more restricted patterns of expression. In general, α2 levels were highest in tissues with large mesodermally derived components (skeletal and cardiac muscle), whereas α3 levels were highest in organs that are rich in epithelia (skin, intestine, and lung). The notion that α2 and α3 are predominantly mesodermal and epithelial products, respectively, has been proposed (Vuolteenaho et al., 1994; Aberdam et al., 1994a ). Finally, expression of laminin α1, the initially described α chain, was the most severely restricted of the five.
Figure 3.
Ribonuclease protection analysis of laminin α chain expression in (A) adult and (B) E17.5 mouse tissues. A probe for elongation factor 1α was used to control for the amount of input RNA in both embryos (B) and adults (not shown). Each α chain is expressed in a distinct pattern in the adult and, in general, these patterns are established by birth. The α5 chain is the most highly expressed, and α1 is the most restricted. B, brain; I, intestine; H, heart; K, kidney; Li, liver; Lu, lung; M, skeletal muscle; S, skin; P, placenta; Y, yeast RNA. The sample of E17.5 placental RNA was included in the panel of adult RNAs to allow comparison between experiments. nd, not done.
We next tested RNAs prepared from the same tissues but at a late fetal (E17.5) stage. In general, patterns of expression seen in the fetus were similar to those seen in adults, although levels of expression were generally higher in the fetus (Fig. 3 B). Laminin α1 was again the least widely expressed α chain, although its RNA was readily detected in fetal kidney as well as in placenta. Likewise, laminins α2–5 were expressed at moderate to high levels in a variety of tissues, consistent with patterns seen in adults. Thus, the distinct patterns of laminin α chain expression found in adult tissues are, for the most part, established before birth.
To map α chain expression in younger embryos (E15.5), we used in situ hybridization. Laminin α1 was detected in the kidney and in the meninges of the central nervous system (Fig. 4 a). Laminin α2 was observed primarily in the developing skeletal musculature, in dorsal root ganglia, and in kidney (Fig. 4 b). Laminin α3A/B was strongly expressed in skin, lung, olfactory epithelium, and the superficial layers of the tongue and palate (Fig. 4 c). Laminin α4 was expressed strongly in mesenchymal tissues of the head, in dorsal root ganglia, and in intestine, and was observed diffusely in skeletal and cardiac muscle (Fig. 4 d). Finally, laminin α5 was expressed in a pattern similar to α3, with additional sites of expression in salivary gland, in intestine, and in the most superficial cells of the liver (Fig. 4 e).
Figure 4.
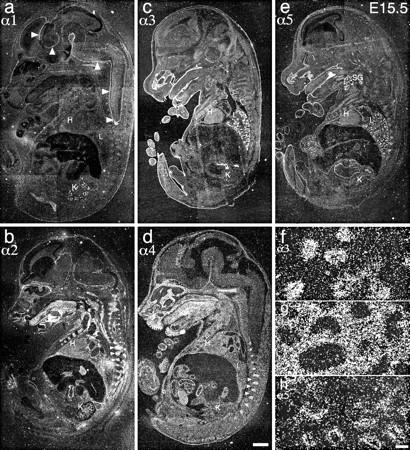
In situ hybridization of laminin α chain probes to E15.5 embryo parasagittal sections. (a) α1, (b) α2, (c) α3, (d) α4, and (e) α5. α1 shows restricted expression in kidney and meninges (arrowheads); α2 and α4 show widespread expression in mesenchymal cells and derivatives as well as in dorsal root ganglia (arrowheads); and α3 and α5 transcripts localize primarily to epithelia. (f–h) High power views of (f) α3, (g) α4, and (h) α5 expression in lung. α3 and α5 are concentrated in the epithelial lung buds, and α4 to the mesenchyme. H, heart; K, kidney; L, lung; SG, salivary gland; T, tongue (muscle). Bars: (d) 1 mm; (h) 50 μm.
When compared with the RNase protection results from E17.5 (Fig. 3 B), it is apparent that the main sites of expression are already established by E15.5: α2 and α4 in muscle, α3 and α5 in skin, and α3–5 in lung. There are, however, a few differences. For example, α2 is barely detectable in heart at E15.5 but is expressed strongly at E17.5; the presence of α4 in heart at E15.5 suggests that there may be a developmental transition in α chain expression in the heart in which α4 is joined by α2. In addition, a particularly interesting pattern of α3–α5 chain expression was observed in the lung, as shown at higher power in Fig. 4, f–h. α3 and α5 were confined to the epithelial lung buds at this stage, while α4 was expressed only in the surrounding mesenchyme. This complementary pattern of expression suggests that these chains may play distinct roles in lung development: α3 and α5 in branching morphogenesis, and α4 in the organization of the mesenchyme.
Identification of Laminin α4 and α5 Proteins
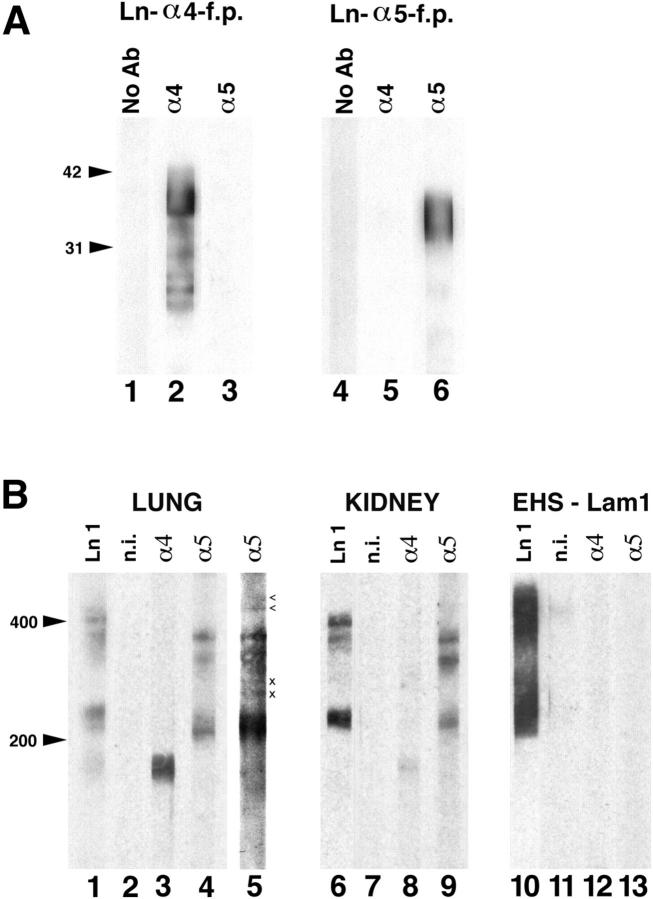
The α1–3 chains were first identified by biochemical and immunochemical methods (Timpl et al., 1979; Leivo and Engvall, 1988; Rousselle et al., 1991; Carter et al., 1991; Verrando et al., 1992). In contrast, α4 and α5 were identified as cDNAs by molecular cloning (Richards et al., 1994; Iivanainen et al., 1995; Miner et al., 1995). Sequence analysis implies that the α4 and α5 cDNAs encode laminin-like proteins, but it is crucial to demonstrate this directly. To this end, we used α4 and α5 cDNAs to produce recombinant proteins in bacteria, and then used the proteins to generate antisera in rabbits. Each antiserum specifically recognized its immunogen on Western blots (Fig. 5 A), and immunoreactivity was removed by incubation with the corresponding fusion protein. Since α3B and α5 sequences are closely related (see above), we also tested anti-α5 on a recombinant fragment from the corresponding domains of α3B. No cross-reaction was detected (data not shown).
Figure 5.
Identification of laminin α4 and α5 proteins in lung and kidney. (A) Characterization of antisera. The α4 and α5 fusion proteins used to immunize rabbits were fractionated by SDSPAGE on 12% gels and transferred to blots. Strips were probed either with no primary antibody (lanes 1 and 4), with the anti-α4 antiserum (lanes 2 and 5), or with the anti-α5 antiserum (lanes 3 and 6). Each antiserum specifically recognized its cognate immunogen. (B) Solubilized and reduced crude membranes from adult rat lung and kidney and purified laminin-1 were fractionated on 7% gels and transferred to blots. Strips were probed either with anti–laminin-1 (lanes 1, 6, and 10), nonimmune (lanes 2, 7, and 11), antilaminin α4 (lanes 3, 8, and 12), or anti-laminin α5 (lanes 4, 5, 9, and 13). The anti-α4 serum recognized a protein of ∼180 kD in lung and kidney. The α5 antiserum recognized several bands in lung and kidney (lanes 4 and 9), the largest of which, ∼450 kD, was observed only after long exposures (lane 5, arrowheads). Neither serum recognized laminin α1 (lanes 12 and 13). n.i., nonimmune serum; x, nonspecific bands seen in all lung lanes with long exposures.
To detect laminin α4 and α5 proteins, we prepared extracts of adult lung and kidney; lung was chosen because it expresses both chains at high levels (Fig. 3 A), and kidney was chosen because it was the focus of immunohistochemical studies detailed below. Tissue BL proteins and purified laminin-1 (α1/β1/γ1) were reduced, fractionated by gel electrophoresis, and transferred to nitrocellulose filters, which were then probed with antisera to laminins α4 or α5 or with an antiserum to laminin-1. Results are shown in Fig. 5 B. Anti-α4 recognized an ∼180-kD protein in both lung and kidney (Fig. 5 B, lanes 3 and 8). Anti-α5 recognized large proteins of ∼380 and ∼350 kD, as well as a smaller protein of ∼210 kD, in both tissues (Fig. 5 B, lanes 4 and 9). Additional specific anti-α5–reactive bands of high M r (∼450 kD) were observed with longer exposure times in several experiments (Fig. 5 B, lane 5). Neither anti-α4 nor anti-α5 reacted with the ∼400-kD α1 chain of laminin-1 (Fig. 5 B, lanes 12 and 13), although this chain was readily detected by a polyclonal antiserum to laminin-1 (lane 10). Nonimmune rabbit serum was not reactive with any of the laminin chains (Fig. 5 B, lanes 2, 7, and 11). Thus, the laminin α4 and α5 chains are present in adult lung and kidney, and this is consistent with results of RNase protection assays (Fig. 3 A).
The M r of the anti-α4–reactive protein, ∼180 kD, is consistent with the size predicted from the open reading frame of the cDNA (190 kD; Iivanainen et al., 1995; Richards et al., 1996). Likewise, the ∼450-kD α5 chain is of the expected size for this presumably glycosylated protein. The observation that smaller α5-immunoreactive bands (380, 350, and 210 kD) are more abundant than the 450-kD species indicates that α5 is subject to posttranslational cleavage. Multiple protease inhibitors were used in preparing tissue, and the relative abundance of the bands in the extracts remained constant for several weeks at 4°C. We therefore suspect that this cleavage occurred in situ, as has been reported for laminins α2 and α3 (Ehrig et al., 1990; Marinkovich et al., 1992a ), rather than during isolation.
Cellular Distribution of Laminin α Chains in Adult Tissues
The laminin α1–3 chains have been shown to be associated with BLs in a variety of tissues, as have the β and γ chains. Here we asked whether α4 and α5 are components of BLs, and whether their expression overlaps those of the α1–3 chains. In addition, we wanted to know whether all BLs contained at least one α chain, as the finding of an apparently α-free BL might suggest the existence of additional α chains. We began by using our α4 and α5 antisera, along with previously characterized antibodies to α1–3 (see Materials and Methods), to investigate the expression patterns of the laminin α chains in kidney, a tissue that contains numerous heterogeneous yet well-characterized BLs (Abrahamson et al., 1989; Sanes et al., 1990; Abrahamson and Leardkamolkarn, 1991; Abrahamson and St. John, 1993; Miner and Sanes, 1994; Virtanen et al., 1995).
Laminin α1 was readily detected in the BLs of a subset of renal tubules (primarily proximal), as shown previously (Horikoshi et al., 1988; Sorokin et al., 1992). The α1 chain was absent from glomerular BL but was present in the glomerular mesangium, an amorphous matrix that is one of the few sites in which laminins are present outside of a formed BL (Fig. 6 a). No α1 was detectable in the BLs of arteries, veins, or capillaries. In the peripheral portion of the kidney, laminin α2 was largely restricted to the mesangium, in agreement with previous studies of human kidney (Fig. 6 b; Sanes et al., 1990; Virtanen et al., 1995). At deeper levels, however, some tubules were weakly α2 positive, particularly in the transitional zone between the cortex and medulla (the corticomedullary junction; Fig. 6 b, inset). Laminin α3 was absent from glomeruli, tubules, and vasculature of the renal cortex (data not shown), but it was present in the epithelial BL that lines the papilla (Fig. 6 c). Laminin α4 was absent from all renal, epithelial, and arterial BLs (Fig. 6 d) but was found in many capillaries of the medulla (not shown). Finally, laminin α5 was detected in virtually all BLs, including those of glomeruli, arteries, and all tubules (Fig. 6 e). Thus, all five of the known α chains are present in adult kidney, but each is expressed in a unique pattern.
Figure 6.
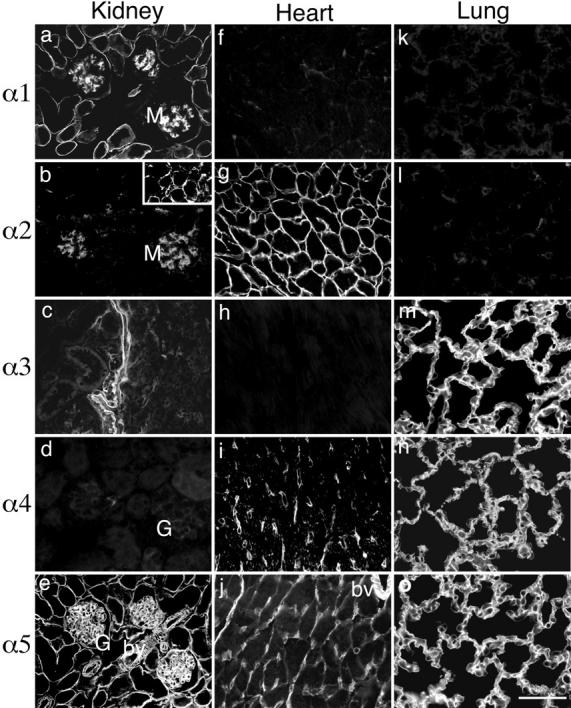
Immunohistochemical localization of laminin α chains in adult mouse kidney (a–e), heart (f–j), and lung (k–o). All five α chains were present in adult BLs, but each chain was distributed in a distinct pattern. α1 was found only in kidney mesangium and in a subset of tubular BLs (primarily proximal) (a). α2 was present in mesangium (b), in a subset of corticomedullary tubular BLs (b, inset), and in cardiomyocyte BLs (g). α3 was detected in kidney papillary BL (c) and in lung alveolar BL (m). α4 was absent from renal cortex (d) but found in capillaries of both the renal medulla (not shown) and heart (i), and in alveolar BLs in lung (n). α5 showed the most widespread expression: in all kidney BLs—glomerular, tubular, and arterial (e); in heart blood vessels and in some cardiomyocyte BLs (j); and in lung alveolar BL (o). G, glomerulus; M, mesangium; bv, blood vessel. Bar, 50 μm.
To extend these studies, we examined two other BL-rich tissues, heart and lung. As in kidney, the laminin α chains were primarily restricted to BLs, and each chain was expressed in a distinct pattern. In the heart, laminins α1 and α3 were undetectable (Fig. 6, f and h). Laminin α2 was abundant in the myocyte BLs (Fig. 6 g), as reported previously (Leivo and Engvall, 1988; Paulsson et al., 1991), while laminin α4, as in the kidney, was restricted to capillaries (Fig. 6 i). Laminin α5 was present in arterioles and capillaries and was also found at low levels in many myocyte BLs (Fig. 6 j). In lung, laminin α3 was present in alveolar BL (Fig. 6 m), consistent with a recent report by Virtanen et al. (1996). The α5 chain was colocalized with α3 in most alveolar BLs (Fig. 6 o), while laminin α4 was detected in a large subset of these BLs (Fig. 6 n). The identity of the α4positive BLs remains to be determined. Interestingly, however, in developing lung, protein localization mirrors the RNA localization documented above: α3 and α5 are concentrated in the epithelial lung buds, with α4 in the mesenchyme (data not shown; Miner, J.H., manuscript in preparation). Laminins α1 and α2 were not detectable in lung (Fig. 6, k and l).
Several conclusions can be drawn from these results. First, all laminin α chains are confined to the extracellular matrix and, with the exception of the glomerular mesangium, to BLs. Cytoplasmic deposits of laminins were not detected, nor were any laminin α chains present in the interstitial collagen– and fibronectin-rich matrix between tubules (in kidney) or myocytes (in heart). Second, each α chain is expressed in a unique pattern. Third, each BL contains at least one α chain. Fourth, BLs can contain either a single α chain (e.g., α5 in glomerular BL) or multiple α chains (e.g., α1 and α5 in proximal tubular BL or α3, α4, and α5 in some alveolar BLs). Fifth, even a single BL can vary in laminin composition along its length (e.g., α1 and α5 in proximal portions of tubular BL, α5 in distal portions, and α2 and α5 at the corticomedullary junction). Sixth, as surmised from studies at the RNA level (see above), α1 is most restricted in its expression, and α5 is the most broadly distributed in adult BLs. Together, these results support the idea that the functional diversity of BLs is achieved in part by laminin α chain diversity.
Finally, it is interesting to compare the distribution of the individual α chains to that previously documented for α1. Many studies, including some from our laboratory, have used the mAb 4C7 (Engvall et al., 1986) to assess the distribution of α1 (Engvall et al., 1990; Sanes et al., 1990; Virtanen et al., 1995, 1996; Sewry et al., 1995; Durham and Snyder, 1995). This antibody was shown to recognize a laminin α chain distinct from α2 at a time when only two α chains had been described (Engvall et al., 1990), but since this antibody does not recognize mouse protein, it could not be tested on bona fide laminin α1 as originally isolated and cloned from the EHS tumor. 4C7-immunoreactive material is more broadly distributed than is α1-immunoreactive material, as recognized by mAbs that bind to mouse laminin α1 (Sorokin et al., 1992) and were used here. Interestingly, the array of BLs recognized by 4C7 most closely resembles that stained by anti-α5 in heart, lung, and kidney (Fig. 6), as well as in skeletal muscle (Patton, B.L., J.H. Miner, and J.R. Sanes, unpublished results). Unfortunately, direct comparisons are not feasible because 4C7 does not recognize mouse or rat antigen, and our antiα5 antiserum does not recognize rabbit or human antigen. However, we speculate that 4C7 may recognize the laminin α5 chain, either instead of or in addition to α1.
Developmental Transitions in Laminin α Chain Expression
We next used our panel of antisera to ask when developing BLs acquire their complement of laminin α chains. Based on the studies of adult organs detailed above, and on the fact that laminins have been implicated as important in renal development and function (Klein et al., 1988; Noakes et al., 1995), we focused on kidney for this analysis. In fact, we found a complex and dynamic pattern of laminin α chain expression in the BLs of the developing nephron. To document these results, it is first necessary to summarize the main stages of nephrogenesis (Fig. 7).
Figure 7.
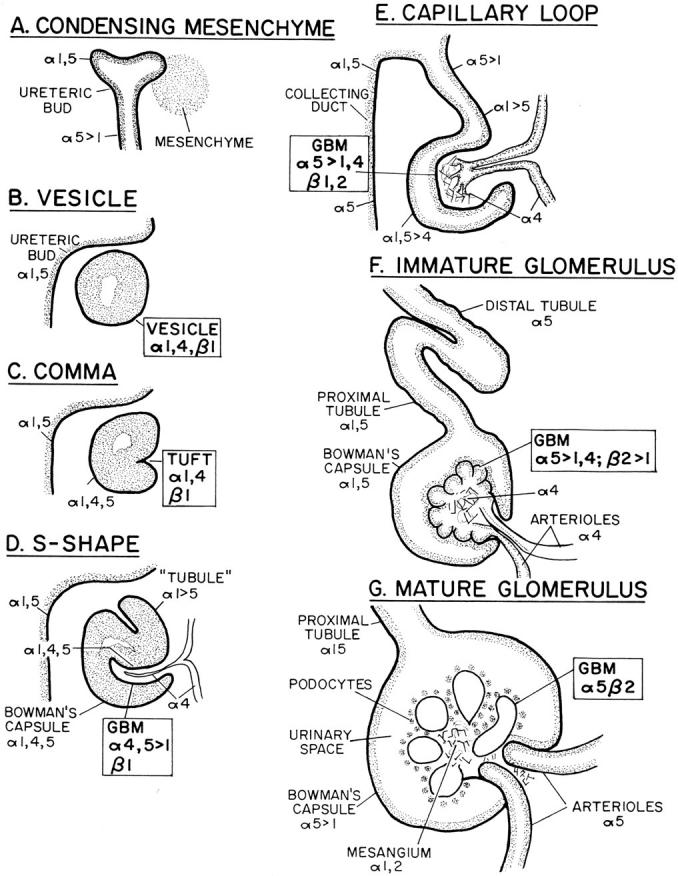
Schematic summary of kidney development, and expression patterns of the laminin α chains in various nephron segment BLs. The laminin α and β chains expressed in the developing glomerular BL (GBM) and its progenitors are boxed. See text for details.
Nephrogenesis begins when the epithelial ureteric bud grows out of the mesonephric duct, invades loose metanephric mesenchyme, and induces it to condense into a sphere (Fig. 7 A). The condensed mesenchyme then epithelializes, forming a vesicle, and secretes a BL around its periphery (Fig. 7 B). One side of the vesicle and then the other invaginates to form, successively, a comma-shaped and an S-shaped figure (Fig. 7, C and D); during this process, a blood vessel invades the primary invagination. Next, the distal portion of the S-shaped body fuses with the ureteric bud to form the tubule, and the invading vessel branches within the widening invagination to form the rudimentary capillary loops of the glomerulus (Fig. 7 E). Further ramification of the capillary loops and their enclosure by glomerular constriction lead successively to the immature and mature glomeruli (Fig. 7, F and G). Multiple waves of induction of cortical mesenchyme by the branching and lengthening ureteric bud make nephrogenesis a graded process that continues from E11 until postnatally, with newly forming nephrons just beneath the cortical surface and more mature stages at increasingly deep levels (Abrahamson, 1991; Sorokin and Ekblom, 1992; Davies, 1993).
To search for potential developmental transitions in laminin α chain expression, we stained sections of E15.5 and neonatal mouse kidney with antibodies to laminins α1–5. Results are summarized in Fig. 7 and examples are shown in Fig. 8. Before vesicle formation, the only BL near the cortical surface was that of the ureteric bud. This BL was rich in laminin α5 (Fig. 8 a) throughout its length and also contained α1 (Fig. 8 g) in the cortical portions. The first-formed BL of the nephron, that of the vesicle, contained laminins α1 (not shown) and α4 (Fig. 8 b). Laminin α1 was detected in the BL of some but not all vesicles, suggesting that it appears after α4 near the end of the vesicle stage. In the comma, α1 and α4 remained (Fig. 8, c and e) and were joined by α5 (Fig. 8 a). At this stage, significant heterogeneity became evident within the single BL that surrounded each comma: laminin α4 was present at higher levels in the tuft than in the periphery (Fig. 8 c), whereas laminin α5 was clearly present in the periphery but was virtually absent from the tuft (Fig. 8 a). Thus, the BL of the tuft, which is the precursor of the glomerular BL, becomes molecularly distinct from continuous but nonglomerular stretches of BL at an early stage of nephrogenesis.
Figure 8.
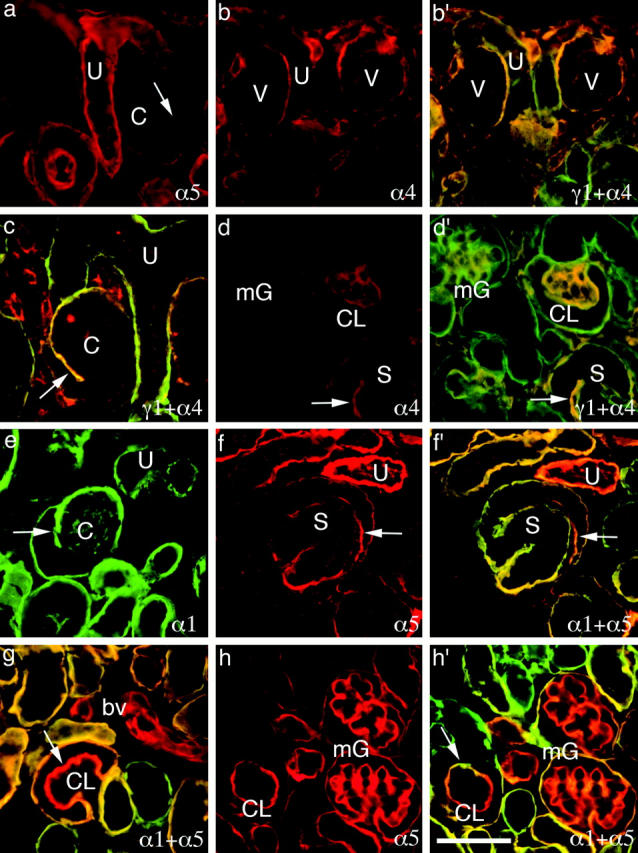
Immunohistochemical analysis of laminins α1, α4, α5, and γ1 in developing kidney shows the dynamic pattern of α chain accumulation depicted schematically in Fig. 7. All sections are from P1 mouse kidney except c, which is from E15.5. b′, c, d′, f′, g, and h′ are double exposures of doubly labeled sections; antibodies listed first and second are shown in green and red, respectively, and regions of overlap are indicated by yellow and light orange. Single exposure companions are shown in b (for b′), d (for d′), f (for f′), and h (for h′). U, ureteric bud; V, vesicle; C, comma-shaped structure; S, S-shaped structure; CL, capillary loop; mG, maturing glomerulus; bv, blood vessel. (Arrows) Progenitors of glomerular BL. Bar, 50 μm.
At the S-shaped stage, distinct portions of the nephron that will give rise to the glomerular filtration apparatus, Bowman's capsule, and the tubule can be distinguished. Interestingly, BLs in each of these regions bore a different complement of laminin α chains. The progenitor of the glomerular BL was rich in α4 and α5 and contained low levels of α1; the progenitor of Bowman's capsule BL was rich in all three chains; and the progenitor of the tubular BL contained abundant α1, low levels of α5, and no detectable α4 (Fig. 8, d and f). In addition, the invading vessel, destined to generate the capillary loops of the glomerulus, was coated by a BL with yet a fourth composition: rich in α4 but with no detectable α1 or α5.
As summarized in Fig. 7, E–G, and documented in Fig. 8, d, g, and h, each stretch of BL underwent further changes in laminin α chain composition as the nephron matured. (1) In the BL of Bowman's capsule, α4 declined in level by the capillary loop stage and disappeared from the capsule in the immature glomerulus. Levels of α1 declined later, leaving the mature capsular BL rich in α5, poor in α1, and without detectable α4. (2) Tubular BL became richer in α5 as development proceeded. Different segments of the tubule either maintained or lost α1, or acquired α2, as described above (Fig. 6, a–e). (3) Arteriolar BL lost α4 and acquired α5 at a late stage of development. (4) The glomerular BL first lost α4 and then lost α1, leaving α5 as the only detectable laminin α chain in the adult. (5) Finally, the mesangial matrix was first detectable at the capillary loop stage, where it was α4 positive. Later, α4 disappeared from this matrix, and α1 and α2 accumulated.
It is interesting to compare the laminin α chain transitions in glomerular BL to those previously documented for the laminin β and collagen IV α chains (Abrahamson and St. John, 1993; Miner and Sanes, 1994; Noakes et al., 1995; Virtanen et al., 1995). Laminin β1 and collagen α1,2(IV) chains are present in the primitive comma and S-shaped figure BLs. At the capillary loop stage, they are joined by laminin β2 and collagen α3-5(IV) in the developing glomerular BL. At the immature glomerulus stage, laminin β1 and collagen α1,2(IV) levels begin to decline, leaving only laminin β2 and collagen α3-5(IV) in the mature glomerular BL. The later appearance of α5 and β2 raises the possibility that these two events are linked. However, the initial accumulation of laminin α5 occurs at the S-shaped stage, while laminin β2 and collagen α35(IV) do not appear until the capillary loop stage (Fig. 7, D and E). Likewise, the roughly parallel disappearance of laminins α1, α4, and β1 raises the possibility that this developmental step reflects loss of α1β1- or α4β1-containing trimers. However, laminin β1 was detectable in maturing glomerular BLs that lacked laminins α1 and α4 (data not shown), suggesting that elimination of these molecules is not obligatorily linked. Surprisingly, therefore, the developmental transitions in laminin α and β chains appear to be regulated independently. If all laminin chains occur in α/β/γ trimers (see below), these results suggest that α1β1-, α1β2-, α4β1-, α4β2-, α5β1-, and α5β2-containing trimers are potentially present, at least transiently, in developing glomerular BL.
To determine whether the isoform transitions detected at the protein level reflected regulation of gene expression, we performed in situ hybridizations on P1 kidney sections using probes for the α1–5 chains. As noted above, nephrons at all stages of development are present in a corticomedullary gradient in neonates, with the most primitive just beneath the cortical surface and the most mature at deeper levels. Laminin α4 transcripts were clustered at the cortex, as expected from its early appearance in vesicular BL (Fig. 9 d). Laminin α1 transcripts were detected in cortical and subcortical clusters, consistent with the expression of this chain in late vesicle, comma, and S-shaped stages (Fig. 9 a). Segments of tubules were also α1 positive, and lower level expression (above background) was found throughout the kidney. Low levels of laminin α5 RNA were present in the superficial layer of the cortex, consistent with the later appearance of this chain in the developing nephron. Occasional clusters that were observed are likely to be the tips of the ureteric buds that have BLs rich in α5 (Fig. 9 e). Deep in the medulla, laminin α5 RNA was abundant in the collecting ducts, which are derived from the α5-positive ureteric bud. α5 labeling was not abundant in all structures that contained the protein (e.g., capillary loop stage glomeruli), suggesting that, in some cases, α5 RNA is unstable or simply present at low levels but translated efficiently. Laminin α2 RNA was concentrated in the deep cortical and medullary portions of the kidney (Fig. 9 b), consistent with its localization in a subset of tubules (Fig. 6 b). Laminin α3 RNA was not detectable within the renal cortex but was found at P15 in the papilla (data not shown), consistent with its protein localization in adult kidney (Fig. 6 c).
Figure 9.
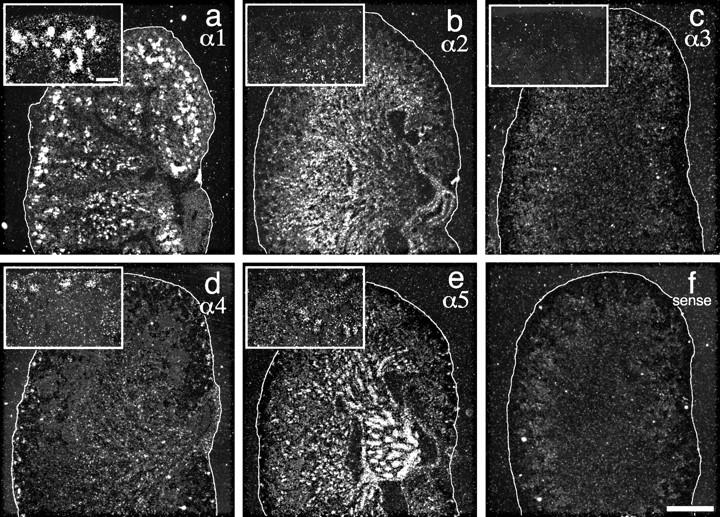
In situ hybridization of laminin α chain probes to P1 kidney sections. α1 was observed in primitive structures in the cortex and in some tubules (a). α2 was absent from the cortical structures but distributed diffusely in the interior (b). α4 was detected primarily in clusters at the cortex (vesicle and comma stage nephrons) but also diffusely in the medulla (d). α5 was mainly in the collecting ducts, but there were also grain clusters in the inner cortex and in the medulla (e). α3 was absent (c), and a sense control was negative (f). These patterns suggest that the developmental transitions demonstrated immunohistochemically in Figs. 7 and 8 reflect, in part, developmental transitions in α chain gene expression. The cortical surface of the kidney is outlined in white. Insets in a–e show higher power views of the cortex. Bars: (f) 0.5 mm; (a, inset) 0.1 mm.
Identification of Heterotrimers Containing the Laminin α4 or α5 Chain
Current understanding of the structure and function of BLs is based on the assumption that all laminins are heterotrimers of α, β, and γ subunits, covalently joined by disulfide bonds (Burgeson et al., 1994). Such trimeric structures have been demonstrated for the α1–3 chains, which form laminins 1–7 (Table I), but not for α4 and α5. We therefore asked whether the α4 and α5 chains also occur as components of trimers. To this end, we fractionated proteins from lung and kidney on SDS gels under nonreducing conditions such that laminins migrate as trimers, the relative sizes of which depend on the constituent α, β, and γ chains. Nitrocellulose blots were then probed with antibodies to α4 or α5 (Fig. 10 A). Both lung and kidney contained high M r complexes that reacted with the α4 (Fig. 10 A, lanes 3 and 7) or α5 (lanes 4 and 8) antisera but not with nonimmune serum (lanes 2 and 6). The α4 complexes were ∼500–600 kD, and the α5 complexes were ∼700–800 kD. These values are consistent with M rs predicted for laminin trimers. None of the α4- or α5-immunoreactive bands (⩽450 kD) that had been seen after reduction (Fig. 5 B) were detectable under these nonreducing conditions, indicating that all of the monomeric species were associated with larger, disulfide-bonded complexes. Moreover, the approximate difference in M r between the α4- and α5-containing complexes (∼200 kD) is consistent with the difference in M r between the full-length α4 and α5 chains themselves. An antiserum to laminin-1, which recognizes the α1, β1, and γ1 chains, blotted material at the M rs of all α4- and α5containing complexes (Fig. 10 A, lanes 1 and 5), whereas anti-α4 and anti-α5 did not recognize laminin-1 (lanes 11 and 12). Together, these results suggest that laminins α4 and α5 occur in heterotrimers with β1 and/or γ1 chains.
Table I.
Composition of Laminin Trimers
| Trimer | Components | References | ||
|---|---|---|---|---|
| Laminin-1 | α1β1γ1 | Beck et al., 1990 | ||
| Laminin-2 | α2β1γ1 | Engvall et al., 1990 | ||
| Laminin-3 | αlβ2γ1* | Engvall et al., 1990 | ||
| Laminin-4 | α2β2γ1 | Engvall et al., 1990 | ||
| Laminin-5 | α3β3γ2 | Rousselle et al., 1991 | ||
| Laminin-6 | α3β1γ1 | Marinkovich et al., 1992b | ||
| Laminin-7 | α3β2γ1 | Champliaud et al., 1996 | ||
| Laminin-8 | α4β1γ1 | This study | ||
| Laminin-9 | α4β2γ1 | This study | ||
| Laminin-10 | α5β1γ1 | This study | ||
| Laminin-11 | α5β2γ1 | This study |
α1 identified immunochemically with the 4C7 mAb.
Figure 10.
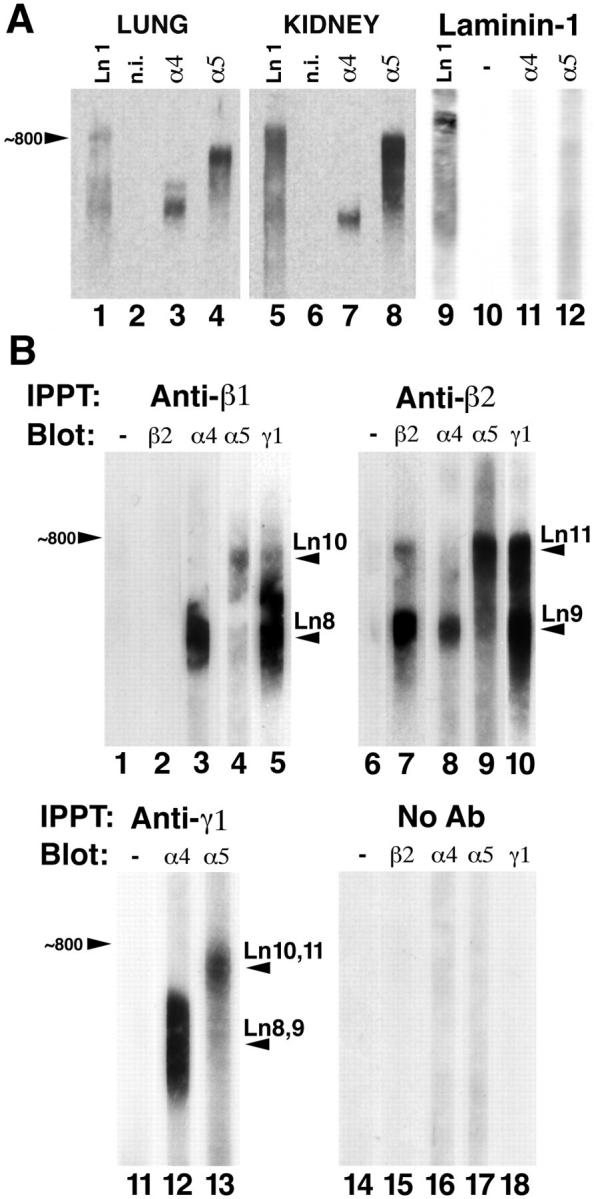
Biochemical identification of laminins-8, -9, -10, and -11. (A) Detection of laminin complexes containing α4 and α5. Crude membrane preparations from adult lung and kidney and purified laminin-1 were solubilized without reducing agent and fractionated by SDS-PAGE on 3.5% polyacrylamide gels. Proteins were transferred to nitrocellulose and probed with nonimmune (n.i.) serum (lanes 2, 6, and 10) or with antibodies to laminin-1 (lanes 1, 5, and 9), α4 (lanes 3, 7, and 11), or α5 (lanes 4, 8, and 12). The size of the laminin-1 trimer (∼800 kD) is indicated. Lung contained at least two complexes containing α4 (lane 3), while kidney contained multiple α5-positive complexes (lane 8). Laminin-1 contained little or no detectable α4 or α5. (B) Identification of laminin trimers from adult lung. Laminins were solubilized, partially purified, and then immunoprecipitated with antibodies to laminin β1 (lanes 1–5), β2 (lanes 6–10), γ1 (lanes 11–13), or without primary antibody (lanes 14–18). The precipitates were then fractionated on nonreducing gels and probed without primary antibody (lanes 1, 6, 11, and 14), or with antibodies to laminins β2 (lanes 2, 7, and 15), α4 (lanes 3, 8, 12, and 16), α5 (lanes 4, 9, 13, and 17), and γ1 (lanes 5, 10, and 18). Four novel laminin trimers were identified: laminin-8 (α4β1γ1), laminin-9 (α4β2γ1), laminin-10 (α5β1γ1), and laminin-11 (α5β2γ1).
To definitively identify the α4- and α5-containing complexes as laminin heterotrimers, we solubilized and partially purified laminins from lung. Lung was chosen for this set of experiments because, unlike kidney, it expresses both α4 and α5 chains at high levels in adulthood (Figs. 3 and 6). Lung homogenates were extracted with EDTA, EGTA, NaCl, and Triton X-100, and the extracts were then fractioned by a combination of ion exchange and size exclusion chromatography (see Materials and Methods). The resulting laminin-rich fraction was then subjected to immunoprecipitation with mAbs specific for laminin β1 or β2. Precipitates were separated on gels under nonreducing conditions, electroblotted onto nitrocellulose, and probed with antibodies specific for individual laminin chains (Fig. 10 B). Antibodies to laminin β1 precipitated a series of complexes of ∼500–800 kD (Fig. 10 B, lanes 1–5). All the complexes comigrated with γ1 (Fig. 10 B, lane 5), but none contained β2 (lane 2). Individual complexes reacted with either anti-α4 (Fig. 10 B, lane 3) or with anti-α5 (lane 4) but not with both. Complexes containing α4 had M rs of ∼500–600 kD, and those with α5 had M rs of ∼700–800 kD, corresponding to M rs of the nonreduced α4 and α5 complexes observed by immunoblotting crude lung samples (Fig. 10 A). Controls in which the primary antibody was omitted did not contain laminins (Fig. 10 B, lanes 14–18). Thus, the laminin α4 and α5 chains are both associated with β1 in distinct heterotrimers.
Parallel results were obtained from material precipitated by antibodies to laminin β2 (Fig. 10 B, lanes 6–10). All of the laminin β2-reactive material precipitated by these antibodies (Fig. 10 B, lane 7) reacted with anti γ1 (lane 10) and with either anti-α4 (lane 8) or anti-α5 (lane 9) but not both. Complexes containing α4 had M rs of ∼500–600 kD, and those with α5 had M rs of ∼700–800 kD, corresponding to those detected in crude lung samples (Fig. 10 A). Electrophoresis of the immunoprecipitates under reducing conditions revealed that the major α4- and α5-reactive bands detected in crude extracts (Fig. 5 B) were complexed with β2 (data not shown). Thus, lung also contains distinct laminin heterotrimers that include α4+β2 and α5+β2.
Two observations indicated that the complexes containing α4β1, α4β2, α5β1, and α5β2 also contained the γ1 chain. First, an mAb to γ1 precipitated α4- and α5-containing complexes (Fig. 10 B, lanes 11–13) that were equivalent in M r to those precipitated by anti-β1 or anti-β2. Second, as noted above, anti-β1 and anti-β2 precipitated γ1-containing complexes that comigrated with all of the α4- and α5immunoreactive trimers (Fig. 10 B, lanes 5 and 10). Together, these results provide strong evidence for the existence of laminin trimers with chain compositions of α4β1γ1, α4β2γ1, α5β1γ1, and α5β2γ1. In accordance with recently adopted rules of nomenclature (Burgeson et al., 1994), these are named laminins 8–11, respectively (Table I).
Chromosomal Mapping of Lama Genes
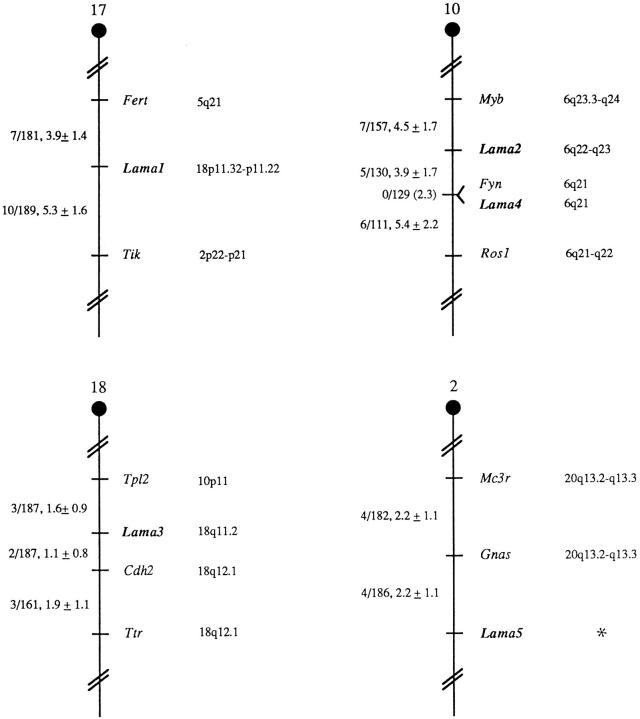
The murine chromosomal locations of Lama1–Lama5 were determined using an interspecific backcross mapping panel derived from crosses of [(C57BL/6J × M. spretus) F1 X C57BL/6J] mice. Mouse genomic or cDNA fragments specific for each locus were used as probes in Southern blot hybridization analysis of C57BL/6J and M. spretus genomic DNA that was separately digested with several different restriction enzymes to identify informative RFLPs (see Materials and Methods). The strain distribution pattern of each RFLP in the interspecific backcross was then determined by following the presence or absence of RFLPs specific for M. spretus in backcross mice.
Backcross mapping refined previously reported chromosomal locations for Lama1–3 and provided the first data on mouse chromosomal locations of Lama4 and Lama5 (Fig. 11). Lama1 has previously been reported to map to the distal region of mouse chromosome 17 in this interspecific backcross (Okazaki et al., 1993; Doyle et al., 1996). Lama2 mapped to the proximal region of mouse chromosome 10, 4.5 centiMorgans (cM) distal of Myb and 3.9 cM proximal of Fyn. This result is in good agreement with previous studies that map Lama2 3.2 ± 1.8 cM distal of Myb on mouse chromosome 10 (Sunada et al., 1994). We have also confirmed the recent results of Aberdam et al. (1994b) and Griffith et al. (1996) by assigning Lama3 to the proximal region of chromosome 18. In our interspecific cross, Lama3 mapped 1.6 cM distal of Tpl1 and 1.1 cM proximal of Cdh2. Lama4 is linked to Lama2 on chromosome 10 and does not recombine with Fyn in 129 animals typed in common, suggesting that the two loci are within 2.3 cM of each other (upper 95% confidence limit). Finally, Lama5 mapped to the very distal region of mouse chromosome 2, 2.2 cM distal of Gnas. Thus, the five Lama loci were distributed on four different mouse autosomes, indicating that most of the Lama genes have become dispersed through evolution.
Figure 11.
Chromosomal locations of Lama loci in the mouse genome, as determined from interspecific backcross analysis. The number of recombinant N2 animals is presented over the total number of N2 animals typed to the left of the chromosome maps between each pair of loci. The recombination frequencies, expressed as genetic distance in centimorgans (± one standard error) are also shown. The upper 95% confidence limit of the recombination distance is given in parentheses when no recombinants were found between loci. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns. The positions of loci on human chromosomes, where known, are shown to the right of the chromosome maps. (Asterisk) The human LAMA5 gene is predicted to reside on chromosome 20q13. References for the human map positions of loci cited in this study can be obtained from GDB (Genome Data Base), a computerized database of human linkage information maintained by The William H. Welch Medical Library of The Johns Hopkins University (Baltimore, MD).
Previous studies have shown that the Lama2 gene encodes the dystrophia muscularis (dy) mouse mutation (Sunada et al., 1994; Xu et al., 1994), but no targeted or spontaneous mouse mutants have been reported for lama1, 3, 4, or 5. Therefore, we compared our maps of mouse chromosomes 2, 10, 17, and 18 with composite mouse maps that report the map locations of many uncloned mouse mutations (obtained from the Mouse Genome Database, maintained at The Jackson Laboratory, Bar Harbor, ME). Mutations in the vicinity of the Lama genes include thin fur (thf) near the Lama1 locus; ataxia (ax), twirler (Tw), and balding (bal) near the Lama3 locus; waltzer (v) and kidney disease (kd) near the Lama4 locus; and ragged (Ra) and wasted (wst) near the Lama5 locus. For most of these mutations, the reported phenotypes are not suggestive of a BL defect. For ragged, however, the phenotype is an intriguing one. The mutation Ra shows semidominance, and the coats of heterozygotes have a thin ragged appearance. Homozygotes are naked, many die in utero, and there are internal visceral defects (Carter and Phillips, 1954; Slee, 1957). Another semidominant allele of ragged, Ra < op > (opossum) (Green and Mann, 1961), is homozygous lethal before E11, which, because of its expression in many tissues, might be expected for a Lama5 mutant. Also, since α5 is expressed in the skin, a mutation may affect the function of hair follicles, which could lead to the ragged coat appearance. Moreover, the disruption of heterotrimeric laminin structure by a defective α chain could explain the semidominant phenotype of both ragged alleles identified to date.
Several of the LAMA genes have also been mapped to human chromosomes (summarized in Fig. 11). LAMA1 maps to human 18p11.32-p11.22, LAMA2 to 6q22-q23, LAMA3 to 18q11.2, and LAMA4 to 6q21. LAMA1 is currently the only gene that maps to human chromosome 18 and mouse chromosome 17. Thus, LAMA1 defines a new region of homology between mouse and human chromosomes. In contrast, the mouse and human map locations of LAMA2–4 confirm and extend the known regions of homology between mouse and human chromosomes (Fig. 11; data not shown). LAMA5 has not been mapped in humans. However, the distal region of mouse chromosome 2 shares a region of homology with human chromosome 20q13, suggesting that LAMA5 will map to 20q13 in humans, as well. As noted in the Introduction, the LAMA2 gene is mutated in some congenital muscular dystrophies, and the LAMA3 gene is mutated in a subset of patients with a skin blistering disease, junctional epidermolysis bullosa. No human mutations that suggest a BL defect map to the vicinity of the human LAMA1 or LAMA4 locus or to the predicted LAMA5 locus.
Conclusion
The laminins are major components of BLs throughout the body, and their α chains are ligands for most cellular laminin receptors identified to date, including at least six integrins, dystroglycan, and several glycoconjugates (see Introduction). Yet the distribution of the laminin α chains has not previously been studied in detail, and the existence of the two most recently identified chains (α4 and α5) has been deduced from cDNA sequence but not shown directly at the protein level. We have therefore generated a panel of antibody and cDNA reagents and have used it to localize the α chain genes on mouse chromosomes and the α chain RNAs and proteins in developing and adult tissues. We have also identified a new isoform of laminin α3 and provided direct evidence for the existence of four novel laminin heterotrimers. Our main conclusions are as follows:
(1) The α subfamily of laminin genes currently consists of five members, which encode six proteins. Four (α1, α2, α3B, and α5) are full-sized chains, and two (α3A and α4) are truncated chains (Fig. 1). The previously hypothesized mid-sized α chain (α3B) may not, in fact, exist (Fig. 2).
(2) All five of the laminin α chain genes are expressed in both embryos and adults, but each is expressed in a distinct pattern (Figs. 3 and 4). Their products are confined to the extracellular matrix and, with a few exceptions (e.g., the glomerular mesangium), to BLs (Fig. 6).
(3) All developing and adult BLs studied to date contain at least one of the known α chains, and some contain two or three distinct α chains. Although additional α chains may well exist, our results provide no evidence for them and no clues as to their nature or distribution.
(4) Laminin α5 is the most widely distributed α chain in adult BLs, and α1 is the most restricted. The α2 chain is particularly abundant in mesodermally derived tissues (e.g., skeletal and cardiac muscle), and α3 is concentrated in epithelial BLs. The restricted distribution of α1 is inconsistent with the broad distribution of immunoreactive material recognized by a widely used mAb, 4C7. This discrepancy, together with data on the distribution of α2–5, raises the possibility that 4C7 recognizes the α5 chain, in addition to or instead of α1.
(5) The main patterns of α chain expression are established embryonically (Fig. 4), but some individual BLs change in α chain composition as development proceeds. In kidney, for example, forming glomeruli express three different α chains in a dynamic progression. Moreover, distinct portions of a continuous BL, such as that of the nephron, can contain distinct complements of α chains, and these change during development as well (Figs. 7 and 8).
(6) Laminins α1–5 all form heterotrimers with β and γ chains. At present, 11 distinct heterotrimers have been identified in mammalian cells or tissues, including at least two containing each α chain (Table I). The four heterotrimers identified in this study (Fig. 10) have been named laminins-8 (α4β1γ1), -9 (α4β2γ1), -10 (α5β1γ1), and -11 (α5β2γ1).
(7) The five α chain genes are distributed on four chromosomes in mouse, and probably on three chromosomes in human. The two α chain genes present on a single chromosome in both species (α2 and α4) are not tightly linked, and two chains present on the same chromosome in humans (α1 and α3) are on different chromosomes in mouse (Fig. 11). Thus, there is no evidence for functionally important linkage of the α chain genes.
The patterns of expression that we have documented indicate that the α chains contribute importantly to the molecular diversity of BLs. Together with evidence that some cellular receptors can distinguish among α chains and that naturally occurring mutations of α2 and α3 have tissuespecific phenotypes (see Introduction), our results suggest that differences among α chains are critical to the multiplicity of roles that BLs play both during development and in maturity.
Note Added in Proof:
A third α3 transcript with a distinct 5′ end and translation start site has now been identified by D. Aberdam and colleagues. It encodes a protein intermediate in size between the α3B isoform reported here and the α3A isoform described by Galliano et al. (1995). Thus, there may be three isoforms of laminin α3: short (A), fulllength (B), and mid-sized (C) (Aberdam, D., personal communication).
Acknowledgments
We are grateful to Drs. Lydia Sorokin, Peter Yurchenco, and Daniel Aberdam for gifts of antisera and to Renate Lewis, Jeanette Cunningham, and Ellen Ryan for technical assistance. J.H. Miner was supported in part by a Damon Runyon-Walter Winchell Cancer Research Fund fellowship.
This work was supported by grants from the National Institutes of Health.
Abbreviations used in this paper
- BL
basal lamina
- cM
centiMorgan
- E
embryonic day
- EHS
Englebreth-Holm-Swarm
- RFLP
restriction fragment length polymorphism
- RT-PCR
reverse transcription coupled–PCR
Footnotes
Please address all correspondence to Joshua R. Sanes, Department of Anatomy and Neurobiology, Washington University School of Medicine, 660 South Euclid Avenue, St. Louis, MO 63110. Tel.: (314) 362-2507. Fax: (314) 747-1150.
J.H. Miner and B.L. Patton contributed equally to this work.
References
- Aberdam D, Aguzzi A, Baudoin C, Galliano M-F, Ortonne J-P, Meneguzzi G. Developmental expression of nicein adhesion protein (laminin-5) subunits suggests multiple morphogenic roles. Cell Adhes Commun. 1994a;2:115–129. doi: 10.3109/15419069409004431. [DOI] [PubMed] [Google Scholar]
- Aberdam D, Galliano MF, Mattei M-G, Pisani-Spadafora A, Ortonne J-P, Meneguzzi G. Assignment of mouse nicein genes to chromosomes 1 and 18. Mamm Genome. 1994b;5:229–233. doi: 10.1007/BF00360551. [DOI] [PubMed] [Google Scholar]
- Abrahamson DR. Glomerulogenesis in the developing kidney. Semin Nephrol. 1991;11:375–389. [PubMed] [Google Scholar]
- Abrahamson DR, Leardkamolkarn V. Development of kidney tubular basement membranes. Kidney Int. 1991;39:382–393. doi: 10.1038/ki.1991.50. [DOI] [PubMed] [Google Scholar]
- Abrahamson DR, St. John PL. Laminin distribution in developing glomerular basement membranes. Kidney Int. 1993;43:73–78. doi: 10.1038/ki.1993.13. [DOI] [PubMed] [Google Scholar]
- Abrahamson DR, Irwin MH, St. John PL, Perry EW, Accavitti MA, Heck LW, Couchman JR. Selective immunoreactivities of kidney basement membranes to monoclonal antibodies against laminin: localization of the end of the long arm and the short arms to discrete microdomains. J Cell Biol. 1989;109:3477–3491. doi: 10.1083/jcb.109.6.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Beck K, Hunter I, Engel J. Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB (Fed Am Soc Exp Biol) J. 1990;4:148–160. doi: 10.1096/fasebj.4.2.2404817. [DOI] [PubMed] [Google Scholar]
- Burgeson RE, Chiquet M, Deutzmann R, Ekblom P, Engel J, Kleinman H, Martin GR, Ortonne J-P, Paulsson M, Sanes J, et al. A new nomenclature for laminins. Matrix Biol. 1994;14:209–211. doi: 10.1016/0945-053x(94)90184-8. [DOI] [PubMed] [Google Scholar]
- Carter TC, Phillips RJS. Ragged, a semidominant coat texture mutant. J Hered. 1954;45:151–154. [Google Scholar]
- Carter WG, Ryan MC, Gahr PJ. Epiligrin, a new cell adhesion ligand for integrin α3β1 in epithelial basement membranes. Cell. 1991;65:599–610. doi: 10.1016/0092-8674(91)90092-d. [DOI] [PubMed] [Google Scholar]
- Champliaud M-F, Lunstrum GP, Rousselle P, Nishiyama T, Keene DR, Burgeson RE. Human amnion contains a novel laminin variant, laminin 7, which like laminin 6, covalently associates with laminin 5 to promote stable epithelial-stromal attachment. J Cell Biol. 1996;132:1189–1198. doi: 10.1083/jcb.132.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Chung AE, Jaffe R, Freeman IL, Vergnes JP, Braginsk JE, Carlin B. Properties of a basement membrane related glycoprotein synthesized by a mouse embryonal carcinoma-derived cell line. Cell. 1979;16:277–287. doi: 10.1016/0092-8674(79)90005-9. [DOI] [PubMed] [Google Scholar]
- Clark EA, Brugge JS. Integrins and signal transduction pathways: the road taken. Science (Wash DC) 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA. Development and applications of a molecular genetic linkage map of the mouse genome. Trends Genet. 1991;7:113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- Davies J. How to build a kidney. Semin Cell Biol. 1993;4:213–219. doi: 10.1006/scel.1993.1025. [DOI] [PubMed] [Google Scholar]
- Doyle J, Hoffman S, Ucla C, Reith W, Mach B, Stubbs L. Locations of human and mouse genes encoding the RFX1 and RFX2transcription factor proteins. Genomics. 1996;35:227–230. doi: 10.1006/geno.1996.0343. [DOI] [PubMed] [Google Scholar]
- Durham PL, Snyder JM. Characterization of α1, β1, and γ1 laminin subunits during rabbit fetal lung development. Dev Dyn. 1995;203:408–421. doi: 10.1002/aja.1002030404. [DOI] [PubMed] [Google Scholar]
- Ehrig K, Leivo I, Argraves WS, Ruoslahti E, Engvall E. Merosin, a tissue-specific basement membrane protein, is a laminin-like protein. Proc Natl Acad Sci USA. 1990;87:3264–3268. doi: 10.1073/pnas.87.9.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblom P. Receptors for laminins during epithelial morphogenesis. Curr Opin Cell Biol. 1996;8:700–706. doi: 10.1016/s0955-0674(96)80112-8. [DOI] [PubMed] [Google Scholar]
- Engvall E, Wewer UM. Domains of laminin. J Cell Biochem. 1996;61:493–501. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C493::AID-JCB2%3E3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Engvall E, Davis GE, Dickerson K, Ruoslahti E, Varon S, Manthorpe M. Mapping of domains in human laminin using monoclonal antibodies: localization of the neurite-promoting site. J Cell Biol. 1986;103:2457–2465. doi: 10.1083/jcb.103.6.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engvall E, Earwicker D, Haaparanta T, Ruoslahti E, Sanes JR. Distribution and isolation of four laminin variants; tissue restricted distribution of heterotrimers assembled from five different subunits. Cell Regul. 1990;1:731–740. doi: 10.1091/mbc.1.10.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MRS, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSF2and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Foster RF, Thompson JM, Kaufman SJ. A laminin substrate promotes myogenesis in rat skeletal muscle cultures: analysis of replication and development using antidesmin and anti-BrdUrd monoclonal antibodies. Dev Biol. 1987;122:11–20. doi: 10.1016/0012-1606(87)90327-7. [DOI] [PubMed] [Google Scholar]
- Galliano M-F, Aberdam D, Aguzzi A, Ortonne J-P, Meneguzzi G. Cloning and complete primary structure of the mouse laminin α3 chain. J Biol Chem. 1995;270:21820–21826. doi: 10.1074/jbc.270.37.21820. [DOI] [PubMed] [Google Scholar]
- Garcia-Alonso L, Fetter RD, Goodman CS. Genetic analysis of Laminin A in Drosophila: extracellular matrix containing laminin A is required for ocellar axon pathfinding. Development (Camb) 1996;122:2611–2621. doi: 10.1242/dev.122.9.2611. [DOI] [PubMed] [Google Scholar]
- Gee SH, Blacher RW, Douville PJ, Provost PR, Yurchenco PD, Carbonetto S. Laminin-binding protein 120 from brain is closely related to the dystrophin-associated glycoprotein, dystroglycan, and binds with high affinity to the major heparin binding domain of laminin. J Biol Chem. 1993;268:14972–14980. [PubMed] [Google Scholar]
- Gerecke SR, Wagman DW, Champliaud MF, Burgeson RE. The complete primary structure for a novel laminin chain, the laminin B1k chain. J Biol Chem. 1994;269:11073–11080. [PubMed] [Google Scholar]
- Green, E.L. 1981. Linkage, recombination and mapping. In Genetics and Probability in Animal Breeding Experiments. Oxford University Press, New York. 77–113.
- Green EL, Mann SJ. Opossum, a semi-dominant lethal mutation affecting hair and other characteristics of mice. J Hered. 1961;52:223–227. [Google Scholar]
- Green TL, Hunter DD, Chan W, Merlie JP, Sanes JR. Synthesis and assembly of the synaptic cleft protein S-laminin by cultured cells. J Biol Chem. 1992;267:2014–2022. [PubMed] [Google Scholar]
- Griffith AJ, Radice GL, Burgess DL, Kohrman DC, Hansen GM, Justice ML, Johnson KR, Davisson MT, Meisler MH. Location of the 9257 and ataxia mutations on mouse Chromosome 18. Mamm Genome. 1996;7:417–419. doi: 10.1007/s003359900124. [DOI] [PubMed] [Google Scholar]
- Hall H, Liu L, Schachner M, Schmitz B. The L2/NHK-1 carbohydrate mediates adhesion of neural cells to laminin. Eur J Neurosci. 1993;5:34–42. doi: 10.1111/j.1460-9568.1993.tb00202.x. [DOI] [PubMed] [Google Scholar]
- Helbling-Leclerc A, Zhang X, Topaloglu H, Cruaud C, Tesson F, Weissenbach J, Tome FM, Schwartz K, Fardeau M, Tryggvason K, Guicheney P. Mutations in the laminin α2-chain gene (LAMA2) cause merosin-deficient congenital muscular dystrophy. Nat Genet. 1995;11:216–218. doi: 10.1038/ng1095-216. [DOI] [PubMed] [Google Scholar]
- Henchcliffe C, Garcia-Alonso L, Tang J, Goodman CS. Genetic analysis of laminin A reveals diverse functions during morphogenesis in Drosophila. . Development (Camb) 1993;118:325–337. doi: 10.1242/dev.118.2.325. [DOI] [PubMed] [Google Scholar]
- Henry MD, Campbell KP. Dystroglycan—an extracellular matrix receptor linked to the cytoskeleton. Curr Opin Cell Biol. 1996;8:625–631. doi: 10.1016/s0955-0674(96)80103-7. [DOI] [PubMed] [Google Scholar]
- Horikoshi S, Koide H, Shirai T. Monoclonal antibodies against laminin A chain and B chain in the human and mouse kidneys. Lab Invest. 1988;58:532–538. [PubMed] [Google Scholar]
- Hunter DD, Porter BE, Bulock JW, Adams SP, Merlie JP, Sanes JR. Primary sequence of a motor neuron-selective adhesive site in the synaptic basal lamina protein S-laminin. Cell. 1989a;59:905–913. doi: 10.1016/0092-8674(89)90613-2. [DOI] [PubMed] [Google Scholar]
- Hunter DD, Shah V, Merlie JP, Sanes JR. A laminin-like adhesive protein concentrated in the synaptic cleft of the neuromuscular junction. Nature (Lond) 1989b;338:229–234. doi: 10.1038/338229a0. [DOI] [PubMed] [Google Scholar]
- Iivanainen A, Sainio K, Sariola H, Tryggvason K. Primary structure and expression of a novel human laminin α4 chain. FEBS Lett. 1995;365:183–188. doi: 10.1016/0014-5793(95)00462-i. [DOI] [PubMed] [Google Scholar]
- Jenkins NA, Copeland NG, Taylor BA, Lee BK. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. . J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice MJ, Siracusa LD, Gilbert DJ, Heisterkamp N, Groffen J, Chada K, Silan CM, Copeland NG, Jenkins NA. A genetic linkage map of mouse chromosome 10: localization of eighteen molecular markers using a single interspecific backcross. Genetics. 1990;125:855–866. doi: 10.1093/genetics/125.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice MJ, Gilbert DJ, Kinzler KW, Vogelstein B, Buchberg AM, Ceci JD, Matsuda Y, Chapman VM, Patriotis C, Makris A, et al. A molecular genetic linkage map of mouse chromosome 18 reveals extensive linkage conservation with human chromosomes 5 and 18. Genomics. 1992;13:1281–1288. doi: 10.1016/0888-7543(92)90047-v. [DOI] [PubMed] [Google Scholar]
- Justice MJ, Morse HC, III, Jenkins NA, Copeland NG. Identification of Evi-3, a novel common site of retroviral integration in mouse AKXD B-cell lymphomas. J Virol. 1994;68:1293–1300. doi: 10.1128/jvi.68.3.1293-1300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki P, Sainio K, Eddy R, Byers M, Kallunki T, Sariola H, Beck K, Hirvonen H, Shows TB, Tryggvason K. A truncated laminin chain homologous to the B2 chain: structure, spatial expression, and chromosomal assignment. J Cell Biol. 1992;119:679–693. doi: 10.1083/jcb.119.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G, Langegger M, Timpl R, Ekblom P. Role of laminin A chain in the development of epithelial cell polarity. Cell. 1988;55:331–341. doi: 10.1016/0092-8674(88)90056-6. [DOI] [PubMed] [Google Scholar]
- Leivo I, Engvall E. Merosin, a protein specific for basement membranes of Schwann cell, striated muscle, and trophoblast, is expressed late in nerve and muscle development. Proc Natl Acad Sci USA. 1988;85:1544–1548. doi: 10.1073/pnas.85.5.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz SI, Miner JH, Sanes JR, Snider WD. Distribution of the ten known laminin chains in the pathways and targets of developing sensory axons. J Comp Neurol. 1997;378:547–561. doi: 10.1002/(sici)1096-9861(19970224)378:4<547::aid-cne9>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Lindblom A, Marsh T, Fauser C, Engel J, Paulsson M. Characterization of native laminin from bovine kidney and comparison with other laminin variants. Eur J Biochem. 1994;219:383–392. doi: 10.1111/j.1432-1033.1994.tb19950.x. [DOI] [PubMed] [Google Scholar]
- Marinkovich MP, Lunstrum GP, Burgeson RE. The anchoring filament protein kalinin is synthesized and secreted as a high molecular weight precursor. J Biol Chem. 1992a;267:17900–17906. [PubMed] [Google Scholar]
- Marinkovich MP, Lunstrum GP, Keene DR, Burgeson RE. The dermal-epidermal junction of human skin contains a novel laminin variant. J Cell Biol. 1992b;119:695–703. doi: 10.1083/jcb.119.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR, Timpl R. Laminin and other basement membrane components. Annu Rev Cell Biol. 1987;3:57–85. doi: 10.1146/annurev.cb.03.110187.000421. [DOI] [PubMed] [Google Scholar]
- McGrath JA, Kivirikko S, Ciatti S, Moss C, Dunnill GS, Eady RA, Rodeck CH, Christiano AM, Uitto J. A homozygous nonsense mutation in the alpha 3 chain gene of laminin 5 (LAMA3) in Herlitz junctional epidermolysis bullosa: prenatal exclusion in a fetus at risk. Genomics. 1995;29:282–284. doi: 10.1006/geno.1995.1246. [DOI] [PubMed] [Google Scholar]
- Mecham, R.P., and A. Hinek. 1996. Non-integrin laminin receptors. In The Laminins. P. Ekblom and R. Timpl, editors. Harwood Academic Publishers, Amsterdam. 159–183.
- Mercurio AM. Laminin receptors: achieving specificity through cooperation. Trends Cell Biol. 1995;5:419–423. doi: 10.1016/s0962-8924(00)89100-x. [DOI] [PubMed] [Google Scholar]
- Miner JH, Sanes JR. Collagen IV α3, α4, and α5 chains in rodent basal laminae: sequence, distribution, association with laminins, and developmental switches. J Cell Biol. 1994;127:879–891. doi: 10.1083/jcb.127.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JH, Wold BJ. c-mycinhibition of MyoD and myogenin-initiated myogenic differentiation. Mol Cell Biol. 1991;11:2842–2851. doi: 10.1128/mcb.11.5.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JH, Lewis RM, Sanes JR. Molecular cloning of a novel laminin chain, α5, and widespread expression in adult mouse tissues. J Biol Chem. 1995;270:28523–28526. doi: 10.1074/jbc.270.48.28523. [DOI] [PubMed] [Google Scholar]
- Noakes PG, Miner JH, Gautam M, Cunningham JM, Sanes JR, Merlie JP. The renal glomerulus of mice lacking s-laminin/laminin β2: nephrosis despite molecular compensation by laminin β1. Nat Genet. 1995;10:400–406. doi: 10.1038/ng0895-400. [DOI] [PubMed] [Google Scholar]
- Okazaki T, Yoshida BN, Avraham KB, Wang H, Wuenschell CW, Jenkins NA, Copeland NG, Anderson DJ, Mori N. Molecular diversity of the SCG10/Stathmin gene family in the mouse. Genomics. 1993;18:360–373. doi: 10.1006/geno.1993.1477. [DOI] [PubMed] [Google Scholar]
- Pall EA, Bolton KM, Ervasti JM. Differential heparin inhibition of skeletal muscle alpha-dystroglycan binding to laminins. J Biol Chem. 1996;271:3817–3821. doi: 10.1074/jbc.271.7.3817. [DOI] [PubMed] [Google Scholar]
- Paulsson M, Saladin K, Engvall E. The 300-kDa chains of murine and bovine heart laminin are related to the human placenta merosin heavy chain and replace the A chain in some laminin variants. J Biol Chem. 1991;266:17545–17551. [PubMed] [Google Scholar]
- Rao CN, Kefalides NA. Identification and characterization of a 43 kilodalton laminin fragment from the “A” chain (long arm) with high-affinity heparin binding and mammary epithelial cell adhesion-spreading activities. Biochemistry. 1990;29:6768–6777. doi: 10.1021/bi00481a004. [DOI] [PubMed] [Google Scholar]
- Reichardt LF, Tomaselli KJ. Extracellular matrix molecules and their receptors: functions in neural development. Annu Rev Neurosci. 1991;14:531–570. doi: 10.1146/annurev.ne.14.030191.002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AJ, Al-Imara L, Carter NP, Lloyd JC, Leversha MA, Pope FM. Localization of the gene (LAMA4) to chromosome 6q21 and isolation of a partial cDNA encoding a variant laminin A chain. Genomics. 1994;22:237–239. doi: 10.1006/geno.1994.1372. [DOI] [PubMed] [Google Scholar]
- Richards A, Al-Imara L, Pope FM. The complete cDNA sequence of laminin α4 and its relationship to the other human laminin α chains. Eur J Biochem. 1996;238:813–821. doi: 10.1111/j.1432-1033.1996.0813w.x. [DOI] [PubMed] [Google Scholar]
- Rousselle P, Lunstrum GP, Keene DR, Burgeson RE. Kalinin: an epithelium-specific basement membrane adhesion molecule that is a component of anchoring filaments. J Cell Biol. 1991;114:567–576. doi: 10.1083/jcb.114.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MC, Tizard R, VanDevanter DR, Carter WG. Cloning of the LamA3 gene encoding the α3 chain of the adhesive ligand epiligrin. J Biol Chem. 1994;269:22779–22787. [PubMed] [Google Scholar]
- Sanes JR, Chiu AY. The basal lamina of the neuromuscular junction. Cold Spring Harbor Symp Quant Biol. 1983;48:667–678. doi: 10.1101/sqb.1983.048.01.070. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Engvall E, Butkowski R, Hunter DD. Molecular heterogeneity of basal laminae: isoforms of laminin and collagen IV at the neuromuscular junction and elsewhere. J Cell Biol. 1990;111:1685–1699. doi: 10.1083/jcb.111.4.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Kleinman HK, Huber H, Deutzmann R, Yamada Y. Laminin, a multidomain protein: the A chain has a unique globular domain and homology with the basement membrane proteoglycan and the laminin B chains. J Biol Chem. 1988;263:16536–16544. [PubMed] [Google Scholar]
- Sewry CA, Chevallay M, Tome FMS. Expression of laminin subunits in human fetal skeletal muscle. Histochem J. 1995;27:497–504. [PubMed] [Google Scholar]
- Slee J. The morphology and development of ragged—a mutant affecting the skin and hair of the house mouse. II. Genetics, embryology and gross juvenile morphology. J Genet. 1957;55:570–584. [Google Scholar]
- Sorokin L, Ekblom P. Development of tubular and glomerular cells of the kidney. Kidney Int. 1992;41:657–664. doi: 10.1038/ki.1992.101. [DOI] [PubMed] [Google Scholar]
- Sorokin LM, Conzelmann S, Ekblom P, Battaglia C, Aumailley M, Timpl R. Monoclonal antibodies against laminin A chain fragment E3 and their effects on binding to cells and proteoglycan and on kidney development. Exp Cell Res. 1992;201:137–144. doi: 10.1016/0014-4827(92)90357-e. [DOI] [PubMed] [Google Scholar]
- Sunada Y, Bernier SM, Kozak CA, Yamada Y, Campbell KP. Deficiency of merosin in dystrophic dy mice and genetic linkage of laminin M chain gene to dylocus. J Biol Chem. 1994;269:13729–13732. [PubMed] [Google Scholar]
- Sung U, O'Rear JJ, Yurchenco PD. Cell and heparin binding in the distal long arm of laminin: identification of active and cryptic sites with recombinant and hybrid glycoprotein. J Cell Biol. 1993;123:1255–1268. doi: 10.1083/jcb.123.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpl R. Macromolecular organization of basement membranes. Curr Opin Cell Biol. 1996;8:618–624. doi: 10.1016/s0955-0674(96)80102-5. [DOI] [PubMed] [Google Scholar]
- Timpl R, Rhode H, Robey PG, Rennard SI, Foidart JM, Martin GR. Laminin—A glycoprotein from basement membranes. J Biol Chem. 1979;254:9933–9937. [PubMed] [Google Scholar]
- Vachon PH, Loechel F, Xu H, Wewer UM, Engvall E. Merosin and laminin in myogenesis; specific requirement for merosin in myotube stability and survival. J Cell Biol. 1996;134:1483–1497. doi: 10.1083/jcb.134.6.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrando P, Partouche O, Pisani A, Ortonne J-P. The 6/2 (AA3) polyclonal antibody identifying a 37 kD keratinocyte protein reacts also with BM-600/nicein, the basement membrane component bound by the monoclonal antibody GB3. Exp Derm. 1992;1:52–58. doi: 10.1111/j.1600-0625.1992.tb00072.x. [DOI] [PubMed] [Google Scholar]
- Virtanen I, Laitinen L, Korhonen M. Differential expression of laminin polypeptides in developing and adult human kidney. J Histochem Cytochem. 1995;43:621–628. doi: 10.1177/43.6.7769233. [DOI] [PubMed] [Google Scholar]
- Virtanen I, Laitinen A, Tani T, Paakko P, Laitinen LA, Burgeson RE, Lehto V-P. Differential expression of laminins and their integrin receptors in developing and adult human lung. Am J Respir Cell Mol Biol. 1996;15:184–196. doi: 10.1165/ajrcmb.15.2.8703474. [DOI] [PubMed] [Google Scholar]
- Vuolteenaho R, Nissinen M, Sainio K, Byers M, Eddy R, Hirvonen H, Shows TB, Sariola H, Engvall E, Tryggvason K. Human laminin M chain (merosin): complete primary structure, chromosomal assignment, and expression of the M and A chain in human fetal tissues. J Cell Biol. 1994;124:381–394. doi: 10.1083/jcb.124.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie TM, Gilbert DJ, Olsen AS, Chen X-N, Amatruda TT, Korenberg JR, Trask BJ, de Jong P, Reed RR, Simon MI, et al. Evolution of the mammalian G protein α subunit multigene family. Nat Genet. 1992;1:85–91. doi: 10.1038/ng0592-85. [DOI] [PubMed] [Google Scholar]
- Xu H, Wu X-R, Wewer UM, Engvall E. Murine muscular dystrophy caused by a mutation in the laminin α2 (Lama2)gene. Nat Genet. 1994;8:297–302. doi: 10.1038/ng1194-297. [DOI] [PubMed] [Google Scholar]
- Yamada KM, Miyamoto S. Integrin transmembrane signaling and cytoskeletal control. Curr Opin Cell Biol. 1995;7:681–689. doi: 10.1016/0955-0674(95)80110-3. [DOI] [PubMed] [Google Scholar]
- Yarnitzky T, Volk T. Laminin is required for heart, somatic muscles, and gut development in the Drosophilaembryo. Dev Biol. 1995;169:609–618. doi: 10.1006/dbio.1995.1173. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD, O'Rear JJ. Basal lamina assembly. Curr Opin Cell Biol. 1994;6:674–681. doi: 10.1016/0955-0674(94)90093-0. [DOI] [PubMed] [Google Scholar]