Abstract
Cell adhesion molecules of the Ig superfamily are implicated in a wide variety of biological processes, including cell migration, axon guidance and fasciculation, and growth control and tumorigenesis. Expression of these proteins can be highly dynamic and cell type specific, but little is known of the signals that regulate such specificity. Reported here is the molecular cloning and characterization of rat CDO, a novel cell surface glycoprotein of the Ig superfamily that contains five Ig-like repeats, followed by three fibronectin type III–like repeats in its extracellular region, and a 256-amino acid intracellular region that does not resemble other known proteins. In rat embryo fibroblasts, cdo mRNA expression is maximal in confluent, quiescent cells. It is rapidly and transiently down-regulated by serum stimulation of such cells, and is constitutively down-regulated in oncogene-transformed derivatives of these cells. CDO protein levels are also dramatically regulated by cell–substratum adhesion, via a mechanism that is independent of cdo mRNA expression. The amount of CDO produced at the surface of a cell may therefore be governed by a complex balance of signals, including mitogenic stimuli that regulate cdo mRNA levels, and substratum-derived signals that regulate CDO protein production. cdo mRNA is expressed at low levels in most adult rat tissues. A closely related human gene maps to chromosome 11q23–24, a region that displays frequent loss of heterozygosity in human lung, breast, and ovarian tumors. Taken together, these data suggest that loss of CDO function could play a role in oncogenesis.
Cell adhesion molecules (CAMs)1 of the Ig superfamily (IgSF) are implicated in cell–cell interactions that mediate, in part, a wide variety of developmental, physiological, and pathological processes (for reviews see Edelman and Crossin, 1991; Hynes and Lander, 1992; Brummendorf and Rathjen, 1995; Cunningham, 1995). In general, CAMs of the IgSF mediate Ca2+-independent, homo- and heterophilic cell–cell adhesion. These molecules typically possess an extracellular region that consists of several Ig-like domains followed, in many cases, by a number of fibronectin type III (FNIII)–like domains. Members of the IgSF that contain both Ig-like and FNIII-like repeats (e.g., neural cell adhesion molecule (N-CAM), L1, F11/contactin, etc.) have been mainly implicated in developmental processes in the nervous system, including cell migration, neurite extension, and axon guidance and fasciculation (Hynes and Lander, 1992; Tessier-Lavigne and Goodman, 1996).
Cell–cell recognition also plays an important role in the proliferative capacity of cells both inside and outside the nervous system, and Ig/FNIII family members may be involved in this phenomenon as well. For example, cell surface N-CAM mediates contact inhibition of growth of a mouse fibroblast cell line (Aoki et al., 1991), and addition of soluble N-CAM to primary cultures of rat astrocytes specifically inhibits their proliferation (Sporns et al., 1995).
Consistent with a role for CAMs in controlling cell proliferation, a large body of evidence suggests that alterations in adhesion-mediated cell regulation may play a role in induction and/or maintenance of the neoplastic phenotype (for review see Hedrick et al., 1993). It has been known since the 1940s that cancer cells display altered adhesiveness when compared to their normal counterparts (Coman, 1944). Moreover, malignant tumors exhibit a loss of appropriate tissue architecture, altered cell shape, cell crowding, and invasiveness. Tumor cell lines, or cells transformed in vitro, show a loss of contact inhibition of growth and proliferate in an anchorage-independent fashion. Although the molecular bases of these phenotypes are still poorly understood, they may arise, in part, because of loss of expression or mutational inactivation of genes that encode CAMs (Hedrick et al., 1993; Berx et al., 1996).
A candidate tumor suppressor gene, DCC, encodes a member of the Ig/FNIII family containing four Ig-like repeats, and six FNIII-like repeats in its extracellular region (Fearon et al., 1990; Hedrick et al., 1994; for reviews see Fearon and Pierceall, 1995; Fearon, 1996). DCC was isolated by positional cloning in a region of chromosome 18q that commonly displays loss of heterozygosity (LOH) in colorectal cancers. DCC was included in the region of allelic loss in >90% of colorectal cancers that exhibited LOH of 18q. Furthermore, loss of DCC protein expression in colorectal tumors is a negative prognostic marker of survival rate in patients with stage II and stage III disease (Shibata et al., 1996). Additional studies of DCC expression and allelic loss indicated that this gene may also be involved in many additional types of cancer and may play a role in cell differentiation (Fearon, 1996). DCC was also recently shown to function as an evolutionarily conserved receptor or component of a receptor for netrin-1, a secreted and membrane-associated protein that acts as a guidance cue for migrating axons in the developing nervous system (Chan et al., 1996; Keino-Masu et al., 1996; Kolodziej et al., 1996). These data raise the possibility that loss of DCC function could alter the motility of cancer cells.
An additional IgSF member implicated in carcinogenesis is cell–cell adhesion molecule (C-CAM)/biliary glycoprotein, which contains four Ig-like domains in its extracellular region. Expression of the C-CAM/Bgp-1 gene was down-regulated in colorectal and prostate malignancies (Kleinerman et al., 1995; Neumaier et al., 1993), and ectopic restoration of its expression suppressed tumorigenicity of cell lines derived from such tumors (Hsieh et al., 1995; Kunath et al., 1995).
It is clear from the studies cited above that CAMs of the IgSF play a role in coordinating normal cellular architecture, motility, and growth control, and that loss of function of such molecules may play an important role in multistage carcinogenesis. We have previously developed and characterized a panel of somatic cell mutant lines derived from Rat 6 embryo fibroblasts that are specifically resistant to the induction of anchorage-independent growth by various oncogenes (Krauss et al., 1992; Feinleib and Krauss, 1996; Kang and Krauss, 1996). Because the oncogene-resistant phenotype of these cells was dominant in somatic cell hybridizations, we sought to isolate genes whose expression is up-regulated in the mutant cell lines relative to a transformation-sensitive control cell line. Reported here is the molecular cloning and characterization of a rat gene designated cdo (CAM-related/down-regulated by oncogenes), which encodes a novel transmembrane member of the Ig/FNIII family. In rat fibroblasts, cdo expression is maximal in confluent, quiescent cells. cdo mRNA and protein levels are strongly, but transiently, reduced when such cells are stimulated to reenter the cell cycle and are constitutively down-regulated in oncogene-transformed derivatives of these cells. Furthermore, CDO protein levels are strikingly regulated by cell–substratum adhesion, with a mechanism that is independent of cdo mRNA expression. A closely related human gene maps to chromosome 11q23–24, a region that displays LOH in human lung, breast, and ovarian tumors. Taken together, these data suggest that loss of CDO function might play a role in oncogenesis.
Materials and Methods
Cell Culture
All Rat 6–derived cell lines were routinely maintained in DME plus 10% bovine calf serum, as previously described (Krauss et al., 1992). NIH 3T3 and NIH 3T3/ras cells were cultured in DME plus 5% FBS. Soft agar assays and preparative methylcellulose cultures were performed as previously described (Kang and Krauss, 1996). Cells were harvested from preparative methylcellulose cultures 48 h after inoculation into the semisolid medium, and were >98% viable as assessed by trypan blue dye exclusion and replating assays (Kang and Krauss, 1996). For serum stimulation experiments, confluent cultures of Rat 6 cells were washed twice with PBS, and fed with DME plus 0.1% calf serum. After 48 h, the cultures were refed with DME plus 20% calf serum and harvested for analysis at various time points thereafter.
RNA Isolation, Library Construction, and Screening
RNA used for cDNA library construction was prepared by the guanidine thiocyanate/cesium chloride centrifugation method (Chirgwin et al., 1979), and the poly(A)+ fraction was isolated by oligo(dT)-cellulose chromatography (Aviv and Leder, 1972). RNA used for Northern blot analyses was isolated either as described above or with the TRIzol reagent (GIBCO BRL, Gaithersburg, MD), which was used according to the manufacturer's instructions.
An oligo(dT)-primed cDNA library was constructed in λZAPII (Stratagene, La Jolla, CA) with mRNA isolated from the transformation-resistant, mutant cell line, R-1a (Krauss et al., 1992). The library was constructed with the SuperScript Choice system kit from GIBCO BRL, and was screened by differential hybridization (Johnson et al., 1987) with 32P-labeled cDNA prepared from R-1a and control, transformation-sensitive PKC3-F4 cells. One clone that exhibited a greater signal with R-1a–derived cDNA, relative to PKC3-F4–derived cDNA, represented a partial cDNA for cdo. Clones representing a nearly full-length cDNA were isolated from a random-primed λZAPII library constructed with mRNA from Rat 6 cells. A human cDNA that is closely related to rat cdo was isolated by screening human fetal lung and fetal brain cDNA libraries (obtained from A. Chan and S. Aaronson, Mount Sinai School of Medicine; and Clontech, Palo Alto, CA, respectively) with the full length rat cdo cDNA.
DNA Sequencing and Computer Analysis
DNA sequencing was carried out by a combination of manual and automated methods. Manual sequencing was performed with a Sequenase 2.0 kit (United States Biochemical Corp., Cleveland, OH). Automated sequencing was performed at the Mount Sinai Molecular Biology Core Facility on an Applied Biosystems 373 computerized DNA sequencer (Foster City, CA) or at the Biotechnology Center at Utah State University (Logan, UT). Searches of the DNA databases were performed with the BLAST server service, and sequence analysis and alignments were carried out with MacVector (International Biotechnologies, Inc., New Haven, CT) and GCG software (Genetics Computer Group, Madison, WI).
RNA Analyses
Northern blot analyses were performed by fractionating total cellular or poly(A)+ RNA through agarose/formaldehyde gels, blotting to nylon membranes, and hybridizing with DNA probes as described by Krauss et al. (1992).
For the study of cdo expression in adult rat tissues, PCR-based “exon-connection” assays were carried out (Fearon et al., 1990). A portion of the cdo gene was isolated from a rat genomic library (a gift of S. Salton, Mount Sinai School of Medicine) and a partial exon/intron structure deduced over a ∼15-kb region. Random primers and reverse transcriptase were used to generate cDNA from total RNA derived from rat tissues. PCR primers (5′-CACACAGTCAGAAGCGTTCCTC-3′ [sense], and 5′-AGGATAAGAGGCTACCACTGGG-3′ [antisense]) directed against sequences from separate exons were then used to amplify a cDNA product that linked these exons. PCR reactions were fractionated on agarose gels, blotted to nylon membranes, and probed with a cdo cDNA probe. To analyze differential splicing of the fifth Ig repeat of cdo in Rat 6 cells, reverse transcription–PCR was performed on total Rat 6 cell RNA with the following primers: 5′-GGAAGCAACTGGAGAAGG-3′ (sense) and 5′-GTCTCGTTCATCGTTCTG-3′ (antisense). The products were analyzed by agarose gel electrophoresis and ethidium bromide staining.
Flow Cytometry
Cells were stained with propidium iodide buffer (50 μg/ml propidium iodide, 0.1% sodium citrate, 0.1% Triton X-100) in the presence of 10 μg of DNase-free RNase and analyzed by flow cytometry on a fluorescence-activated cell sorter (FACS®can; Becton and Dickinson, Co., Mountain View, CA). The data were analyzed by the CellFIT software program (Becton and Dickinson, Co., Mountain View, CA).
Antibodies and Western Blot Analyses
A portion of the intracellular region of rat cdo (amino acids 1057–1241) was ligated into pGEX-5X-1 (Pharmacia LKB Biotechnology Inc., Piscataway, NJ) to produce a glutathione-S-transferase fusion protein. Fusion protein produced in Escherichia coli was purified by SDS-PAGE and electroelution. Rabbit polyclonal antibodies to CDO protein were generated with a proprietary immunization regimen referred to as PolyQuikTM (Zymed Laboratories Inc., South San Francisco, CA). Crude antisera were used for Western blot analyses.
Immunoblot analyses were performed essentially as described in Kang et al. (1996). Cells from monolayer or methylcellulose suspension cultures were harvested in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl, pH 7.2,150 mM NaCl/1% Triton X-100/1% sodium deoxycholate/ 0.1% SDS/1 mM EGTA) containing 1 mM PMSF, 10 ng/ml leupeptin, 50 mM NaF, and 1 mM sodium orthovanadate. Total proteins were then separated by SDS-PAGE, transferred to nitrocellulose membranes (Amersham Corp., Arlington Heights, IL), and probed with anti-CDO antiserum. The blots were washed and reprobed with HRP-conjugated secondary antibody and specific protein bands visualized with the ECL chemiluminescent detection system (Amersham Corp.) as described previously (Kang, et al., 1996).
Surface Labeling and Tunicamycin Treatment
To surface biotinylate Rat 6–derived cultures, cells were detached in 0.04% EDTA in PBS, washed with serum-free DME, and incubated on ice with serum-free DME plus 50 μM sulfo-NHS-LC-biotin (Pierce Chemical Co., Rockford, IL) for 0.5 h. Cells were then washed again and lysed in RIPA buffer, and CDO was immunoprecipitated. Western blotting was then performed as described above, except that HRP-linked streptavidin was used for detection. To assess N-glycosylation of CDO, Rat 6 cells were treated with tunicamycin (5 μg/ml) for 48 h, lysed, and analyzed by Western blotting.
Ectopic Expression of cdo
A complete rat cdo open-reading frame, including 95 bp of the 5′ untranslated region and 633 bp of the 3′ untranslated region, was reconstructed from overlapping bacteriophage inserts and ligated into the retroviral expression vector, pBabePuro (Morganstern and Land, 1990). Production of recombinant retroviruses and infection of Rat 6–derived cell lines was performed as previously described (Krauss et al., 1992). Infected cultures were selected in medium that contained 5 μg puromycin per ml, and >1,000 independent, drug-resistant colonies were pooled and analyzed as described in Results.
Chromosomal Localization of Human cdo
A portion of the human cdo gene was PCR amplifed from human monochromosomal somatic cell hybrids. The National Institute of General Medical Sciences human/rodent somatic cell hybrid mapping panel No. 2, version 3 (Coriell Institute for Medical Research, Camden, NJ), was used for chromosomes 1, 6–9, 11, 12, 14, 16, 17, 20, 22, X, and Y. The remaining chromosomes were represented by monochromosomal hybrids: GM11686 (chr. 2); GM11713 (chr. 3); GM11687 (chr. 4); GM11714 (chr. 5); GM11688 (chr. 10); GM11689 (chr. 13); GM11715 (chr. 15); GM12082 (chr. 18); GM10612 (chr. 19); and GM08854 (chr. 21) (Coriell Institute for Medical Research). PCR amplifications were performed with the following primers: 5′-CACAACAGGCTGGGAGCGA-3′ (sense), and 5′-GGGGAAGAAAACATTTAAG-3′ (antisense). The identity of the chromosome 11 monochromosomal hybrid was confirmed by PCR of the previously mapped locus catalase (11p13) (Theune et al., 1991). Human cdo was further localized with a chromosome 11 regional mapping panel (Coriell Institute for Medical Research).
Results
Isolation, Sequence Analysis, and Tissue Distribution of cdo
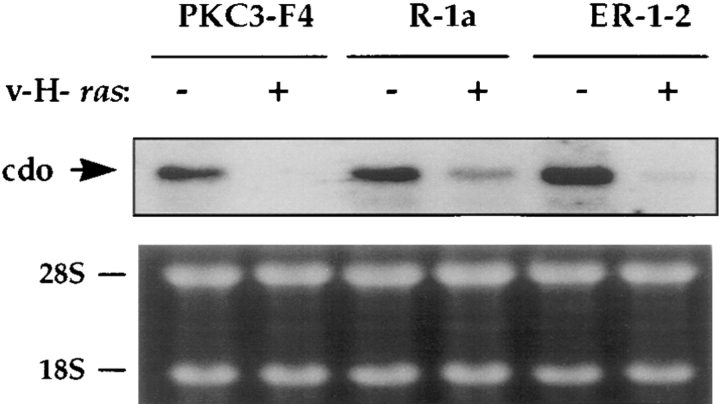
The R-1a and ER-1-2 mutant rat embryo fibroblast cell lines are resistant to induction of anchorage-independent growth by various oncogenes, and this property is dominant in somatic cell hybridizations (Krauss et al., 1992). Because this dominant resistance could have arisen by the stable expression (or overexpression) of a transformation suppressor gene, a cDNA library constructed from R-1a cells was screened by differential hybridization to isolate genes whose expression was enhanced in R-1a cells, relative to a transformation-sensitive control cell line, PKC3-F4. One cDNA clone isolated in this screen, later named cdo, hybridized to an ∼8.5-kb mRNA species that reproducibly showed a mild (1.5–2-fold) up-regulation in both R-1a and ER-1-2 cells, relative to the control PKC3-F4 cells (Fig. 1). Interestingly, cdo mRNA levels were dramatically down-regulated in PKC3-F4 cells that had been infected with a v-H-ras–expressing retrovirus. This down-regulation was incomplete in similarly infected cultures of the ras-resistant R-1a and ER-1-2 cell lines (Fig. 1).
Figure 1.
Northern blot analysis of cdo mRNA expression in the PKC3-F4, R-1a, and ER-1-2 cell lines, plus or minus infection with a v-H- ras–expressing retrovirus. + and −, expressing or not expressing v-H-ras, respectively. The ethidium bromide–stained gel (displaying the 28S and 18S ribosomal RNA bands), is shown as a loading control.
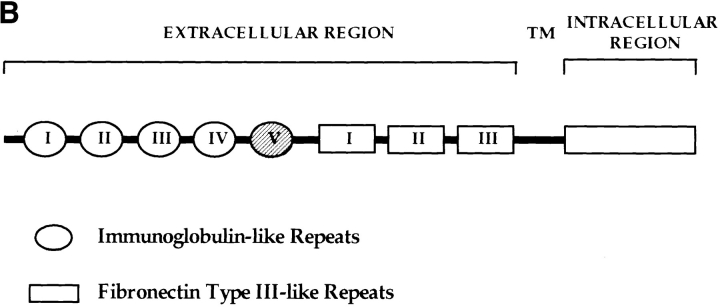
Sequence analysis of overlapping cDNAs revealed cdo to be a member of the Ig/FNIII repeat family. Fig. 2 A shows the predicted amino acid sequence of rat cdo; also shown is the amino acid sequence of a closely related human gene (see below). The rat cdo mRNA is predicted to encode a 1,242–amino acid protein, comprised of a 24– amino acid hydrophobic signal sequence followed by an extracellular region of five Ig V–like repeats (Vaughn and Bjorkman, 1996) and three FNIII-like repeats, a 25-residue, membrane-spanning region, and a putative intracellular region of 256 amino acids. A hydropathy plot (not shown) is consistent with a single membrane-spanning region, suggesting that cdo encodes a transmembrane protein with a structure similar to members of the N-CAM family. A schematic representation of the predicted CDO structure is shown in Fig. 2 B. While there are no other IgSF members with this numerical combination of Ig- and FNIII-like repeats in the current databases, individual Ig and FNIII repeats in cdo appeared to be most closely related to such repeats found in rat NB-2 and NB-3 (Ogawa et al., 1996), members of the F11/contactin/F3 subfamily of CAMs (up to 50% amino acid similarity; data not shown). Interestingly, however, the putative intracellular region of cdo showed no significant resemblance to any gene currently in the standard databases. A proline-rich stretch in the intracellular region that might serve as a potential SH3 domain binding site is indicated in Fig. 2 A. 10 potential N-linked gycosylation sites present in the extracellular region are also indicated.
Figure 2.
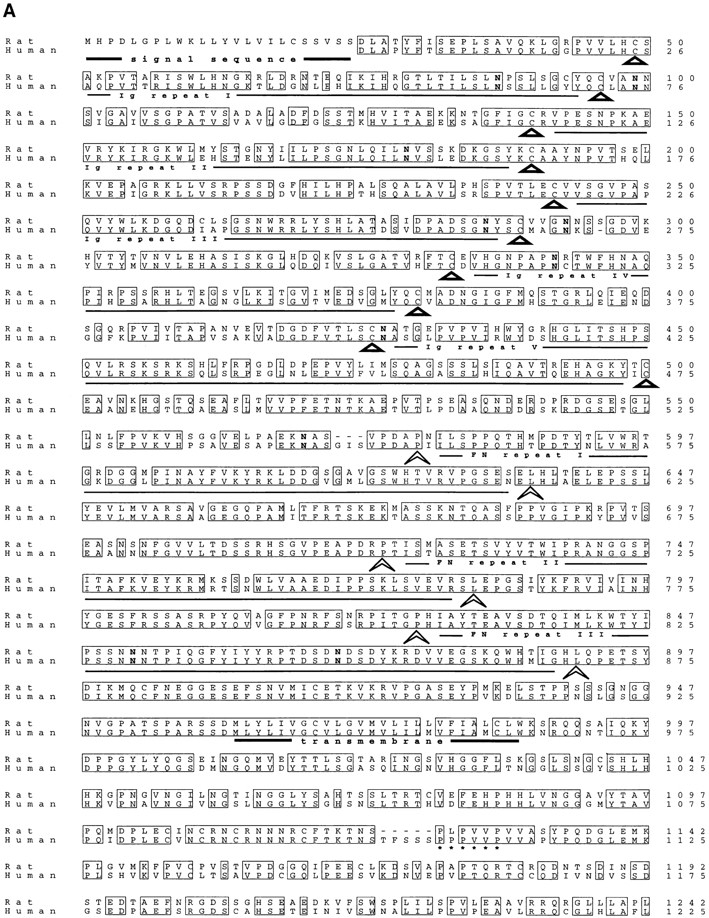
(A) Predicted amino acid sequence of rat and human CDO. Identical residues are boxed. The five Ig-like repeats are indicated by the triangles, which correspond to the two conserved cysteines present in each domain (Williams and Barclay, 1988). The three FNIII-like repeats are indicated by the arrowheads, which correspond to the conserved prolines and leucines at the end of each domain (Patthy, 1990). The putative signal sequence and transmembrane regions are underlined in bold. 10 asparagines in the extracellular region that represent potential sites of N-linked glycosylation are in bold print. A proline-rich stretch in the intracellular region is underscored with asterisks. These sequence data are available from EMBL/ GenBank/DDBJ, under accession numbers AF004840 (rat), and AF004841 (human). (B) Schematic representation of CDO structure, which contains five Ig-like repeats, three FNIII-like repeats, a transmembrane region (TM), and a 256–amino acid intracellular region. Ig repeat 5 (cross-hatched) is alternatively spliced.
Sequencing of multiple, independent cDNAs revealed a potential for alternative splicing of the fifth Ig repeat, leading to removal of amino acids 400–516. Reverse transcriptase–PCR analysis of total Rat 6 cell RNA with primers that flank this region produced a major product corresponding to mRNA that encodes the fifth Ig domain, and a minor product corresponding to mRNA that lacks this domain (data not shown). Consistent with the notion that this is a minor splice variant in Rat 6 cells, only one major form of cdo mRNA and protein was detected by Northern and Western blot analyses of these cells (see below).
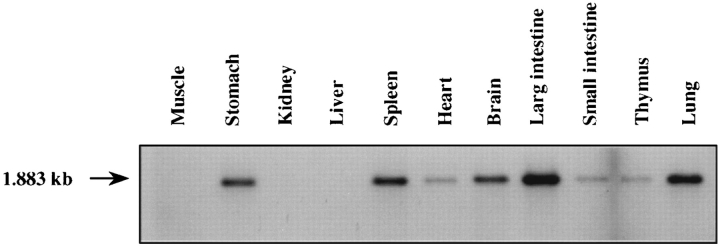
Expression of cdo in adult rat tissues was analyzed initially by Northern blotting of poly(A)+ RNA derived from brain, liver, kidney, heart, large and small intestines, spleen, thymus, lung, stomach, and skeletal muscle, but only a very faint signal could be detected. This result was reminiscent of that observed for the related gene, DCC. We chose, therefore, to use the “exon-connection” strategy originally used by Fearon et al. (1990) to analyze DCC expression. A product of the predicted size was found in all of the tissues examined except liver, kidney, and skeletal muscle (Fig. 3). Thus, cdo is expressed at low levels in most adult tissues.
Figure 3.
Expression of cdo in adult rat tissues. PCR-based “exon-connection” assays for detection of cdo transcripts were performed as described in the text.
Regulation of cdo mRNA by Oncogenes and Serum
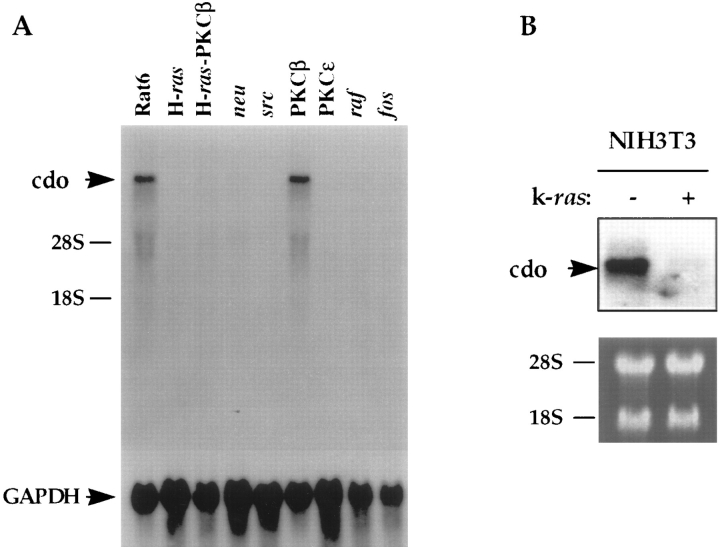
The PKC3-F4, R-1a, and ER-1-2 cell lines all share the same original parental cell line, Rat 6. Expression of cdo mRNA in Rat 6 cells, as well as in Rat 6 cells transformed by different oncogenes, was therefore assessed. Transformation of Rat 6 cells by the H-ras, neu, v-src, v-raf, protein kinase C ε, or v-fos oncogenes led to a dramatic loss of expression of cdo (Fig. 4 A). Each of these oncogene-expressing cell lines was grossly morphologically transformed and formed large colonies in soft agar (Borner et al., 1992). In contrast, overexpression of protein kinase C β1, which led to enhanced mitogen-induced signaling but was a very weak transforming gene (Housey et al., 1988; Borner et al., 1995), had little effect on cdo expression. Furthermore, hybridization of a rat cdo cDNA probe to Northern blots of RNA from the murine embryo fibroblast line, NIH 3T3, revealed an mRNA species that was indistinguishable in size from that seen in Rat 6 cells, and this mRNA was nearly undetectable in NIH 3T3 cells transformed by a K-ras oncogene (Fig. 4 B). Thus, down-regulation of cdo mRNA expression correlated with establishment of the transformed phenotype by several different oncogenes in Rat 6 cells and by ras oncogenes in two different cell lines.
Figure 4.
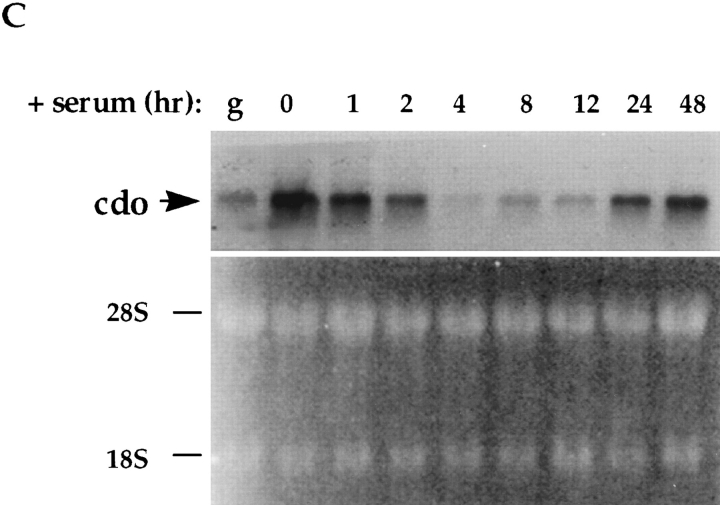
Effect of oncogenes and serum on cdo mRNA expression. (A) Northern blot analysis of cdo expression in Rat 6 cells and various oncogene-transformed Rat 6 cell derivatives. The lane designations refer to the oncogene expressed in the given Rat 6 cell derivative. The same blot was rehybridized with a rat glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA probe as a loading control. (B) Northern blot analysis of cdo expression in NIH 3T3 cells, and NIH 3T3 cells transformed by the k-ras oncogene. + and −, expressing or not expressing k-ras, respectively. The ethidium bromide–stained gel, displaying the 28S and 18S ribosomal RNA bands, is shown as a loading control. (C) Northern blot analysis of the time course of cdo mRNA expression in serum-stimulated Rat 6 cells. Confluent cultures of wild-type Rat 6 cells were serum starved (0 h) and then fed with fresh, serum-containing medium. Lane designations represent the time, in hours, after serum stimulation; lane g represents cells growing logarithmically in serum-containing medium. The ethidium bromide–stained gel, displaying the 28S and 18S ribosomal RNA bands, is shown as a loading control.
The possibility that cdo expression might be down-regulated by mitogen treatment was examined next. Fig. 4 C demonstrates that when confluent, serum-starved Rat 6 cells were stimulated by refeeding with fresh, serum-containing medium, cdo expression was nearly extinguished within 4 h. Expression began to return within 8–12 h, and by 24–48 h, it had been restored to the level of the original cultures. Parallel cultures that were analyzed for cell cycle progression by flow cytometry indicated that >70% of the cells were in S phase 16 h after serum stimulation, and that the cultures were fully quiescent again 24–48 h after-stimulation (data not shown). Thus, steady-state levels of cdo mRNA were transiently down-regulated as Go-arrested cells reentered the cell cycle and were restored as the cultures regained quiescence. Fig. 4 C also shows that logarithmically growing cells had lower levels of cdo mRNA than did confluent, serum-starved cells. This appeared not to be due simply to cell proliferation, per se, or solely to the fact that these growing cultures were in the presence of serum, because growth arrest of such cultures by removal of serum did not significantly increase cdo expression (data not shown). These data indicate that cell density and/or cell–cell contact might also play a role in cdo mRNA regulation.
Analysis of CDO Protein in Rat 6 Cells
A complete cdo open reading frame was inserted into the retroviral expression vector, pBabePuro. This vector was transiently transfected into the human embryonic kidney cell line, 293, and Western blot analysis performed on total cell lysates with polyclonal rabbit antisera raised against the intracellular region of cdo. A protein of ∼190 kD was detected in transfected cells, but not in mock-transfected controls (Fig. 5 A). An immunoreactive protein with similar mobility was detected in Rat 6 cell lysates (Fig. 5 A). A nonspecific, cross-reactive band of lower molecular weight is also visible in mock-transfected 293 cells and, frequently, in Rat 6 cells. Consistent with the RNA analyses described above, CDO protein levels were dramatically down-regulated in ras-transformed Rat 6 cells (C1-T24 cells; Fig. 5 B). Likewise, serum stimulation of quiescent Rat 6 cultures led to transient down-regulation of CDO protein levels (Fig. 5 C). It was not possible to perform similar studies with NIH 3T3 cells, because the antibodies to CDO appear to be specific for rat.
Figure 5.
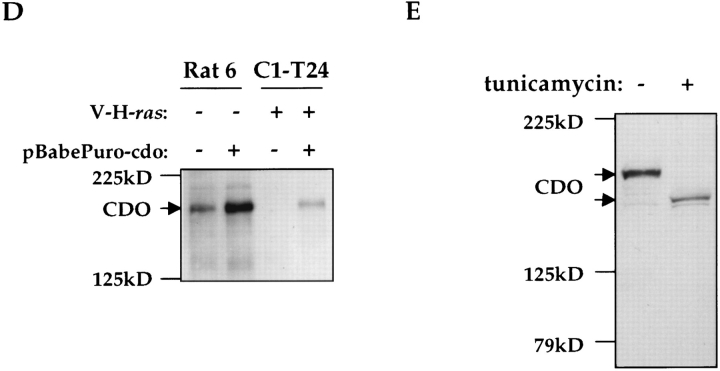
Western blot analyses of CDO protein expression with antisera against the CDO intracellular region. (A) Transient transfection of the pBabePuro/cdo vector into 293 cells. + and −, cells were transfected or mock transfected with the expression vector, respectively. (B) Ras-mediated down-regulation of CDO protein levels in Rat 6 cells. C1-T24 cells are a ras-transformed Rat 6 cell subclone. (C) Serum-mediated down-regulation of CDO protein levels in Rat 6 cells. Confluent cultures of wild-type Rat 6 cells were serum starved (0 h) and then fed with fresh, serum-containing medium. Lane designations represent the time, in hours, after serum stimulation; note that the times after stimulation at which CDO protein was analyzed are not identical to those used in analysis of cdo mRNA in Fig. 2. (D) Cell surface expression of CDO. Rat 6 and C1-T24 cell derivatives were surface biotinylated and detected on Western blots with HRP-linked streptavidin. + and −, cells did, or did not, stably express the pBabePuro/cdo vector. (E) N-linked glycosylation of CDO. + and −, Rat 6 cells treated or not treated with tunicamycin, respectively, and then analyzed by immunoblotting with CDO antisera.
The amino acid sequence of cdo predicts a transmembrane protein. To confirm that CDO was expressed on the cell surface, various Rat 6 cell derivatives were surface biotinylated. CDO was then immunoprecipitated and detected on Western blots with HRP-coupled streptavidin. As shown in Fig. 5 D, a band of the appropriate molecular weight was observed in Rat 6 cells, but was not present in ras-transformed C1-T24 cells. Furthermore, the signal intensity of the band was increased in Rat 6 cells that overproduce CDO, and the band was restored in C1-T24 cells that ectopically produce this protein (see below for a description of these cell lines). It is concluded that CDO is expressed on the cell surface.
Members of the IgSF are typically modified in their extracellular region by N-linked glycosylation (Brummendorf and Rathjen, 1995). To test whether CDO is also N-glycosylated, Rat 6 cells were treated with tunicamycin, an inhibitor of this process. CDO from treated cells displayed a significant decrease in apparent molecular weight (Fig. 5 E), consistent with the notion that it is a glycoprotein.
Effect of Cell–Substratum Adhesion on cdo RNA and Protein Expression
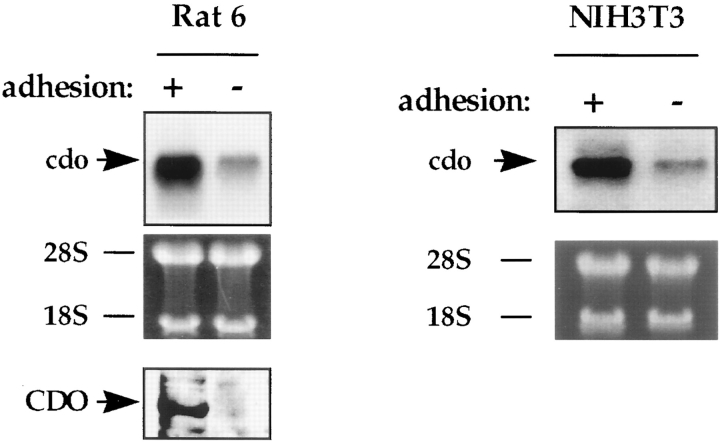
The expression pattern of cdo in Rat 6 and NIH 3T3 cells (Fig. 4) suggests that cdo might encode a protein with growth inhibitory or transformation suppressor function. Because the primary defect in the R-1a and ER-1-2 mutant cell lines is an inability to proliferate in an anchorage-independent manner in response to ras (Krauss et al., 1992; Kang and Krauss, 1996), it was of interest to determine the effects of cell–substratum adhesion on the expression of cdo. Rat 6 cells suspended in methylcellulose-containing medium had significantly lower steady state levels of cdo mRNA than did parallel adherent cultures. A similar effect was observed in NIH 3T3 cells (Fig. 6). cdo mRNA levels were consistently lower in suspension cultures than in adherent cultures of low cell density (data not shown). It seems likely, however, that the decrease in steady state cdo mRNA levels seen in the methylcellulose cultures of Rat 6 cells may reflect a combination of two phenomena: (a) loss of cell anchorage; and (b) the low cell density typically used for these suspension cultures, since decreased cell density in adherent cultures also led to partially reduced levels of cdo mRNA (Fig. 4 C, and see above).
Figure 6.
Regulation of cdo mRNA and protein by cell–substratum adhesion in Rat 6 and NIH 3T3 cells. + and −, cells were cultured on plastic dishes or suspended in methylcellulose-containing medium, respectively. CDO protein and cdo mRNA were analyzed by Western and Northern blotting techniques, respectively. For the Northern blots, the ethidium bromide–stained gel, displaying the 28S and 18S ribosomal RNA bands, is shown as a loading control.
Although cdo mRNA levels were down-regulated in nonadherent Rat 6 cells, some level of expression was always detectable in these cultures. In contrast, CDO protein could not be detected in suspension cultures (Fig. 6). These data raised the possibility that CDO protein levels might be regulated by cell–substratum adhesion by a mechanism that is independent of cdo mRNA expression. Additional experiments that address this point are described below.
Ectopic Expression of cdo
To test the hypothesis that cell–substratum adhesion can regulate CDO protein levels independently of cdo mRNA expression, we sought to express a cdo cDNA from a promoter whose activity was not influenced by cell culture conditions. To achieve this, parental Rat 6 cells and a ras-transformed derivative, C1-T24, were infected with a recombinant retrovirus that drives expression of cdo from the viral long terminal repeat. The cells were infected with either the cdo-expressing virus or, as a control, a virus that lacked a cDNA insert. Infectants were then pooled and the polyclonal populations were analyzed.
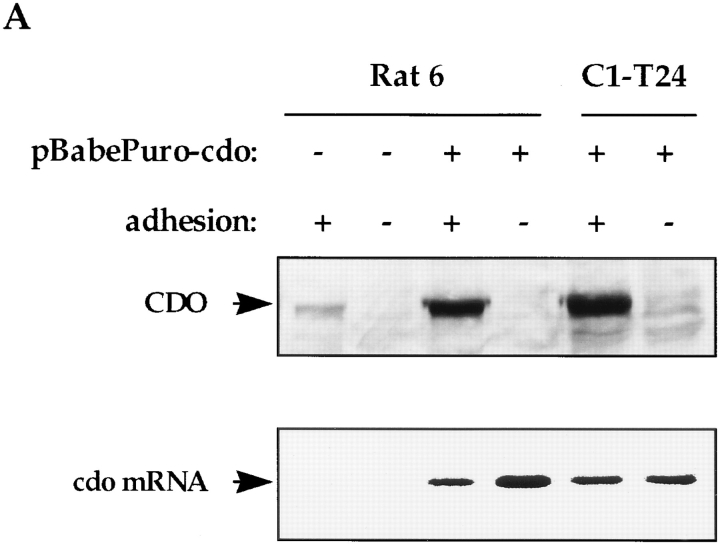
Ectopic expression of cdo was assessed by Northern and Western blot analyses of infected cells cultured under both adherent and nonadherent conditions. As shown in Fig. 7 A, Rat 6 and C1-T24 cells that were infected with the cdo virus (Rat 6/cdo and C1-T24/cdo cells, respectively) expressed abundant, vector-derived mRNA whether they were attached to plates or suspended in methylcellulose. When adherent cultures were tested, Rat 6/cdo cells produced significantly more immunoreactive CDO protein than control vector-infected cells (Fig. 7 A). C1-T24/cdo cells also produced large quantities of CDO protein when grown on tissue culture plates (Fig. 7 A). Control vector-infected C1-T24 cells expressed no detectable CDO, similar to the parental C1-T24 cell line shown in Fig. 5 B (see also Fig. 7 B). Interestingly, no CDO protein was produced when any of these cell lines was cultured under nonadherent conditions, even though they stably expressed exogenous, vector-derived cdo mRNA (Fig. 7 A). We have previously demonstrated that production of several cell cycle proteins is not altered by culturing these cells in methylcellulose-containing medium (Kang and Krauss, 1996). Furthermore, immunoblotting of these same cell extracts with antibodies to a different cell surface protein, the insulin-like growth factor I receptor, did not reveal significant differences between adherent and nonadherent cultures in the expression of this protein (data not shown). Loss of CDO production in nonadherent cultures therefore was not due to a nonspecific effect, nor was it due to undetected damage to the cells. These data are consistent with the data described in Fig. 5, and indicate a requirement, at least in Rat 6 cells, for cell–substratum adhesion for stable production of CDO protein.
Figure 7.
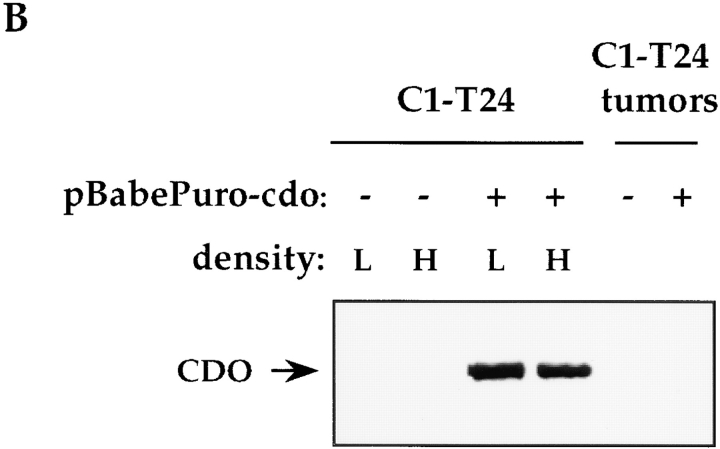
Regulation of ectopic CDO protein production by cell– substratum adhesion. (A) Rat 6 and C1-T24 cells were infected with recombinant pBabePuro/cdo virus (+) or pBabePuro virus lacking a cDNA insert (−), and were cultured on plastic dishes (+ adhesion) or suspended in methylcellulose-containing medium (− adhesion). CDO protein expression and mRNA expression were analyzed by Western and Northern blotting techniques, respectively. Prolonged exposure of the Northern blot revealed the endogenous cdo mRNA species in the first two lanes, similar to the data presented in Fig. 6. (B) C1-T24 cells were infected with recombinant pBabePuro/cdo virus (+) or pBabePuro virus lacking a cDNA insert (−), and were then cultured on plastic dishes at low (L) or high (H) cell density. Also shown are extracts from tumors derived from C1-T24/puro (−) and C1-T24/cdo (+) cells. Samples were analyzed for CDO protein expression by Western blotting techniques. Western blots were also stained with Ponceau S to confirm that protein samples were not degraded.
The failure of Rat 6 cells to produce either endogenous or ectopically expressed CDO protein in the absence of cell–substratum adhesion might have arisen, at least in part, as a result of the low cell density in the methylcellulose cultures used for the nonadherent conditions, rather than as an absolute need for cell anchorage. To distinguish between these possibilities, we cultured C1-T24/cdo cells at low and high cell densities on tissue culture plates and measured CDO levels by immunoblotting. C1-T24/cdo cells are most appropriate for addressing this question because they express only the vector-derived cdo mRNA which, unlike the endogenous cdo gene, is not regulated by cell density or cell anchorage. As shown in Fig. 7 B, adherent cultures of C1-T24/cdo cells harvested at low and high cell densities each produced abundant CDO protein, indicating that loss of cell substratum adhesion was most likely to be responsible for the effects observed in Fig. 7 A.
As would be expected of cells that failed to produce the exogenously derived protein, there was no difference between C1-T24/cdo cells and control vector-infected C1-T24 cells in their frequency of colony formation in soft agar (∼20% cloning efficiency). The tumorigenicity of C1-T24/cdo and control vector-infected C1-T24 cells was also tested. When 106 cells of each cell line were injected into the flanks of nude mice, both produced tumors of 1-cm diam in 10 d. The tumors were then excised, and tested for expression of CDO protein. Similar to what was observed with the nonadherent cultures, tumors derived from C1-T24/cdo cells produced barely detectable levels of CDO (Fig. 7 B), further indicating a need for engagement of the cells with an appropriate substratum for stable production of this protein.
Finally, ectopic expression of CDO protein in adherent cultures of Rat 6/cdo and C1-T24/cdo cells did not alter the gross morphology or growth properties of either cell type when compared with their respective vector control or parental cell lines (data not shown). Notably, no increase in cell–cell aggregation was observed, as might be expected for ectopic expression of a cell surface molecule that mediated homophilic cell adhesion.
It should be noted that the ras-resistant mutant cells also did not produce CDO protein when cultured in suspension (data not shown). Thus, the failure of the mutant cell lines to down-regulate cdo mRNA levels to the same extent observed in transformation-sensitive control cells presumably does not account for their resistance to anchorage-independent growth. These data do not, however, exclude a role for CDO as a transformation suppressor in other systems where stable expression of the protein may occur.
Sequence and Chromosomal Localization of a Human Gene Closely Related to cdo
A Southern “zoo” blot containing DNA from various species revealed that mouse and human DNA displayed strong cross-species hybridization to a rat cdo cDNA probe, while chicken, toad (Xenopus laevis), and zebrafish DNA displayed a clearly detectable, but weaker signal (data not shown). Screening of human fetal brain and fetal lung libraries yielded clones that corresponded to a single closely related gene. These human cDNAs hybridized to an mRNA species present in several human neuroblastoma cell lines that was indistinguishable in size from cdo mRNA in Rat 6 cells (data not shown).
The predicted amino acid sequence of this human gene is shown in comparison to that of rat cdo in Fig. 2 A. The human and rat genes have identical domain structures in their extracellular regions, and their intracellular regions, which do not resemble other known proteins, are also highly related. Across their full lengths (except for the signal sequence which was missing from the human cDNA clones obtained), the two proteins are ∼81% identical and ∼96% similar. The relative positions of amino acids that define the Ig- and FNIII-like domain consensus sequences are conserved between the two predicted proteins, as are all 10 potential N-linked glycosylation sites. Two potential microexons are apparent in the human sequence: one is a three–amino acid insertion between Ig-like repeat 5 and FNIII-like repeat 1; the second is a five–amino acid insertion that is immediately adjacent to the proline-rich sequence found in the cytoplasmic tail. Although we cannot rule out the existence of human genes that are even more closely related to rat cdo, in this article, this gene will be referred to as human cdo.
To identify a chromosomal location for human cdo, a panel of human/rodent monochromosomal hybrids was screened by PCR with primers that amplify human, but not rodent, cdo DNA. These studies mapped human cdo to chromosome 11 (data not shown). PCR analysis of a chromosome 11 regional mapping panel showed that cdo mapped to the interval 11q23–24 as defined by the breakpoints of the hybrids GM10482 (11q23) and GM11222C (11q24). Hybrid GM11222C was positive for cdo, whereas GM10482 was negative (Fig. 8). This region has substantial overlap with similarly sized regions that display LOH in a high percentage of human breast, ovarian, and lung cancers (Negrini et al., 1995; Rasio et al., 1995; Gabra et al., 1996). PCR for the catalase locus (11p13) was used as a control to confirm the identity of the hybrids (Fig. 8).
Figure 8.
Regional localization of human cdo to chromosome 11q23–24. PCR amplification of human cdo and, as a control, the catalase locus (CAT; 11p13) in a regional mapping panel containing the displayed portions of human chromosome 11 in a rodent cell background. Ethidium bromide–stained gels are shown. CHO, genomic DNA from CHO cells; Mus, genomic DNA from mouse.
Discussion
Proteins of the Ig/FNIII family play important roles in a variety of developmental and physiological processes. Additionally, expression of Ig/FNIII proteins can be highly dynamic and cell type specific, although little is known of the signaling pathways that regulate such expression (Edelman and Crossin, 1991; Brummendorf and Rathjen, 1995; Cunningham, 1995). We report here the molecular cloning and characterization of rat CDO, a novel cell surface glycoprotein of the Ig/FNIII family. A cdo cDNA was isolated by screening a genetically dominant, oncogene-resistant mutant cell line for genes whose expression was enhanced relative to a transformation-sensitive control cell line. cdo mRNA and protein were dramatically down-regulated in rat and mouse embryo fibroblast cell lines that were transformed by various oncogenes. Additionally, cdo mRNA and protein levels were strongly, but transiently, reduced when confluent, quiescent cells were stimulated to reenter the cell cycle. These observations raise the possibility that CDO may serve as a negative regulator of cell proliferation, transformation, and/or tumorigenesis. Consistent with this possibility, a human gene that is very closely related to rat cdo mapped to chromosome 11q23–24, a region that exhibits LOH in a high percentage of human breast, ovarian, and lung cancers (Negrini et al., 1995; Rasio et al., 1995; Gabra et al., 1996). cdo therefore joins a growing list of genes that are not only down-regulated at the mRNA level in transformed cell lines or tumors, but may also have transformation suppressor function. Such genes include those that encode components of the extracellular matrix (fibronectin, collagen); cell surface proteins (E- and H-cadherin, connexins 26 and 43, certain integrins, and maspin); and proteins that organize the actin cytoskeleton (vinculin, α-actinin, tropomyosin 1) (Plantefaber and Hynes, 1989; Giancotti and Ruoslahti, 1990; Frixen et al., 1991; Vleminckx et al., 1991; Fernandez et al., 1992; Gluck et al., 1993; Prasad et al., 1993; Akamatsu et al., 1996; Hirschi et al., 1996; Lee, 1996; Sheng et al., 1996; Travers et al., 1996).
In addition to regulation of cdo mRNA levels by oncogenes and serum, CDO protein levels were powerfully controlled by cell–substratum adhesion, via a mechanism that was independent of cdo mRNA expression. Rat 6 cells and the Ras-transformed derivative cell line C1-T24 each displayed a cell anchorage requirement for stable production of CDO, even when this protein was derived from an ectopically expressed mRNA, whose levels were not altered by the culture conditions. Preliminary experiments indicate that when suspended C1-T24/cdo cells were allowed to attach and spread on fibronectin-coated dishes, CDO was restored to maximal levels 2 h after replating. Furthermore, attachment-mediated production of CDO was partially blocked by cytochalasin D (Kang, J.-S., and R.S. Krauss, unpublished observations). These data suggest a possible role for integrin-mediated signaling and cytoskeletal organization in this process. They also raise the possibility that stable production of CDO may require an appropriate extracellular matrix microenvironment. Such a requirement would have important implications for cell type specificity in the production of CDO and other proteins that might be regulated in a similar fashion. It will be very interesting to determine whether anchorage or specific extracellular matrix proteins are required for stable production of other Ig/FNIII family members. It is not known whether the loss of CDO protein in suspension cultures occurred at the level of translation or stability. Zhu et al. (1996) have reported that translation of cyclin D1 mRNA is dependent on cell–substratum adhesion, but unlike the observations described here for CDO, this effect of cell anchorage was lost when ectopic cyclin D1 transcripts lacked untranslated regions.
Most CAMs of the IgSF can mediate homophilic binding, although DCC may be an exception (Fearon, 1996). However, Rat 6 and C1-T24 cells that ectopically expressed CDO did not exhibit enhanced cell–cell aggregation. Furthermore, preliminary studies indicate that a soluble fusion protein that contains the entire CDO extracellular region is unable to bind to Rat 6 or Rat 6/cdo cells, each of which has abundant CDO on their cell surface (Kang, J.-S., and R.S. Krauss, unpublished observations). These observations indicate that Rat 6 cells do not express a ligand for CDO on their cell surface, and also that CDO is probably not a homophilic CAM. Therefore, it will also be interesting to determine the effects of cell anchorage on CDO production in a cell system in which a known ligand for CDO is present. The presence of a ligand could conceivably stabilize or stimulate production of CDO, even in the absence of optimal cell anchorage. The levels of CDO at the cell surface may therefore be governed by complex interactions between (a) signals that can regulate cdo gene expression, such as mitogenic stimuli; and (b) signals that can regulate CDO protein production, such as appropriate cell–substratum interactions and, possibly, the presence of ligands.
Members of the Ig/FNIII repeat family can be organized into subfamilies by the number of Ig- and FNIII-like repeats they contain in their extracellular regions (Brummendorf and Rathjen, 1995; Cunningham, 1995). We are not aware of any previously reported members of this family that have the five Ig plus three FNIII combination of repeats. CDO's unique intracellular region also distinguishes it from other Ig/FNIII proteins. cdo is therefore a founding member of a new subfamily within the Ig/FNIII repeat family. Given the structural resemblance of CDO to N-CAM, DCC, and other Ig/FNIII family members, it is tempting to speculate that CDO might also play a role in cell migratory events during development of the nervous system and/or other organ systems. Consistent with this notion, cdo mRNA is expressed at high levels in several human neuroblastoma cell lines (Gao, M., and R.S. Krauss, unpublished observations); in addition, preliminary in situ hybridization studies indicate that cdo mRNA is expressed at high levels in particular regions of the developing rat spinal cord and, interestingly, limb bud (Brose, K., J.-S. Kang, R.S. Krauss, and M. Tessier-Lavigne, unpublished studies). Because under some conditions Rat 6 cells express cdo mRNA but not its protein product, it will be important to identify cell surface production of CDO protein by particular cell types during development to understand its function. Understanding CDO's possible functions in development and oncogenesis will also require analysis of the protein's long cytoplasmic tail, which bears no significant resemblance to other known proteins. The dramatic regulation of cdo mRNA and protein production by mitogenic and anchorage-related stimuli, combined with CDO's unique intracellular region, indicate that further studies of this protein should yield interesting new insights into cell proliferation and/or motility, and how such processes may go awry during tumorigenesis. Such studies are in progress.
Acknowledgments
We thank C. Goodman, T. Kidd, and M. Tessier-Lavigne for communicating results before publication; E. Spanopoulou for cell lines; and L. Ossowski, J. Hirsch, and D. Colman for comments on the manuscript.
This work was supported by grants from the National Intstitutes of Health (CA59474) and the Elsa U. Pardee Foundation, as well as a scholarship from the Alexandrine and Alexander L. Sinsheimer Fund to R.S. Krauss. J.L. Feinleilb is supported by a U.S. Army Medical Research and Materiel Command Breast Cancer predoctoral training grant (DAMD17-94-J-4111). R.S. Krauss is a Career Scientist of the Irma T. Hirschl Trust.
Footnotes
1. Abbreviations used in this paper: CAM, cell adhesion molecule; C-CAM, cell–cell adhesion molecule; FNIII, fibronectin type III; N-CAM, neural cell adhesion molecule; IgSF, Ig superfamily; LOH, loss of heterozygosity; FNIII, fibronectin type III.
Please address all correspondence to Robert S. Krauss, Department of Biochemistry, Box 1020, Mount Sinai School of Medicine, New York 10029. Tel.: (212) 241-2177. Fax: (212) 996-7214. e-mail: rkrauss@smtplink.mssm.edu
References
- Akamatsu H, Ichihara-Tanaka K, Ozono K, Kamiike W, Matsuda H, Sekiguchi K. Suppression of the transformed phenotypes of human fibrosarcoma cells by overexpression of recombinant fibronectin. Cancer Res. 1996;56:451–456. [PubMed] [Google Scholar]
- Aoki J, Umeda M, Takio K, Titani K, Utsemi H, Sasaki M, Inoue K. Neural cell adhesion molecule mediates contact-dependent inhibition of growth of near-diploid mouse fibroblast cell line m5S/1M. J Cell Biol. 1991;115:1751–1761. doi: 10.1083/jcb.115.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H, Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci USA. 1972;69:1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berx G, Cleton-Jansen A-M, Strumane K, de Leeuw WJF, Nollet F, van Roy F, Cornelisse C. E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene. 1996;13:1919–1925. [PubMed] [Google Scholar]
- Borner C, Guadagno SN, Hsiao WW-L, Fabbro D, Barr M, Weinstein IB. Expression of four protein kinase C isoforms in rat fibroblasts. Differential alterations in ras-, src-, and fos- transformed cells. J Biol Chem. 1992;267:12900–12910. [PubMed] [Google Scholar]
- Borner C, Ueffing M, Jaken S, Parker PJ, Weinstein IB. Two closely related isoforms of protein kinase C produce reciprocal effects on the growth of rat fibroblasts. J Biol Chem. 1995;270:78–86. doi: 10.1074/jbc.270.1.78. [DOI] [PubMed] [Google Scholar]
- Brummendorf T, Rathjen FG. Cell adhesion molecules 1: immunoglobulin superfamily. Protein Profile. 1995;2:963–1108. [PubMed] [Google Scholar]
- Chan SS-Y, Zheng H, Su M-W, Wilk R, Killeen MT, Hedgecock EM, Culotti JG. UNC-40, a C. eleganshomologue of DCC (deleted in colorectal cancer), is required in motile cells responding to UNC-6 netrin cues. Cell. 1996;87:187–195. doi: 10.1016/s0092-8674(00)81337-9. [DOI] [PubMed] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Coman DR. Decreased mutaul adhesiveness, a property of cells from squamous cell carcinomas. Cancer Res. 1944;4:625–629. [Google Scholar]
- Cunningham BA. Cell adhesion molecules as morphoregulators. Curr Opin Cell Biol. 1995;7:628–633. doi: 10.1016/0955-0674(95)80103-0. [DOI] [PubMed] [Google Scholar]
- Edelman GM, Crossin KL. Cell adhesion molecules: implications for a molecular histology. Annu Rev Biochem. 1991;60:155–190. doi: 10.1146/annurev.bi.60.070191.001103. [DOI] [PubMed] [Google Scholar]
- Fearon ER. DCC: is there a connection between tumorigenesis and cell guidance molecules? . Biochim Biophys Acta. 1996;1288:M17–M23. doi: 10.1016/0304-419x(96)00023-6. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Pierceall WE. The deleted in colorectal cancer (DCC)gene: a candidate tumor suppressor gene encoding a cell surface protein with similarity to the neural cell adhesion molecules. Cancer Surv. 1995;24:3–17. [PubMed] [Google Scholar]
- Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler K, et al. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science (Wash DC) 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- Feinleib JL, Krauss RS. Dissociation of rasoncogene-induced gene expression and anchorage-independent growth in a series of somatic cell mutants. Mol Carcinog. 1996;16:139–148. doi: 10.1002/(SICI)1098-2744(199607)16:3<139::AID-MC4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Fernandez JLR, Geiger B, Salomon D, Sababay H, Zoller M, Ben-Ze'ev A. Suppression of tumorigenicity in transformed cells after transfection with vinculin cDNA. J Cell Biol. 1992;119:427–438. doi: 10.1083/jcb.119.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Lochner D, Birchmeir W. E-cadherin–mediated cell–cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113:173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabra H, Watson JEV, Taylor KJ, Mackay J, Leonard RCF, Steel CM, Porteous DJ, Smyth JF. Definition and refinement of a region of loss of heterozygosity at 11q23.3-q24.3 in epithelial ovarian cancer associated with poor prognosis. Cancer Res. 1996;56:950–954. [PubMed] [Google Scholar]
- Giancotti FG, Ruoslahti E. Elevated levels of the α5β1fibronectin receptor suppress the transformed phenotype of Chinese hamster ovary cells. Cell. 1990;60:849–859. doi: 10.1016/0092-8674(90)90098-y. [DOI] [PubMed] [Google Scholar]
- Gluck U, Kwiatkowski DJ, Ben-Ze'ev A. Suppression of tumorigenicity in simian virus 40-transformed 3T3 cells transfected with α-actinin cDNA. Proc Natl Acad Sci USA. 1993;90:383–387. doi: 10.1073/pnas.90.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick L, Cho KR, Vogelstein B. Cell adhesion molecules as tumour suppressors. Trends Cell Biol. 1993;3:36–39. doi: 10.1016/0962-8924(93)90148-t. [DOI] [PubMed] [Google Scholar]
- Hedrick L, Cho KR, Fearon ER, Wu T-C, Kinzler KW, Vogelstein B. The DCC gene product in cellular differentiation and colorectal tumorigenesis. Genes & Dev. 1994;8:1174–1183. doi: 10.1101/gad.8.10.1174. [DOI] [PubMed] [Google Scholar]
- Hirschi KK, Xu C-E, Tsukamoto T, Sager R. Gap junction genes Cx26 and Cx43individually suppress the cancer phenotype of human mammary carcinoma cells and restore differentiation potential. Cell Growth Differ. 1996;7:861–870. [PubMed] [Google Scholar]
- Housey GM, Johnson MD, Hsiao W-LW, O'Brian CA, Murphy JP, Kirschmeir P, Weinstein IB. Overproduction of protein kinase C causes disordered growth control in rat fibroblasts. Cell. 1988;52:343–354. doi: 10.1016/s0092-8674(88)80027-8. [DOI] [PubMed] [Google Scholar]
- Hsieh J-T, Luo W, Song W, Wang Y, Kleinerman DI, Van NT, Lin S-H. Tumor suppressive role of an androgen-regulated epithelial cell adhesion molecule (C-CAM) in prostate carcinoma cell revealed sense and anti-sense approaches. Cancer Res. 1995;55:190–197. [PubMed] [Google Scholar]
- Hynes RO, Lander AD. Contact and adhesive specificities in the associations, migrations, and targeting of cells and axons. Cell. 1992;68:303–322. doi: 10.1016/0092-8674(92)90472-o. [DOI] [PubMed] [Google Scholar]
- Johnson MD, Housey GM, Kirschmeir PT, Weinstein IB. Molecular cloning of gene sequences regulated by tumor promoters and mitogens through protein kinase C. Mol Cell Biol. 1987;7:2821–2829. doi: 10.1128/mcb.7.8.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-S, Krauss RS. Ras induces anchorage-independent growth by subverting multiple adhesion-regulated cell cycle events. Mol Cell Biol. 1996;16:3370–3380. doi: 10.1128/mcb.16.7.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS-Y, Culotti JG, Tessier-Lavigne M. Deleted in colorectal cancer (DCC)encodes a netrin receptor. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- Kleinerman DI, Troncoso P, Lin S-H, Pisters LL, Sherwood ER, Brooks T, von Eisenbach AC, Hsieh J-T. Consistent expression of an epithelial cell adhesion molecule (C-CAM) during human prostate development and loss of expression in prostate cancer: implication as a tumor suppressor gene. Cancer Res. 1995;55:1215–1220. [PubMed] [Google Scholar]
- Kolodziej PA, Timpe LC, Mitchell KJ, Fried SR, Goodman CS, Jan LY, Jan YN. frazzledencodes a Drosophila member of the DCC immunoglobulin subfamily and is required for CNS and motor axon guidance. Cell. 1996;87:197–204. doi: 10.1016/s0092-8674(00)81338-0. [DOI] [PubMed] [Google Scholar]
- Krauss RS, Guadagno SN, Weinstein IB. Novel revertants of H-ras oncogene-transformed R6-PKC3 cells. Mol Cell Biol. 1992;12:3117–3129. doi: 10.1128/mcb.12.7.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Ordonez-Garcia C, Turbide C, Beauchemin N. Inhibition of colonic tumor cell growth by biliary glycoprotein. Oncogene. 1995;11:2375–2382. [PubMed] [Google Scholar]
- Lee SW. H-cadherin, a novel cadherin with growth inhibitory functions and diminished expression in human breast cancer. Nat Med. 1996;2:776–782. doi: 10.1038/nm0796-776. [DOI] [PubMed] [Google Scholar]
- Morganstern JP, Land H. Advanced mammalian gene transfer: high titer retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini M, Rasio D, Hampton GM, Sabbioni S, Rattan S, Carter SL, Rosenberg AL, Schwartz GF, Shiloh Y, Cavenee WK, et al. Definition and refinement of chromosome 11 regions of loss of heterozygosity in breast cancer: identification of a new region at 11q23.3. Cancer Res. 1995;55:3003–3007. [PubMed] [Google Scholar]
- Neumaier M, Paululat S, Chan A, Matthaes P, Wagener C. Biliary glycoprotein, a potential human cell adhesion molecule, is down-regulated in colorectal carcinomas. Proc Natl Acad Sci USA. 1993;90:10744–10748. doi: 10.1073/pnas.90.22.10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa J, Kaneko H, Masuda T, Nagata S, Hosoya H, Watanabe K. Novel neural adhesion molecules in the contactin/F3 subgroup of the immunoglobulin superfamily: isolation and characterization of the cDNAs from rat brain. Neurosci Lett. 1996;218:173–176. doi: 10.1016/s0304-3940(96)13156-6. [DOI] [PubMed] [Google Scholar]
- Patthy L. Homology of a domain of the growth hormone/prolactin receptor with type III modules of fibronectin. Cell. 1990;61:13–14. doi: 10.1016/0092-8674(90)90208-v. [DOI] [PubMed] [Google Scholar]
- Plantefaber LC, Hynes RO. Changes in integrin receptors on oncogenically transformed cells. Cell. 1989;56:281–290. doi: 10.1016/0092-8674(89)90902-1. [DOI] [PubMed] [Google Scholar]
- Prasad GL, Fuldner RA, Cooper HL. Expression of transduced tropomyosin 1 cDNA suppresses neoplastic growth of cells transformed by the rasoncogene. Proc Natl Acad Sci USA. 1993;90:7039–7043. doi: 10.1073/pnas.90.15.7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasio D, Negrini M, Manetti G, Dragani TA, Croce CM. Loss of heterozygosity at chromosome 11q in lung adenocarcinoma: identification of three independent regions. Cancer Res. 1995;55:3988–3991. [PubMed] [Google Scholar]
- Sheng S, Carey J, Seftor EA, Dias L, Hendrix MJC, Sager R. Maspin acts at the cell membrane to inhibit invasion and motility of mammary and prostatic cancer cells. Proc Natl Acad Sci USA. 1996;93:11669–11674. doi: 10.1073/pnas.93.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata D, Reale MA, Lavin P, Silverman M, Fearon EF, Steele G, Jessup JM, Loda M, Summerhayes IC. The DCC protein and prognosis in colorectal cancer. N Engl J Med. 1996;335:1727–1732. doi: 10.1056/NEJM199612053352303. [DOI] [PubMed] [Google Scholar]
- Sporns O, Edelman GM, Crossin KL. The neural cell adhesion molecule (N-CAM) inhibits proliferation in primary cultures of rat astrocytes. Proc Natl Acad Sci USA. 1995;92:542–546. doi: 10.1073/pnas.92.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science (Wash DC) 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- Theune S, Fung J, Todd S, Sakaguchi AY, Naylor SL. PCR primers for human chromosomes: reagents for the rapid analysis of somatic cell hybrids. Genomics. 1991;9:511–516. doi: 10.1016/0888-7543(91)90418-e. [DOI] [PubMed] [Google Scholar]
- Travers H, French NS, Norton JD. Suppression of tumorigenicity in ras-transformed fibroblasts by α2(I) collagen. Cell Growth Differ. 1996;7:1353–1360. [PubMed] [Google Scholar]
- Vaughn DE, Bjorkman PJ. The (Greek) key to structures of neural adhesion molecules. Neuron. 1996;16:261–273. doi: 10.1016/s0896-6273(00)80045-8. [DOI] [PubMed] [Google Scholar]
- Vleminckx K, Vakaert J, Mareel LM, Fiers W, Van Roy F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66:107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- Williams AF, Barclay AN. The immunoglobulin superfamily: domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- Zhu X, Ohtsubo M, Bohmer RM, Roberts JM, Assoian RK. Adhesion-dependent cell cycle progression linked to the expression of cyclin D1, activation of cyclin E-cdk2, and phosphorylation of the retinoblastoma protein. J Cell Biol. 1996;133:391–403. doi: 10.1083/jcb.133.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]