Abstract
The yeast vacuolar protein aminopeptidase I (API) is synthesized as a cytosolic precursor that is transported to the vacuole by a nonclassical targeting mechanism. Recent genetic studies indicate that the biosynthetic pathway that transports API uses many of the same molecular components as the degradative autophagy pathway. This overlap coupled with both in vitro and in vivo analysis of API import suggested that, like autophagy, API transport is vesicular. Subcellular fractionation experiments demonstrate that API precursor (prAPI) initially enters a nonvacuolar cytosolic compartment. In addition, subvacuolar vesicles containing prAPI were purified from a mutant strain defective in breakdown of autophagosomes, further indicating that prAPI enters the vacuole inside a vesicle. The purified subvacuolar vesicles do not appear to contain vacuolar marker proteins. Immunogold EM confirms that prAPI is localized in cytosolic and in subvacuolar vesicles in a mutant strain defective in autophagic body degradation. These data suggest that cytosolic vesicles containing prAPI fuse with the vacuole to release a membrane-bounded intermediate compartment that is subsequently broken down, allowing API maturation.
The yeast vacuole plays a key role in cellular protein metabolism. During periods of nitrogen starvation, amino acid supplies are maintained by protein degradation reactions that take place in this organelle (Klionsky et al., 1990; Hilt and Wolf, 1992). Accordingly, the vacuole contains numerous hydrolytic enzymes, compartmentalized to prevent nonspecific cellular damage. Substrate proteins are delivered to the vacuole from the cytosol by autophagy (Takeshige et al., 1992; Noda et al., 1995), and from the cell surface by endocytosis (Riezman, 1993). Precise subcellular transport reactions are necessary to ensure both the faithful compartmentalization of hydrolytic enzymes and the recognition and timely delivery of protein substrates for degradation. The majority of vacuolar hydrolases are delivered to the organelle via the secretory pathway (Conibear and Stevens, 1995; Stack et al., 1995). Aminopeptidase I (API)1, however, does not use the secretory system and is instead targeted by a nonclassical pathway from the cytoplasm to the vacuole (Klionsky et al., 1992).
API is synthesized as a cytosolic precursor containing an amino-terminal propeptide that mediates its vacuolar delivery (Segui-Real et al., 1995; Oda et al., 1996). The proregion consists of two predicted α helices separated by a proline residue. The first helix is amphipathic and directs the binding of API to a target membrane (Oda et al., 1996). Upon arrival in the vacuolar lumen, the proregion is cleaved in a proteinase B (PrB)–dependent reaction (Klionsky et al., 1992). Thus, maturation of API is a convenient marker for correct vacuolar delivery.
Two lines of evidence suggest that API transport may be a vesicular event. First, analysis of cytoplasm to vacuole targeting mutants (cvt; Harding et al., 1995) indicates that there is substantial overlap between genes involved in API transport and those required for autophagy (Harding et al., 1996; Scott et al., 1996). In addition, phenotypic examination of cvt and autophagy mutants (apg, aut) revealed that the majority of these mutants are both autophagy defective and accumulate API precursor (prAPI) (Harding et al., 1996; Scott et al., 1996). The apg mutants (Tsukada and Ohsumi, 1993) and, presumably, aut mutants (Thumm et al., 1994) are defective in macroautophagy, a process by which cytosolic proteins and cytoplasmic organelles are engulfed by double-membraned vesicles and delivered to the vacuole for degradation (Takeshige et al., 1992; Baba et al., 1994).
The second indication that API transport is vesicular comes from recent analysis of the biogenesis of this protein. These studies indicate that prAPI is transported as a 732-kD dodecamer (Kim et al., 1997). Since this complex is much too large to penetrate a typical translocation channel, and large porelike structures cannot be present on an energetically active membrane such as the vacuole, API must be transported by a vesicular mechanism.
The degree to which the Cvt pathway is analogous to bulk macroautophagy remains to be determined. There are several fundamental differences between these two pathways. API transport is a specific biosynthetic event that is constitutive, occurring even under nutrient-rich conditions (Klionsky et al., 1992; Scott et al., 1996). Bulk autophagy is nonselective, degradative, and only detectable during starvation for nutrients such as nitrogen or carbon (Takeshige et al., 1992). Also, prAPI has a half-time for vacuolar delivery of 30–45 min (Klionsky et al., 1992), while the half-time for delivery of cytosolic proteins by macroautophagy is at least 12 h when induced by nitrogen starvation (Scott et al., 1996). One possible model that accommodates these differences is that the Cvt pathway uses essentially the same molecular mechanism as macroautophagy to deliver API to the vacuole. The transport of API could be made specific, rapid, and constitutive if API is concentrated into vesicles by a receptor-mediated mechanism, and the constitutive level of autophagic vesicle formation is sufficient for delivery of API during exposure to various nutrient conditions.
To provide direct evidence that API in fact uses a vesicular mechanism for vacuolar delivery, we have examined the API transport process biochemically and morphologically. We have identified two yeast mutants that trap prAPI inside a vesicular compartment and used one of these mutant strains to purify an API transport intermediate. In addition, these data are supported by immuno-EM images demonstrating that API is concentrated in vesicles during transport to the vacuole.
Materials and Methods
Strains and Media
Saccharomyces cerevisiae strains used in this study were: JSR18Δ1 MATα vps18Δ1::TRP1 leu2-3,112 ura3-52 his3-Δ200 trp1Δ901 lys2-801 suc2-Δ9 with a plasmid bearing a temperature-sensitive allele of vps18, pJSR9 (Robinson et al., 1991); SEY6210 MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL (Robinson et al., 1988); SEY6211 MATa leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 ade2-101 suc2-Δ9 GAL (Robinson et al., 1988); THY32 cvt17-1 derivative of SEY6211 (Harding et al., 1995, 1996); and DYY101 MATα leu2,3-112 ura3-52 his3-Δ200 trp1-Δ901 ade2-101 suc2-Δ9 GAL ape1Δ::LEU2 with a single copy plasmid containing the P22L API construct (Oda et al., 1996). Unless otherwise indicated, cells were grown in synthetic minimal medium (SMD; 0.067% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as needed). Synthetic minimal medium without amino acids or ammonium sulfate containing 2% glucose (SD-N) was used for nitrogen starvation experiments, and YPD (containing 1% yeast extract, 2% peptone, and 2% glucose) was used for cell growth before immuno-EM.
Reagents
Gold-conjugated goat anti–rabbit IgG was from Biocell (Cardiff, UK). LR white resin was from London Resin (Hampshire, UK). Optiprep was from Accurate Chemical and Scientific Corp. (Westbury, NY); Oxalyticase from Enzogenetics (Corvallis, OR); Expre35S35S was from Dupont-New England Nuclear Research Products (Boston, MA); proteinase K and Pefabloc were from Boehringer Mannheim Biochemicals (Indianapolis, IN); SYPRO orange was from BioRad Laboratories (Hercules, CA); and all other reagents were from Sigma Chemical Co. (St. Louis, MO). Antiserum against peptides in the mature region of API (Klionsky et al., 1992), API propeptide (Harding et al., 1995), carboxypeptidase Y (CPY) and proteinase A (PrA) (Klionsky et al., 1988), and alkaline phosphatase (ALP) (Scott et al., 1996) were as described previously. The following antisera were generously supplied as indicated: phosphoglycerate kinase (PGK) (J. Thorner, Department of Biochemistry, University of California, Berkeley), Sec13p (C. Barlow, Dartmouth Medical School, Hanover, NH), Kex2p (G. Payne, Department of Biological Chemistry, University of California, Los Angeles), and Sec61p (R. Schekman, Department of Molecular and Cellular Biology, University of California, Berkeley).
Pulse-Chase Analysis and Immunoprecipitation
For whole cell labeling experiments, yeast strains were grown to 1 OD600/ml in SMD. Cells were harvested and labeled with 100 μCi/ml Expre35S35S in SMD at a cell density of 100 OD600/ml for 5 min. Cold cysteine and methionine were added to 2 mM and 1 mM, respectively. The labeled cells were chased at a cell density of 1 OD600/ml in SMD for the indicated times. Where indicated, SD-N was used for chase reactions. Spheroplast labeling was performed as described (Scott and Klionsky, 1995). Samples were immunoprecipitated as described (Klionsky et al., 1992).
Differential Lysis Fractionation
Fractionation experiments were performed using previously described methods based on differential osmotic lysis (Harding et al., 1995; Oda et al., 1996). Spheroplasts were permeabilized in 200 mM sorbitol, 20 mM K-Pipes, pH 6.8 (PS200), and soluble (S200) and organelle-containing (P200) fractions were collected by centrifugation at 5,000 g for 5 min. This treatment consistently releases >95% of the cytosolic marker PGK into the supernatant while maintaining ∼70% of the vacuole lumen marker PrA in the pellet fraction (Harding et al., 1995, 1996; Oda et al., 1996). Where indicated, a second osmotic lysis step was performed by resuspending the P200 fraction in 20 mM K-Pipes, pH 6.8 (PS0), and separating the S0 and P0 fractions by centrifugation at 10,000 g for 5 min.
Subvacuolar Vesicle Isolation
Vacuoles were isolated from 700–800 OD600 U of spheroplasts prepared from the cvt17 strain. Differential lysis was performed in PS200 containing 1 mM Pefabloc and 2 μg/ml pepstatin A, and the resulting P200 fraction was resuspended in 3 ml of 12% Ficoll. This was overlaid with 3 ml of 10% Ficoll, 3 ml of 4% Ficoll, and 1 ml of PS200. All Ficoll solutions were made in PS200 with protease inhibitors. Vacuoles were isolated from the 4% Ficoll/PS200 interface after centrifugation at 100,000 g for 90 min at 8°C. The vacuoles were diluted with 2 vol of PS200 and collected by centrifugation at 60,000 g for 15 min. An aliquot of the vacuole fraction was saved for analysis. The remaining vacuoles were resuspended in 200 μl PS0 containing protease inhibitors and loaded on top of an Optiprep step gradient consisting of 500 μl of 37% Optiprep in PS0 (density 1.12) overlaid with 1.3 ml of 19% Optiprep in PS0 (density 1.06). The resulting step gradient was subjected to centrifugation at 160,000 g for 60 min at 12°C. Four fractions were collected: the PS0/1.06 interface (vacuolar vesicles [VV]), an equal aliquot of the 1.06 density region (low density [LD]), the 1.06/1.12 interface (subvacuolar vesicles [SV]), and the 1.12 density region (high density [HD]).
Vacuole Isolation
Vacuoles were isolated from small scale preparations for the experiments in Figs. 2 and 3. Spheroplasts (50 OD600 U) were lysed in PS200 buffer and centrifuged at 5,000 g for 5 min to generate the P200 fraction. This fraction was resuspended in 900 μl 10% Ficoll, and then overlaid with 1 ml 4% Ficoll and 200 μl PS200. The resulting step gradient was centrifuged at 166,000 g for 60 min at 8°C. The vacuole fraction (V) was collected from the 0/4 % Ficoll interface, the intermediate fraction (I) from the 4/10% Ficoll interface, and the pellet fraction (P) from the gradient pellet.
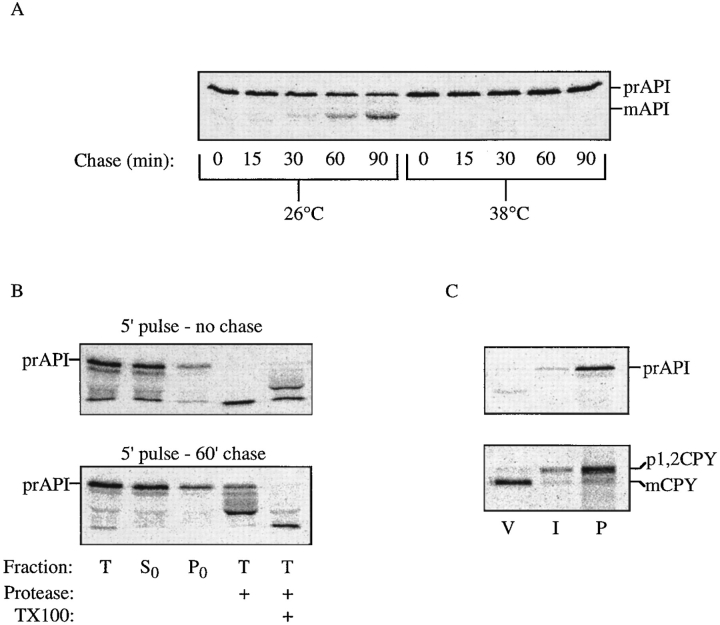
Figure 2.
Precursor API is trapped in a prevacuolar compartment in vps18 cells. (A) Yeast strain JSR18Δ1 with plasmid pJSR9 (vps18 ts) was grown at the permissive temperature of 26°C. Before labeling, the cells were incubated at either 26° or 38°C for 10 min. Pulse labeling was for 5 min, followed by the indicated chase times. (B) vps18 spheroplasts were shifted to 38°C for 5 min, pulse labeled for 5 min, and then chased for either 0 or 60 min. At each time point the spheroplasts were collected by centrifugation and resuspended in PS0 buffer. The Total (T) fraction either received no treatment, was treated with 50 μg/ml proteinase K, or received proteinase K in the presence of 0.2% Triton X-100. The supernatant (S0) and pellet (P0) fractions were collected after centrifugation at 10,000 g for 5 min. (C) vps18 spheroplasts were pulse labeled for 10 min, chased for 10 min at 26°C, and then shifted to 38°C and chased for an additional 60 min. These cells were then lysed in PS200 buffer, and the S200 and P200 fractions were collected by centrifugation at 5,000 g for 5 min. The P200 fraction was resuspended in 10% Ficoll solution made in PS200, and vacuoles were isolated by flotation through 4% Ficoll. V, vacuole fraction; I, 4/10% Ficoll interface fraction; P, gradient pellet. Proteins of interest were recovered by immunoprecipitation, followed by SDS-PAGE, and detected by a Molecular Dynamics STORM phosphorimager. The positions of precursor and mature API and CPY are indicated.
Figure 3.
A mutant in the API propeptide causes accumulation of prAPI on a nonvacuolar compartment. (A) Spheroplasts from an ape1Δ strain containing a plasmid bearing P22L API were subjected to differential lysis in PS200 buffer. The Total (T) fraction (after differential lysis) either received no treatment, was treated with 50 μg/ ml proteinase K, or received proteinase K in the presence of 0.2% Triton X-100. (B) The supernatant (S200) and pellet (P200) fractions were collected after centrifugation at 5,000 g for 5 min. The P200 fraction was resuspended in 10% Ficoll, and vacuoles were isolated by flotation through 4% Ficoll. V, vacuole; I, 4/10% Ficoll interface; P, gradient pellet. Proteins were detected by Western blotting. The positions of prAPI, mAPI, and mPrA are indicated. (C) ape1Δ cells containing P22L API on a plasmid were pulse labeled for 5 min, harvested by centrifugation, and subjected to a nonradioactive chase in either nitrogen-containing (SMD) or nitrogen starvation (SD-N) medium. Samples were collected at the times indicated and immunoprecipitated with API antiserum. The resulting SDS-PAGE gels were quantified using a Molecular Dynamics STORM phosphorimager.
Protease treatment was with 50 μg/ml proteinase K for 15 min on ice. Where indicated, Triton X-100 was added at a final concentration of 0.2%. Protease treatment results in the degradation of precursor API to intermediate- and mature-sized bands; mature API is resistant to digestion by proteinase K (Oda et al., 1996). Immunoblotting was performed as before (Harding et al., 1996).
Immuno-EM
Samples were prepared for immuno-EM as described (Baba et al., 1994) with modifications. Cells were sandwiched between an aluminum and a copper disk, frozen in liquid propane, and transferred to 0.1% formaldehyde in absolute acetone at −80°C. After 1–2 d, they were transferred to −20°C, washed with absolute acetone, and then replaced stepwise with cold absolute ethanol. LR white resin was applied and allowed to polymerize for 48 h with UV irradiation at −20°C. Ultrathin sections were collected onto Formvar-coated nickel grids, blocked with 2% BSA-PBS with purified goat serum (1:50 dilution), and immunolabeled with affinity-purified API antibody by floating the grids on a 1:200 or 1:500 dilution of antiserum for 1 h at room temperature. The grids were then washed, incubated with 10 nm gold–conjugated goat anti–rabbit IgG, washed again, and then fixed with 1% glutaraldehyde for 3 min. Sections were stained for 7 min with 4% uranyl acetate and with 0.2% lead citrate for 30 s, and then examined with an electron microscope (H-800; Hitachi Ltd., Tokyo, Japan) at 125 kV.
Results
API Maturation Is Severely Inhibited by Low Temperatures
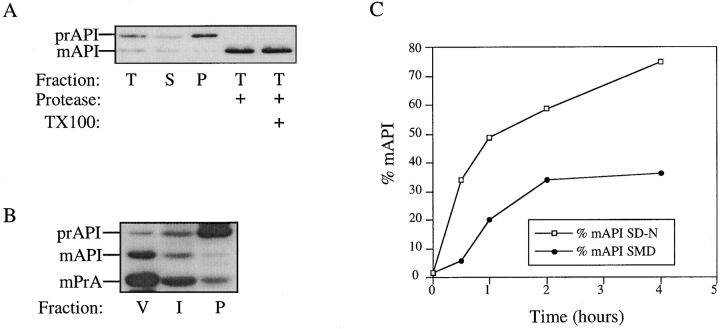
One way to distinguish between membrane translocation through a proteinaceous channel and vesicular transport reactions is by sensitivity to low temperature. For example, at 10°C translocation of CPY into the ER lumen occurs efficiently, while the packaging and transport of CPY from the ER to the Golgi (a vesicular process) is blocked (Baker and Schekman, 1989). We have previously shown that API maturation in vitro is sensitive to low temperature (Scott and Klionsky, 1995). To examine the effect of low temperature on the kinetics of API transport in cells, pulse-chase experiments were performed at 10°, 14°, and 30°C. At 30°C API has a half-time for vacuolar delivery of 30–45 min (Klionsky et al., 1992); the reaction is essentially complete after 2 h of chase (Fig. 1). However, at 10°C no mature API was detected, after incubation for 4 h (Fig. 1). When CPY transport was also monitored in the same samples, the ER form of CPY (p1CPY) accumulated, indicating that membrane translocation into the ER continued while vesicular reactions that carry cargo from the ER to the Golgi were blocked (data not shown). When the reactions were carried out at 14°C, a trace of mature API was apparent at the 4-h chase point, indicating that even at this intermediate temperature API delivery is severely inhibited. This requirement for relatively high temperatures is typical of vesicle-mediated transport but not membrane translocation through a proteinaceous channel.
Figure 1.
API delivery is blocked at low temperatures. SEY6210 cells were pulse labeled for 5 min and chased for the times indicated. Identical experiments were performed at 10°, 14°, and 30°C. API was recovered by immunoprecipitation followed by SDS-PAGE and detected by a STORM phosphorimager (Molecular Dynamics, Sunnyvale, CA). The positions of prAPI and mAPI are indicated.
Identification of Prevacuolar API-containing Vesicles
If API is transported to the vacuole by a vesicular intermediate, it may require some of the same components as the vacuolar protein sorting (Vps) pathway for correct compartment identification, docking, or fusion. Similarly, if API-containing vesicles fuse at the endosome or late endosome instead of at the vacuole, then proteins that are required for late steps in the Vps pathway would also be required for API targeting. Previously, we examined vps mutants for their API phenotype. The vps mutants display differential effects on API processing; some vps mutants are essentially wild type for API transport, while others are strongly blocked (Klionsky et al., 1992; Scott and Klionsky, 1995). To reduce the possibility of secondary defects and to allow a more direct assessment of the role of a vps gene product in API import, we examined API delivery in a vps18 temperature-conditional mutant.
vps18Δ cells containing a temperature-sensitive allele of vps18 on a plasmid were subjected to pulse-chase experiments at the permissive and nonpermissive temperatures. The vps18 strain was completely blocked for API delivery at the nonpermissive temperature (Fig. 2 A). Previous studies of Vps18p (Pep3p) suggest that it is involved in transport between the endosome and the vacuole (Robinson et al., 1991), and that it may be located on the vacuolar membrane surface (Preston et al., 1991). The fact that API transport is tightly blocked in this mutant suggests that API-containing vesicles may either join vacuole-destined proteins somewhere within the endosomal system, or may use some of the same molecular components used by vesicles traveling from the endosome to the vacuole to fuse at the vacuolar membrane.
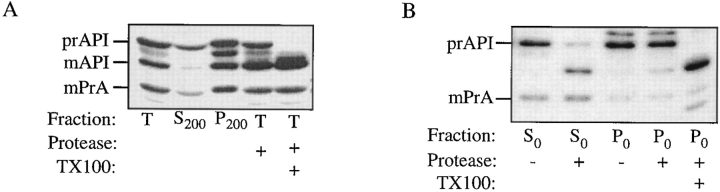
To determine the step in the API transport pathway that is affected by vps18, subcellular fractionation experiments were performed. Spheroplasts were pulse labeled and lysed in PS0, a buffer of low osmotic strength that lyses both the plasma membrane and the vacuole membrane, while smaller vesicles remain intact (see Fig. 4). After a 5-min pulse at the nonpermissive temperature, the majority of prAPI remained in the supernatant fraction and was sensitive to added protease, consistent with a cytosolic localization (Fig. 2 B). After 60 min of chase, a fraction of prAPI was contained in the pellet fraction and was protease protected after lysis in PS0 buffer. Since this treatment lyses both the plasma membrane and the vacuole, this result suggests that prAPI is trapped in a prevacuolar vesicular compartment.
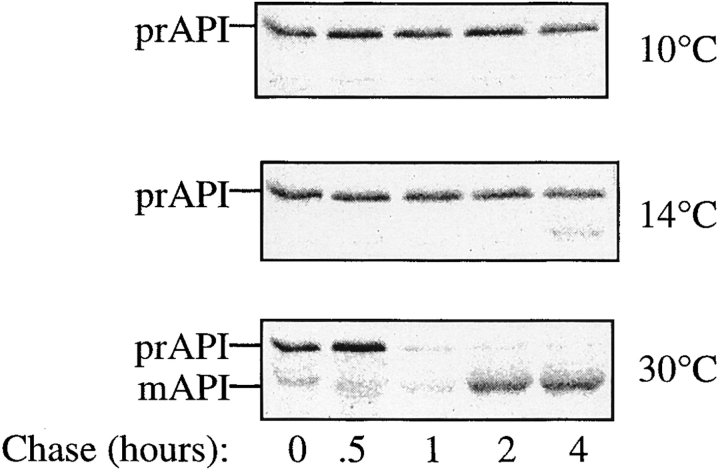
Figure 4.
Precursor API is trapped within a vesicle in cvt17 mutants. (A) Precursor API is in a protease-protected compartment in cvt17 mutants. THY32 (cvt17) spheroplasts were subjected to differential lysis and protease treatment in PS200 buffer as in Fig 3. (B) Precursor API is not free in the vacuolar lumen in cvt17 mutants. Differential fractionation was continued by resuspending the P200 fraction in PS0 buffer and separating the supernatant (S0) and pellet (P0) fractions by centrifugation at 10,000 g. The S0 and P0 fractions were treated with proteinase K in the presence and absence of Triton X-100 as indicated. All fractions were precipitated with 10% TCA and resolved by SDS PAGE; proteins of interest were detected by Western blotting. The positions of prAPI, mAPI, and mPrA are indicated.
To confirm that the population of prAPI contained within the pellet fraction is not within the vacuole, further fractionation experiments were performed. These experiments were carried out using a differential lysis technique based on resuspension of spheroplasts in PS200 buffer. Marker protein analysis shows that this treatment lyses the plasma membrane, while maintaining the integrity of the majority of vacuoles (Harding et al., 1996, 1995). Cells containing the vps18 mutation were pulse labeled for 10 min, and then chased for 10 min at the permissive temperature before shifting to the nonpermissive temperature for 60 min. This pulse-chase regime allowed for transport of CPY to the vacuole to occur before the onset of the vps18 block. Since delivery of API is slower than CPY, maturation of API was still prevented. As in the PS0 buffer, a population of prAPI was maintained in the pellet fraction after lysis in PS200 buffer. Vacuoles were then isolated from the P200 fraction by flotation through 4% Ficoll. After this treatment, mCPY was recovered in the vacuole fraction, while trapped p2CPY was recovered in the gradient pellet. In the same samples, none of the trapped prAPI was collected in the vacuole fraction (Fig. 2 C). These results indicate that the vps18 block causes the accumulation of prAPI in a nonvacuolar compartment, and suggest that API may be transported to the vacuole via a vesicular intermediate.
A Targeting Mutant in the API Propeptide Accumulates in a Nonvacuolar Compartment
To extend our analysis of the cytoplasmic intermediate compartment, we examined the location of API with a mutation in the propeptide region. Mutations in the API proregion interfere with the membrane binding or subsequent steps of the transport process. In particular, changing proline 22 to leucine (P22L) causes prAPI to accumulate in a membrane-bound state that undergoes further transport at a very minimal rate (Oda et al., 1996). This mutation is predicted to disrupt a β turn separating two α helices that make up the API propeptide and results in a mutant protein with increased membrane affinity compared with the wild-type protein (Oda et al., 1996). Differential lysis fractionation experiments were performed in PS200 buffer using cells containing P22L API. P22L API was primarily precursor sized, but it was recovered in the membrane pellet fraction where it was protease sensitive, indicating that, although it is membrane bound, it is not compartmentalized (Fig. 3 A). Under these same conditions, wild-type precursor is located exclusively in the supernatant fraction (Harding et al., 1995, 1996). Vacuoles were then isolated from the pellet containing P22L API. As was the case with the trapped prAPI in the vps18 mutant strain, precursor P22L API did not cofractionate with vacuolar markers such as PrA, but was instead recovered from the gradient pellet (Fig. 3 B), suggesting that prAPI is first localized to a nonvacuolar membrane.
Since P22L API is transport defective (Oda et al., 1996), it is possible that prAPI that accumulates in this mutant may not be on the correct transport pathway. To examine this question, the severity of the P22L defect was followed by pulse-chase analysis. P22L API-containing cells were pulse labeled for 5 min, and then subjected to a nonradioactive chase reaction in either nitrogen-containing or nitrogen starvation medium. Nitrogen starvation increases the capacity of the API transport machinery in wild-type cells (Scott et al., 1996). In nitrogen-containing medium, the transport of P22L API is slow but not completely defective. However, when the chase was performed in SD-N, the import efficiency was significantly improved (Fig. 3 C). The rate of API delivery in this experiment is too rapid to be explained by bulk autophagy alone. These results indicate that the deficiency of P22L API can be overcome in conditions where the capacity for API transport is increased, suggesting that the defective API is maintained on the import pathway.
Identification and Purification of Subvacuolar Vesicles Containing API
Genetic and phenotypic analyses indicate that the majority of the cvt, apg, and aut mutants are defective in both API maturation and autophagy (Harding et al., 1996; Scott et al., 1996). Our results with vps18 and P22L API suggested that API enters a cytoplasmic vesicle. If API uses the same machinery as that used by the autophagic pathway, the cytoplasmic intermediate compartment may be an autophagosome (Baba et al., 1994, 1995). Autophagosomes are double-membraned structures that fuse with the vacuole, resulting in the release of a subvacuolar single-membraned vesicle termed an autophagic body (Takeshige et al., 1992; Baba et al., 1994). This vesicle is subsequently degraded in a PrB-dependent manner. Accordingly, mutations in either PrA or PrB cause the accumulation of autophagic bodies inside the vacuole (Takeshige et al., 1992), as well as causing a processing defect in API (Klionsky et al., 1992). Lack of API maturation in pep4 or prb1 mutant strains could be the result of a direct processing block and/or inaccessibility of precursor API that is contained within intact subvacuolar vesicles. To investigate whether API can be trapped within a subvacuolar vesicle, additional fractionation studies were performed using the cvt17 mutant. Like PrA and PrB mutants, cvt17 also accumulates autophagic bodies in the vacuole (Harding et al., 1996) but has the additional advantage that it is not defective in PrA, so that this protein can be used as a marker protein for the vacuolar lumen.
When cvt17 cells were subjected to differential osmotic lysis in PS200 buffer, the majority of prAPI and PrA was maintained within the pellet fraction (Fig. 4 A), while cytosolic proteins such as PGK were released into the supernatant fraction (data not shown). The prAPI in these cells was protease protected, indicating its localization to a membrane-enclosed compartment (Fig. 4 A). Additional fractionation experiments were performed to determine whether this population of API was localized in the vacuolar lumen or in a separate protease-protected compartment.
For these studies, cvt17 spheroplasts were resuspended first in PS200 to lyse the plasma membrane while maintaining vacuolar integrity. After separating the cytosolic fraction from the vacuole-containing pellet fraction by centrifugation, the resulting pellet (P200) was subjected to a second lysis step with a very low osmotic strength buffer (PS0). After separation by centrifugation at 10,000 g, this second lysis step resulted in the S0 and P0 fractions. While the majority of the vacuolar lumen marker PrA was recovered in the P200 fraction, it was released into the S0 fraction upon resuspension in PS0, indicating vacuolar lysis. However, a considerable amount of prAPI is still maintained within the pellet fraction, indicating that it is not free in the vacuolar lumen (Fig. 4 B). The pelletable prAPI is released into a soluble fraction by treatment with Triton X-100, indicating that it is enclosed within a membrane rather than an aggregate (data not shown). In addition, prAPI is collected in the pellet fraction by the relatively low force of centrifugation at 10,000 g, consistent with its association with a large compartment such as an autophagic body. Microscopy studies indicate that autophagic bodies are ∼ 300–900 nm in diam (Baba et al., 1994, 1995), much larger than secretory vesicles that exist in 50- and 100-nm classes (Novick et al., 1980).
The S0 and P0 fractions were subjected to protease treatment with proteinase K. The prAPI that was released into the S0 fraction was digested to the mature size, while prAPI in the P0 fraction was protease protected unless membranes were solubilized by the addition of detergent (Fig. 4 B), suggesting that the prAPI in the P0 fraction remains in a vesicular compartment. This result, together with the fact that the prAPI is in a less osmotically sensitive compartment than the vacuolar lumen, suggests that it may instead be within a subvacuolar vesicle such as the autophagic bodies previously identified microscopically.
Purification of Subvacuolar Vesicles from cvt17 Cells
Since purified vacuoles contain little contamination from other organelles, we used isolated vacuoles from cvt17 cells as the starting material for purification of prAPI-containing subvacuolar vesicles. Isolated vacuoles were permeabilized by resuspension in PS0 buffer to release API-containing vesicles. Optiprep step gradients were then used to separate the released subvacuolar vesicles from remaining vacuolar membranes, which may reseal into vesicles after lysis and release of luminal contents. Appropriate densities for separation of these two vesicle populations in our buffer system were determined empirically.
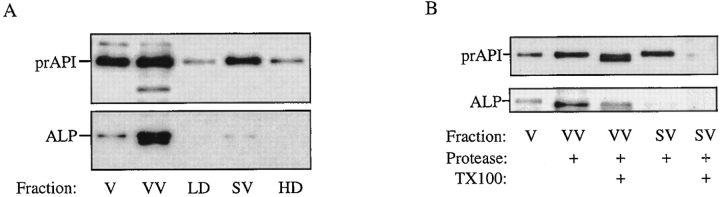
We found that vacuolar vesicles are unable to enter an Optiprep solution with a density of 1.06, while some of the prAPI-containing subvacuolar vesicles sediment through the 1.06 density step and become concentrated at the 1.06/ 1.12 interface (Fig. 5 A). The majority of vacuolar membranes in this gradient, marked by ALP, is in the VV fraction collected from the 0/1.06 density interface. This is in contrast with prAPI, which is localized in both the VV fraction and the SV fraction (the 0/1.06 and the 1.06/1.12 density interfaces). A portion of the VV fraction and the SV fraction was subjected to protease treatment. Precursor API in both fractions was protease protected in the absence of detergent, indicating that it is still localized within a membrane-bound compartment (Fig. 5 B). ALP contained in the VV fraction was digested to a slightly lower molecular weight form both in the presence and absence of detergent. As ALP is a type II transmembrane protein that contains a cytosolic domain (Klionsky and Emr, 1989), this is the expected result, indicating that the added protease is working efficiently.
Figure 5.
Precursor API is contained within subvacuolar vesicles in cvt17 mutants. (A) Isolated vacuoles (V) from cvt17 cells were lysed in PS0 buffer, loaded on top of an Optiprep step gradient, and centrifuged at 170,000 g for 60 min as described in Materials and Methods. Fractions VV, 0/1.06 interface; LD, 1.06 region; SV, 1.06/1.12 interface; HD, 1.12 region. (B) An aliquot of fractions VV and SV was treated with proteinase K either in the presence or absence of Triton X-100 as indicated. Because of the presence of protease inhibitors in the vesicle isolation procedure, an intermediate-sized API degradation product results from proteinase K treatment. The V fraction represents one-tenth of the load of the Optiprep gradient. Proteins were detected by Western blotting. The positions of prAPI and mALP are indicated.
API-containing Vesicles Can Be Detected by Immuno-EM
To provide additional evidence for a vesicular mode of API transport, cells were examined using immuno-EM with antibodies against API. Wild-type and cvt17 cells were grown in YPD to late log phase, harvested, and rapidly frozen. When wild-type cells were probed with preimmune serum, or ape1Δ cells were probed with API antiserum, no immunogold signal was detected (data not shown). Wild-type cells probed with API antiserum contained dispersed gold particles in the vacuole lumen, confirming that mature API is a soluble luminal hydrolyase and indicating that API itself does not appear to aggregate, despite its dodecameric structure (Fig. 6 A). When sections from cvt17 cells were examined, both cytosolic and subvacuolar API-containing vesicles were evident (Fig 6, B and C). The cytosolic vesicles appeared near the surface of the vacuole. These are most likely double-membraned vesicles that are approaching the vacuole surface for docking and fusion. Subvacuolar vesicles were detected in cvt17 cells, but not in wild-type cells, confirming that the accumulation of these vesicles is the result of mutation in the CVT17 gene.
Figure 6.
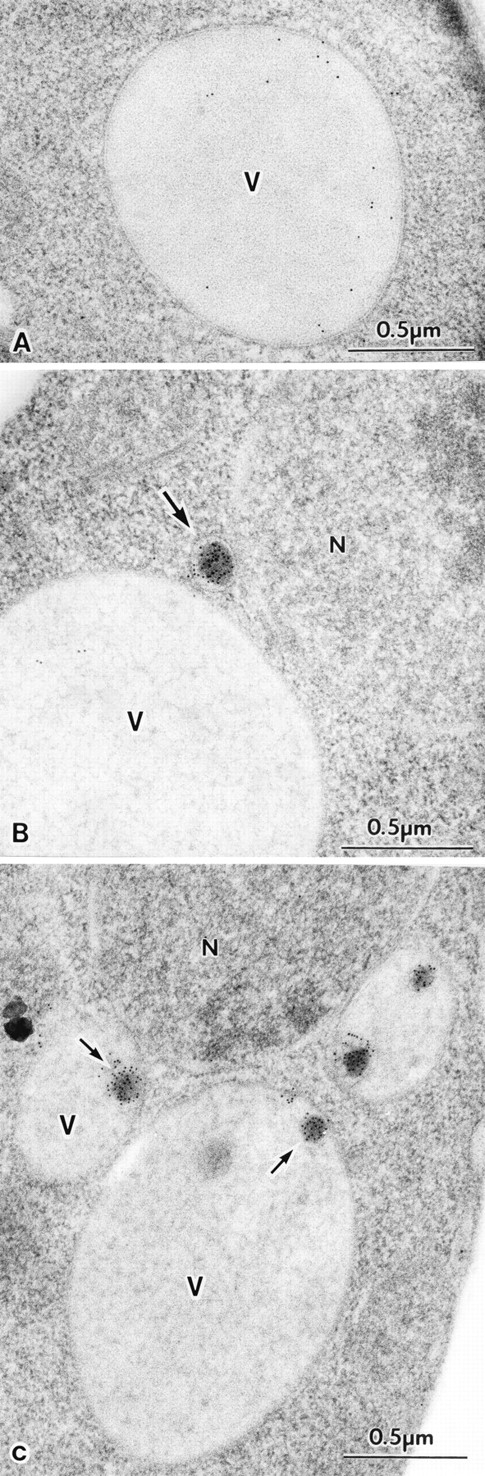
Immuno-EM of cvt17 cells. Yeast strains SEY6210 (parental wild type) and THY32 (cvt17) were grown in YPD and prepared for microscopy as described in Materials and Methods. (A) Wild-type cells probed with antibody against mature API. (B) A cvt17 section showing a cytosolic vesicle containing prAPI probed with antibody against the API proregion. (C) A cvt17 section showing subvacuolar vesicles containing API probed with antibody against mature API. N, nucleus; V, vacuole; (arrows) API containing vesicles. Bars, 0.5 μm.
Characterization of prAPI-containing Vesicles
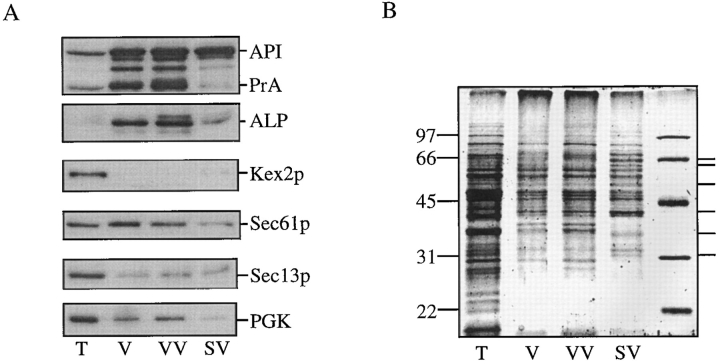
Additional analysis of the subvacuolar vesicles isolated from cvt17 cells was performed to evaluate the degree of purity attained by this fractionation procedure. After differential osmotic lysis and recovery on Optiprep step gradients, fractions were analyzed by immunoblot analysis with antibodies against marker proteins for various yeast organellar compartments. As in the experiment shown in Fig. 5, the SV fraction was nearly devoid of PrA and ALP, but still contained prAPI (Fig. 7 A). When these same fractions were probed with antibodies raised against Kex2p, a Golgi-resident protein (Redding et al., 1991), it was only detected in the total fraction. Similarly, Sec13p, a component of COPII-coated secretory vesicles (Barlowe et al., 1994), was primarily detected in the total fraction. In addition, components of the coatomer complex were detected in the total fraction, but not in the VV and SV fractions (data not shown). Sec61p, an ER membrane protein (Stirling et al., 1992), was found in all of the recovered fractions. Sec61p is not enriched in the vacuole fraction when compared with the total fraction, however, suggesting that it is contaminating this preparation, rather than specifically purified. The cytosolic marker protein PGK also contaminates the vacuole and subvacuolar fractions to a certain degree, but is not specifically enriched. These fractions were also examined by staining with SYPRO orange. The protein profiles of the vacuolar vesicle and subvacuolar vesicle fractions were clearly distinct. A number of proteins appeared specifically enriched in the subvacuolar vesicle fraction (Fig. 7 B).
Figure 7.
Characterization of API-containing vesicles. (A) Fractionations were performed as in Fig. 5. The total (T), vacuole (V), vacuolar vesicle (VV), and subvacuolar vesicle (SV) fractions are shown. Proteins were detected by immunoblotting as indicated in the figure. (B) SYPRO orange staining detected by fluorescent scanning mode on a Molecular Dynamics STORM phosphorimager. The relative mobility of the molecular mass standards are indicated on the left. Polypeptides enriched in the subvacuolar vesicles are indicated on the right.
Discussion
Recent evidence that the majority of mutants in the autophagy and Cvt pathways display the same phenotypes suggested that API transport is vesicle mediated (Harding et al., 1996; Scott et al., 1996). Additionally, studies of the oligomeric state of API during transit indicate that this protein is assembled into a dodecamer in the cytosol before transport (Kim et al., 1997). These results, coupled with the fact that API transport is extremely sensitive to low temperatures (Fig. 1) and requires a GTP-binding protein (Scott and Klionsky, 1995), indicated that the Cvt pathway may be vesicular. In this report we demonstrate biochemically and morphologically that prAPI is transported by a vesicle-mediated process.
We performed fractionation analysis on various protein targeting mutants to identify those that might accumulate transport intermediates on the API targeting pathway. Examination of prAPI that accumulates in the vps18 mutant revealed that a population of prAPI is trapped in a nonvacuolar vesicular compartment in this strain (Fig. 2). Although the precise role of Vps18p is not known, it is involved at a late step in vacuolar protein sorting, presumably after the endosome (Preston et al., 1991; Robinson et al., 1991). This result, together with the fact that the P22L API mutant accumulates on a nonvacuolar membrane (Fig. 3), indicates that prAPI first binds and then becomes encapsulated in a vesicle before delivery to the vacuole. These results are supported by immunoelectron microscope analysis, which indicates the presence of API-containing vesicles in the cytosol (Fig. 6 B).
We predicted that, analogous to macroautophagy, API is transported inside double-membraned vesicles (autophagosomes) that are formed in the cytosol. The autophagosomes would be subsequently targeted to the vacuole where the outer membrane would fuse with the vacuolar membrane, releasing the inner vesicle (autophagic body) into the vacuolar lumen. This hypothesis is supported by subcellular fractionation experiments using cvt17 that demonstrate the presence of API-containing subvacuolar transport vesicles (Figs. 4 and 5). In addition, immuno-EM of cvt17 confirmed the presence of API-containing subvacuolar vesicles (Fig. 6). Purification and characterization of the complement of proteins contained in this compartment suggest that proteins unique from those of the vacuole make up these subvacuolar transport vesicles (Fig. 7).
Together, these data indicate that API is constitutively packaged into cytosolic vesicles that are then transported to the vacuole. These vesicles could either be targeted directly to the vacuole, or could first fuse with an endosomal compartment. In the event that they fuse at the vacuole, they may use common recognition and fusion components such as a vacuolar t-SNARE and v-SNARE, as well as the sec17/sec18 complex, to facilitate this reaction. If fusion instead occurs at the endosome, then all of the components necessary for late steps in vacuolar targeting or maturation would be required for vacuolar delivery. This is similar to the situation that has been proposed in mammalian cells, where lysosomes develop through the fusion of various different classes of endosomal compartments (Dunn, 1995). Possibly the yeast endosomal system operates similarly: autophagic vesicles and secretory vesicles carrying vacuole-destined cargo could both fuse and deliver their contents to the endosome. Through continued fusion reactions, this compartment could develop into an enzymatically mature vacuole. Further studies will provide information on the mechanism of API import and in particular the stage at which it integrates with vacuolar delivery through the secretory pathway and endosomal systems.
These results clearly demonstrate that API transport is a vesicular process that has many similarities to the degradative macroautophagic pathway. Despite this evidence, the relationship between the constitutive API delivery pathway and macroautophagy remains unclear. In addition, many uncertainties remain regarding protein trafficking by autophagic vesicles. These include such basic mechanistic questions as the membrane of origin of autophagic vesicles and what triggers the formation, movement, fusion, and, finally, breakdown of these transport vesicles. The availability of purified vesicular compartments and the use of API as a marker protein will be valuable tools for these studies.
Acknowledgments
We thank Dr. Masako Osumi, Japan Women's University for the use of EM facilities, John Kim for helpful discussion and critical reading of the manuscript, and Drs. C. Barlow, G. Payne, R. Schekman, and J. Thorner for providing antisera.
This work was supported by Public Health Service grant GM53396 from the National Institutes of Health to D.J. Klionsky.
Footnotes
1. Abbreviations used in this paper: ALP, alkaline phosphatase; API, aminopeptidase I; prAPI, API precursor; CPY, carboxypeptidase Y; HD, high density; LD, low density; PGK, phosphoglycerate kinase; PrA, proteinase A; PrB, proteinase B; SV, subvacuolar vesicle; VV, vacuolar vesicle; V, vacuole fraction; I, intermediate fraction; P, pellet fraction; Vps, vacuolar protein sorting.
Please address all correspondence to Daniel J. Klionsky, Section of Microbiology, University of California, Davis, CA 95616. Tel.: (916) 752-0277. Fax: (916) 752-9014.
References
- Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- Baker D, Schekman R. Reconstitution of protein transport using broken yeast spheroplasts. Methods Cell Biol. 1989;31:127–141. doi: 10.1016/s0091-679x(08)61605-2. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vacuolar biogenesis in yeast: sorting out the sorting proteins. Cell. 1995;83:513–516. doi: 10.1016/0092-8674(95)90088-8. [DOI] [PubMed] [Google Scholar]
- Dunn WA. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1995;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Hilt W, Wolf DH. Stress-induced proteolysis in yeast. Mol Microbiol. 1992;6:2437–2442. doi: 10.1111/j.1365-2958.1992.tb01419.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Scott SV, Oda M, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biogenesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO (Eur Mol Biol Organ) J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Banta LM, Emr SD. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol Cell Biol. 1988;8:2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiaeis localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. . Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD, Klionsky DJ. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J Cell Biol. 1996;132:999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston RA, Manolson MF, Becherer K, Weidenhammer E, Kirkpatrick D, Wright R, Jones EW. Isolation and characterization of PEP3, a gene required for vacuolar biogenesis in Saccharomyces cerevisiae. . Mol Cell Biol. 1991;11:5801–5812. doi: 10.1128/mcb.11.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. . J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H. Yeast endocytosis. Trends Cell Biol. 1993;3:273–277. doi: 10.1016/0962-8924(93)90056-7. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Graham TR, Emr SD. A putative zinc finger protein, Saccharomyces cerevisiaeVps18p, affects late Golgi functions required for vacuolar protein sorting and efficient α-factor prohormone maturation. Mol Cell Biol. 1991;11:5813–5824. doi: 10.1128/mcb.11.12.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Klionsky DJ. In vitro reconstitution of cytoplasm to vacuole protein transport in yeast. J Cell Biol. 1995;131:1727–1735. doi: 10.1083/jcb.131.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm to vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segui-Real B, Martinez M, Sandoval IV. Yeast aminopeptidase I is post-translationally sorted from the cytosol to the vacuole by a mechanism mediated by its bipartite N-terminal extension. EMBO (Eur Mol Biol Organ) J. 1995;14:5476–5484. doi: 10.1002/j.1460-2075.1995.tb00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Horazdovsky B, Emr SD. Receptor-mediated protein sorting to the vacuole in yeast: roles for a protein kinase, a lipid kinase and GTP-binding proteins. Annu Rev Cell Dev Biol. 1995;11:1–33. doi: 10.1146/annurev.cb.11.110195.000245. [DOI] [PubMed] [Google Scholar]
- Stirling CJ, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch M, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. . FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. . FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]