Abstract
The development of an induced transcript environment was investigated at the supramolecular level through comparative localization of the human cytomegalovirus immediate early (IE) transcripts and specific nuclear domains shortly after infection. Compact aggregates of IE transcripts form only adjacent to nuclear domain 10 (ND10), and the viral protein IE86 accumulates exclusively juxtaposed to the subpopulation of ND10 with transcripts. The stream of transcripts is funneled from ND10 into the spliceosome assembly factor SC35 domain through the accumulation of IE86 protein, which recruits some components of the basal transcription machinery. Concomitantly the IE72 protein binds to ND10 and later disperses them. The domain containing the zinc finger region of IE72 is essential for this dispersal. Positional analysis of proteins IE86 and IE72, IE transcripts, ND10, the spliceosome assembly factor SC35, and basal transcription factors defines spatially and temporally an immediate transcript environment, the basic components of which exist in the cell before viral infection, providing the structural environment for the virus to usurp.
The nucleus is increasingly recognized as a highly compartmentalized structure. The spatial distribution of different chromosomes into discrete territories (Cremer et al., 1982; Lichter et al., 1988; Pinkel et al., 1988) is but one level of organization. In addition, an increasing number of extrachromosomal domains are being described and assigned functions in the overall integrated system of differential gene expression. Individual genes have not been located to specific sites in the nucleus except to those which they induce, in the case of ribosomal genes. Recently however, some genes have been shown to be located adjacent to sites containing pre-mRNA splicing components (Fu and Maniatis, 1990; Spector 1990) and poly(A)+ RNA (Carter et al., 1991; Visa et al., 1993; Xing et al., 1993, 1995). These nonchromosomal sites are distributed in 30–40 major nuclear domains to which the spliceosome assembly factor, SC35, localizes (Fu and Maniatis, 1990) and correspond to interchromatinic granule clusters and possibly perichromatin fibrils (Fakan and Bernard, 1971; Fakan and Puvion, 1980; Spector et al., 1991; Visa et al., 1993). These SC35 domains appear important for the processing of transcripts, as several highly transcribed genes line their periphery and as their transcripts as well as transcripts from transiently transfected genes are funneled through this compartment (Huang and Spector, 1991, 1996; Xing et al., 1995). Nontranscribed genes showed no such preferential location (Xing et al., 1995). We wondered where genes induced to high transcriptional rates would be located if allowed to choose the site of transcription. The double requirement of movement and strong induction of transcription is given for DNA viruses. We therefore used the human cytomegalovirus as a model to investigate, at the supramolecular level, the nuclear environment of an induced gene.
The viral particle is very large, and disassembly before the entry into the nucleus is likely, since no infectious core particle is ever observed in the nucleus. Viral DNA or some DNA–protein complex enters the nucleus, and it might be expected that immediate early gene transcription could start anywhere in the nucleus, since polymerase II activity is distributed throughout the nucleus (Wansink et al., 1993; Dirks et al., 1995). However, we recently showed that viruses from three virus families begin replication in the immediate vicinity of a nuclear domain designated ND10 (Ishov and Maul, 1996; Maul et al., 1996). ND10 (Ascoli and Maul, 1991; Maul and Everett, 1994), also referred to as POD or promyelocytic leukemia protein (PML)1 bodies (Dyck et al., 1994; Weiss et al., 1994) are multiprotein complexes, and most proteins found in these domains are upregulated by interferons (Guldner et al., 1992; Koken et al., 1994; Lavau et al., 1995). The function of these proteins is not known, although one of them, PML, has been shown to have growth repressive properties (Mu et al., 1994; Le et al., 1996). ND10 have previously been shown to be modified after herpes simplex type 1 (HSV-1), human cytomegalovirus (HCMV), and adenovirus 5 (Ad5) infection (Maul et al., 1993; Maul and Everett, 1994; Carvalho et al., 1995; Doucas et al., 1996). The large DNA viruses appear to require the dispersion of ND10-associated proteins.
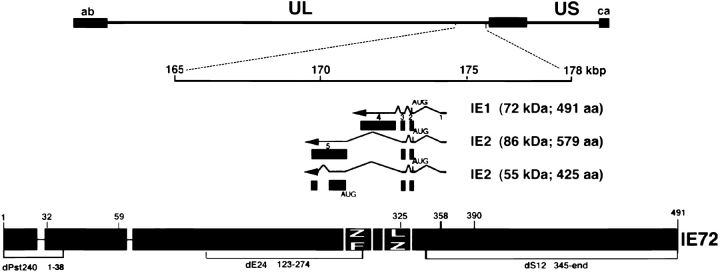
Herpes viruses start their replication cycle in the nucleus by a coordinated expression of viral genes, beginning with immediate early (IE) gene transcription, which is required for subsequent early and late gene transcription. Transcription of the major IE gene region (UL 122-123) for HCMV results in a single 18-kb pre-mRNA, which forms several mRNA species through differential splicing and polyadenylation site usage (Stenberg et al., 1984, 1985, 1989; Stinski et al., 1983). These transcripts include the mRNA for the IE1 (IE72) and the IE2 (IE86) proteins. They synergistically activate viral and cellular gene expression (for reviews see Mocarski, 1996; Stenberg, 1996). The IE72 protein can upregulate the IE promoter (Cherrington and Mocarski, 1989; Stenberg et al., 1990); and the IE86 protein acts through a cis-repression signal to repress this promoter (Pizzorno and Hayward, 1990; Cherrington et al., 1991; Liu et al., 1991). IE86 interacts with a number of cellular proteins such as the TATA-binding protein (TBP; Hagemeier et al., 1992; Jupp et al., 1993; Sommer et al., 1994), the basal transcription factor TFIIB (Caswell et al., 1993), the retinoblastoma protein Rb (Sommer et al., 1994; Hagemeier et al., 1994), the cellular transcription factor CREB (Lang et al., 1995), and transcription factors SP1 and Tef-1 (Lukac et al., 1994). Both IE72 and IE86 are accumulated in the nucleus (Kelly et al., 1995; Korioth et al., 1996).
In the present study, we sought to determine the spatial and temporal changes in the nucleus shortly after HCMV infection. We determined ND10 to be the site of viral transcription from which the transcripts directionally move to the SC35 domain and documented the positional changes in the IE proteins of HCMV over time relative to ND10 and SC35 domains. In addition, we established the molecular domain of the IE72 protein that affects ND10 integrity and demonstrated that an IE86 cloud surrounding the emerging transcripts accumulates some components of the basal transcription machinery. The specific positional arrangement of a number of intranuclear compartments suggests that the virus develops an immediate transcript environment constrained by preexisting nuclear structures.
Materials and Methods
Antibodies and Cell Culture
ND10 were visualized using the following antibodies: mAb 1150, which recognizes Sp100 (Maul et al., 1995), mAb 5E10 (Stuurman et al., 1992) which recognizes PML (Maul et al., 1995), polyclonal rabbit antiserum against PML (Dyck et al., 1994), polyclonal rabbit anti-peptide antibodies 8575 and 1218, which interact with IE72 and IE86, respectively (Stenberg et al., 1989). Polyclonal anti-TFIIB rabbit antiserum (Santa Cruz Biotechnology, Santa Cruz, CA), mAb that recognizes TBP (Promega Biotech, Madison, WI, and from L. Hernandez, Cold Spring Harbor Laboratory (Cold Spring Harbor, NY); Ruppert et al., 1996), anti-p53 mAb (from Oncogene), mAbs anti-IE72 (HA 3.118) and anti-IE86 (SMX; Lang et al., 1995), and mAb anti-SC35 (Fu and Maniatis, 1990) were also used. Human antibody 1745, which recognizes Sp100 and PML (Maul et al., 1995), was used in triple-labeling experiments. Isotype-matched mAbs of unrelated specificity were used as controls.
Human WI38 fibroblasts were maintained in minimal essential medium supplemented with 10% FCS and antibiotics. Cells were plated on round coverslips in 24-well plates (Corning Glass, Inc., Corning, NY), grown at 37°C in a humidified atmosphere containing 5% CO2 until ∼80% confluent, and infected.
Virus Infection
2 d after plating, WI38 cells were infected with HCMV (Towne strain) at a multiplicity of one per cell, resulting in 95% infected cells as determined by staining with IE72 antibodies at 5 h after infection. Use of higher and lower multiplicities resulted in different percentages of cells infected and changes in the temporal progression of infection but did not affect the relative positional location of any of the tested components. Cells were fixed at different time intervals after infection and were assayed with different antibodies or by in situ hybridization. HSV1 infection was carried out as described by Maul et al. (1996).
Immunolocalization of Virus and Host Proteins
Cells were fixed at room temperature for 15 min with freshly prepared 1% paraformaldehyde in PBS, washed with PBS, and permeabilized for 20 min on ice with 0.2% (vol/vol) Triton X-100 (Sigma Chemical Co., St. Louis, MO) in PBS. Antigen localization was determined after incubation of permeabilized cells with rabbit antiserum, mAb, or human antiserum diluted in PBS for 1 h at room temperature. Avidin–fluorescein or avidin– Texas red was complexed with primary antibodies through biotinylated secondary antibodies (Vector Laboratories, Burlingame, CA). Cells were double or triple labeled with the respective second antibodies conjugated with FITC, Texas red, or Cy-5 using biotin–avidin (Rockland, Gilbertsville, PA) enhancement and FITC for structures with the lowest staining intensity. Cells were then stained for DNA with 0.5 μg/ml of bis-benzimide (Hoechst 33258; Sigma Chemical Co.) in PBS and mounted with Fluoromount G (Fisher Scientific, Pittsburgh, PA). Cells were analyzed using a confocal scanning microscope (Leica Inc., Deerfield, IL). The two channels were recorded simultaneously when no cross-talk was detectable. In the case of intense FITC labeling, sequential images were acquired with more restrictive filters to prevent possible breakthrough of the FITC signal into the red channel. Both acquisition modes gave the same results. Image enhancement software (Leica Inc.) was used solely in balancing signal strength. Because of the variability of the infectious cycle progression in any given culture, the most prevalent and representative images were photographed and presented as limited number of nuclear images to retain high magnification. At least 500 cells were studied in each sample. The experiments were repeated at least three times and evaluated by two investigators.
In Situ Hybridization of Viral DNA and RNA
Fluorescence in situ hybridization techniques were used for the localization of virus DNA and RNA as described (Lawrence et al., 1989; Huang and Spector, 1991; Ishov and Maul, 1996). The hybridization probe for HCMV was prepared using pSVH which encodes the major IE gene region of the HCMV genome (Stenberg et al., 1990). The hybridization probe for HSV1 has been described (Maul et al., 1996). Plasmid DNA was nick translated in the presence of biotinylated 11-dUTP (Sigma Chemical Co.). The DNase concentration and the duration of nick translation were adjusted to obtain probe fragments 200–500 bp in size. Cells infected with HCMV for various time periods were fixed with freshly prepared 1% paraformaldehyde and permeabilized with 0.2% Triton X-100. For DNA visualization, cellular and probe DNA were denatured simultaneously for 3 min at 90°C on a heat block. For RNA visualization, only probe DNA was denatured. Mock infected cells were used as controls. Hybridization was performed for 2 h at 37°C in a hybridization mixture containing 50% deionized formamide, 10% dextran sulfate, 1× SSC, and 10 ng of biotinylated viral DNA, 1 μg of salmon sperm DNA, and 1 μg of human DNA (for DNA hybridization) or 1 μg yeast tRNA (for RNA hybridization) per μl. After hybridization, cells were washed three times in 50% formamide/ 2× SSC prewarmed to 42°C followed by three washes for 5 min each in 0.1× SSC prewarmed to 60°C. Washed cells were stained with fluoresceinated avidin (Vector Laboratories) in 3% BSA/2× SSC. The signal was enhanced with an additional round of biotinylated anti-avidin antibodies followed by fluoresceinated avidin treatment. All preparations were stained with Hoechst 33258. For DNase treatment, coverslips were incubated with RNase-free DNase I (10 μg/ml) in the presence of 5 mM Mg2+ and RNase inhibitor (1 U/μl RNasin; Promega Biotech) for 2 h at room temperature. The effectiveness of DNase treatment was judged based on residual Hoechst 33258 staining. RNase digestion was carried out using 20 μg/ml RNase (DNase free; Boehringer Mannheim, Indianapolis, IN) for 2 h at room temperature. For simultaneous immunohistochemical procedures, cells were stained with the respective antibodies as described above after the in situ hybridization.
Transfection
Cells were transiently transfected by calcium phosphate precipitation (Chen and Okayama, 1987) and assayed at various times later using antibodies to various ND10 antigens and HCMV proteins. pIE86, IE55, pSVH, and the insertion mutants m1, m2, and m16–m19 have been described (Stenberg et al., 1990). The IE72 and IE72 mutants encoding plasmids pSVCC3, dE24, and dS12 were previously described (Stenberg and Stinski, 1985). Plasmid dPst240, derived from pSVCC3, is an in-frame deletion of the 240-bp PstI fragment in the amino terminus of IE72 (see Fig. 3). Plasmid pSVOd, which contains no HCMV-specific sequences (Stenberg et al., 1990), was used as a negative control.
Figure 3.
Outline of the HCMV genome depicting the IE transcript with mutations used in transfection experiments. Short lines with numbers at the upper part of the IE bar show the location of insertion mutants and brackets at the lower part show deletions in the IE1 gene.
Results
IE Transcripts Are Not Randomly Localized
To visualize viral RNA at various times after infection, we used in situ hybridization with an IE probe of HCMV under nondenaturing conditions in HCMV-infected WI38 cells. At 2 h after infection, immediate early transcripts were detected in a low number of specific short tracks (Fig. 1 a). A control image of mock-infected cells is shown in Fig. 1 b. The combined use of in situ hybridization with immunohistochemical localization of different nuclear domains was used to determine the spatial relationship of the transcript tracks with various nuclear domains. Nearly all transcript tracks overlapped part of the SC35 domain (Fig. 1 c), although the number of transcript signals was lower than the number of SC35 domains. The signal derived from the transcript neither filled the entire SC35 domain, nor was all of the in situ hybridization signal contained in the SC35 domain. These images did not allow us to distinguish the transcription start site along the elongated tracks, i.e., whether the transcript stream enters or leaves the SC35 domain.
Figure 1.
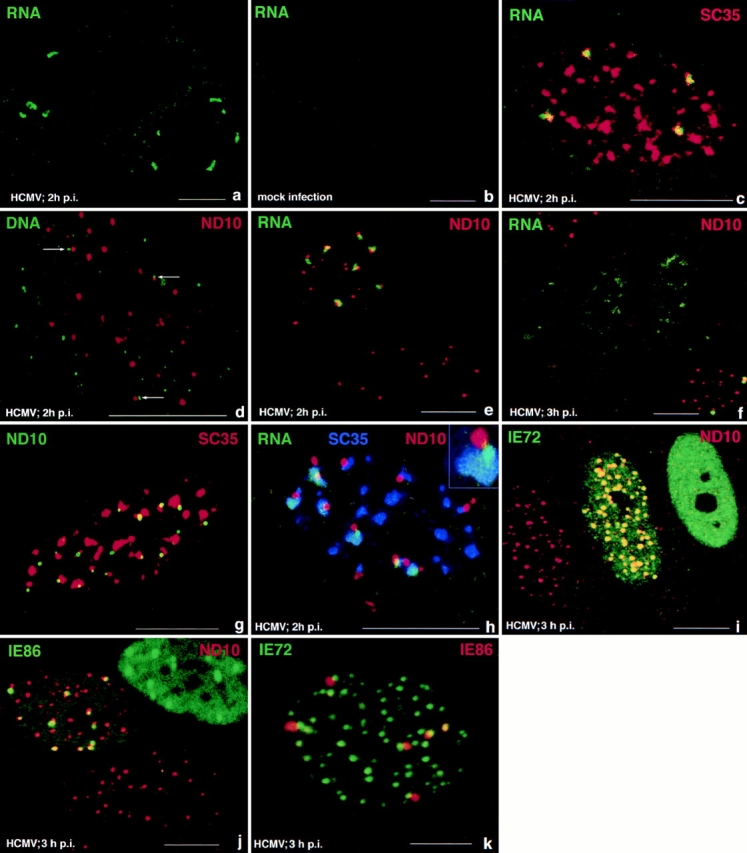
Correlation of HCMV IE gene transcripts with various nuclear domains as visualized by combined immunohistochemistry and in situ hybridization. Confocal microscopy was used to document single optical sections of WI38 cells, which are presented at high magnification. Pseudocolors were chosen to reflect either FITC labeling (green), Texas red (red), or cy5 (blue) as indicated in the upper part of each panel for the respective macromolecules or structures labeled. (a) Two infected cells showing a limited number of short and compact IE transcript-derived signals; (b) control, mock-infected cell to demonstrate probe specificity. (c) Nucleus double labeled for IE transcripts and the SC35 domain. The transcripts partially overlap with the SC35 domain. (d) In situ hybridization using the IE gene probe under denaturing conditions and after RNase treatment. Small punctate hybridization signals indicate input viral genomes, and some are found juxtaposed to ND10 (arrows). (e) Double labeling of IE transcripts and ND10. The lower cell is uninfected, and the upper cell shows the transcript signals juxtaposed to ND10. (f) Same as e but at 3 h after infection. A cell in the lower right corner shows the same configuration as at 2 h after infection, but in three nuclei seen in the center, the RNA signal is less compact, and no ND10 are seen. (g) Uninfected cell double labeled for ND10 and the SC35 domain. A large number of ND10 appear associated with the rim of the SC35 domain. (h) Infected cell triple labeled for ND10, IE transcripts, and SC35. Transcript signal appears to locate with highest concentration at ND10 and is also found in the SC35 domain. Inset shows the arrangement as expected when all three components appear in the same plane. (i) Cells double labeled for ND10 and IE72. The left nucleus is apparently not infected, the middle nucleus shows IE72 and ND10 colocalization at all ND10, and the right nucleus shows only dispersed IE72 and no ND10. (j) Cells double labeled for ND10 and IE86. The lower cell appears uninfected, the top left nucleus shows IE86 located adjacent to a few ND10, and the top right nucleus reveals IE86 throughout, with some sites of higher concentration. (k) Double labeling of IE72 and IE86. All IE86 accumulations are located next to a few IE72 sites. Bars, 10 μm.
We had previously determined that several DNA viruses are deposited at ND10 (Ishov and Maul, 1996). To confirm that HCMV input virus was also deposited at ND10 like HSV-1 (Maul et al., 1996), we tested, by in situ hybridization under denaturing conditions and RNase digestion, the location of the input viral DNA. Labeling ND10 in the same preparation showed at 2 h after infection that a high percentage of viral input DNA was still in the cytoplasm, and from those located in the nucleus only a few were positioned adjacent to ND10 (Fig. 1 d, arrows). Presumably transcription may start on any of the virus genomes in the nucleus. However, the experimental determination of immediate early transcript sites relative to ND10 location at 2 h after infection showed clearly that although not all ND10 contained a transcript signal, all transcripts were associated with ND10 (Fig. 1 e). Thus only genomes deposited at ND10 began to transcribe. Later, beginning at 3 h after infection, we found that in some cells ND10 were dispersed and the transcript signal became diffuse. An intermittent stage is shown in the three nuclei in the center of Fig. 1 f, where ND10 are dispersed, and the transcripts are not strictly contained but partially dispersed. Within the same microenviroment (right lower side of Fig. 1 f) some cells retained ND10 and contained the transcript in the same tight configuration as shown in Fig. 1 e. Such cells may be infected with a lower viral load and progress more slowly through the IE transcription program.
If the virus begins transcription at ND10 and the transcripts are partially overlapping the SC35 domain, ND10 should be positioned close to the SC35 domain. Whether this was the consequence of viral infection was tested by double labeling of uninfected cells to determine the relative position of ND10 and the SC35 domain. By quantitative assessment we found that in uninfected cells the rim of the SC35 domain was juxtaposed to ∼80% of all ND10 (Fig. 1 g). The close physical proximity between ND10 and the SC35 domain exists therefore in uninfected cells.
We used triple labeling to determine whether the transcript emerged randomly from ND10 or whether the SC35-ND10 apposition site determined the transcript emergence from ND10. Transcripts were not found opposite the ND10-SC35 apposition site; rather they appeared to emanate from ND10 closest to the SC35 domain. This point also showed the highest concentration of transcripts (Fig. 1 h). The insert most clearly shows this arrangement. Together with the deposition of input viral genomes at ND10, this image suggests polarization of the transcription track with the beginning at ND10 and a gradual spreading into the SC35 domain.
Differential Nuclear Localization of IE72 and IE86
Since both IE72 and IE86 are involved in the regulation of HCMV IE transcription we decided to test if specific intranuclear accumulation of these proteins accompanies IE transcript accumulation. In addition we had observed that during the early stages of HCMV infection, ND10-associated proteins became dispersed. Questions arising were which of the viral IE proteins determines this effect and whether a preliminary accumulation of this protein at ND10 is necessary for modification of ND10. Double immunolabeling to localize ND10 and IE72 proteins simultaneously in HCMV-infected cells (3 h after infection) revealed (Fig. 1 i) a variety of different images. The cell at the left appears uninfected, as it does not contain the green-labeled IE72 protein. The middle cell shows yellow-labeled nuclear domains and a greenish background due to IE72 staining. Therefore, in this cell, IE72 and ND10-associated proteins are colocalized. In the cell at the right only a rather homogeneously distributed IE72 is recognized, no specific accumulation of IE72 is seen, and ND10 staining is absent. This indicates that as early as 3 h after infection either a colocalization of ND10 and IE72 or the loss of ND10 and homogeneous nuclear distribution of IE72 can be seen.
Since IE72 and IE86 contain the same NH2-terminal region from the first two exons, we determined whether the targeting domain that brings IE72 to ND10 is also present in IE86. At 3 h after infection, IE86 localized at ND10 but did not occupy the exact same space, i.e., it was located juxtaposed to ND10 (Fig. 1 j, top left cell). Again as with IE72, a few cells at this time after infection had no ND10, but besides diffuse IE86 distribution, some specific accumulation of this protein remained (Fig. 1 j, top right cell). However, the major difference between IE72 and IE86 is that IE72 was always found at all ND10, whereas IE86 accumulated at only a few and was positioned juxtaposed to ND10, suggesting that IE72 and IE86 are positioned next to each other and that all IE86 should be situated at IE72 sites. When infected cells were double labeled for IE72 and IE86, all IE86 were seen located in domains beside a few IE72 domains (Fig. 1 k). This finding implies that both proteins are targeted to the same nuclear structure, ND10, but bind to different components of this nuclear subcompartment.
Since the distribution of IE72 and IE86 differed at a given time after infection, we determined by time-course analysis whether this reflected sequential changes or was possibly due to a difference in amount of virus input. Fig. 2 shows the changes in the distribution of IE72 and IE86 from a single experiment, i.e., from cells grown and infected at the same time. Cells fixed at different time points after infection were double labeled for ND10 and IE72 or for ND10 and IE86, counted from random fields, and categorized as cells with ND10 only, cells with ND10 and with IE72 or IE86 (Fig. 1 i, middle and top left), cells with either IE72 or IE86 domains with no ND10 staining (Fig. 1 j, right), and those cells with either IE72 or IE86 diffuse staining (Fig. 1 i, top right). For each time point, 500 cells were analyzed by two investigators. As early as 1.5 h after infection, both IE72 and IE86 domains are recognizable at ND10 in some cells. IE72 colocalized with ND10 for more than 1 h, possibly longer, before the first cells displaying IE72 domains only or diffuse IE72 without ND10 were detected. By 4 h after infection, most cells showed only IE72 dispersed throughout the nucleus. This time course suggests that IE72 needs to accumulate in ND10 for a substantial time and disperses together or shortly after ND10 have dispersed. An observation that can not be gleaned from the counts is that within the same cell, ND10 appeared to disperse differentially, i.e., most ND10 disappear early, but a few remained longer.
Figure 2.
Time course of IE72 and IE86 localization relative to ND10. 500 cells were evaluated for each time point for ND10 and IE72 or ND10 and IE86. Each cell was placed in either of four categories: with ND10 only, with ND10 and either IE72 or IE86, with IE72 or IE86 in specific domains without ND10, or with only homogeneous IE72 or IE86 staining. These figures are from a single experiment with >95% infected cells at 6 h after infection (3 PFU). Minor variations in this time course were found when lower or higher PFU were used.
Although IE86 and IE72 accumulation appear at ND10 at the same time, IE86 remained there for a substantially longer time as most of the cells show IE86 domains even at 6 h after infection. Thus the distribution of IE72 and IE86 differ not only in position and number relative to ND10, but also in the retention of IE86 at specific sites over time. A population of ND10 which remained longer in a given cell was found in association with IE86 (see below and Fig. 4 h) suggesting that the presence of IE86 correlates with ND10 preservation.
Figure 4.
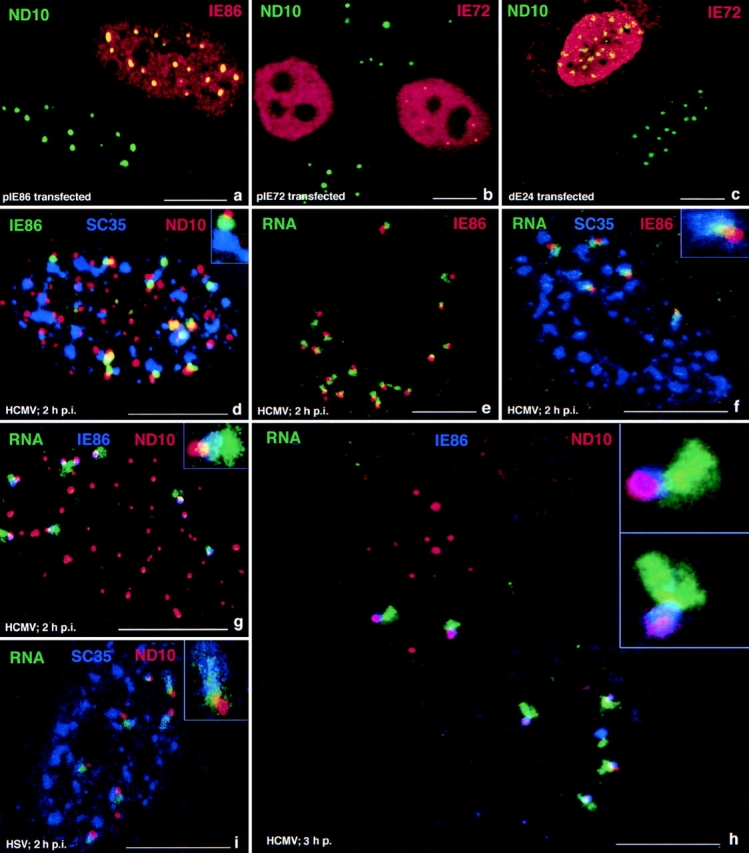
IE72, IE86, and IE transcript localization relative to other nuclear domains in human WI38 fibroblasts after transfection or infection with HCMV. (a) Cells transfected with pIE86 expressing IE86 and double labeled for ND10 and IE86. The bottom left nucleus is untransfected. The top nucleus shows a high concentration of IE86 and colocalization with ND10, which remain intact in IE86- expressing cells. (b) Cells transfected with pIE72 and double labeled for ND10 and IE72. The left transfected nucleus shows no ND10, whereas the right transfected cell is one of the few that had retained some ND10 16 h after transfection. (c) Cell transfected with dE24 expressing the putative zinc finger region deletion mutant of IE72 and double labeled for ND10 and IE72. The bottom nucleus is untransfected, and the top one shows retention of ND10 in the transfected cell. (d) HCMV-infected cell triple labeled for ND10, IE86, and the SC35 domain. IE86 domains associated with ND10 are also associated with the SC35 domain. The inset illustrates the presumed positioning if all components were in two dimensions. (e) Single nucleus double labeled for the IE86 protein and transcripts. Both signals overlap partially or appear next to each other. (f) Cell triple labeled for IE86, IE transcripts, and the SC35 domain. The IE86 cloud partly overlaps with the transcript signal, most of which is in the SC35 domain. (g) Cell triple labeled for ND10, IE transcripts, and IE86 protein. IE86 appears as a cloud surrounding the transcript exemplified in the inset. (h) Same as g but showing two cells at slightly different stages of ND10 removal. The top left nucleus still has all ND10 and only two transcript signals. The lower right nucleus has very few and smaller ND10, all of which are associated with transcript signals. The insets show one such immediate transcript environment from each nucleus. (i) HSV-1–infected cell labeled for ND10, IE1 gene transcripts, and SC35 domains, demonstrating the same general development as for HCMV. Inset shows at higher magnification that the transcript signals have the same position to ND10 and SC35 domains. Bars, 10 μm.
Interaction of IE72 with ND10
The different localization of IE72 and IE86 raised the question whether this difference was due to the individual ND10 or whether the presence of virus determined the ND10 at which IE86 was deposited. We transiently transfected vectors that expressed both IE72 and IE86 or both proteins individually and determined the location of either proteins at various times after transfection. Expression from pSVH resulted in both proteins localized at ND10 at early times after transfection (data not shown) and loss of ND10 ∼12 h after transfection (Table I). Transient expression of IE86 did not disperse ND10, and more importantly, IE86 fully colocalized with all ND10 (see Fig. 4 a, and Table I). This suggests that both the localization of IE86 juxtaposed to ND10, and its localization to only a few ND10 occurs only in the context of viral infection and that IE86 does not disperse ND10. IE72 was also initially deposited at ND10 as expected but later dispersed ND10- associated proteins. Its high concentration in a few nuclei that retained ND10 until 16 h after transfection (see Fig. 4 b, right nucleus with yellow dots) suggests that the fast elimination of ND10 takes place only in the context of viral infection.
Table I.
Determination of the IE Domain Necessary for ND10 Dispersal
| Plasmid | Protein expressed | Insertion points or deletion, aa | ND10 elimination | |||
|---|---|---|---|---|---|---|
| pSVH | IE72+86 | wt | + | |||
| pSVCC3 | IE72 | wt | + | |||
| pIE86 | IE86 | wt | − | |||
| pIE55 | IE55 | wt | − | |||
| Insertion mutants | ||||||
| pSVH m1 | IE72+86 | 32 | + | |||
| pSVH m2 | IE72+86 | 59 | + | |||
| pSVH m16 | IE72+86 | 325 | + | |||
| pSVH m17 | IE72+86 | 325 | + | |||
| pSVH m18 | IE72+86 | 358 | + | |||
| pSVH m19 | IE72+86 | 390 | + | |||
| Deletion mutants | ||||||
| pSVCC3 dE24 | IE72 | 132-274 | − | |||
| pSVCC3 dS12 | IE72 | 345-end | + | |||
| pSVCC3 dPst240 | IE72 | 1-38 | + |
aa, amino acid; wt, wild type
We have established that proteins that are encoded by the IE region express a protein that eliminates ND10. We also showed that neither the expression of the IE86 nor the IE55 protein (alternatively spliced form of IE86) can do so and that IE72 alone can disperse ND10-associated proteins (Table I). We therefore used insertion mutations or deletion mutations to determine which part of the IE72 protein is essential to disperse ND10 (see map of the IE region, Fig. 3). As shown in Fig. 4 c and Table I, only transfection with an expression plasmid where amino acids 132–274 (dE24) were deleted, retained ND10. All other mutants, including the deletion in dPst240 (deleted from amino acids 1–38) retained ND10 dispersion capability. These results suggest that the putative zinc finger region located between amino acids 266–285 may be necessary for ND10 dispersion and thus a likely candidate for protein–protein interaction that influences ND10 integrity.
IE86 Connects ND10 and the SC35 Domain
As shown, the IE86 protein domain attaches to the rim of ND10 and ND10 localized predominantly at the periphery to the SC35 domain. Triple labeling was therefore necessary to determine whether IE86 was positioned randomly around ND10 or whether the ND10-SC35 apposition influenced the position of IE86. Surprisingly, IE86 was not positioned away from the ND10-SC35 connection but rather appeared to be either interposed between ND10 and SC35 or immediately beside the apparent ND10-SC35 attachment (Fig. 4 d). The appearance of a polarized structural unit emerges and is exemplified in the Fig. 4 d inset, where IE86 is placed between ND10 and SC35.
The deposition and colocalization of IE86 to all ND10, if expressed from transfected plasmids, contrasts strongly with the selective position of this protein between a few ND10 and the SC35 domain shortly after infection. The general question of what causes this selective IE86 deposition was approached by asking whether the sites of immediate early HCMV transcription could determine IE86 localization. The transcripts were clearly found in close association with IE86, and all IE86 domains have transcripts associated with them (Fig. 4 e). Triple labeling showed that IE86, the transcript and the SC35 domain partially overlapped (Fig. 4 f, inset). Fig. 4 g shows the spatial relationship of ND10, transcripts, and IE86. In the inset a cloud of IE86 appears to surround the transcript emanating from ND10 but only for a certain distance, after which the transcript appears devoid of IE86. Of the four elements correlated, ND10, IE86 domains, IE transcripts, and the SC35 domain, only three can be visualized simultaneously (Figs. 1 h and 4, d, f, and g). These four images, however, allow aggregation into a schematic representation which is shown as Fig. 6 (see Discussion).
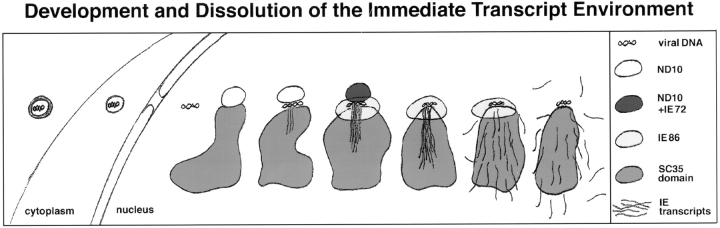
Figure 6.
Schematic sequence of events during the immediate early steps of HCMV infection. Key to structures and shading is shown at the right.
At any time interval after infection we find cells that present different stages in the infectious cycle (Fig. 4 h). The cell at the top left has only two in situ signals, and ND10 have not yet been eliminated. The cell on the bottom right has lost all those ND10 not associated with transcription tracks and IE86 domains and has more than double the number of in situ signals. A similar situation is seen in Fig. 1 f, bottom right, where the cell with only two transcript signals still has ND10, whereas the three nuclei in the center with more transcript signals have lost all ND10. The earlier loss of ND10 in neighboring cells correlates with a higher number of transcript sites. Fig. 4 h (bottom nucleus) illustrates an additional observation. Those ND10 that are in association with IE86 are retained longer than those that are not. We presented in Fig. 6 a schematic sequence of the changes in the immediate transcript environment as observed at defined time intervals after infection and supported by the quantitative evaluation over 6 h after infection (Fig. 2).
Whether our observations represented an HCMV-specific situation or could be a more generalized form of arrangement in the infected nucleus was tested using HSV-1. As shown in Fig. 4 i, the same relative localization of ND10, HSV-1 IE transcripts, and the SC35 domains was found as had been documented for HCMV. The same arrangement of intranuclear components, therefore, exists also for other herpes viruses.
Factors of the Basal Transcription Machinery Are Recruited into IE86 Domains
IE86 interacts directly with at least two general transcription factors: TBP (Hagemeier et al., 1992; Jupp et al., 1993; Sommer et al., 1994) and TFIIB (Caswell et al., 1993). Because of the specific correlation between IE86 domains and viral transcripts, we asked whether the specific accumulation of IE86 at ND10 resulted in a redistribution or recruitment of these transcription factors. Cells infected for 3 h were tested for the localization of TBP or TFIIB relative to IE86. In the IE86-negative (and apparently uninfected) cell, a dispersed TBP localization is found, but the infected cells display a substantial increase in TBP concentrations in the IE86 domains (Fig. 5, a–c). These data were confirmed with another mAb against TBP (data not shown). The same evidence for recruitment was obtained with antibodies to TFIIB (Fig. 5, d–f). Both basal transcription factors colocalized with the IE86 cloud. p53, the third protein tested, did not change intranuclear distribution under the same infection conditions and was not found to be relocated into IE86 domains (Fig. 5, g–i).
Figure 5.
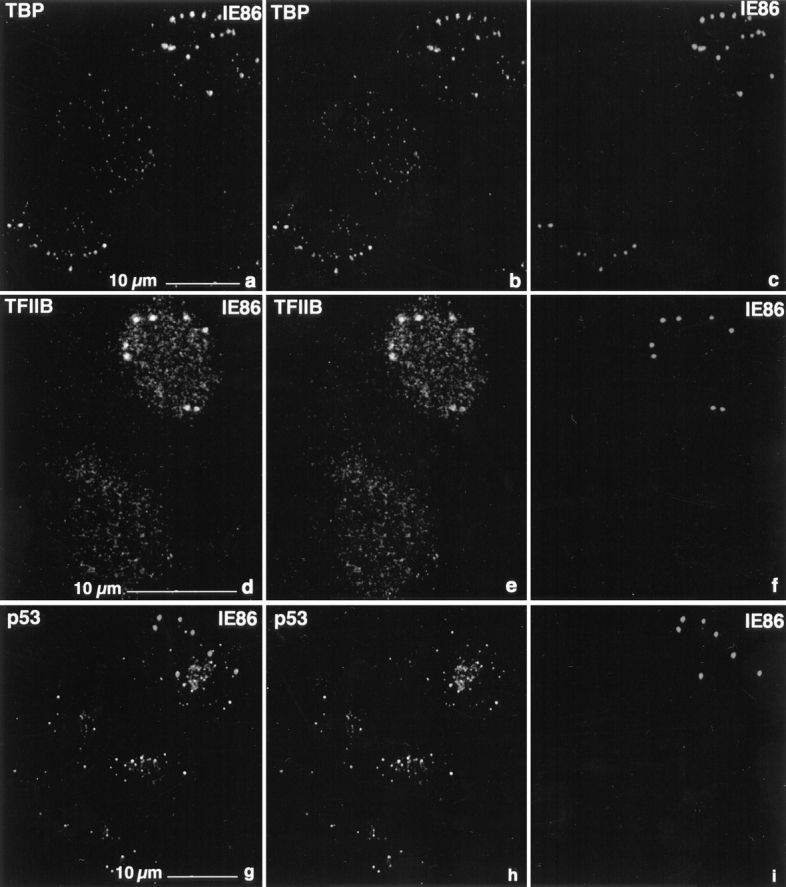
Localization of the cellular proteins relative to IE86 by double labeling of fibroblasts 3 h after infection. (a–c) Separate scans of FITC signal derived from TBP (b) and IE86 (c). Superimposition (a) shows colocalization. (d–f) Separate scans of FITC signal derived from TFIIB (e) and IE86 (f). Superimposition (d) shows colocalization. (g–i) Separate scans of FITC signal derived from p53 (h) and IE86 (i). Superimposition (g) shows no enrichment of p53 at the IE86 domains. Bars, 10 μm.
Discussion
Cellular transcription, processing, and transport of transcripts takes place in a highly structured nuclear environment consisting of various compartments and on genes spatially fixed to the nuclear matrix (for review see Pienta et al., 1991). A principal requirement of incoming virus is to position itself either in a preexisting transcriptional environment or to enable recruitment of factors essential for initial transcription of immediate early genes (Jimenez-Garcia and Spector, 1993). Our study provides a picture at the supramolecular level of the spatial and temporal intranuclear organization during the initial stages of viral transcription from the time HCMV input DNA arrives at the specific site where transcription begins (assumption made by viral transcripts accumulation), followed by the development of a structurally recognizable immediate transcript environment and the subsequent disassembly of the immediate transcript environment. How the rather large viral genome enters the nucleus and how it moves through the highly viscous nucleoplasm and the maze of the nuclear matrix and chromatin remains enigmatic. However, the number of viral genomes entering the nucleus seems larger than the number actually transcribed. This discrepancy may reflect the early stage of our observation. It may also reflect the presence of infectious virus that are competent enough to penetrate the cell and move into the nucleus but incompetent to transcribe, or to the failure of some virus to arrive at a nuclear position favorable for transcription. The latter possibility implies the inability to recruit essential transcription factors or to use preassembled transcription environments. On the other hand, the finding that transcription takes place in association with ND10 suggests the existence of a specific environment where HCMV can transcribe. We previously demonstrated that viruses as diverse as HSV-1, Ad5, and SV40 begin their replication at ND10 (Ishov and Maul, 1996; Maul et al., 1996). A mechanism appears to exist that deposits or retains transcriptionally competent viral genomes at this specific nuclear site. It is also possible that certain factors or binding components that allow viral transcription are concentrated at ND10. The demonstration that HCMV starts its transcription only at ND10 establishes these nuclear matrix specializations as part of a specific transcription environment.
For cellular genes with a high rate of transcription, the rim of the spliceosome assembly factor SC35 domain emerges as the specific site of transcription. The collagen gene, the actin gene in human fibroblasts, and the fibronectin gene in rat fibroblasts have been detected at the periphery of the SC35 domain (Xing et al., 1993, 1995). The highly spliced transcripts are then found in the SC35 domain either at its edge (actin) or throughout the SC35 domain-like collagen Iα1 (Xing et al., 1995) and the transcripts of transiently transcribed plasmids (Huang and Spector, 1996). The highly transcribed and extensively spliced HCMV IE gene transcripts are also funneled into the SC35 domains. This may be solely due to the association of ∼80% of ND10 with the SC35 domain. However, a random release of transcripts from ND10 should show some transcripts accumulating at sites other than the ND10-SC35 apposition point. Triple labeling of ND10, the transcripts, and the SC35 domain clearly demonstrated that the transcripts emerge from ND10, where they are at their highest concentration, and move into the SC35 domain. This observation suggests that the ND10-SC35 domain association constitutes a specific vectorial transcript environment for the IE transcripts, although it does not suggest what or whether any processing of these transcripts takes place.
Newly synthesized IE72 and IE86 proteins can be recognized as early as 1.5 h after infection. Their surprisingly specific localization to ND10 early after the beginning of synthesis suggests that they are targeted to these sites and become part of the evolving transcript environment. However, there are subtle differences between IE72 and IE86 that may reflect on their respective functions. Spatially, IE72 colocalizes completely with all ND10, and transfection experiments show that this localization is inherent in the protein. This protein is also responsible for the dispersion of ND10-associated proteins. Mutations and deletions of many functional domains have no influence on the ability to disperse ND10. Only the removal of a region that contains the putative zinc finger domain abolished the ND10 dispersing activity, suggesting that binding to an as yet unknown cellular protein by the zinc finger region may be the triggering effect that initiates dispersion. In this context, it may not be surprising that IE72 appeared to accumulate apparently throughout ND10 for a substantial time before ND10 was dispersed.
Like IE72, the IE86 protein has the property to accumulate at all ND10 as shown by transfection experiments. However, in the context of viral infection, IE86 was present only at the subpopulation of ND10 with viral transcripts, and it remained at this position even after ND10 and IE72 were dispersed. The cloud of IE86 around the transcripts and situated between ND10 and the SC35 domain is considered an integral part of the evolving transcript environment. At the immediate early time investigated here, the IE86 protein may also have structural functions. We may speculate that the IE86 cloud protects ND10 temporarily, since those ND10 that are part of an immediate transcript environment are retained for some time after the others in the same nucleus are eliminated. The IE86 cloud may be involved in the sequestration and accumulation of basal transcription factors as demonstrated for TBP and TFIIB, although the quantity of these transcription factors accumulated seems disproportionately large relative to a single viral transcription unit.
The Immediate Transcript Environment
On the supramolecular level, the transcript environment of some constitutively transcribed genes appears as a specific intranuclear space that includes the gene itself, the SC35 domain, and transcripts in transit through the SC35 domain. An analogous transcript environment forms after the induction of viral transcription. The virus genome, moving through the nucleus, apparently finds the space at the periphery of the SC35 domain conductive for its own transcription but attaches to the SC35 domain only at sites where ND10 apposes it. Alternatively, the virus transcribing at ND10 that are close to the SC35 domain may recruit components from the SC35 domains to establish the direct pathway into the preexisting larger SC35 domain. The immediate transcript environments developing may then be defined by the position of viral genomes at a few preexisting sites with high transcriptional potential. These sites develop into a structurally organized space through which transcripts are directionally moved in a compact form from their immediate site of synthesis. Whether the move through the SC35 domain signifies any additional processing or is essential for transport out of the nucleus can not be established with the methodology used. The recognition of adjacent nuclear domains through which macromolecular complexes move, with the implication that additional processing can take place, seems to be conceptually important. The virus may utilize environments that preexist for cellular transcripts, or these environments may evolve in a temporal fashion, as described for HCMV IE transcripts (Fig. 6). The viral immediate transcript environment is further characterized by the IE86 cloud which holds or recruits an excess of transcription factors. We envision IE86 and the transcription factors positioned like a collar around the tightly bundled transcripts forming the connecting piece between the transcription site and the SC35 domain. The viral immediate transcript environment may become a paradigm for a cell's individual transcript environments for induced genes, since two of the observed structural components of the immediate transcript environment, ND10 and SC35 domains, are very frequently associated in uninfected cells. We predict that identification of the protein that retains IE86 around the IE transcripts will help to establish new properties of the immediate transcript environment which should have equivalents in the uninfected cell.
Acknowledgments
We thank Mr. Qin Wu Lin for excellent technical assistance, Mrs. A. Schuttfort for anti-Sp100 and PML antibodies, Dr. B. Plachter for the mAb to each of the individual IE antigens, and Dr. N. Hernandez for anti-TBP antibodies. We thank Dr. E Gonzol for providing enough HCMV to carry out these experiments.
This study was supported by The Wistar Institute, National Institutes of Health Core grant CA-10815 (G.G. Maul, and A.M. Ishov), and National Institutes of Health grant AI 23313 (R.M. Stenberg).
Abbreviations used in this paper
- IE
immediate early
- HCMV
human cytomegalovirus
- PML
promyelocytic leukemia protein
- TBP
TATA-binding protein
Footnotes
Please address all correspondence to Gerd G. Maul, The Wistar Institute, 3601 Spruce Street, Philadelphia, PA 19104. Tel.: (215) 898-3817; Fax: (215) 898-3868; E-mail: Maul@wista.wistar.upenn.edu
References
- Ascoli CA, Maul GG. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KS, Taneja KL, Lawrence JB. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J Cell Biol. 1991;115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T, Seeler J-S, Ohman K, Jordan P, Petterson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell R, Hagemeier C, Chou C-J, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcription regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington JM, Mocarski ES. Human cytomegalovirus ie1 transactivates the promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington JM, Khoury EL, Mocarski ES. Human cytomegalovirus ie2 negatively regulates a gene expression via a short target sequence near the transcription start site. J Virol. 1991;65:887–896. doi: 10.1128/jvi.65.2.887-896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C, Schneider T, Baumann H, Hens L, Kirsch-Volders Analysis of chromosome positions in the interphase nucleus of Chinese hamster cells by laser-UV-microirradiation experiments. Hum Genet. 1982;62:201–209. doi: 10.1007/BF00333519. [DOI] [PubMed] [Google Scholar]
- Dirks RW, Daniel KC, Raap AK. RNAs radiate from gene to cytoplasm as revealed by fluorescence in situ hybridization. J Cell Sci. 1995;108:2565–2572. doi: 10.1242/jcs.108.7.2565. [DOI] [PubMed] [Google Scholar]
- Doucas V, Ishov AM, Romo A, Juguilon J, Weitzman M, Evans RM, Maul GG. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- Dyck JA, Maul GG, Miller WH, Jr, Chen JD, Kakizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Fakan S, Bernhard W. Localization of rapidly and slowly labeled nuclear RNA as visualized by high resolution autoradiography. Exp Cell Res. 1971;67:129–141. doi: 10.1016/0014-4827(71)90628-8. [DOI] [PubMed] [Google Scholar]
- Fakan S, Puvion P. The ultrastructural visualization of nuclear and extranucleolar RNA synthesis and distribution. Int Rev Cytol. 1980;65:255–299. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature (Lond) 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Guldner HH, Szostecki C, Grotzinger T, Will H. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J Immunol. 1992;149:4067–4073. [PubMed] [Google Scholar]
- Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO (Eur Mol Biol Organ) J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 1991;5:2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Garcia LF, Spector DL. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- Jupp R, Hoffman S, Stenberg RM, Nelson JA, Ghazal P. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J Virol. 1993;67:7539–7546. doi: 10.1128/jvi.67.12.7539-7546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C, Van Driel R, Wilkinson WG. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- Koken MH, Puvion-Dutilleul F, Guillemin MC, Viron V, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, et al. The t(15; 17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO (Eur Mol Biol Organ) J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korioth F, Maul GG, Plachter B, Stamminger T, Ferry J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi PP, Pelicci PG, Dejean A. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871–876. [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Le X-F, Yang P, Chang K-S. Analysis of growth and transformation suppressor domains of the promyelocytic leukemia gene, PML. J Biol Chem. 1996;271:130–135. doi: 10.1074/jbc.271.1.130. [DOI] [PubMed] [Google Scholar]
- Lichter P, Cremer T, Borden L, Ward DC. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Liu B, Hermiston TW, Stinski MF. A cis-acting element in the major immediate early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Manuppello JR, Alwine JC. Transcription activation by human cytomegalovirus immediate-early proteins: recruitments for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG, Everett RD. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- Maul GG, Guldner HH, Spivack JG. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- Maul GG, Yu E, Ishov AM, Epstein AL. Nuclear domain 10 (ND10) associated proteins are present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:499–514. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- Maul GG, Ishov AM, Everett RD. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- Mocarski, E.S., Jr. 1996. Cytomegaloviruses and their replication. In Fields Virology. B.N. Fields, D.M. Knipe, and P.M. Howley, editors. Lippincott-Raven Publishers, Philadelphia. 2447–2492.
- Mu ZM, Chin KV, Liu JH, Lozano G, Chang KS. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pienta KJ, Getzenberg RH, Coffey DS. Cell structure and DNA organization. Crit Rev Eukaryotic Gene Expr. 1991;1:355–385. [PubMed] [Google Scholar]
- Pinkel D, Landegent C, Collins C, Fuscoe J, Seagraves R, Lucas J, Gray JW. Fluorescence in situ hybridization with human chromosome-specific libraries: Detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci USA. 1988;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno MC, Hayward GS. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target located near the cap site. J Virol. 1990;64:6154–6165. doi: 10.1128/jvi.64.12.6154-6165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert SM, McCulloch V, Meyer M, Bautista C, Falcowski M, Stunnenberg HG, Hernandez N. Monoclonal antibodies directed against the amino-terminal domain of human TBP cross-reacted with TBP from other species. Hybridoma. 1996;15:55–68. doi: 10.1089/hyb.1996.15.55. [DOI] [PubMed] [Google Scholar]
- Sommer MH, Scully AL, Spector DH. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Higher order nuclear organization: three-dimensional distribution of small nuclear ribonucleoprotein particles. Proc Natl Acad Sci USA. 1990;87:147–152. doi: 10.1073/pnas.87.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Fu X-D, Maniatis T. Association between distinct pre-mRNA splicing components and the cell nucleus. EMBO (Eur Mol Biol Organ) J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg RM. The human cytomegalovirus major immediate-early gene. Intervirology. 1996;39:343–349. doi: 10.1159/000150505. [DOI] [PubMed] [Google Scholar]
- Stenberg RM, Stinski MF. Autoregulation of the human cytomegalovirus major immediate-early gene. J Virol. 1985;56:676–682. doi: 10.1128/jvi.56.3.676-682.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg RM, Thomsen DR, Stinski MF. Structural analysis of the major immediate early gene of human cytomegalovirus. J Virol. 1984;49:190–199. doi: 10.1128/jvi.49.1.190-199.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg RM, Witte PR, Stinski MF. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg RM, Depto AS, Fortney J, Nelson JA. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J Virol. 1989;63:2699–2708. doi: 10.1128/jvi.63.6.2699-2708.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg RM, Fortney J, Barlow SW, Magrane BP, Nelson JA, Ghazal P. Promoter-specific trans-activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski MF, Thomsen DR, Stenberg RM, Goldstein LC. Organization and expression of the immediate early genes of human cytomegalovirus. J Virol. 1983;46:1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- Visa N, Puvion-Dutilleul F, Harper F, Bachellerie J-P, Puvion E. Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res. 1993;208:19–34. doi: 10.1006/excr.1993.1218. [DOI] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Long L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR α in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Dobner P, Lawrence JB. Higher level organization of individual gene transcription and RNA splicing: integration of nuclear structure and function. Science (Wash DC) 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Moen PT, Jr, McNeil JA, Lawrence JB. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]