Abstract
Neocentromere activity is a classic example of nonkinetochore chromosome movement. In maize, neocentromeres are induced by a gene or genes on Abnormal chromosome 10 (Ab10) which causes heterochromatic knobs to move poleward at meiotic anaphase. Here we describe experiments that test how neocentromere activity affects the function of linked centromere/kinetochores (kinetochores) and whether neocentromeres and kinetochores are mobilized on the spindle by the same mechanism. Using a newly developed system for observing meiotic chromosome congression and segregation in living maize cells, we show that neocentromeres are active from prometaphase through anaphase. During mid-anaphase, normal chromosomes move on the spindle at an average rate of 0.79 μm/min. The presence of Ab10 does not affect the rate of normal chromosome movement but propels neocentromeres poleward at rates as high as 1.4 μm/min. Kinetochore-mediated chromosome movement is only marginally affected by the activity of a linked neocentromere. Combined in situ hybridization/immunocytochemistry is used to demonstrate that unlike kinetochores, neocentromeres associate laterally with microtubules and that neocentromere movement is correlated with knob size. These data suggest that microtubule depolymerization is not required for neocentromere motility. We argue that neocentromeres are mobilized on microtubules by the activity of minus end–directed motor proteins that interact either directly or indirectly with knob DNA sequences.
C urrent models suggest that chromosomes move by a combination of forces generated by microtubule disassembly (Inoue and Salmon, 1995; Waters et al., 1996) and the activity of molecular motors (Vernos and Karsenti, 1996; Yen and Schaar, 1996). Microtubule disassembly generates a constant poleward force; while molecular motors can generate force in either poleward or away-from-pole directions, depending on the characteristics of the motor protein. Both plus and minus end–directed microtubule-based motors are localized to kinetochores (Hyman and Mitchison, 1991). Immunolocalization experiments indicate that mammalian kinetochores contain the minus end– directed motor dynein throughout metaphase and anaphase (Pfarr et al., 1990; Steuer et al., 1990). The kinesin-like proteins CENP-E, which has a transient kinetochore localization in animals, and MCAK, which is localized between the kinetochore plates of mammalian chromosomes, are also thought to generate and/or regulate chromosome movement (Yen et al., 1992; Lombillo et al., 1995; Wordeman and Mitchison, 1995).
In addition to the molecular motors on kinetochores, several kinesin-like proteins are localized to chromosome arms (Vernos and Karsenti, 1996). Two subfamilies of arm-based motors have been identified in animals: the NOD subfamily (Afshar et al., 1995; Tokai et al., 1996) and the Xklp1/chromokinesin subfamily (Vernos et al., 1995; Wang and Adler, 1995). Both Nod and Xklp1 are required for positioning chromosomes on the metaphase plate, suggesting that they encode plus end–directed motors (Afshar et al., 1995; Vernos et al., 1995). Other evidence suggests that minus end–directed motors interact with chromosome arms. In the plant Haemanthus, a poleward force acts along chromosome arms during metaphase (Khodjakov et al., 1996), and forces propelling chromosome arms poleward have been detected during anaphase in crane fly spermatocytes (Adames and Forer, 1996). Little is known about how poleward arm motility at metaphase–anaphase affects the fidelity or rate of chromosome segregation.
The neocentromeres of maize (Rhoades and Vilkomerson, 1942) provide a particularly striking example of poleward chromosome arm motility. In the presence of Abnormal chromosome 10 (Ab10),1 heterochromatic DNA domains known as knobs are transformed into neocentromeres and mobilized on the spindle (Rhoades and Vilkomerson, 1942; Peacock et al., 1981; Dawe and Cande, 1996). Knobs are primarily composed of a tandem 180-bp repeat (Peacock et al., 1981) which shows homology to a maize B centromere clone (Alfenito and Birchler, 1993). A characteristic feature of neocentromeres is that they arrive at the spindle poles in advance of centromeres; in extreme cases the neocentromere-bearing chromosome arms stretch towards the poles (Rhoades and Vilkomerson, 1942; Rhoades, 1952). A recently identified mutation (smd1) demonstrates that a trans-acting factor(s) encoded on Ab10 is essential for converting the normally quiescent heterochromatic knobs into active neocentromeres (Dawe and Cande, 1996).
Here we use neocentromeres as a model for understanding the mechanisms and importance of nonkinetochore chromosome movement. As a part of our analysis, we developed a four-dimensional system for observing chromosome segregation in living meiocytes. Our experiments were designed to determine (a) how poleward arm motility affects the rate and fidelity of chromosome segregation; and (b) whether the mechanism of neocentromere motility is comparable to the mechanism of kinetochore motility.
Materials and Methods
Plant Material
The inbred maize line W23, with four knobs, was the primary source of wild-type meiocytes. A line with Ab10 linked to R (by two map units) was backcrossed five times to a derivative of the W23 inbred that carries all the factors required for kernel pigmentation except r. These heterozygous Ab10 plants were selfed to obtain plants that were homozygous for Ab10. The Ab10 and W23 strains are near-isogenic: knob counts on homozygous Ab10 plants revealed six knobs, including the knob on Ab10 and an extra knob not present in W23. We also used a high-knob strain, with knob numbers ranging from 6 to 10, that was segregating wild-type and heterozygous Ab10 plants. The presence or absence of Ab10 in the high-knob strain was determined using linked R alleles.
Analysis of Living Meiocytes
Tassels from greenhouse-grown plants were harvested at a stage when anthers were 1–2 mm in length. The tips were excised from anthers using a No. 15 scalpel blade, and the meiocytes were gently extruded into a Petri dish filled with liquid culture medium. The culture medium was essentially that published by Pena (1986), except that the sucrose concentration was reduced to 0.1 M and the medium was supplemented with 0.1 M maltose, 1% Guillard's antibiotic concentrated solution (G5535; Sigma Chemical Co., St. Louis, MO), and 0.25 mM n-propyl gallate. The medium was filter sterilized. Meiocytes in roughly 65 μl of culture medium were transferred from the petri dish to a depression slide (12-560A; Fisher Scientific, Pittsburgh, PA). The fluorescent DNA stain Syto 12 was added to the medium at a final concentration of 2 μM (10 other fluorescent DNA stains were found not to stain the chromosomes in cultured maize meiocytes: DAPI, Hoechst 33258, Hoechst 33342, propidium iodide, and Syto 11 and 13–17). A coverslip was placed over the concavity, leaving a ∼2 mm air bubble and sealed using rubber cement. The temperature on the microscope stage was maintained at 25 ± 1°C and continuously monitored using a digital recording thermometer. The Syto 12–stained chromosomes were visualized using an FITC filter set.
Cell viability after 6 or 9 h under various treatments (Table I) was determined by adding 5 μm Calcein AM to the cultured cells (Molecular Probes, Eugene, OR). Calcein AM is converted into a fluorescent product in living cells but does not stain dead cells. The 5-min green light treatment (450–490 nm) reported in Table I was delivered using a fluorescence microscope (Axiophot, Zeiss, Inc., Thornwood, NY) with the objective removed so that the entire concavity was irradiated.
Table I.
Effects of n-Propyl Gallate, Syto 12, and FITC-Channel Excitation Light on Cultured Meiocytes*
| Treatment | NPG‡ | Syto 12§ | Light‖ | Viability (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exp. 1 (6 h) | Exp. 2 (9 h) | |||||||||
| t = 0 | − | − | − | 71.2 | 73.1 | |||||
| A | − | − | − | 49.4 | 45.0 | |||||
| B | − | − | + | 38.9 | 27.5 | |||||
| C | − | + | − | 51.0 | 39.6 | |||||
| D | − | + | + | 32.6 | 32.5 | |||||
| F | + | + | + | 45.1 | 45.8 | |||||
Two different experiments are reported. Viability was measured immediately after extruding W23 meiocytes from anthers (t = 0) and after 6 h in the first experiment (Exp. 1) and after 9 h in the second experiment (Exp. 2). The average number of cells counted to determine percent viability was 213.
0.25 mM n-propyl gallate.
2 μm Syto12.
5-min exposure to 450- to 490-nm light at t = 0.
Combined In Situ Hybridization/Immunocytochemistry
Knob/Spindle Staining.
Meiocytes were fixed with 4% formaldehyde in Phems as described previously (Dawe and Cande, 1996). To avoid cell flattening during adhesion of cells to coverslips, cells were dried onto poly-l-lysine–coated coverslips without centrifugation. The coverslips were inverted onto a slide with the four corners supported by small broken pieces of coverslip. Ten different FITC-labeled 18-mers were prepared (Bioserve Biotechnologies, Laurel, MD) that are homologous to different and nonoverlapping portions of the 180-bp knob repeat. The oligonucleotides were combined in equimolar amounts and diluted to 10 μg/ml in PBS. Approximately 75 μl of the oligonucleotide solution was injected beneath the coverslip and sealed with rubber cement. The slide was heated to 95°C for 5 min and rinsed three times for 5 min at room temperature in PBS. After in situ hybridization, an α-tubulin monoclonal antibody (generously supplied by D. Asai, Purdue University, Layfayette, Indiana; Asai et al., 1982), diluted 1:500 in antibody dilution buffer (1× PBS, 3% BSA, 0.02% sodium azide), was applied to the coverslip. The cells were incubated with the tubulin primary antibody in a moist chamber overnight at 30°C. After three 5-min rinses with PBS, the cells were treated for 2 h with 20% goat serum in antibody dilution buffer at 30°C. Rhodamine-conjugated goat anti–mouse secondary antibody (115-025-146; Jackson ImmunoResearch, West Grove, PA) at a dilution of 1:30 in antibody dilution buffer was applied to the coverslip and incubated at 30°C for another 2 h. The cells were stained with 0.01 μg/ml DAPI, mounted in Mowiol mounting medium (Harlow and Lane, 1988), and observed using three-dimensional light microscopy.
Centromere/Spindle Staining.
PCR primers (GATTGGAAACAGTTAAAGAAC and CATGCTTAAAATGGACTCTGT) were used to amplify the pSau3A9 centromere repeat (Jiang et al., 1996) from Sorghum bicolor DNA. The ∼700-bp PCR product was cloned using the TA cloning kit (Invitrogen Corp., San Diego, CA). The gel-purified insert from this clone was labeled using a random priming kit (Prime-It Fluor; Stratagene, LaJolla, CA) and tetramethylrhodamine-6-dUTP (Boehringer Mannheim, Indianapolis, IN). Unincorporated nucleotides were removed using microcolumns (ProbeQuant G-50; Pharmacia Biotech, Piscataway, NJ) before in situ hybridization. W23 meiocytes were spun down onto poly-l-lysine–coated coverslips and hybridized with the labeled probe essentially as described previously (Dawe and Cande, 1996). Unlike the knob labeling protocol, this protocol involves the use of 30% deionized formamide. The protocol for spindle staining was the same as for knob/ spindle staining, except that all solutions were in 2× PBS, incubations were at 27°C, and the secondary antibody was FITC-conjugated goat anti– mouse (14274020; Boehringer Mannheim).
Microscopy and Image Analysis
All microscopy and image analysis was carried out using a multidimensional microscopy system (DeltaVision SA3.1; Applied Precision Inc., Issaquah, WA) equipped with a thermoelectrically cooled CCD (TEA/ CCD-768K-1UV; Princeton Instruments, Princeton, NJ). Slides were mounted on an inverted microscope (IMT2; Olympus Corp., Lake Success, NY) and observed with a 1.4 oil immersion objective (60× NA; Nikon, Inc., Melville, NY). For observing living meiocytes, the exposure time required for each optical section was reduced to 0.1 s by summing each 2 × 2 array of pixels and assigning the resulting values to one pixel (a process called binning; see Chen et al., 1995). This process increases sensitivity by fourfold but reduces resolution by twofold (pixel size = 0.294 μm). Optical sections from live cells were taken at either 0.5- or 1.0-μm intervals. For fixed specimens, optical sections were taken using a 1.5× Optivar without binning (pixel size = 0.0981 μm) at 0.2-μm intervals.
The out-of-focus information in the raw data was removed by three- dimensional constrained iterative deconvolution (Chen et al., 1995) using software supplied with the DeltaVision system. The resulting images were scaled to optimize contrast but were not processed further. The stereo pair in Fig. 8 was generated by volume rendering (Chen et al., 1995). When making measurements, the cursor was placed over the approximate center of the chromosome or neocentromere being measured. To determine the rate of anaphase chromosome movement in living cells (Table II), two or three pairs of chromosomes (presumably disjoining sister chromatids) were chosen for measurement. The distance between the chromosomes was recorded for each pair at each time point. These distances were divided in half (to obtain the rates for the individual chromosomes), averaged with the distances from the one or two other pairs of chromosomes from the same cell, and plotted with respect to elapsed time. When measuring the distance of neocentromeres from the equatorial plate (see Fig. 10), the equatorial plate was calculated as the midway point between the spindle poles (or when the poles were not well defined, the cell walls closest to the poles).
Figure 8.
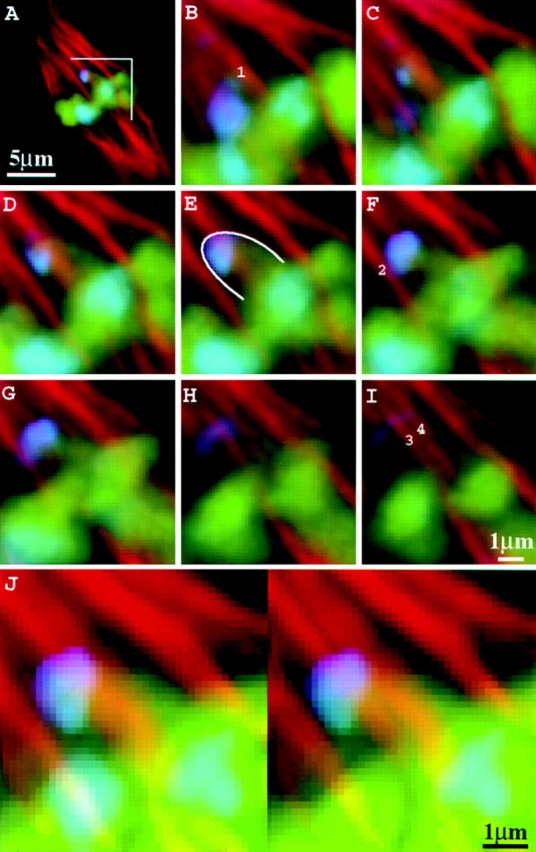
Microtubules interact laterally with neocentromeres. Images are from a heterozygous Ab10 meiocyte (high-knob background) at early metaphase II. (A) Optical section of the entire cell; (B–I) consecutive optical sections at 0.2-μm intervals over the region indicated in A; (J) images in B through I presented as a volume-rendered stereo pair. In B, F, and I, two spindle fibers that contact the knob (1 and 2) and two others that may contact it (3 and 4) are indicated. Chromosomes are shown in green; neocentromeres in blue; microtubules in red. The blue staining in the spindle region of B is derived from knobs that are out of the plane of focus in A. The neocentromere is outlined in E.
Table II.
Rates of Chromosome and Neocentromere Motility during Anaphase II*
| Genotype‡ | Chromosome motility (μm/min ± SD) | Neocentromere motility (μm/min ± SD) | No. cells measured | |||
|---|---|---|---|---|---|---|
| High-knob | 087 ± 0.15 | − | 5 | |||
| W23 | 0.79 ± 0.17 | − | 9 | |||
| Ab10§ | 0.78 ± 0.13 | 1.08 ± 0.23‖ | 10 |
Data report the average rate of movement between consecutive time points at mid-anaphase, which was defined as the stage when chromosomes were 3–6 μm from the metaphase plate.
The high-knob and W23 are wild-type strains (and did not show neocentromeres).
In the W23 background.
Derived from measurements of 13 neocentromeres in 10 cells.
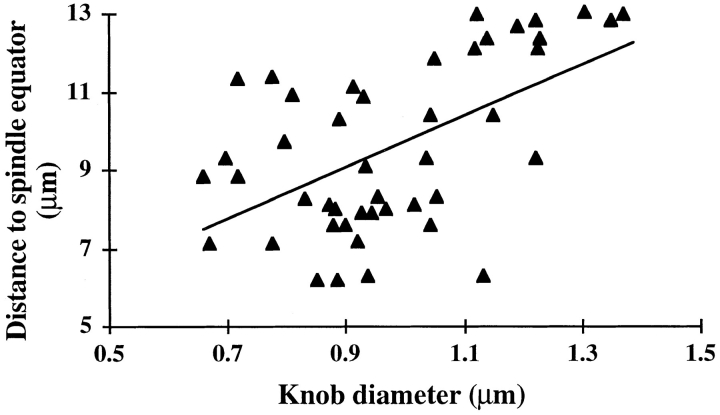
Figure 10.
The position of neocentromeres in the anaphase II spindle correlates with knob size. Knob size is plotted with respect to distance from the spindle equator (all the knobs from eight anaphase II cells with extreme neocentromere activity are shown). The regression line shown has a coefficient of 3.11 (P < 0.01, r 2 = 0.31).
Results
Chromosome Segregation in Living Maize Meiocytes
Using a modified rye meiocyte culture medium (Pena, 1986) to support growth, we were able to record chromosome movement in 53 maize meiocytes undergoing meiosis II. Modifications in the medium included a reduction of sucrose to minimize the toxic effects of sucrose decomposition by living cells (Scott and Lyne, 1994), and the addition of the antioxidant n-propyl gallate, which is known to increase the longevity of maize protoplasts in culture (Cutler et al., 1988). Approximately 45% of the meiocytes extruded into this medium were viable for 9 h or longer (Table I).
To identify a vital chromosome stain, 11 commercially available fluorescent DNA dyes were screened (see Materials and Methods). Only Syto 12, a green-fluorescent DNA dye, selectively stained living cells. Syto 12 did not affect the viability of maize meiocytes (Table I). However, aberrant division planes were observed when some meiocytes, particularly those going through prometaphase, were exposed to excessive light (490 nm). We were able to eliminate this problem by keeping the total light exposure during time-lapse experiments to <1 min (see Materials and Methods). Four-dimensional data (three dimensions over time) were collected using wide field three-dimensional light microscopy (Chen et al., 1995). To minimize light exposure, images were acquired from four to six optical sections at 1 to 5 min intervals. A series of optical sections from a living meiocyte undergoing meiosis II can be seen in Fig. 1. These and other data indicate that meiosis II in maize takes ∼5 h to complete: the interkinesis between telophase I and prophase II takes ∼150 min; prometaphase– metaphase takes ∼90 min; and anaphase–telophase II takes ∼60 min.
Figure 1.
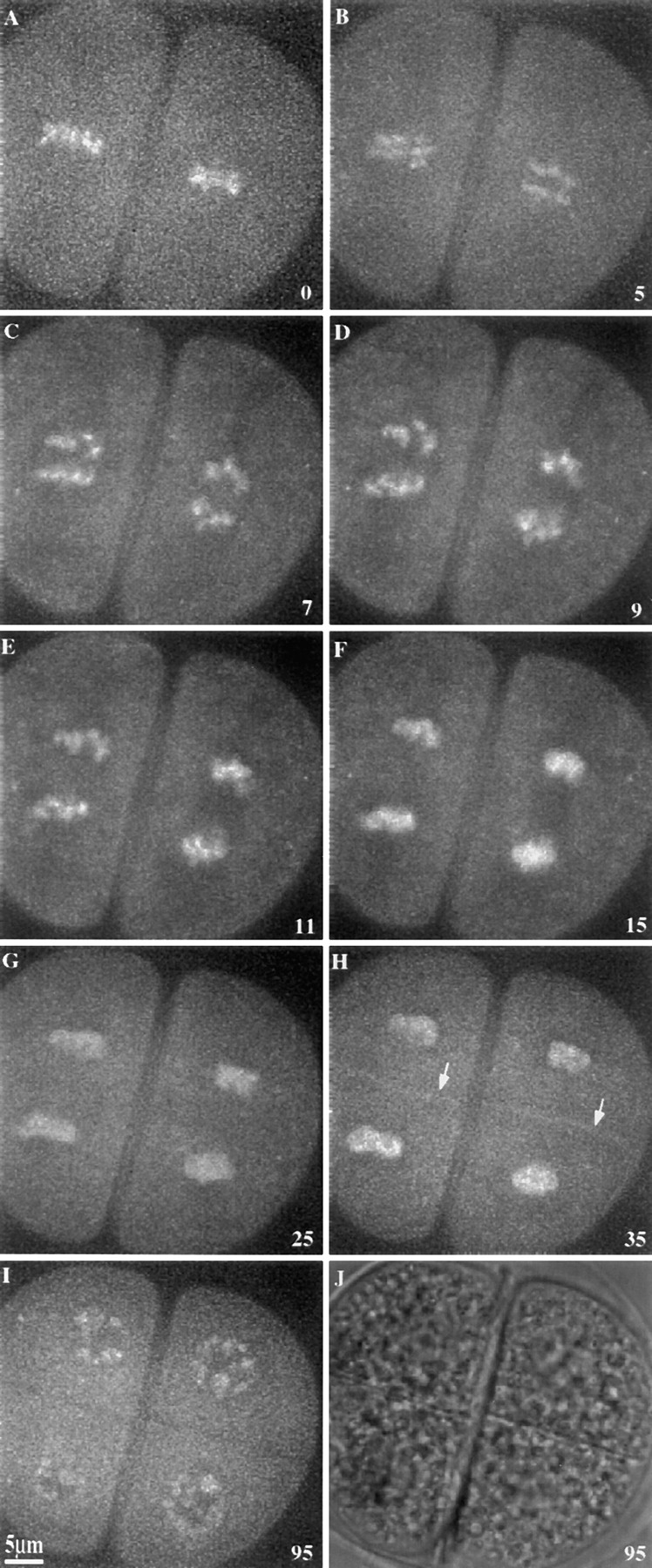
Time-lapse sequence showing a wild-type meiocyte undergoing meiosis II. Images are single optical sections of Syto12-stained chromosomes in a high-knob strain, except J, which is a phase contrast image. (A) Metaphase; (B–E) anaphase; (F–H) telophase; and (I and J) final interphase. Arrows in H indicate the new cell plates. Time in minutes is shown in the lower right corner of each frame.
Neocentromeres Form as Early as Prometaphase
Neocentromeres are most easily detected at metaphase– anaphase II in plants homozygous for Ab10 (Rhoades and Vilkomerson, 1942) but have been reported in heterozygous Ab10 plants and at meiosis I (Rhoades, 1952). According to Rhoades (1952), neocentromere activity is also occasionally observed in late prophase II. We observed, as have others (e.g., Emmerling, 1959), that the penetrance of the neocentromere phenotype is highly variable and environmentally influenced. Neocentromeres were observed only during the winter months in greenhouse-grown plants, with less than half of the plants showing neocentromere activity.
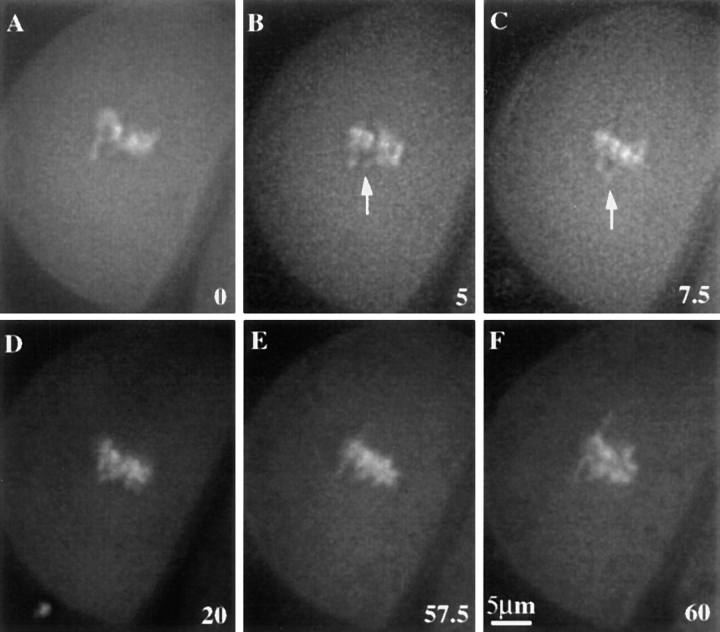
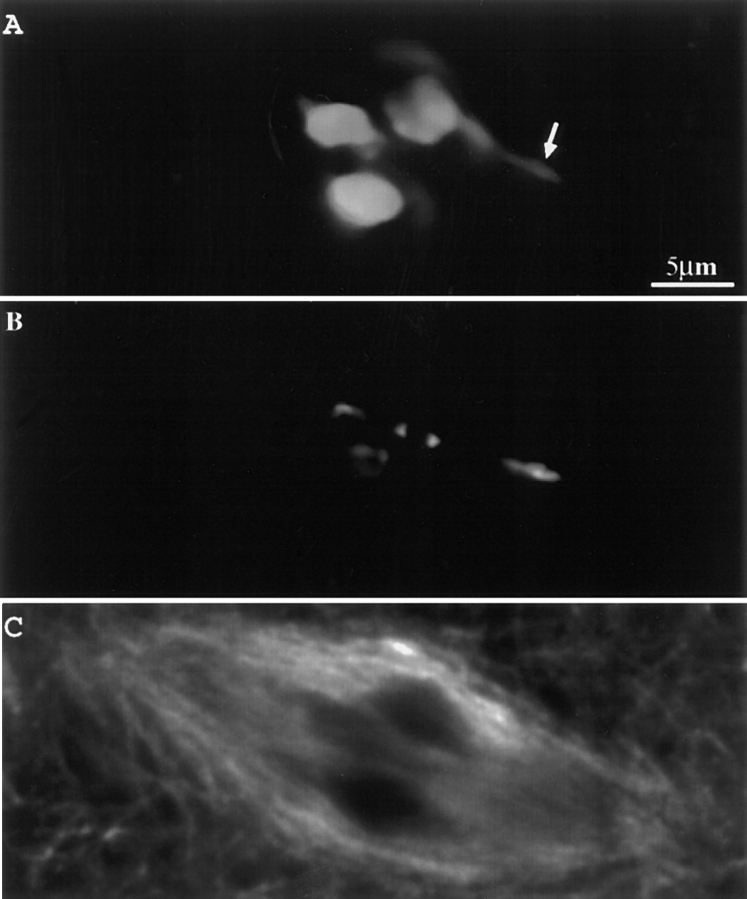
During prometaphase II in both wild-type and Ab10 plants, the chromosomes move towards the spindle and then slowly oscillate in the midzone of the spindle. We defined metaphase as the stage when the chromosomes ceased to oscillate, or alternatively, as the stage when a nonstaining gap appeared between the chromatids at the metaphase plate (presumably due to tension from the half spindles; Fig. 1 A). Significant poleward arm movement was not observed in any of the 38 living wild-type meiocytes observed during prometaphase. However, in the presence of Ab10, distinct poleward chromatin extensions were observed. Fig. 2 shows a living meiocyte from a plant that was homozygous for Ab10 and five additional knobs on other chromosomes (there are 10 chromosomes in maize). Poleward chromatin extensions were first observed 30 min before metaphase (Fig. 2, arrows) and continued throughout prometaphase, metaphase, and anaphase. Evidence that the chromatin extensions observed in live cells are the result of neocentromere activity was obtained by hybridizing fixed cells with fluorescent oligonucleotide probes that identify the knob repeat. Knob-terminated chromatin extensions were clearly observed in one fixed prometaphase II cell (not shown) and two fixed prometaphase I cells carrying Ab10. A meiosis I cell that was fixed and stained for chromosomes, knobs, and microtubules is illustrated in Fig. 3. This cell was heterozygous for Ab10 and at the time of fixation was in the process of aligning the chromosomes on the metaphase plate. As shown, a noticeably elongated neocentromere is positioned midway between the spindle equator and pole (Fig. 3, arrow).
Figure 2.
Prometaphase II neocentromere activity in a living meiocyte. Images are single optical sections of Syto12-stained chromosomes in a homozygous Ab10 strain (W23 background). (A–D) Prometaphase; (E) metaphase; and (F) metaphase–anaphase transition. Arrows in B and C indicate a neocentromere stretching towards a pole. Metaphase occurred at 30 to 35 min after the first data set (A) was taken. Time in minutes is shown in the lower right corner of each frame.
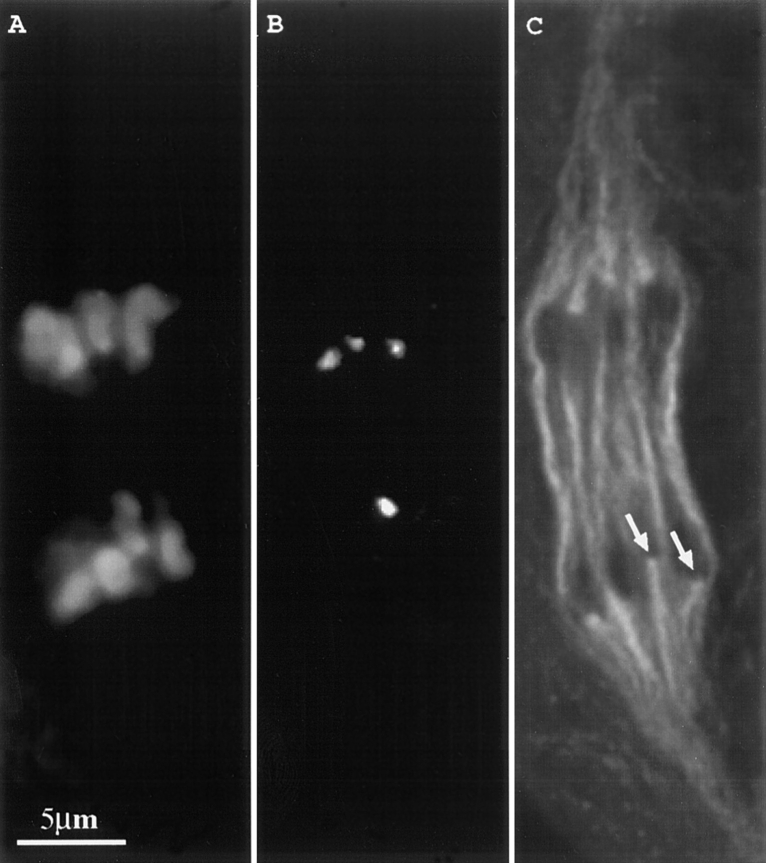
Figure 3.
Prometaphase I neocentromere activity in a fixed and triple-stained meiocyte. Ab10 was heterozygous in the high-knob background. Three images from a single optical section are shown. (A) Chromosomes; (B) knobs; and (C) spindle. Arrow identifies the neocentromere.
Neocentromeres Move Faster than Kinetochores at Anaphase
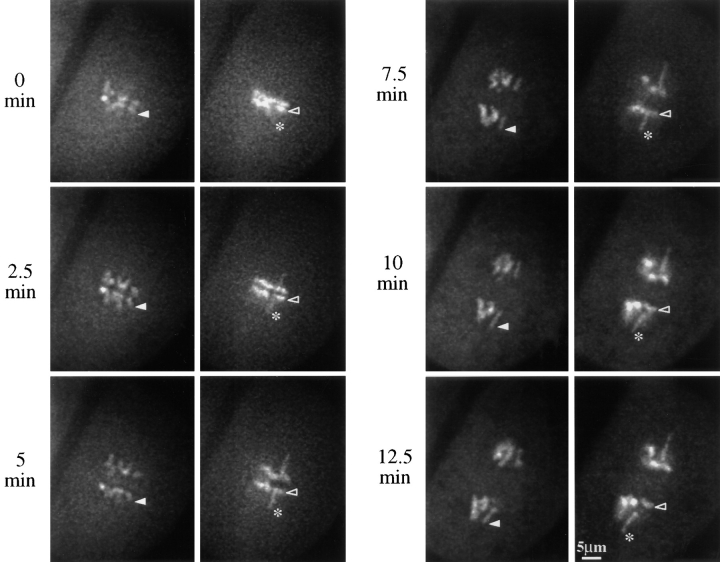
During anaphase, kinetochores and neocentromeres proceed poleward at different rates (Fig. 4). The rate of knobless chromosome movement in Ab10 strains, at 0.78 μm/ min, was not significantly different from the rate of 0.79 μm/min observed in the near-isogenic W23 wild-type strain (Table II). In contrast, neocentromeres moved at an average maximum rate of 1.08 μm/min and were observed moving at rates as high as 1.4 μm/min. Fig. 4 shows two optical sections at each of six time points from an Ab10 meiocyte undergoing anaphase II. Several neocentromeres can be seen moving towards the poles at rates faster than either the knobless chromosomes in the same cell or the kinetochores on the same chromatids (causing stretching). Knobless chromosomes and neocentromeres both undergo periods of acceleration in early anaphase and periods of deceleration in late anaphase (Fig. 5), as has been observed in previous studies (e.g., Ryan, 1983). In mid-anaphase, the rate of neocentromere movement can be twice as fast as the maximum rate of normal chromosome movement (Fig. 5). In cases where a neocentromere had started to stretch a chromosome arm poleward in metaphase, the arm continued to elongate by 1 to 3 μm during anaphase (e.g., Fig. 4, asterisk). In other cases the neocentromeres were not visible at metaphase, and the chromosome arms stretched by as much as 4 μm during anaphase (e.g., Fig. 4, solid arrowhead). Most neocentromeres did not affect the rate of linked kinetochore movement, but some neocentromeres appeared to cause the entire chromosome, including the kinetochore, to accelerate towards a pole (Fig. 4, asterisk).
Figure 4.
Anaphase II neocentromere activity in a living meiocyte. Images are from a homozygous Ab10 cell in the W23 background. Two optical sections (across) are shown at each of six time points. Arrowheads indicate the neocentromere (closed) and chromosome (open) used for the data presented in Fig. 5. The asterisk identifies a neocentromere that appeared to accelerate the movement of its linked kinetochore (the neocentromere is slightly out of the focal plane in the image taken at 0 min).
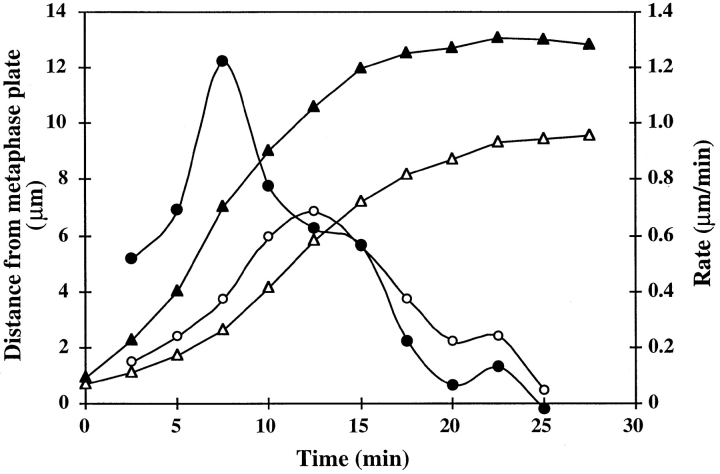
Figure 5.
Chromosome and neocentromere movement during anaphase–telophase. Data were derived from the cell shown in Fig. 4. Time 0 = anaphase onset. (▴) Distance of a neocentromere from the metaphase plate; (•) rate of movement for the neocentromere; (▵) distance of a normal chromosome from the metaphase plate; (○) rate of movement for the normal chromosome. Rates shown occurred over the preceding time interval; e.g., the rate shown at 7.5 min is the rate that occurred between 5.0 and 7.5 min after anaphase onset.
Microtubules Interact Laterally with Neocentromeres
A combined in situ hybridization/immunocytochemistry approach was used to analyze the interaction of knobs with microtubules. Knobs were localized using a collection of 10 FITC-labeled oligonucleotides which together represent the entire 180-bp repeat. Because the knobs are brightly labeled with this probe, we were able to carry out the hybridization in the absence of the denaturing agent formamide and obtain well preserved microtubules for immunocytochemistry.
As observed in studies with other plants (e.g., Inoue, 1953), we observed that maize microtubules are evenly distributed throughout the spindle in prometaphase I and II. The uniform spindle staining typical of prometaphase can be seen in Fig. 3 C. As the cell progresses into metaphase and anaphase (I or II), the spindle staining shifts to a pattern primarily composed of bundled fibers. The fibers were divided into two classes: the nonkinetochore fibers, which pass from pole-to-pole or terminate without contacting chromosomes; and the kinetochore fibers, which terminate at chromosomes. The kinetochore fibers have the following characteristics: (a) they terminate ∼0.3 μm from the DAPI-stained chromatin, leaving a nonstaining gap that presumably represents the kinetochore; (b) they are more brightly stained adjacent to the chromosomes (appearing thicker; Fig. 6, arrows); and (c) they interact with centromere/kinetochores as judged by their colocalization with centromere sequences. A conserved cereal centromere sequence that was identified in sorghum by Jiang et al. (1996) was used as a probe for maize chromosomes. The centromere signal was associated with the ends of spindle fibers, as expected if the fibers interact with the centromere/kinetochore complex (Fig. 7). To detect hybridization of the sorghum sequence on maize chromosomes it was necessary to add formamide in the hybridization step (see Materials and Methods). Under these conditions, the kinetochore fibers at metaphase I were resolved into two distinct fibers that presumably interact with the sister kinetochores of the chromosome (Fig. 7, inset). A separation of sister spindle fibers in metaphase I has not been observed previously, but it is consistent with observations from other plants that sister kinetochores can be resolved at late metaphase I (Lima-de-Faria, 1956).
Figure 6.
Fixed and triple-stained W23 wild-type cell at anaphase II. Three images from a single optical section are shown. (A) Chromosomes; (B) knobs; and (C) spindle. Arrows indicate the thickening of kinetochore fibers adjacent to the chromatin.
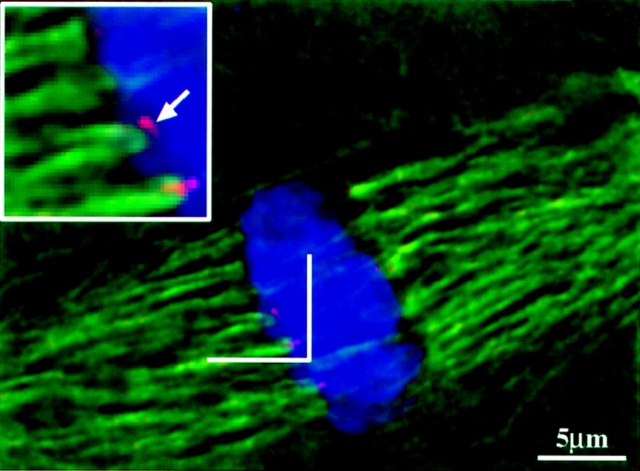
Figure 7.
Colocalization of the centromeres with the ends of kinetochore fibers. The image was taken at a single optical section from a W23 wild-type cell. Inset is a twofold magnification of the region indicated by a bracket. Arrow indicates a centromere. Chromosomes are shown in blue; centromeres in red; microtubules in green.
In contrast to kinetochores, neocentromeres interact laterally with spindle fibers. The knobs in maize are located within the distal half of their respective chromosome arms (Longley, 1939) and usually appear to occupy telomeric positions at metaphase and anaphase (Peacock et al., 1981; Dawe and Cande, 1996). In the absence of Ab10, knobs invariably lag behind the bulk of the chromatin at anaphase (Fig. 6). Knobs are converted into neocentromeres in the presence of Ab10 and with few exceptions are pulled poleward. In 46 three-dimensional data sets containing 160 neocentromeres, an end-on interaction of a spindle fiber with a neocentromere was never detected. Fig. 8 shows a single neocentromere and its associated microtubules at early metaphase II. At least two distinct spindle fibers are laterally associated with the neocentromere with no evidence of a fiber terminating in the knob-stained region. Of 39 other neocentromeres studied in detail, 20 were laterally associated with at least two spindle fibers, 16 were laterally associated with one spindle fiber, and 3 were in a region of the spindle where they appeared to be interacting with nonbundled microtubules. Although spindle fibers are frequently associated with neocentromeres, it is unlikely that bundled fibers are required for motility, because neocentromeres are active in prometaphase cells where the microtubules are evenly distributed (Fig. 3). Given the resolution of immunofluorescence microscopy, we cannot rule out the possibility that a small number of microtubules make end-on contact with neocentromeres.
Rhoades (1952) suggested that the true centromere was inactivated when a neocentromere on the same chromosome was formed. Because of the tight grouping of chromosomes at anaphase, we found it difficult to identify single chromosomes and their respective kinetochore fibers. However, in two cases we were able to identify chromosomes with both an active neocentromere and a distinct kinetochore fiber (one is shown in Fig. 9).
Figure 9.
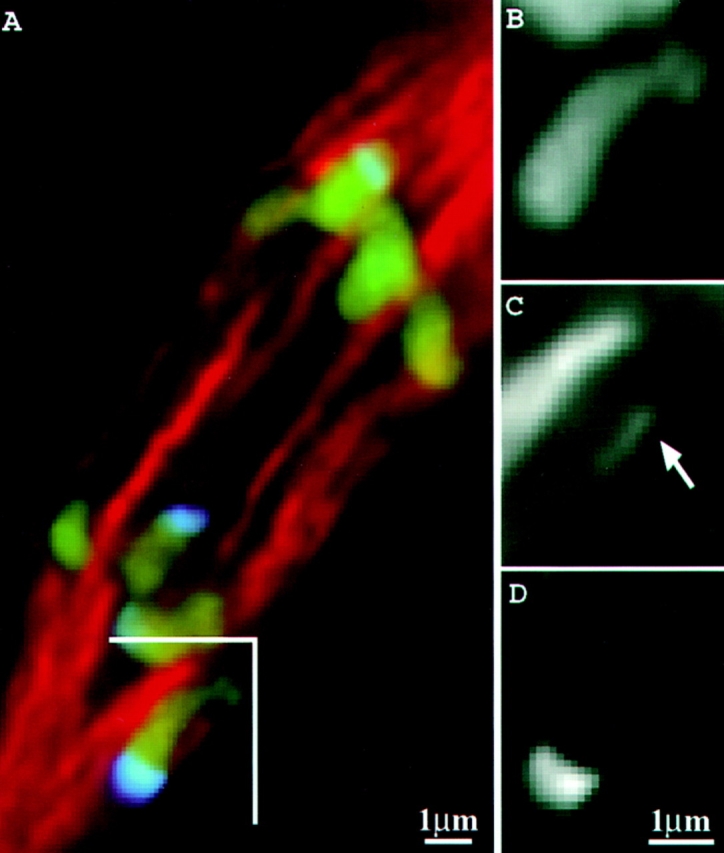
Kinetochore fiber interacting with a neocentromere-carrying chromosome. Chromosomes are shown in green; neocentromeres in blue; microtubules in red. Region indicated with a bracket is reproduced in B (chromosomes only), C (spindle only), and D (knobs only). Arrow indicates the kinetochore fiber associated with the chromosome in B.
The Rate of Neocentromere Motility is a Function of Knob Size
In normal maize meiocytes the chromosomes tend to move at roughly the same speed during anaphase (Fig. 1). This contrasts with the rates of neocentromere movement, which are highly variable. Rates of neocentromere movement as fast as 1.4 μm/min and as slow as 0.91 μm/min were documented in a single Ab10 meiocyte. Although it is difficult to tell the size of a knob in living cells, differences in knob size are apparent after in situ hybridization with a knob probe. To address the question of whether the size of a knob correlated with its motility on the spindle, the knobs in eight anaphase cells with extreme neocentromere activity were scored for diameter and location relative to the spindle equator. A significant positive correlation between knob size and position in the spindle was observed (Fig. 10), indicating that the poleward force exerted by a knob is correlated with the number of 180-bp knob repeats.
Discussion
Using a live-cell system and three-dimensional light microscopy, we have been able to accurately measure the rate of chromosome movement in maize. The analysis of chromosome movement in living plant cells has previously been possible in only species with chromosomes large enough to be viewed using transmission light microscopy, such as Lilium, Allium, and Haemanthus (Inoue, 1953; Ryan, 1983; Khodjakov et al., 1996). While maize chromosomes are large, it has not been possible to resolve them in living cells using phase or differential interference contrast optics. We were able to compensate for the relatively small size of maize chromosomes with high-resolution three-dimensional microscopy and the fluorescent DNA stain Syto 12. Multiply-labeled fixed specimens were used to confirm and extend the observations of living meiocytes.
Neocentromere Activity Has Little Effect on the Overall Rate of Chromosome Segregation: Kinetochores Govern Chromosome Motion at Anaphase
It is well documented that chromosomes segregate synchronously during anaphase even when the chromosomes differ dramatically in size or when forces are applied that restrain poleward motion. For instance, when two segregating chromosomes are joined by a chromatin bridge, chromosome movement continues nearly unabated (Bajer, 1963). Using glass needles to impede chromosome motion, Nicklas estimated that the available force at the kinetochore is 10,000 times more than is actually needed. Nicklas proposed that there is a “governor” at the centromere regulating chromosome motion and indicated that the best candidate for the rate governor is the kinetochore fiber (Nicklas, 1988).
We have shown that neocentromeres can cause a chromosome arm to lengthen by as much as 4 μm during the course of anaphase (Fig. 4). Assuming maize chromosomes are elastic (as are Haemanthus chromosomes; Bajer, 1963), anaphase neocentromere activity generates a poleward force at the kinetochore. However, kinetochores rarely respond by moving faster towards the pole (Fig. 4), providing evidence of a “brake” at the kinetochore. We attribute the braking effect of the kinetochore to structural constraints associated with end-on microtubule interactions at the kinetochore and the slow process of microtubule depolymerization (Nicklas, 1988). Neocentromeres, by interacting with microtubules laterally (Fig. 8), can move rapidly poleward without these constraints.
The importance of nonkinetochore chromosome motility is a matter of debate (Östergren, 1960; Bajer et al., 1987; Fuge, 1990; Adames and Forer, 1996). Recent data show that chromosome arms carry molecular motors that are involved in positioning chromosomes on the metaphase plate (Afshar et al., 1995; Vernos et al., 1995). However, more subtle arm motility events are also observed at anaphase. It has been shown that poleward arm motility at anaphase is a normal part of crane fly spermatogenesis (Fuge, 1975; Adames and Forer, 1996). Similar poleward forces at anaphase have been observed in mammalian cells (Liang et al., 1993). However, it is more commonly observed that chromosome arms move towards the spindle equator during anaphase (e.g., Rieder and Salmon, 1994; Khodjakov et al., 1996). In maize, chromosome arms normally lag behind the kinetochores, presumably applying an away-from-the-pole force to the kinetochore (a chromosome with lagging arms can be seen in the upper right of Fig. 6 A; Rhoades, 1952). Neocentromeres reverse this orientation and apply a poleward force to the kinetochore that only marginally affects the rate of movement (Fig. 4). We have also failed to detect any obvious effects on the fidelity of chromosome segregation. This is surprising because neocentric chromosomes effectively have two centromeres: the true centromere and the neocentromere (Fig. 9). The data of McClintock (1943) and Novitski (1952) suggest that chromatin bridges should be produced from such dicentric chromosomes due to the occasional segregation of linked centromeres to opposite poles. A single Ab10 meiocyte may have a large number of chromosomes with both a centromere and a neocentromere; nevertheless, no chromatin bridges were observed in this study and are apparently very rare (Rhoades, 1952). One possible explanation for the absence of bridges is that the early prometaphase activity of neocentromeres (Figs. 2 and 3; Rhoades, 1952) causes kinetochores to preferentially orient with the poleward-stretching chromosome arms.
The Mechanism of Neocentromere Motility in Maize
As a first step towards understanding the mechanism of neocentromere movement, we determined at which point during the cell cycle neocentromeres are active. The data in Figs. 2 and 3 substantiate the claim by Rhoades (1952) that neocentromeres are first observed in meiotic prometaphase and continue their poleward movement into metaphase. Before our study, it remained possible that all or most of the neocentromere activity was completed in these stages. If the neocentromeres had already moved close to the poles in metaphase, they could have conceivably ceased their poleward motion at anaphase and still arrived early at the poles. However, the data in Fig. 4 showing that chromosome arms continue to stretch during anaphase rule out this possibility. These data, combined with the observation that Ab10 has no effect on the movement of chromosomes lacking knobs (Table II), suggest that Ab10 encodes a product that acts specifically at neocentromeres from prometaphase through anaphase.
Two major forces have been implicated in chromosome movement: the disassembly of spindle fibers at kinetochores (Gorbsky et al., 1988; Waters et al., 1996) and the forces generated by molecular motors at the centromere/ kinetochore complex (Vernos and Karsenti, 1996). Recent data suggest that the two forces are tethered together by molecular motors at the kinetochore, such as CENP-E, which remain coupled to shortening microtubules (Lombillo et al., 1995; Desai and Mitchison, 1995). We believe it is unlikely that microtubule depolymerization plays a major role in neocentromere motility for two reasons. First, neocentromeres interact with spindle microtubules primarily in a lateral manner, not the end-on fashion required to use the energy of depolymerization (Fig. 8). Second, the rate of neocentromere movement varies with knob size (Fig. 10). Tubulin polymerization/depolymerization, while varying as a function of the cell cycle, is relatively uniform within the spindle (Gorbsky et al., 1988; Mitchison and Salmon, 1992). The spindle-wide uniformity of depolymerization is inconsistent with the asynchronous movement of neocentromeres.
Our neocentromere data are best explained by the activity of a motor protein that associates with microtubules laterally. The asynchronous movement of knobs (Fig. 10) is similar to a known motor-driven process: the placement of achiasmate (nonrecombinant) chromosomes in the Drosophila meiotic spindle. Achiasmate chromosomes are seen within metaphase half spindles at locations that correspond to size. Smaller chromosomes are very close to the pole, while larger chromosomes are positioned farther from the pole. The placement of achiasmate chromosomes is regulated by the Drosophila NOD protein, which is a kinesin-like motor that is localized to chromosome arms (Afshar et al., 1995). It has been argued that the larger chromosomes contain more of the NOD protein and as a result are pushed closer to the metaphase plate (Afshar et al., 1995). Similarly, by binding more motor proteins, the larger neocentromeres may be more effective at overcoming the elastic (drag) forces associated with chromosome stretching. In vitro, kinesin-mediated motility is positively correlated with the number of kinesin molecules and negatively correlated with drag forces (e.g., Hunt et al., 1994; Hall et al., 1996). Unlike NOD (Afshar et al., 1995), the motor responsible for neocentromere motility is presumably minus end directed because the minus ends of the microtubules are predominantly localized at the spindle poles (Euteneuer et al., 1982).
Lateral microtubule interactions were observed with nearly all of the 160 neocentromeres analyzed here and appear to be typical of a wide variety of nonkinetochore transport phenomena (for review see Fuge, 1990). For example, rows of microtubules interact laterally with poleward-pointing chromosome arms, as well as with poleward-moving acentric fragments in the crane fly Pales ferruginea (Fuge, 1972, 1975). Microtubules also interact laterally with animal kinetochores during prometaphase (Rieder et al., 1990; Merdes and De May, 1990), and this interaction generates rates of chromosome movement up to 10 times faster than anaphase chromosome movement (Rieder et al., 1990). Similarly, neocentromeres move at rates significantly faster than normal anaphase chromosomes (Fig. 4 and Table II). The similarities to animal prometaphase kinetochores may indicate that neocentromeres are “normal” maize kinetochores that only display prometaphase-like movement. However, the mechanism of chromosome alignment in animal mitotic (astral) spindles and maize meiotic (anastral) spindles may differ significantly (Rieder et al., 1993).
An additional property of kinetochores is that they function as microtubule organizing centers (e.g., Galatis and Apostolakos, 1991). Whether neocentromeres can also nucleate microtubules was addressed indirectly by Rhoades and Dempsey (1966). Using inversion heterozygotes in an Ab10 background, they generated a series of chromatin fragments that carried a knob but lacked a centromere. The knobbed acentric fragments migrated to a pole when they were brought to the equator in meiosis I (by association with a bivalent), but after the neocentromeres had separated from their linked kinetochores at meiosis II, they lost their ability to interact with the spindle. We have reproduced these results and further observed that acentric neocentromeres in meiosis II are nearly always isolated in the cytoplasm and devoid of any association with microtubules (data not shown). Plant spindles are thought to be organized in large part by microtubule nucleation at the kinetochores and side-by-side associations between microtubules (for review see Smirnova and Bajer, 1992; Palevitz, 1993). Given this, one interpretation of the acentric fragment data is that neocentromeres cannot nucleate microtubules. A neocentromere on a chromosome fragment will be ejected from the spindle by its minus end–directed motor activity and once isolated, will have no means of nucleating the microtubules that would allow it to reassociate with the main spindle (Smirnova and Bajer, 1992; Vernos and Karsenti, 1995). An inability to nucleate microtubules is also consistent with the observation that neocentromeres do not interact in an end-on fashion with spindle fibers (Figs. 8 and 9).
In contrast to kinetochores, neocentromeres interact with microtubules laterally, appear to be incapable of regulating their rates of movement, and are probably incapable of congressing on a metaphase plate. The primary similarity to kinetochore motility appears to be the involvement of a microtubule-based motor(s); indeed, all the available data on neocentromeres can be explained by a meiosis-specific, minus end–directed motor activity that interacts either directly or indirectly with knobs from prometaphase to anaphase. No chromosome-associated motors have been identified in plants, although several kinesin-like proteins have recently been described (Liu et al., 1996; Reddy et al., 1996; Wang et al., 1996). To identify the proteins responsible for neocentromere motility, and ultimately the evolutionarily related proteins required for normal chromosome segregation, we are taking a genetic approach. Neocentromeres are one component in a complex meiotic drive system that causes aberrant segregation ratios (for review see Rhoades and Dempsey, 1985). Because neocentromeres are required for meiotic drive, meiotic drive can be used as a phenotype to score for mutations in neocentromere activity (Dawe and Cande, 1996). One mutation derived from such a screen, suppressor of meiotic drive 1 (smd1), showed a ∼69% reduction in neocentromeres by a quantitative assay (Dawe and Cande, 1996). Smd1, or other genes identified in our ongoing mutant screens, may encode the postulated knob-associated minus end–directed motor.
Acknowledgments
We thank B. Palevitz for critically reading this manuscript.
This work was supported by a grant from the National Science Foundation. A. Chan was supported by a National Institutes of Health grant to W.Z. Cande.
Footnotes
Address all correspondence to R. Kelly Dawe, Department of Botany, Miller Plant Sciences Building, University of Georgia, Athens, GA 30602. Tel.: (706) 542-1658. Fax: (706) 542-1805. E-mail: kelly@dogwood.botany.uga.edu
1. Abbreviation used in this paper: Ab10, abnormal chromosome 10.
References
- Adames KA, Forer A. Evidence for polewards forces on chromosome arms during anaphase. Cell Motil Cytoskeleton. 1996;34:13–25. doi: 10.1002/(SICI)1097-0169(1996)34:1<13::AID-CM2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Afshar K, Barton NR, Hawley RS, Goldstein LSB. DNA binding and meiotic chromosomal localization of the DrosophilaNod kinesin-like protein. Cell. 1995;81:129–138. doi: 10.1016/0092-8674(95)90377-1. [DOI] [PubMed] [Google Scholar]
- Alfenito MR, Birchler JA. Molecular characterization of a maize B chromosome centric sequence. Genetics. 1993;135:589–597. doi: 10.1093/genetics/135.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai DJ, Brokaw CJ, Thompson WC, Wilson L. Two different monoclonal antibodies to tubulin inhibit the bending of reactivated sea urchin spermatozoa. Cell Motil. 1982;2:599–614. doi: 10.1002/cm.970020608. [DOI] [PubMed] [Google Scholar]
- Bajer A. Observations of dicentrics in living cells. Chromosoma. 1963;14:18–30. [Google Scholar]
- Bajer A, Vantard M, Mole-Bajer J. Multiple mitotic transports expressed by chromosome and particle movement. Fortschr Zool. 1987;34:171–186. [Google Scholar]
- Chen, H., J.R. Swedlow, M. Grote, J.W. Sedat, and D.A. Agard. 1995. The collection, processing, and display of digital three-dimensional images of biological specimens. In Handbook of Biological Confocal Microscopy. J.B. Pawley, editor. Plenum Press, New York. 197–210.
- Cutler AJ, Saleem M, Coffey MA, Loewen MK. Role of oxidative stress in cereal protoplast recalcitrance. Plant Cell Tiss Organ Cult. 1989;18:113–128. [Google Scholar]
- Dawe RK, Cande WZ. Induction of centromeric activity in maize by suppressor of meiotic drive 1. Proc Natl Acad Sci USA. 1996;93:8512–8517. doi: 10.1073/pnas.93.16.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. A new role for motor proteins as couplers to depolymerizing microtubules. J Cell Biol. 1995;128:1–4. doi: 10.1083/jcb.128.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerling MH. Preferential segregation of structurally modified chromosomes in maize. Genetics. 1959;44:625–645. doi: 10.1093/genetics/44.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U, Jackson WT, McIntosh JR. Polarity of spindle microtubules in Haemanthusendosperm. J Cell Biol. 1982;94:644–653. doi: 10.1083/jcb.94.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuge H. Morphological studies on the structure of univalent sex chromosomes during anaphase movement in spermatocytes of the crane fly. Chromosoma. 1972;39:403–417. doi: 10.1007/BF00326175. [DOI] [PubMed] [Google Scholar]
- Fuge H. Anaphase transport of akinetochoric fragments in tipulid spermatocytes. Chromosoma. 1975;52:149–158. doi: 10.1007/BF00326264. [DOI] [PubMed] [Google Scholar]
- Fuge H. Non-kinetochore transport phenomena, microtubule-chromosome associations, and force transmission in nuclear division. Protoplasma. 1990;158:1–9. [Google Scholar]
- Galatis B, Apostolakos P. Patterns of microtubule reappearance in root cells of Vigna sinensisrecovering from a colchicine treatment. Protoplasma. 1991;160:131–143. [Google Scholar]
- Gorbsky GJ, Sammak PJ, Borisy GG. Microtubule dynamics and chromosome motion visualized in living anaphase cells. J Cell Biol. 1988;106:1185–1192. doi: 10.1083/jcb.106.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall K, Cole D, Yeh Y, Baskin RJ. Kinesin force generation measured using a centrifuge microscope sperm-gliding motility assay. Biophys J. 1996;71:3467–3476. doi: 10.1016/S0006-3495(96)79542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. p. 418.
- Hunt AJ, Gittes F, Howard J. The force exerted by a single kinesin molecule against a viscous load. Biophys J. 1994;67:766–781. doi: 10.1016/S0006-3495(94)80537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Mitchison TJ. Two different microtubule-based motor activities with opposite polarities in kinetochores. Nature. 1991;351:206–211. doi: 10.1038/351206a0. [DOI] [PubMed] [Google Scholar]
- Inoue S. Polarization optical studies of the mitotic spindle. I. The demonstration of spindle fibers in living cells. Chromosoma. 1953;5:487–500. doi: 10.1007/BF01271498. [DOI] [PubMed] [Google Scholar]
- Inoue S, Salmon ED. Force generation by microtubule assembly/ disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Nasuda S, Dong F, Scherrer CW, Woo S, Wing RA, Gill BS, Ward DC. A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc Natl Acad Sci USA. 1996;93:14210–14213. doi: 10.1073/pnas.93.24.14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Bajer AS, Rieder CL. The force for poleward chromosome motion in Haemanthuscells acts along the length of the chromosome during metaphase but only at the kinetochore during anaphase. J Cell Biol. 1996;132:1093–1104. doi: 10.1083/jcb.132.6.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-de-Faria A. The role of the kinetochore in chromosome organization. Hereditas. 1956;42:85–160. [Google Scholar]
- Liang H, Wright WH, Cheng S, He W, Berns MW. Micromanipulations of chromosomes in PtK2 cells using laser microsurgery (optical scalpel) in combination with laser-induced optical force (optical tweezers) Exp Cell Res. 1993;204:110–120. doi: 10.1006/excr.1993.1015. [DOI] [PubMed] [Google Scholar]
- Liu B, Cyr R, Palevitz BA. A kinesin-like protein, KatAp, in the cells of Arabidopsisand other plants. Plant Cell. 1996;8:119–132. doi: 10.1105/tpc.8.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley AE. Knob positions on corn chromosomes. J Agric Res. 1939;59:475–490. [Google Scholar]
- Merdes A, De May J. The mechanism of kinetochore-spindle attachment and polewards movement analyzed in PtK2cells at the prophase-prometaphase transition. Eur J Cell Biol. 1990;53:313–325. [PubMed] [Google Scholar]
- Mitchison TJ, Salmon ED. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J Cell Biol. 1992;119:569–582. doi: 10.1083/jcb.119.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Maize genetics. Yearbook Carnegie Inst Wash. 1943;42:148–152. [PubMed] [Google Scholar]
- Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- Novitski E. The genetic consequences of anaphase bridge formation in Drosophila. . Genetics. 1952;37:270–287. doi: 10.1093/genetics/37.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östergren G, Mole-Bajer J, Bajer A. An interpretation of transport phenomena at mitosis. Ann NY Acad Sci. 1960;90:381–408. doi: 10.1111/j.1749-6632.1960.tb23258.x. [DOI] [PubMed] [Google Scholar]
- Palevitz BA. Morphological plasticity of the mitotic apparatus in plants and its developmental consequences. Plant Cell. 1993;5:1001–1009. doi: 10.1105/tpc.5.9.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock WJ, Dennis ES, Rhoades MM, Pryor AJ. Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc Natl Acad Sci USA. 1981;78:4490–4494. doi: 10.1073/pnas.78.7.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena ADL. “In vitro” culture of isolated meiocytes of rye, Secale cereale L. Environ Exp Bot. 1986;26:17–23. [Google Scholar]
- Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990;345:263–266. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Narasimhulu SB, Safadi F, Golovkin M. A plant kinesin heavy chain-like protein is a calmodulin-binding protein. Plant J. 1996;10:9–21. doi: 10.1046/j.1365-313x.1996.10010009.x. [DOI] [PubMed] [Google Scholar]
- Rhoades, M.M. 1952. Preferential segregation in maize. In Heterosis. J.W. Gowen, editor. Iowa State College Press, Ames, Iowa. 66–80.
- Rhoades MM, Dempsey E. The effect of abnormal chromosome 10 on preferential segregation and crossing over in maize. Genetics. 1966;53:989–1020. doi: 10.1093/genetics/53.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.M., and Dempsey, E. 1985. Structural heterogeneity of chromosome 10 in races of maize and teosinte. In Plant Genetics. M. Freeling, editor. Alan R. Liss, Inc. New York. 1–18.
- Rhoades MM, Vilkomerson H. On the anaphase movement of chromosomes. Proc Natl Acad Sci USA. 1942;28:433–443. doi: 10.1073/pnas.28.10.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP, Rupp G. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in Newt lung cells. J Cell Biol. 1990;110:81–90. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., J. Ault, U. Eichenlaub-Ritter, and G. Sluder. 1993. Morphogenesis of the mitotic and meiotic spindle: conclusions from one system are not necessarily applicable to the other. In Chromosome Segregation and Aneuploidy. B.K. Vig, editor. Springer-Verlag, Berlin. 183–197.
- Ryan KG. Prometaphase and anaphase chromosome movements in living pollen mother cells. Protoplasma. 1983;116:24–33. [Google Scholar]
- Scott P, Lyne RL. The effect of different carbohydrate sources upon the initiation of embryogenesis from barley microspores. Plant Cell Tiss Organ Cult. 1994;36:129–133. [Google Scholar]
- Smirnova EA, Bajer A. Spindle poles in higher plant mitosis. Cell Motil Cytoskel. 1992;23:1–7. doi: 10.1002/cm.970230102. [DOI] [PubMed] [Google Scholar]
- Steuer ER, Wordeman L, Schroer TA, Sheetz MP. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature. 1990;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- Tokai N, Fujimoto-Nishiyama A, Toyishima Y, Yonemura S, Tsukita S, Inoue J, Yamamoto T. Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO (Eur Mol Biol Organ) J. 1996;15:457–467. [PMC free article] [PubMed] [Google Scholar]
- Vernos I, Karsenti E. Chromosomes take the lead in spindle assembly. Trends Cell Biol. 1995;5:297–301. doi: 10.1016/s0962-8924(00)89045-5. [DOI] [PubMed] [Google Scholar]
- Vernos I, Karsenti E. Motors involved in spindle assembly and chromosome segregation. Curr Opin Cell Biol. 1996;8:4–9. doi: 10.1016/s0955-0674(96)80041-x. [DOI] [PubMed] [Google Scholar]
- Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C. Xklp1, a chromosomal Xenopuskinesin-like protein essential for spindle organization and chromosome positioning. Cell. 1995;81:117–127. doi: 10.1016/0092-8674(95)90376-3. [DOI] [PubMed] [Google Scholar]
- Wang S, Adler R. Chromokinesin: a DNA-binding, kinesin-like nuclear protein. J Cell Biol. 1995;128:761–768. doi: 10.1083/jcb.128.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Takezawa D, Narasimhulu SB, Reddy ASN, Poovaiah BW. A novel kinesin-like protein with a calmodulin-binding domain. Plant Mol Biol. 1996;31:87–100. doi: 10.1007/BF00020609. [DOI] [PubMed] [Google Scholar]
- Waters JC, Mitchison TJ, Rieder CL, Salmon ED. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol Biol Cell. 1996;7:1547–1558. doi: 10.1091/mbc.7.10.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Schaar BT. Kinetochore function: molecular motors, switches and gates. Curr Opin Cell Biol. 1996;8:381–388. doi: 10.1016/s0955-0674(96)80014-7. [DOI] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]