Abstract
To understand the role of microtubule-associated proteins (MAPs) in the regulation of microtubule (MT) dynamics we have characterized MAPs prepared from Xenopus laevis eggs (Andersen, S.S.L., B. Buendia, J.E. Domínguez, A. Sawyer, and E. Karsenti. 1994. J. Cell Biol. 127:1289–1299). Here we report on the purification and characterization of a 310-kD MAP (XMAP310) that localizes to the nucleus in interphase and to mitotic spindle MTs in mitosis. XMAP310 is present in eggs, oocytes, a Xenopus tissue culture cell line, testis, and brain. We have purified XMAP310 to homogeneity from egg extracts. The purified protein cross-links pure MTs. Analysis of the effect of this protein on MT dynamics by time-lapse video microscopy has shown that it increases the rescue frequency 5–10-fold and decreases the shrinkage rate twofold. It has no effect on the growth rate or the catastrophe frequency. Microsequencing data suggest that XMAP230 and XMAP310 are novel MAPs. Although the three Xenopus MAPs characterized so far, XMAP215 (Vasquez, R.J., D.L. Gard, and L. Cassimeris. 1994. J. Cell Biol. 127:985–993), XMAP230, and XMAP310 are localized to the mitotic spindle, they have distinct effects on MT dynamics. While XMAP215 promotes rapid MT growth, XMAP230 decreases the catastrophe frequency and XMAP310 increases the rescue frequency. This may have important implications for the regulation of MT dynamics during spindle morphogenesis and chromosome segregation.
The function of the mitotic spindle is to ensure even segregation of the duplicated chromosomes to the two daughter cells (Kirschner and Mitchison, 1986; McIntosh, 1994; Inoué and Salmon, 1995; Hyman and Karsenti, 1996; Waters and Salmon, 1997). Assembly of the mitotic spindle involves a dramatic reorganization of the radial interphase microtubule (MT)1 network into an elliptical, bipolar shape. This reorganization results from a significant increase in MT turnover at the onset of mitosis (Olmsted et al., 1989; Belmont et al., 1990; Verde et al., 1990, 1992; Vale, 1991; Shiina et al., 1992a ; Sawin and Mitchison, 1994; Hyman and Karsenti, 1996; McNally et al., 1996; Zhai et al., 1996; Tournebize et al., 1997). MT turnover is largely due to the dynamic instability of MTs (Kirschner and Mitchison, 1986), meaning that MTs are not stable rods but instead alternate between phases of growth and shrinkage. Therefore a given turnover state for MTs will be determined by the respective values of four parameters: the growth rate (vg), the shrinkage rate (vs), and the transition between growth and shrinkage, a catastrophe which occurs with a certain frequency (fcat). The opposite event, when a MT transit from shrinkage to growth, a rescue also occurs with a certain frequency (fres) (Walker et al., 1988). Spindle MTs are stabilized and have reduced dynamicity compared with their cytoplasmic counterparts (Karsenti et al., 1984; Saxton et al., 1984; Toso et al., 1993; Vasquez et al., 1994; Dogterom et al., 1996; Heald et al., 1996; Zhai et al., 1996). Microtubule- associated proteins (MAPs) may have an important function in the stabilization of spindle MTs, since they in general dampen MT dynamics and since their activity can be regulated by phosphorylation. However, little is known about the properties of non-neuronal MAPs in general (for reviews see Olmsted, 1986; Matus, 1990; Wiche et al., 1991; Bulinski, 1994; Hirokawa, 1994; Mandelkow and Mandelkow, 1995; Hyman and Karsenti, 1996). Most probably, several different MAPs are involved in regulation of MT turnover during the cell cycle and in the spindle. However, only XMAP230, MAP4, and recently XMAP215 have been localized to the spindle and functionally characterized in vitro (Gard and Kirschner, 1987; Andersen et al., 1994; Vasquez et al., 1994; Ookata et al., 1995; Charrasse, S., M. Schroeder, C. Gauthier-Rouviere, L. Cassimeris, D.L. Gard and C. Larroque. 1996. Mol. Biol. Cell. 7:222a).
To proceed with the identification and characterization of non-neuronal MAPs, we have raised mAbs against MT proteins bound to taxol stabilized MTs isolated from interphase Xenopus laevis egg extracts (Andersen et al., 1994). Several studies indicate that the MT binding activity of MAPs is finely tuned during the cell cycle, rather than modulated by a strict on- off- switch (Zieve and Solomon, 1982; Olmsted et al., 1989; Shiina et al., 1992b ; Andersen et al., 1994; Masson and Kreis, 1995). Therefore, from the interphase MAP fraction, it should be possible to identify MAPs associated with the interphasic MT network, the mitotic spindle MTs, or both. Here we report on the structural and functional in vitro characterization of a new 310-kD Xenopus MAP (XMAP310) that localizes to the mitotic spindle. Interestingly, this MAP turns out to be a strong rescue factor.
Materials and Methods
Extracts, Antibodies, and Immunofluorescence
Xenopus egg extracts were prepared as described by Murray (1991). Interphase and mitotic extracts (CSF-arrested) had H1 kinase activities of 2–5 and 15–25 pmol/μl/min (Félix et al., 1993), respectively. Total MAPs for mouse mAb production were produced as described by Andersen et al. (1994). The screening procedure to obtain the Q4 mAb consisted of immunofluorescence and western blotting in parallel; the mouse polyclonal antiserum gave the same immunofluorescence pattern as the final anti-XMAP310 Q4 mAb (subtyped as IgG1; Mouse-Hybridoma-Subtyping Kit, Boehringer Mannheim GmbH, Mannheim, Germany). Treatment of Western blots with alkaline phosphatase did not result in loss of reactivity with the Q4 mAb, indicating that Q4 does not recognize a phosphorylated epitope (data not shown). For the experiment described here either ascites or culture supernatants of Q4 were used at a 1/500 and 1/3 dilution, respectively, for Westerns. The L7 anti-XMAP230 mAb was as described by Andersen et al. (1994). Immunofluorescence was performed as described by (Andersen et al., 1994). The Q4 ascites was used at a 1/200 dilution. Preextraction of cells was performed twice for 3 s in BRB80, 5 mM EGTA, 0.5% Tx-100, pH 6.8. Confocal fluorescence microscopy was performed as described by Reinsch and Karsenti (1994).
Purification of XMAP310 from Xenopus Egg Extracts
Interphase egg extracts were prepared by activation of dejellied eggs with calcium ionophore A23187 (Sigma Chem. Co., St. Louis, MO) at a 1/10,000 dilution (stock 2 mg/ml in DMSO) in MMR plus 200 μg/ml cycloheximide for 5 min (see also Murray, 1991). Concentrated extracts were frozen in liquid nitrogen in 200-μl aliquots and stored at −80°C. Concentrated extracts (5–7 ml/purification) were rapidly thawed at 37°C, transferred to ice, and diluted with 1 vol 50 mM sucrose, 10 mM K-Hepes (pH 7.7), 1 mM EGTA, 1 mM MgCl2, 0.1% β-mercaptoethanol, protease inhibitors. This mix was centrifuged at 40,000 rpm (192,000 g) for 40 min at 4°C in a SW50 rotor. 20 μM (final) taxol (10 mM stock in DMSO; Molecular Probes) was added to this clarified extract on ice, together with 1/10 vol prepolymerized MTs: 400 μl cycled tubulin at 9 mg/ml was mixed with 400 μl 66% glycerol, BRB80 (80 mM K-Pipes, 1 mM EGTA, 1 mM MgCl2, pH 6.8), 10 mM MgCl2, and 1 mM GTP final and incubated for 30 min at 37°C; Taxol was then added to 20 μM (final) and incubation continued for 5 min before addition to the extract. The extract with the prepolymerized MTs was incubated on ice for 10 min, and then loaded on 40% sucrose cushions, BRB80, 20 μM taxol, and protease inhibitors at a 1:1 volume ratio. The MTs were spun through the cushion at 40,000 rpm (70,000 g) for 20 min at 4°C in a TLA100.2 rotor. The supernatant was discarded and the MT pellets resuspended in 1/4 the volume of the clarified extract (2 ml) using buffer A (20 mM K-Pipes (pH 7), 50 mM NaCl) plus 5 mM CaCl2, protease inhibitors and 0.1% β-mercaptoethanol. This mix was incubated for 10 min on ice to allow depolymerization of the taxol stabilized MTs and then centrifuged twice for 10 min, 50,000 rpm (110,000 g), 4°C. The supernatant (see Fig. 3, lane 3), enriched in XMAP310 and tubulin, was applied onto a PC 1.6/5 MonoS column using the SMART system (Pharmacia LKB Biotechnology, Uppsala, Sweden) at 4°C and 100 μl/min; After loading the column was washed with 1 ml 5% B buffer (20 mM K-Pipes (pH 7), 800 mM NaCl), a step to 20% B resulted in elution of XMAP310 along with other proteins; XMAP310 was enriched in the early fractions after shift to 20% B; pooled fractions (100–150 μl) from the MonoS column (see Fig. 3, lane 5) were loaded on a PC 3.2/30 Superose 6 column equilibrated in 100 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10% glycerol, 10 mM K-Hepes (pH 7.7) using the SMART system at 4°C and 40 μl/min. Fractions containing pure XMAP310 (see Fig. 3, lane 6) were stored separately or pooled with later fractions (see Fig. 3, lanes 7–8), aliquoted by 20 μl and stored in liquid nitrogen. The concentration of XMAP310 in the extract is ∼200 nM.
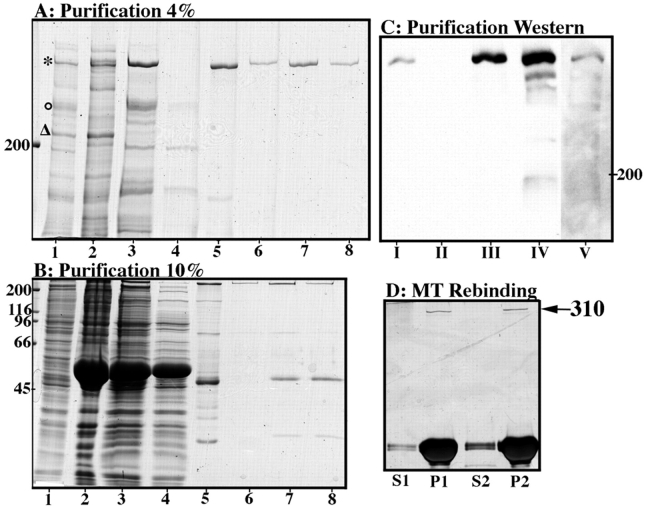
Figure 3.
Purification of XMAP310. A and B show the different steps of the purification by Coomassie-stained 4% (A) and 10% (B) SDS-PAGE: lane 1, total MAPs eluted with salt (Andersen et al., 1994) from taxol stabilized MTs; lane 2, MT pellet of Ca2+ undepolymerized MTs; lane 3, Supernatant of Ca2+ depolymerized MTs/Sample loaded on the MonoS column; lane 4, Flow-through from the MonoS column; lane 5, pooled fractions from the MonoS column (the major 49-kD protein was determined as EF1-γ); lanes 6–8, fractions from the Superose 6 column. (C) Steps of the purification monitored with the Q4 mAb: lane I, start extract; lane II, extract after depletion with MTs; lane III, total MTs (V = 2 ml); lane IV, Ca2+-stable MT fraction (V = 500 μl); lane V, pooled fractions from the MonoS column (compare with A and B, lanes 5). (D) Purified XMAP310 rebinds to and pellets (P) with taxol stabilized MTs. Binding to MTs was for 10 min on ice. In lane 1 the MT concentration (in tubulin equivalents) was 2.5 μM and in 2 0.6 μM. However, the total amount of MTs used was the same in lanes 1 and 2 (S, supernatant after pelleting MTs).
Video Microscopy and MT Cross-linking Assay
Chambers with a volume of ∼4 μl were used. Two strips of double-sided tape were fixed on a microscope slide (76 × 22mm, 0.8 mm thick, Select Micro Slide model BS3836-1975; Chance Propper Ltd., UK) ∼3 mm apart and perpendicular to the long axis of the slide. A 22 × 22mm coverslip (gold seal cover glass model 3306; Clay Adams, UK) was then placed on top of the two tape strips and firmly pressed to attach it well. Slide and coverslip were ethanol and water washed before use. The channel formed by the tape strips, the slide and the coverslip made up the chamber. Before use, centrosomes (Bornens et al., 1987) were diluted to 3 × 103/μl with working concentration of tubulin, and injected into the chamber and left for 5 min on ice. 10 μl 5 mg/ml Casein in BRB80 was then by capillary forces flowed through the chamber and left for 5 min on ice. The chamber was then washed with 15 μl BRB80 and the sample was injected. The volume of the sample was always 30 μl and mixed immediately before use. For most of the preparations the sample consisted of: 10.0 μl Superose 6 column buffer with XMAP310 or without XMAP310 (control), 15.1 μl BRB80, 4.83 μl cycled tubulin at 87 μM (final tubulin concentration 14 μM [using BSA as Standard]), 0.3 μl 0.1 M GTP. After application of the sample the chamber was sealed at both ends with grease and placed on the stage of a Zeiss Axiovert 10 photomicroscope (Zeiss GmbH, Oberkochen, Germany) equipped with a Zeiss 100× achrostigmat 100×/1.25 oil objective and a Hamamatsu Phototonics (Japan) C3077 CCD camera. Objective, condenser, and stage were heated to 37°C. The temperature in the chamber was 35°C. Measurements of MT dynamics were performed as described (Andersen et al., 1994) using 4-s time interval between individual video frames and NIH 1.55 and Microsoft Excel 5 programs. No samples were observed for more than 40 min.
Tubulin at 40 μM plus 2 μM rhodamine-labeled tubulin (Hyman et al., 1991) in BRB80 and 1 mM GTP was mixed on ice with 1/2 vol MAP or control buffer (same dilution of MAP as used in the video experiments) and incubated for 15 min at 37°C. The sample was then processed for negative stain electron microscopy. Alternatively, the sample was diluted 200-fold into BRB80 plus 20 μM taxol at room temperature (without fixation), loaded on top of a 3 ml 15% glycerol cushion, BRB80, 20 μM taxol in modified Corex tubes (Evans et al., 1985) and then centrifuged at 12,000 rpm for 20 min at 20°C using a HB4 rotor. Coverslips were fixed for 5 min in −20°C methanol and mounted in 90% glycerol, 50 mM Tris-HCl (pH = 8), and analyzed. For negative stain EM, 3 μl MT-MAP mix was absorbed onto a glow-discharged 300 mesh carbon grid for 1 min at room temperature, followed by three 5-μl washes and staining with 1% uranyl acetate for 1 min and analysis with a Zeiss 10C electron microscope.
Miscellaneous
For microsequencing XMAP230 and XMAP310 were prepared in preparative amounts by immunoprecipitation. Dynabeads (model 110.12; Dynal GmbH, Hamburg, Germany) with the Q4 or L7 mAb precoupled were incubated in extracts for 30 min at 4°C. Beads were washed 10 times for 1 min, each time with 1 ml PBS, 150 mM NaCl, 0.5% TX-100, 1 mM PMSF at 4°C before analysis on 4% SDS-PAGE. The bands corresponding to XMAP230 and XMAP310 were excised and digested with trypsin. Peptides were separated as described by Tetaz et al. (1993). Edman degradation was performed using an Applied Biosystems Machine (model 477A; Foster City, San Francisco, CA). These microsequences as well as the identity of the Q4 immunoprecipitations, the purified XMAP310 protein (see Fig. 3, lane 6), and EF1-γ (see Fig. 3, lane 5) were confirmed by Matrix-assisted laser desorption ionization mass spectroscopy (MALDI MS). MALDI MS consists of tryptic cleavage of the protein followed by mass spectroscopy of the peptides (Jensen et al., 1996). Therefore, using MALDI MS one gets a fingerprint spectrum of the tryptic cleavage sites of the protein. The spectrum can then be compared with hypothetical tryptic cleavage spectra of proteins in the data bases. Very conserved proteins will show homology by this method because trypsin sites will be conserved in a conserved sequence. Calf brain tubulin was prepared as described by Ashford et al. (1998). The relative molecular mass for XMAP310 of 310 kD was estimated on the basis of the migration on 4% SDS-PAGE of bovine brain MAP1B (M r 325 kD). The hydrodynamic data for XMAP230 and XMAP310 were determined as described by Wilhelm et al. (1997). Gel electrophoresis and Western blotting were performed as described by Andersen et al. (1994). Western blots were developed using alkaline phosphatase conjugated secondary antibodies or ECL (Amersham Corp.). Protein concentrations were measured with Bradford (Bio-Rad Labs., Hercules, CA) or BCA (Pierce, BA oud-Beijerland, Holland) reagents. Tissue extracts were prepared as described in Andersen et al., (1994). Unless otherwise indicated, chemicals were from Sigma or Boehringer Mannheim GmbH.
Results
Identification and Tissue Distribution of XMAP310
Three major MAPs larger than 200 kD cosediment with taxol stabilized MTs from interphasic Xenopus egg extracts: XMAP215, XMAP230, and a 310-kD MAP (XMAP310) that was not previously characterized (see Fig. 1 A in Andersen et al., 1994 and Fig. 3 A, lane 1 in this study: XMAP215 (Δ), XMAP230 (o), XMAP310 (*). We raised mAbs against XMAP310, and obtained one clone (Q4) that recognized XMAP310 in extracts of the Xenopus tissue culture cell line XL177 (Miller and Daniel, 1977), in total Xenopus egg extracts, and in the MAP fraction (data not shown). When Xenopus tissue extracts were probed with the Q4 mAb, XMAP310 was very abundant in brain and testis tissues but could not be detected in heart, lung, stomach, or muscle tissues (data not shown).
Figure 1.
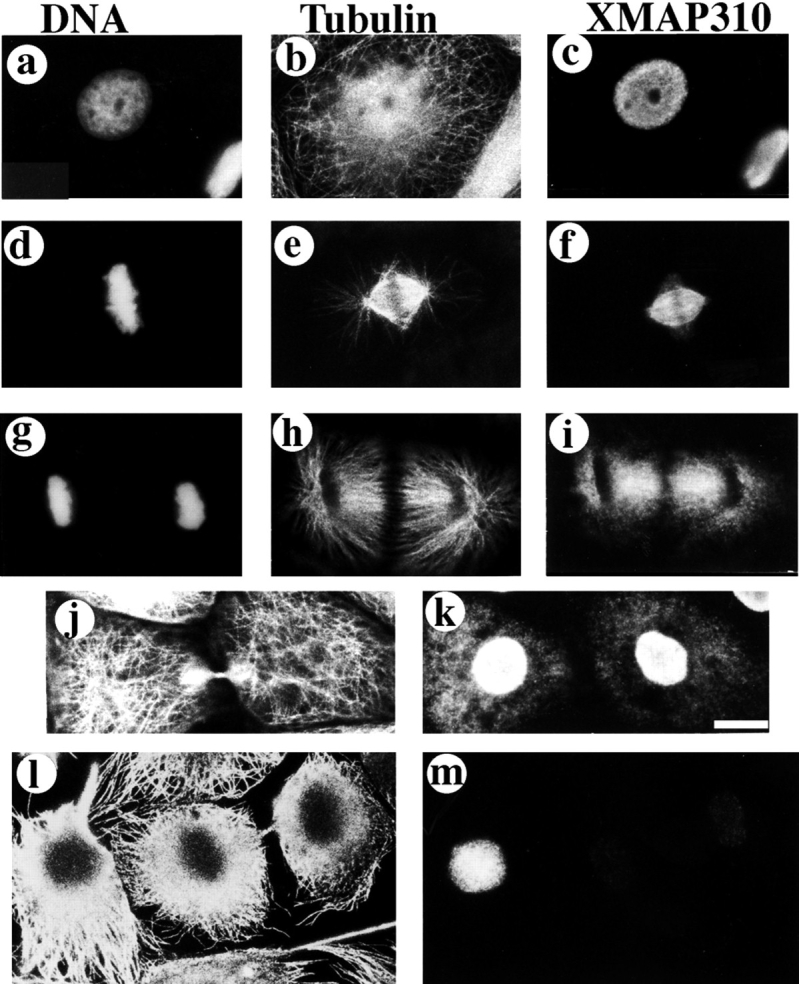
XMAP310 localization in XL177 cells (Miller and Daniel, 1977) during the cell cycle. XL177 cells were stained with: Hoechst (a, d, g), a rabbit anti-tubulin pAb (b, e, h, j, l), and the Q4 mAb (c, f, i, k, m). Interphase (a–c), metaphase/early anaphase after pr-extraction with 0.5% TX-100 (d–f), late anaphase (g–i), late telophase without preextraction (j and k), late telophase with preextraction (l and m). Bar: (a–k) 10 μm; (l and m) 12.5 μm.
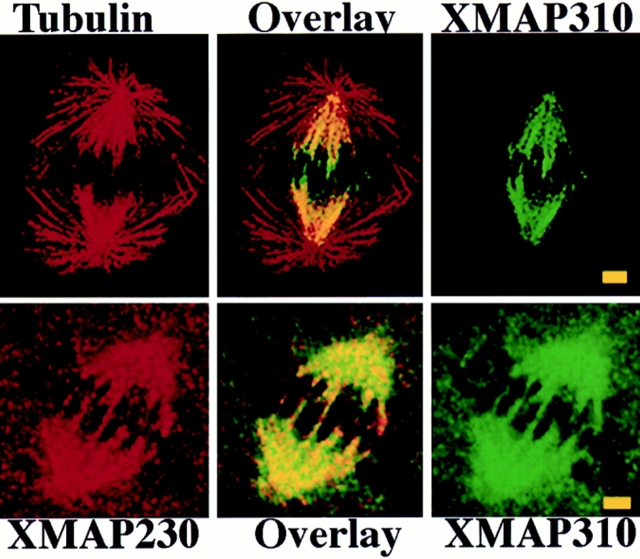
Since XMAP310 had been identified biochemically due to its affinity for MTs, we wanted to determine whether it localized to MTs in vivo. The Q4 mAb was used to probe the localization of XMAP310 at different stages during the cell cycle of Xenopus XL177 tissue culture cells (Fig. 1). In interphase cells XMAP310 was present only in the nucleus (Fig. 1, a–c). The analysis of confocal sections showed that XMAP310 was homogeneously localized throughout the nucleus (data not shown). This localization was maintained through prophase, but XMAP310 relocalized to spindle MTs at the exclusion of astral MTs during metaphase (data not shown and Fig. 1, d–f). The metaphase localization was resistant to preextraction of the cells with detergents (Fig. 1, d–f). During anaphase, XMAP310 was concentrated to areas of high MT density (Fig. 1, g–i), but this staining could be removed by preextraction. During cytokinesis XMAP310 began to reaccumulate in the nucleus (Fig. 1, j and k). Some of the protein that remained diffusely localized in the cytoplasm, was completely removed by preextraction (Fig. 1, l and m, note the faint staining of the nucleus after preextraction). The localization of XMAP310 in the nucleus during interphase and to the spindle during mitosis was confirmed by immunostaining of Xenopus embryos at the mid blastulae stage (data not shown). Double immunostaining of XL177 cells with XMAP230 and XMAP310 mAbs and observation by confocal microscopy revealed that the two proteins colocalize in the mitotic spindle (Fig. 2). However, immunoprecipitation and native molecular weight estimation showed that XMAP230 and XMAP310 do not form a complex in the extract, suggesting that the co-localization in the spindle MTs is due to independent regulation of binding for these two proteins (data not shown; hydrodynamic data for XMAP230 and XMAP310: stokes radius (ao), 13.3 nm and 16.5 nm, respectively; sedimentation coefficients, (S20w) of 5.2 × 10−13 s−1 and 10.1 × 10−13 s−1, respectively; native molecular masses, 285 and 685 kD, respectively).
Figure 2.
(Top) XMAP310 localization in the mitotic spindle: MTs stained with a rabbit anti-tublin pAb in the rhodamine channel (Tubulin), XMAP310 in the fluorescein channel (XMAP310) and in overlay (Overlay). (Bottom) XMAP230 and XMAP310 colocalize (Overlay) to the central region of the mitotic spindle in XL177 cells. Images were obtained by confocal microscopy. Bar, 2.5 μm.
Purification of XMAP310 from Xenopus Egg Extracts
To determine the effect of XMAP310 on MTs it was purified from interphase Xenopus egg extracts. The first purification step consisted in the preparation of MAPs using taxol stabilized MTs, as shown in Fig. 3 A and B, lane 1. XMAP310 could be eluted from MTs by 400 mM salt together with XMAP230 and XMAP215 (Fig. 3, lane 1). However, instead of eluting the MAPs with salt, we used Ca2+ ions to depolymerize taxol stabilized MTs on ice (Collins, 1991; Bulinski and Bossler, 1994). This results in the depolymerization of ∼50% of the taxol MTs (Fig. 3 B, lanes 2 and 3; the large band at ∼50 kD is tubulin), and most of XMAP310 was released from the MT pellet by this treatment, although a heterogeneous fraction remained bound to MTs (Fig. 3 A, *, lanes 3 and 2, note the doublet in lane 2). XMAP230 was also eluted after Ca2+ depolymerization (Fig. 3 A o, lane 3). However, XMAP215 was highly enriched in the Ca2+-stable MT fraction (Fig. 3 A, Δ, lane 2). The Ca2+-eluted XMAP310 was then loaded on a MonoS column, and eluted with a salt gradient (Fig. 3 A, lanes 4 and 5). The MonoS fractions enriched in XMAP310 (Fig. 3 A, lane 5) contained one major contaminant of ∼49 kD (Fig. 3 B, lane 5), that was identified as EF1-γ by MALDI MS (data not shown, Cormier et al., 1991; Hershey, 1991; Jensen et al., 1996). EF1-γ, was eliminated from the preparation by gel filtration through a Superose 6 column (Fig. 3 A and B, lane 6 and later fractions, lanes 7 and 8). The purification was monitored by western blotting with the Q4 mAb. A fraction appeared to remain bound to MTs after Ca2+-depolymerization (Fig. 3 C, lane IV). However, analysis of the same fractions by Coomassie staining (Fig. 3 A, lane 3) revealed that most of the protein was eluted from the MTs. This could be due to the presence of different proteins of the same molecular weight, the presence of isoforms (see Fig. 3 A, lane 2; occasionally three bands were resolved) or the loss of the epitope during the purification. To resolve this issue we checked by MALDI MS whether the protein recognized by the Q4 mAb and the purified protein were the same. The MALDI MS analysis showed that the protein immunoprecipitated by the Q4 mAb and the protein purified on the Superose 6 column were the same (data not shown). We conclude that the poor recognition of pure XMAP310 by the Q4 mAb reflects the presence of isoforms (as previously reported for XMAP230) and perhaps reduced recognition of the epitope on the pure XMAP310.
We checked that XMAP310 is a bona fide MAP by assaying the capacity of the pure protein to bind back to taxol stabilized MTs. The experiment showed that XMAP310 bound back quantitatively to MTs (Fig. 3 D, arrow). These experiments also showed that EF1-γ does not rebind to MTs (data not shown), and that the apparent dissociation constant of XMAP310 for MTs was ∼200 nM (data not shown).
XMAP310 Causes Microtubule Cross-linking
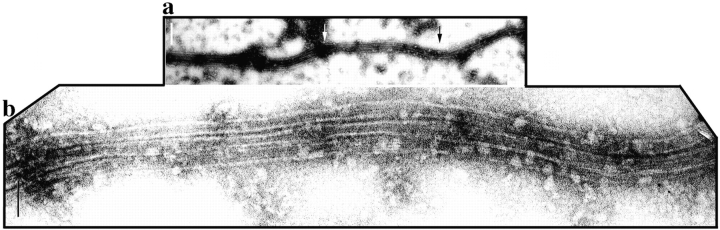
As a first step in the characterization of the effect of pure XMAP310 on MTs, we polymerized rhodamine labeled tubulin, either in the presence or absence of XMAP310. The morphology of the MTs obtained suggested that they formed bundles in the presence of XMAP310 (data not shown). This was confirmed by electron microscopy, which showed long bundles of 2–5 MTs, in the presence of XMAP310 (Fig. 4). The short distance between the individual MTs resembles the cross-links obtained with 280 kD Syncolin (Feick et al., 1991), but is different from the larger spacing reported for brain MAPs (Hirokawa et al., 1988; Sato-Yoshitake et al., 1989; Chen et al., 1992).
Figure 4.
Electron micrographs showing cross-linked MTs in bundles formed in the presence of XMAP310. After spontaneous assembly in the presence XMAP310, MTs were spotted onto carbon grids and visualized by negative stain and electron microscopy. (a) A long bundle consisting of three cross-linked MTs (b) magnification of the arrow-outlined area in a, note the uniform short spacing of the MTs. Bars: (a) 200 nm; (b) 60 nm.
XMAP310 Primarily Promotes Rescues
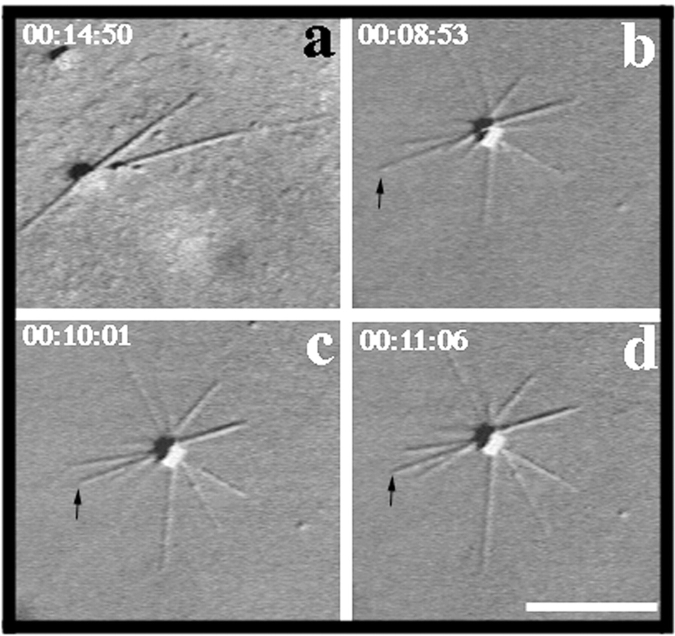
To determine how XMAP310 affected the parameters of MT dynamic instability, we examined MT growth in the presence and absence of XMAP310 by time lapse video microscopy (Fig. 5). Centrosomes and tubulin were preabsorbed to the glass of a microscope chamber on ice, followed by further blocking of the glass with casein. The sample containing soluble tubulin with or without XMAP310 was then injected into the chamber and observed under a microscope at 35°C. The polymerization of individual MTs was then followed in real time (Fig. 5). The average growth rate of MTs (vg) was not significantly influenced by XMAP310, whereas the shrinkage rate (vs) was reduced twofold compared with the control (Table I). Moreover, XMAP310 promoted rescues 5–10-fold (fres) without having any effect on the catastrophe frequency (fcat, Table I). The dynamicity of MTs in the presence of XMAP310 relative to the control was calculated to 0.6, showing that XMAP310 dampens MT dynamics (Toso et al., 1993; Vasquez et al., 1994). In these experiments the concentration of XMAP310 in the chamber varied between 50 and 150 nM depending on the batch used, and the total tubulin concentration was 14 μM. Assuming a stoichiometry <1:5, we calculated (Andersen et al., 1994) that MTs should be saturated with XMAP310, even at a concentration of 50 nM (data not shown). At higher concentrations (>150 nM) catastrophes were rare (data not shown). We believe that this is caused by a stronger promotion of rescues which would cause a MT that undergoes a catastrophe to be rescued immediately. This idea is supported by the observation of pauses, that only occurred in the presence of XMAP310. We verified that the activity of pure XMAP310 from Superose 6 fractions without (Fig. 3, lane 6) and with (Fig. 3, lane 8) EF1-γ was the same, indicating that EF1-γ does not influence MT polymerization (data not shown). These data show that XMAP310 stabilizes MTs by moderately decreasing the shrinkage rate and increasing the rescue frequency 5–10-fold.
Figure 5.
MT dynamics visualized by video microscopy. MTs are nucleated by a centrosome (center of the MT asters). (a) MTs polymerized in the absence of XMAP310. (b–d) MTs polymerized with XMAP310. The MT indicated with an arrow experienced a catastrophe shortly after the frame shown in b, and shrank to the length shown in c where it was rescued and continued to grow, as shown in d. Time since transfer to 37°C is indicated by xx:xx:xx, which represents hours:minutes:seconds, respectively. Bar, 10 μm.
Table I.
Effects on MT Dynamics of XMAP310
| Control | +XMAP310 | |||
|---|---|---|---|---|
| vg ± sd (μm/min) | 1.44 ± 0.97 | 1.13 ± 0.91 | ||
| vs ± sd (μm/min) | 21.26 ± 16.09 | 10.61 ± 9.31 | ||
| fcat (s−1)/No of cat | 0.0024/16 | 0.0024/66 | ||
| fres (s−1)/No of res | 0.0042/3 | 0.0231/61 | ||
| ΣPauses (min)/No pauses | 0.00/0 | 7.93/12 | ||
| ΣTime in growth (min) | 109.32 | 459.73 | ||
| ΣTime in shrinkage (min) | 11.80 | 44.00 | ||
| ΣΣTime of observation (min) | 121.12 | 511.66 | ||
| No of MTs measured | 28 | 39 | ||
| Σof measurements | 173 | 567 |
The experiments were performed in a chamber at 35°C with BRB80 + 1 mM GTP (Materials and Methods). The final tubulin concentration was 14 μM, and 1/3 of the final sample volume consisted of control buffer (Control) or XMAP310 (+XMAP310) from the Superose 6 column (Fig. 8, lanes 6, 7, or 8). The data in the table represent pooled data from three independent purifications of XMAP310. No samples were observed for more than 40 min. ν g is the average growth rate, ν s the average shrinkage rate, f cat is the catastrophe frequency, No of cat is the total number of catastrophes observed, f res is the rescue frequency, No of res is the total number of rescues observed, ΣPauses is the accumulated time during which vg was lower than 0.1 μm/min, No pauses is the total number of pauses observed, ΣTime in growth is the accumulated time during which vg was above 0.1 μm/min, ΣTime in shrinkage is the accumulated time spent in the shrinkage phase, ΣΣTime of observation is the total time of observation, No of MTs measured is the total number of MTs measured for the shown data, Σof measurements is the total number of individual rate measurements, ±sd denotes the standard deviation, time is expressed in min (min) or seconds (s).
Discussion
XMAP310, A Novel Mitotic Spindle MAP
Like XMAP310, other MAPs localized to the nucleus in interphase and to spindle MTs during mitosis have been described previously (i.e., Izant et al., 1982; Pepper et al., 1984; Bonifacino, 1985; and NuMA: Compton et al., 1991; Cleveland, 1995; Merdes et al., 1996). This could suggest that XMAP310 is homologous to known proteins. However, by Edman degradation we obtained the following amino acid sequences for XMAP310: IVQPFGGK, LETAY(P or V)MI and VLPINAA, and these sequences did not show any significant sequence homology with other known proteins. Moreover, MALDI MS (Jensen et al., 1996; Materials and Methods) analysis also did not reveal any homology between XMAP310 or other proteins in the data bases. Moreover, Edman degradation of XMAP230 gave the following sequences that did not show homology with any proteins in the data bases: APLAKPSTALAL, LPTA(S or T)NVTNAA, T(Q or K)EAKRTNQGE, QSIVVEE, NGEPAAAPTEGDNQR, (S or A)- VKIVE, and ATVPAE. MALDI MS of XMAP230 also did not reveal any homologous proteins in the data bases. However, a MALDI MS analysis of XMAP215 revealed that it has strong homology with the human protein TOGp, as previously reported (Charrasse, S., M. Schroeder, C. Gauthier-Rouviere, L. Cassimeris, D.L. Gard and C. Larroque. 1996. Mol. Biol. Cell. 7:222a; Podtelejnikov, A., and M. Mann, unpublished observation). Thus, XMAP310 and XMAP230 appear to be new MAPs whereas XMAP215 is a TOGp homologue (Charasse et al., 1995).
Interestingly, XMAP230 and E-MAP-115 (Masson and Kreis, 1995) are localized to spindle MTs although they do not have maximal binding affinity for MTs during mitosis. Preliminary results indicate that XMAP310 is a phosphoprotein and that its binding to MTs, like XMAP230 and E-MAP-115, is reduced by phosphorylation during mitosis (Andersen, S.S.L., unpublished observation). We previously hypothesized that MAPs may be locally less phosphorylated around chromosomes during mitosis, allowing their specific binding to spindle MTs (Andersen et al., 1994). It has been shown that mitotic chromatin can regulate the phosphorylation level of the MT polymerization inhibitor Stathmin/Op18, and it seems likely that a similar mechanism could locally regulate the binding activity/ phosphorylation of MAPs like XMAP310, XMAP230, and E-MAP-115 to spindle MTs (Sobel, 1991; Belmont and Mitchison, 1996; Andersen et al., 1997). However, more work will be required to determine how preferential binding of MAPs to spindle MTs is achieved during metaphase.
Effects of XMAP310 and other MAPs on MT Dynamics In Vitro
Brain MAPs and XMAP230 strongly reduce fcat and increase vg whereas XMAP310 primarily increases rescues (Drechsel et al., 1992; Pryer et al., 1992; Andersen et al., 1994; Itoh and Hotani, 1994; Trinczek et al., 1995). MAP4, a nonneuronal MAP resembling MAP2 and tau at the molecular level, strongly promotes rescues without having any effect on any of the other dynamic instability parameters (Ookata et al., 1995). However, the experimental conditions between our study and the former are so different that a direct comparison is difficult. Moreover, an important function of MAP4 seems to be the targeting of cdc2 kinase to MTs (Ookata et al., 1995). XMAP215 promotes MT turnover by strongly increasing vg at the MT plus end, increasing vs, and decreasing fres without affecting fcat (Gard and Kirschner, 1987; Vasquez et al., 1994). Comparatively, the brain MAPs together with XMAP230 are the most potent MAP MT stablizers, followed by XMAP310, MAP4, and then XMAP215.
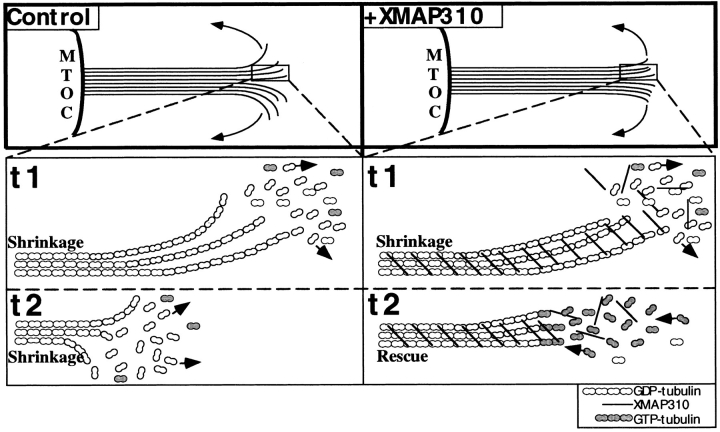
The effect of XMAP310 on fres is unusual and raises interesting questions about the mechanism of MT polymerization. How could XMAP310 increase rescues and not growth? Protofilaments tend to change from a straight configuration during the assembly phase to a curved one during disassembly. This change in curvature is thought to drive MT depolymerization (Simon and Salmon, 1990; Mandelkow et al., 1991; Chrétien et al., 1995). We propose that XMAP310 does not increase vg because it does not bind along the length of tubulin dimers as brain MAPs are thought to do (Drechsel et al., 1992; Pryer et al., 1992). Based on our observations, XMAP310 rather cross-links MTs and protofilaments. As shown in the model (Fig. 6), XMAP310 could, by cross-linking protofilaments, reduce the tendency of protofilaments to change to a curved conformation during MT depolymerization. Together with a reduction in vs, this may increase the rescue frequency by allowing new tubulin-GTP to recap depolymerizing MTs (Fig. 6). This model fits also with the unusually high frequency of pauses observed in MTs assembled in the presence of XMAP310. The plus end of MTs with laterally cross-linked protofilaments could stay in a metastable state, some subunits being added to the end of some protofilaments while other subunits would be removed from other protofilaments.
Figure 6.
Model for how XMAP310 may promote rescues and MT cross-linking. (Top) Plane-projection of a depolymerizing (shrinking) MT in the absence (Control) or presence of XMAP310 (+XMAP310), with protofilaments (lines) peeling off the MT by out-wards curvature (arrows). Note the more blunt MT end in the presence of XMAP310 which could be promoted if cross-linking of protofilaments by XMAP310 prevents the outwards curvature of protofilaments. (Bottom) Magnifications of the boxed areas, showing part of the MTs at two different time points, t1 and t2. In the Control, the MT is shrinking both at t1 and t2, by loss of GDP-tubulin (arrows). In the presence of XMAP310 the MT is shrinking at t1. At t2 the MT has been rescued. A depolymerizing MT (t1) will meet resistance towards depolymerization due to the cross-linking of protofilaments/MTs, but XMAP310 does not dramatically prevent the loss of the tubulin dimers from the depolymerizing ends (little effect on vs). Eventually depolymerization of the MT is blocked long enough to maintain the depolymerizing MT-GDP-lattice in a straight conformation allowing new GTP-tubulin subunits to be added to the end, and the MT has been rescued (arrows indicate the direction of the flux of tubulin subunits).
Role of MAPs in Regulation of Spindle MT Dynamics
At present the role of MAPs in spindle assembly is unclear. Some reports suggest that MAPs are redundant (Pereira et al., 1992; Wang et al., 1996), whereas others report that they are involved in spindle assembly, cell growth, and differentiation (Mangan and Olmsted, 1996; Nguyen et al., 1997; Saunders et al., 1997, and discussion of the MAP4 literature therein). On the basis of what is known about the subcellular localization and the effect of Xenopus MAPs on MT dynamics in vitro, it seems that XMAP215 may produce the rapid polymer growth and turnover observed for spindle MTs. Such a MAP would significantly increase the search efficiency of MTs searching for a kinetochore during spindle assembly (Kirschner and Mitchison, 1986; Holy and Leibler, 1994). Since XMAP310 is localized to the nucleus during interphase and does not stably localize to spindle MTs until metaphase, this MAP may, like XMAP215, be involved in enhancing the searching efficiency of future kinetochore MTs. XMAP230 and XMAP310 may be involved in generating stable kinetochore MTs (XMAP230; Andersen et al., 1994), and other types of stable spindle MTs, such as cross-linked interpolar MTs (XMAP310; Mastronarde et al., 1993). Thus, the abundance of XMAP310 in brain tissue, together with the functional in vitro data, suggest that an important in vivo function of this protein is to cross-link and stabilize MTs. Our immunofluorescence results show that XMAP310 is tightly associated to metaphase MTs but released from anaphase MTs, indicating that XMAP310 could specifically stabilize metaphase MTs and may not be required during anaphase. The release of XMAP310 from anaphase MTs may indicate that MT rescues are undesirable events during anaphase, or perhaps other MAPs, like XMAP230, become reactivated, suppressing catastrophes and therefore the occurrence of rescues.
In mitotic Xenopus egg extracts lacking chromosomes, MTs display a high growth rate, a high catastrophe frequency and rescues and pauses are very infrequent (Belmont et al., 1990; Verde et al., 1992; Tournebize et al., 1997). One could wonder why, when a protein like XMAP310 is present in such extracts. The absence of rescues in mitotic extracts could be explained if during mitosis XMAP310 is inactivated globally by phosphorylation in the cytoplasm. In contrast, due to the presence of XMAP310 on spindle MTs, the rescue frequency of spindle MTs could be high. Therefore, it seems likely that XMAP310 and XMAP230 could be part of the machinery that ensures preferential growth of MTs towards chromosomes during spindle assembly (Andersen et al., 1997), XMAP310 by locally increasing fres and XMAP230 by locally reducing fcat (Andersen et al., 1994). In contrast, XMAP215 seems to be globally active during mitosis, since the MT growth rate remains high and XMAP215 probably is the main regulator of vg in the extract.
It is interesting to see that different MAPs affect different parameters of MT dynamic instability in a complex system like Xenopus. When dynamic instability was discovered, one of the immediate predictions was that different molecules could affect each parameter specifically, and now this is becoming reality. In the next phase, it will be very exciting to determine precisely the function of XMAP215, XMAP230, and XMAP310 in the global and local regulation of MT dynamics during mitosis and in spindle assembly. Before this becomes feasible, these molecules have to be cloned and efficient tools developed.
Acknowledgments
We are grateful to Byeong Cha, Emily Harwood, and David L. Gard for the gifts of the XMAP215 and XMAP230 pAbs and IF of Xenopus embryos. We are indebted to Thomas Kreis for the antitubulin pAb. Special thanks to Alan Sawyer who was of great help in the preparation of the mAbs. Thanks to Alexandre Podtelejnikov, Ole Jensen, and Juri Rappsilber for the MALDI MS analysis, to Keith Ashman for Edman data, and to Anja Habermann for assistance with the EM. Thanks to Rebecca Heald, and Torsten Wittmann for comments on the manuscript.
Footnotes
1. Abbreviations used in this paper: fcat, catastrophe frequency; fres, rescue frequency; mAb, monoclonal antibody; MALDI MS, Matrix Assisted Laser Desorption Ionization Mass Spectroscopy; MAP, microtubule-associated protein; MT, microtubule; pAb, polyclonal antibody; vg, growth rate; vs, shrinkage rate.
Søren Andersen was partially supported by an individual European Union Marie Curie Grant (ERB-CHBIT 930755) and an EU network Grant to Tony Hyman; Eric Karsenti by a Human Frontier Science Program Grant (RG-350/94) as well as a European Union Grant (ERB-CHRXCT 940568).
References
- Andersen SSL, Buendia B, Domínguez JE, Sawyer A, Karsenti E. Effect on microtubule dynamics of XMAP230, a microtubule-associated protein present in Xenopus laeviseggs and dividing cells. J Cell Biol. 1994;127:1289–1299. doi: 10.1083/jcb.127.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SSL, Ashford AJ, Tournebize R, Gavet O, Sobel A, Hyman AA, Karsenti E. Mitotic chromatin regulates phosphorylation of Stathmin/Op18. Nature. 1997;389:640–643. doi: 10.1038/39382. [DOI] [PubMed] [Google Scholar]
- Ashford, A.J., S.S.L. Andersen, and A.A. Hyman. 1998. Purification of bovine brain tubulin. In Cell Biology: A Laboratory Handbook. J.E. Celis, editor. Academic Press, San Diego. Vol. II. 205–212.
- Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–589. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS. A widely distributed nuclear protein immunologically related to the microtubule associated protein MAP1 is associated with the mitotic spindle. Proc Natl Acad Sci USA. 1985;82:1146–1150. doi: 10.1073/pnas.82.4.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M, Paintrand M, Berges J, Marty MC, Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Motil Cytoskel. 1987;8:238–249. doi: 10.1002/cm.970080305. [DOI] [PubMed] [Google Scholar]
- Bulinski, J.C. 1994. MAP4. In Microtubules. J.H. Hyams, and C.W. Lloyd, editors. Wiley-Liss, Inc., New York. 167–182.
- Bulinski JC, Bossler A. Purification and characterization of ensconsin, a novel microtubule stabilizing protein. J Cell Sci. 1994;107:2839–2849. doi: 10.1242/jcs.107.10.2839. [DOI] [PubMed] [Google Scholar]
- Charasse S, Mazel M, Taviaux S, Berta P, Chow T, Larroque C. Characterization of the cDNA and pattern of expression of a new gene over-expressed in human hepatomas and colonic tumors. Eur J Biochem. 1995;234:406–413. doi: 10.1111/j.1432-1033.1995.406_b.x. [DOI] [PubMed] [Google Scholar]
- Chen J, Kanai Y, Cowan NJ, Hirokawa N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature. 1992;360:674–677. doi: 10.1038/360674a0. [DOI] [PubMed] [Google Scholar]
- Chrétien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW. NuMA: a protein involved in nuclear structure, spindle assembly, and nuclear re-formation. Trends Cell Biol. 1995;5:60–64. doi: 10.1016/s0962-8924(00)88947-3. [DOI] [PubMed] [Google Scholar]
- Collins C. Reversible assembly purification of taxol-treated microtubules. Methods Enzymol. 1991;196:246–253. doi: 10.1016/0076-6879(91)96023-k. [DOI] [PubMed] [Google Scholar]
- Compton DA, Yen TJ, Cleveland DW. Identification of novel centromere/kinetochore-associated proteins using monoclonal antibodies against human mitotic chromosome scaffolds. J Cell Biol. 1991;112:1083–1097. doi: 10.1083/jcb.112.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier P, Osborne HB, Morales J, Bassez T, Poulhe R, Mazabraud A, Mulner-Lorillon O, Bellé R. Molecular cloning of Xenopuselongation factor 1g, major M-phase promoting factor substrate. Nucleic Acids Res. 1991;19:6644. doi: 10.1093/nar/19.23.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M, Felix MA, Guet CC, Leibler S. Influence of M-phase chromatin on the anisotropy of microbule asters. J Cell Biol. 1996;133:125–140. doi: 10.1083/jcb.133.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel DN, Hyman AA, Cobb MH, Kirschner MW. Modulation of dynamic instability by the microtubule-associated-protein tau. Mol Biol Cell. 1992;3:1141–1154. doi: 10.1091/mbc.3.10.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L, Mitchison TJ, Kirschner MW. Influence of the centrosome on the structure of nucleated microtubules. J Cell Biol. 1985;100:1185–1191. doi: 10.1083/jcb.100.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feick P, Foisner R, Wiche G. Immunolocalization and molecular properties of a high molecular weight microtubule-bundling protein (Syncolin) from chicken erythrocytes. J Cell Biol. 1991;112:689–699. doi: 10.1083/jcb.112.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix, M.A., P. Clarke, J. Coleman, F. Verde, and E. Karsenti. 1993. Frog egg extracts as a system to study mitosis. In The Cell Cycle: A Practical Approach. IRL Press, New York. 253–283.
- Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around chromatin beads in Xenopusegg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hershey JWB. Translational control in mammalian cells. Ann Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. Microtubule organization and dynamics dependent on microtubule-associated proteins. Curr Opin Cell Biol. 1994;6:74–81. doi: 10.1016/0955-0674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Shiomura Y, Okabe S. Tau proteins: the molecular structure and mode of binding on microtubules. J Cell Biol. 1988;107:1449–1459. doi: 10.1083/jcb.107.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holy TE, Leibler S. Dynamic instability of microtubules as an efficient way to search in space. Proc Natl Acad Sci USA. 1994;91:5682–5685. doi: 10.1073/pnas.91.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Karsenti E. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–411. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Drexel D, Kellog D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison TJ. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–485. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- Inoué S, Salmon ED. Force generation by microtubule assembly/ disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh TJ, Hotani H. Microtubule-stabilizing activity of microtubule-associated proteins (MAPs) is due to increase in frequency of rescue in dynamic instability: shortening length decreases with binding of MAPs onto microtubules. Cell Struct Funct. 1994;19:279–290. doi: 10.1247/csf.19.279. [DOI] [PubMed] [Google Scholar]
- Izant JG, Weatherbee JA, McIntosh JR. A microtubule-associated protein in the mitotic spindle and the interphase nucleus. Nature. 1982;295:248–250. doi: 10.1038/295248a0. [DOI] [PubMed] [Google Scholar]
- Jensen ON, Podtelejnikov A, Mann M. Delayed extraction improves specificity in database searches by MALDI peptide maps. Rapid Commun Mass Spec. 1996;10:1371–1378. doi: 10.1002/(SICI)1097-0231(199608)10:11<1371::AID-RCM682>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Karsenti E, Newport J, Kirschner M. Respective roles of centrosomes and chromatin in the conversion of microtubule arrays from interphase to metaphase. J Cell Biol. 1984;99:47s–57s. doi: 10.1083/jcb.99.1.47s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Mandelkow E, Mandelkow EM. Microtubules and microtubule- associated proteins. Curr Opin Cell Biol. 1995;7:72–81. doi: 10.1016/0955-0674(95)80047-6. [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan ME, Olmsted JB. A muscle-specific variant of microtubule-associated protein 4 (MAP4) is required in myogenesis. Development. 1996;122:771–781. doi: 10.1242/dev.122.3.771. [DOI] [PubMed] [Google Scholar]
- Masson D, Kreis TE. Binding of E-MAP-115 to microtubules is regulated by cell cycle-dependent phosphorylation. J Cell Biol. 1995;131:1015–1024. doi: 10.1083/jcb.131.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A. Microtubule-associated proteins. Curr Opin Cell Biol. 1990;2:10–14. doi: 10.1016/s0955-0674(05)80024-9. [DOI] [PubMed] [Google Scholar]
- McIntosh, R.J. 1994. The role of microtubules in chromosome movement. In Microtubules. J.H. Hyams and C. W. Lloyd, editors. Wiley-Liss, Inc., New York. 413–434.
- McNally FJ, Okawa K, Iwamatsu A, Vale RD. Katanin, the microtubule-severing ATPase, is concentrated at centrosomes. J Cell Sci. 1996;109:561–567. doi: 10.1242/jcs.109.3.561. [DOI] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Miller L, Daniel JC. Comparison of in vivo and in vitro ribosomal RNA synthesis in nucleolar mutants of Xenopus laevis. . In Vitro. 1977;13:557–567. doi: 10.1007/BF02627851. [DOI] [PubMed] [Google Scholar]
- Murray, A. 1991. Cell cycle extracts. In Xenopus laevis: Practical Uses in Cell and Molecular Biology, Vol. 36. B.K. Kay and H.B. Peng, editors. Academic press, San Diego. 581–605.
- Nguyen H-L, Chari S, Gruber D, Lue C-M, Chapin SJ, Bulinski JC. Overexpression of full- or partial-length MAP4 stabilizes microtubules and alters cell growth. J Cell Sci. 1997;110:281–294. doi: 10.1242/jcs.110.2.281. [DOI] [PubMed] [Google Scholar]
- Olmsted JB. Microtubule associated proteins. Ann Rev Cell Biol. 1986;2:421–457. doi: 10.1146/annurev.cb.02.110186.002225. [DOI] [PubMed] [Google Scholar]
- Olmsted JB, Stemple DL, Saxton WM, Neighbors BW, McIntosh JR. Cell cycle-dependent changes in the dynamics of MAP 2 and MAP 4 in cultured cells. J Cell Biol. 1989;109:211–223. doi: 10.1083/jcb.109.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookata K, Hisanaga S, Bulinski JC, Murofushi H, Aizawa H, Itoh TJ, Hotani H, Okumura E, Tachibana K, Kishimoto T. Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34cdc2 kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J Cell Biol. 1995;128:849–862. doi: 10.1083/jcb.128.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper DA, Kim HY, Berns MW. Studies of a microtubule-associated protein using a monoclonal antibody elicited against mammalian mitotic spindles. J Cell Biol. 1984;99:503–511. doi: 10.1083/jcb.99.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A, Doshen J, Tanaka E, Goldstein LSB. Genetic analysis of a Drosophilamicrotubule-associated protein. J Cell Biol. 1992;116:377–383. doi: 10.1083/jcb.116.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer NK, Walker RA, Skeen VP, Bourns BD, Soboeiro MF, Salmon ED. Brain microtubule-associated proteins modulate microtubule dynamic instability in vitro. Real-time observations using video microscopy. J Cell Sci. 1992;103:965–976. doi: 10.1242/jcs.103.4.965. [DOI] [PubMed] [Google Scholar]
- Reinsch S, Karsenti E. Orientation of spindle axis and distribution of plasma membrane proteins during cell division in polarized MDCKII cells. J Cell Biol. 1994;126:1509–1526. doi: 10.1083/jcb.126.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Yoshitake R, Shiomura Y, Miyasaka H, Hirokawa N. MAP1B: molecular structure, localization and phosphorylation-dependent expression in developing neurons. Neuron. 1989;3:229–238. doi: 10.1016/0896-6273(89)90036-6. [DOI] [PubMed] [Google Scholar]
- Saunders RDC, Avides MC, Howard T, Gonzalez C, Glover DM. The Drosophila gene abnormal spindleencodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J Cell Biol. 1997;137:881–890. doi: 10.1083/jcb.137.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Mitchison T J. Microtubule flux in mitosis is independent of chromosomes, centrosomes, Mol Biol Cell. 1994;5:217–226. doi: 10.1091/mbc.5.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Stemple DL, Leslie RJ, Salmon ED, Zavortink M, McIntosh JR. Tubulin dynamics in cultured mammalian cells. J Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N, Gotoh Y, Nishida E. A novel homo-oligomeric protein responsible for an MPF-dependent microtubule severing activity. EMBO (Eur Mol Biol Organ) J. 1992a;11:4723–4731. doi: 10.1002/j.1460-2075.1992.tb05577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N, Moriguchi T, Ohta K, Gotoh Y, Nishida E. Regulation of a major microtubule-associated protein by MPF and MAP kinase. EMBO (Eur Mol Biol Organ) J. 1992b;11:3977–3984. doi: 10.1002/j.1460-2075.1992.tb05491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JR, Salmon ED. The structure of microtubule ends during the elongation and shortening phases of dynamic instability examined by negative-stain electron microscopy. J Cell Sci. 1990;96:571–582. doi: 10.1242/jcs.96.4.571. [DOI] [PubMed] [Google Scholar]
- Sobel A. Stathmin: a relay phosphoprotein for multiple signal transduction? . Trends Biochem Sci. 1991;16:301–305. doi: 10.1016/0968-0004(91)90123-d. [DOI] [PubMed] [Google Scholar]
- Tetaz, T., E. Bozas, J. Kanellos, I. Walker, and I. Smith. 1993. In situ tryptic digestion of proteins separated by sds-page: improved procedures for extraction of peptides before microsequencing. In Techniques in Protein Chemistry IV, Vol. IV. R.H. Angeletti, editor. Academic Press, Inc., San Diego. 389–397.
- Toso RJ, Jordan M A, Farrell KW, Matsumoto B, Wilson L. Kinetic stabilization of microtubule dynamic instability in vitro by vinblastine. Biochemistry. 1993;32:1285–1293. doi: 10.1021/bi00056a013. [DOI] [PubMed] [Google Scholar]
- Tournebize R, Andersen SSL, Verde F, Dorée M, Karsenti E, Hyman AA. Distinct roles of PP1 and PP2A-like phosphatases in control of microtubule dynamics during mitosis. EMBO (Eur Mol Biol Organ) J. 1997;16:5537–5549. doi: 10.1093/emboj/16.18.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinczek B, Biernat J, Baumann K, Mandelkow E-M, Mandelkow E. Domains of tau protein, differential phosphorylation and dynamic instability of microtubules. Mol Biol Cell. 1995;6:1907–1918. doi: 10.1091/mbc.6.12.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. Severing of stable microtubules by a mitotically activated protein in Xenopus egg extracts. Cell. 1991;64:827–839. doi: 10.1016/0092-8674(91)90511-v. [DOI] [PubMed] [Google Scholar]
- Vasquez RJ, Gard DL, Cassimeris L. XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J Cell Biol. 1994;127:985–993. doi: 10.1083/jcb.127.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Labbe J-C, Doree M, Karsenti E. Regulation of microtubule dynamics by cdc2 protein kinase in cell-free extracts of Xenopus eggs. Nature (Lond) 1990;343:233–238. doi: 10.1038/343233a0. [DOI] [PubMed] [Google Scholar]
- Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S. Control of microtubule dynamics and length by cyclin A and cyclin B dependent kinases in Xenopus egg extracts. J Cell Biol. 1992;118:1097–1108. doi: 10.1083/jcb.118.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RA, O'Brien ET, Pryer NK, Sobeiro MF, Voter WA, Erickson HP, Salmon ED. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol. 1988;107:1437. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XM, Peloquin JG, Zhai Y, Bulinski JC, Borisy GG. Removal of MAP4 from microtubules in vivo produces no observable phenotype at the cellular level. J Cell Biol. 1996;132:345–357. doi: 10.1083/jcb.132.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Salmon ED. Pathways of spindle assembly. Curr Opin Cell Biol. 1997;9:37–43. doi: 10.1016/s0955-0674(97)80149-4. [DOI] [PubMed] [Google Scholar]
- Wiche G, Oberkanins C, Himmler A. Molecular structure and function of microtubule-associated proteins. Int Rev Cytol. 1991;124:217–273. doi: 10.1016/s0074-7696(08)61528-4. [DOI] [PubMed] [Google Scholar]
- Wilhelm H, Anderson SSL, Karsenti E. Purification of recombinant cyclin B1/cdc2 kinase from Xenopus egg extracts. Methods Enzymol. 1997;283:12–28. doi: 10.1016/s0076-6879(97)83004-2. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Simon PM, Borisy GG. Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J Cell Biol. 1996;135:201–214. doi: 10.1083/jcb.135.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieve G, Solomon F. Protein specifically associated with the microtubules of the mammalian mitotic spindle. Cell. 1982;28:233–242. doi: 10.1016/0092-8674(82)90341-5. [DOI] [PubMed] [Google Scholar]