Abstract
Localization of dynein–green fluorescent protein (GFP) to cytoplasmic microtubules allowed us to obtain one of the first views of the dynamic properties of astral microtubules in live budding yeast. Several novel aspects of microtubule function were revealed by time-lapse, three-dimensional fluorescence microscopy. Astral microtubules, about four to six in number for each pole, exhibited asynchronous dynamic instability throughout the cell cycle, growing at ≅0.3–1.5 μm/min toward the cell surface then switching to shortening at similar velocities back to the spindle pole body (SPB). During interphase, a conical array of microtubules trailed the SPB as the nucleus traversed the cytoplasm. Microtubule disassembly by nocodozole inhibited these movements, indicating that the nucleus was pushed around the interior of the cell via dynamic astral microtubules. These forays were evident in unbudded G1 cells, as well as in late telophase cells after spindle disassembly. Nuclear movement and orientation to the bud neck in S/G2 or G2/M was dependent on dynamic astral microtubules growing into the bud. The SPB and nucleus were then pulled toward the bud neck, and further microtubule growth from that SPB was mainly oriented toward the bud. After SPB separation and central spindle formation, a temporal delay in the acquisition of cytoplasmic dynein at one of the spindle poles was evident. Stable microtubule interactions with the cell cortex were rarely observed during anaphase, and did not appear to contribute significantly to spindle alignment or elongation into the bud. Alterations of microtubule dynamics, as observed in cells overexpressing dynein-GFP, resulted in eventual spindle misalignment. These studies provide the first mechanistic basis for understanding how spindle orientation and nuclear positioning are established and are indicative of a microtubule-based searching mechanism that requires dynamic microtubules for nuclear migration into the bud.
The ultimate developmental program of any organism relies on the proper coordination of mitosis with cytokinesis. In many organisms, coordination is accomplished when the mitotic apparatus signals for the position of the cleavage plane, which later bisects the segregated chromosomes. This is graphically observed in many asymmetric cell divisions where microtubule-dependent mechanisms align the spindle along the axis of polarity (White and Strome, 1996). Spindle positioning is of particular importance in the budding yeast Saccharomyces cerevisiae where the cleavage plane is established at the start of the cell cycle independent of the mitotic apparatus. The nucleus and its intranuclear spindle must migrate to the bud neck and orient along the mother/bud axis to segregate chromosomes into mother and bud before cell cleavage. Though microtubules are required for nuclear positioning and spindle alignment in budding yeast (Jacobs et al., 1988; Sullivan and Huffaker, 1992), the role of microtubule organization, interactions, and dynamics in these processes is not well understood.
Cytoplasmic dynein and astral microtubules are required for spindle orientation along the mother/bud axis before chromosome segregation and for timely nuclear migration into the bud during anaphase (Palmer et al., 1992; Sullivan and Huffaker, 1992; Eshel et al., 1993; Li et al., 1993). In the absence of the cytoplasmic dynein heavy chain gene, DHC1/DYN1, nuclear migration is delayed, resulting in binucleate cells, while spindle formation, initial elongation, and chromosome segregation to opposite poles continue (Yeh et al., 1995). A conditional, lethal mutation in β-tubulin (tub2-401) that preferentially destabilizes the astral microtubules prevents nuclear migration into the daughter cell, and leads to bi- and anucleated cells after cytokinesis (Sullivan and Huffaker, 1992). The similarity in phenotypes suggests that dynein's role in nuclear orientation and spindle alignment occurs via astral microtubules.
The requirement for astral microtubules and cytoplasmic dynein for proper nuclear movement into the bud, together with dynein's ability to move toward the minus end of a microtubule lattice, has led to the notion that dynein at the cell cortex exerts a pulling force on the astral microtubules (Eshel et al., 1993; Li et al., 1993; Schroer, 1994). In the absence of a central spindle, ndc1, spindle pole bodies (SPBs)1 (only one of which is associated with chromosomes) are segregated to the mother and bud, indicating the presence of forces on the SPBs independent of the central spindle (Thomas and Botstein, 1986; Winey et al., 1993). Dynein's role in the generation of such forces was demonstrated by Yeh et al. (1995), who showed that only the defective SPB without attached chromatin could be translocated into the bud in cells lacking dynein and the central spindle (ndc1, dhc1 double mutants). These data, together with the localization of cytoplasmic dynein to astral microtubules (Yeh et al., 1995) indicate that dynein plays a dominant role in pulling the SPB into the bud via astral microtubules. Cells lacking cytoplasmic dynein and force generators in the central spindle (Cin8p and Kip1p) are deficient in spindle elongation (Saunders et al., 1995), substantiating the view that dynein exerts a pulling force.
Astral microtubules are reported to be oriented toward the site of bud growth before bud emergence. Coimmunofluorescence of microtubules and Spa2p have indicated that microtubules at the cell cortex preferentially localize with proteins that mark the incipient site of bud growth (Snyder et al., 1991). Similarly, in electron microscopic studies, the SPB is reported to be oriented toward the bud in cells with small buds (Byers and Goetsch, 1975; Byers, 1981). Thus the microtubule cytoskeleton has been hypothesized to orient the nucleus toward the bud site, and then dock the nucleus to the future neck of the budded cell. An apparent paradox between the immunofluorescence studies with fixed cells and imaging of the nucleus in live cells is the finding that the nucleus moves with a high degree of freedom in unbudded cells, and is restricted in its movement only as it migrates to the neck of budded cells and not before (Koning et al., 1993; Yeh et al., 1995).
In this study we have constructed a dynein fusion with green fluorescent protein (dynein-GFP) that allows visualization of astral microtubule dynamics throughout the cell cycle in yeast. Time-lapse imaging indicates dramatic microtubule dynamic instability providing both the mechanical force for nuclear movement, as well as a searching mechanism for the nascent bud. Astral microtubules remain dynamic as the spindle elongates, indicating that transient interactions of microtubules within the bud play a predominant role in the alignment of the mitotic spindle. These transient interactions may also provide an inherent mechanism for error correction in segregation of spindle poles to mother and daughter cells.
Materials and Methods
Strains
All strains used in this study were derived from strain 15C (Mata, leu2-3, 112, ura3-52, his4-580, trp1Δ, pep4-3) and MAY591 (Matα, leu2-3,112, lys2-801, his3-200, ura3-52) (Saunders and Hoyt, 1992). Haploid derivatives 8d (Mata, lys2-801, trp1Δ, ura3-52, leu2-3,112) and 9d (Matα, lys2-801, his3-200, ura3-52, leu2-3,112) were mated to generate the diploid strain (8d×9d) (Mata/α, lys2/lys2, ura3/ura3, leu2/leu2, his3/+, +/trp1). A complete deletion of the 12,276-bp dynein heavy chain was obtained by fragment-mediated transformation into 9dΔdhc (dhc1::LEU2). The GAL1–glutathione-S-transferase (GST)–dynein-GFP–containing plasmid (pKBY701; see Fig. 1) was introduced into 9d, 9dΔdhc, or the diploid, 8d×9d by standard transformation.
Figure 1.
Construction of the dynein-GFP–containing plasmid, pKBY701. The parent plasmid is based on a high copy, galactose-inducible shuttle vector previously described (Baldari et al., 1987) that contains the 2-μm origin of replication, URA3, and Leu2-d. Mitchell et al. (1993) modified the vector for the conditional expression of GST fusions in S. cerevisiae. We inserted the dynein heavy chain gene (11,333-bp NheI fragment, amino acids 303–4,081) into the XbaI site of pEGKG. The resulting plasmid, pJU1 contains a galactose-inducible (black), GST (blue)/dynein (gray) fusion protein. The S65T derivative of GFP (green) (716 bp) was fused to the COOH terminus of dynein, at the unique SalI site generating the plasmid pKBY701 (22.3 kbp).
Construction and Expression of a Dynein-GFP Fusion Protein
A dynein-GFP fusion protein was constructed by cloning GFP (S65T) into the COOH terminus of the cytoplasmic dynein heavy chain (DHC1, DYN1; see Fig. 1). The resulting protein is ∼500 kD, containing the COOH-terminal 300–4,081 amino acids of cytoplasmic dynein fused to GFP. The Ser-65-Thr GFP mutation was used for the enhanced solubility and fluorescence of the fusion protein (Heim et al., 1995). Expression of the fusion protein was controlled by the GAL1 promoter to allow varied induction/ repression regimes for regulation of protein levels (galactose, induction, and glucose, repression). Cells were grown to mid-logarithmic growth phase in glucose, followed by a short induction on galactose (∼2 h), and aliquots were removed and placed onto a 5-μm–thick gelatin slab containing glucose (Yeh et al., 1995). Expression was repressed for the duration of recording to restrict analysis to existent dynein.
Immunofluorescence Staining
Immunofluorescence staining of yeast cells was performed essentially as described (Pringle et al., 1989). Cells were fixed overnight at 4°C in 3.7% formaldehyde. Microtubules were visualized with the rat anti–α-tubulin antibody YOL1/34 (1:200 dilution) (Accurate Chemical and Scientific Corp., Westbury, NY). Rabbit anti-GST antibodies (gift of E. Bi, University of North Carolina at Chapel Hill) (1:5 dilution), were used to visualize the dynein-GFP fusion protein (see Fig. 1). FITC-conjugated goat anti–rat and rhodamine-conjugated goat anti–rabbit (Cappel Labs, Cochranville, PA) were used as secondary antibodies (1:200 dilution).
Microscopy and Image Processing
Cells were pipetted onto slabs of 25% gelatin containing minimal media +2% glucose as described by Yeh et al. (1995). Imaging was performed using the microscope system and acquisition protocol described in Shaw et al. (1997). The microscope (Salmon et al., 1994) was modified for automated switching between fluorescence and differential interference contrast (DIC) by replacement of the camera mount with a filter wheel (BioPoint 99B100; Ludl Electronic Products Ltd., Hawthorne, NY) containing the analyzer component of the DIC optics. A ×60 1.4 numerical aperture Plan-Apochromatic objective lens and ×2 + ×1.25 intermediate projection magnified the specimen image ×150 directly onto the cooled, slow-scan CCD imaging device (C4880; Hamamatsu Photonics, Bridgewater, NJ). The computer-controlled (MetaMorph 2.5 software; Universal Imaging Corp., West Chester, PA) microscope executed an acquisition protocol taking fluorescence images at 1-μm axial steps and a single DIC image corresponding to the central fluorescence image. Fluorescence excitation through a 490 ± 10-nm filter was normally attenuated to 1–10% of the available light from the 100 W mercury arc lamp. Fluorescence emission was collected through a 530 ± 15-nm band pass filter. DIC images were made by rotating the analyzer into the light path and taking a 0.6-s exposure.
A single background image (average of 24, 3-s exposures with no illumination) was subtracted from each GFP image. The images corresponding to a single time-lapse point were projected to a single image by using only the brightest pixel at any one location. Registration of DIC and fluorescence images was verified by imaging of 1-μm fluorescent beads in DIC and fluorescence modes. For presentation, dynein-GFP and corresponding DIC images were overlaid or placed side by side and contrast enhanced using MetaMorph (Universal Imaging Corp., West Chester, PA) or Photoshop (Adobe Systems Corp., San Jose, CA). Images for publication were scaled and interpolated to 300 dots per inch.
Analysis and Quantitation
Variable levels of dynein-GFP were observed in all transformed strains necessitating a standard procedure for the selection of cells for imaging. The average and maximum fluorescence (arbitrary units) from a 3-s exposure of a single cell was assessed, using the quantitation tools in MetaMorph, and compared to the identical measurements made on a plastic fluorescent reference slide (Applied Precision Inc., Issaquah, WA). Cells having a maximum fluorescence <20% of the reference were used for further analysis and quantitation. Two image acquisition protocols were used, one for the analysis of cells over long periods of time (>40 min) and a second for determining rates.
For time-lapse analysis of cells (>40 min), five fluorescence exposures of 3-s duration and a single DIC image were taken at 1-min intervals. Excitation was attenuated to between 1–10% to prevent phototoxicity. A total of 1,493 time points were compiled as two-dimensional images from 15 cells for analysis. Anaphase was observed in 11 out of 15 cells and initial spindle pole separation was discretely followed in 4 of 15 cells.
Rates of microtubule elongation and shortening were determined by measurement of microtubule length in time-lapse image sequences from 35 cells. Three exposures of 1-s duration were taken at 100% fluorescence excitation at 1 μm axial steps and projected to a single plane for analysis. The acquisition regime was repeated every 15 s for a maximum of 15 min and represents >1,800 time points. Microtubule length was determined by measuring individual microtubules from the center of the SPB fluorescence to the microtubule end and converting from pixels to microns using the image of a stage micrometer.
Nocodazole Treatment
Exponentially growing cultures were treated with 1% DMSO (control) or 1% DMSO with 20 μg/ml nocodazole (Sigma Chemical Co. St. Louis, MO). After 2 h, >80% of nocodazole-treated cells were large budded.
Results
Dynamic Instability of Astral Microtubules
The function of a plasmid-borne, cytoplasmic dynein-GFP fusion protein (Fig. 1) was assessed by the fidelity of nuclear segregation in individual cells whose sole source of dynein was the GFP fusion. Anucleate and binucleate cells were not generated in individuals expressing an average fluorescence of <20% of a fluorescence reference (described in Materials and Methods). Orientation of the nucleus along the mother–bud axis was not affected by replacing the wild-type dynein with the dynein-GFP fusion. The rates of spindle elongation were in accordance with previous observations (Yeh et al., 1995). Only a slight delay (<5 min) was observed in the penetration of the nucleus through the bud neck at the time of anaphase onset (see below). Thus the fusion protein was able to complement the spindle orientation and nuclear movement defects of dynein null mutants (Yeh et al., 1995).
The dynein-GFP fusion protein decorated both the SPB and astral microtubules exclusively throughout the cell cycle. In cells fixed for microtubule immunofluorescence by standard procedures in yeast (Pringle et al., 1989), dynein-GFP staining was concentrated at SPBs and distributed along the length of the great majority of astral microtubules preserved in cells expressing the dynein-GFP (Fig. 2). Occasionally (23 out of 153 microtubules), a region of a microtubule or a whole microtubule lacked detectable dynein-GFP staining (Fig. 2). The distribution of dynein-GFP staining along the microtubules in the fixed preparations was very uneven (Fig. 2 B) in comparison to the uniform linear distributions seen in the live cell images (Figs. 3 and 4). This indicates that the microtubule fixation procedures are not able to completely preserve the native distribution of dynein-GFP along microtubules. For the live cell studies, a single fluorescence image was inadequate to follow dynamic microtubules, which often extended in and out of the plane of focus. To solve this problem, a two-dimensional representation of their three-dimensional distribution was obtained by the projection of five fluorescent images taken at 1-μm focal steps through the cell onto one plane. A single DIC image was taken at the middle focal plane to provide accurate definition of the cortex and nuclear boundaries (Shaw et al., 1997). At no time was any significant concentration of dynein-GFP observed at the cell cortex or associated with membranous structures. A general background fluorescence was observed in the cytosol presumably representing unbound dynein-GFP. Dynein-GFP did not enter the nucleus at any stage, and consequently, the nucleus was observed as a dark area within the slightly brighter cytosol. The SPB was identified as a focus of fluorescence at the periphery of the nuclear envelope (Figs. 3 and 4). The accompanying DIC image allowed visualization of the nucleus, and confirmation that the SPB was indeed the site of intranuclear spindle assembly, as well as the point of origin for the cytoplasmic astral microtubules (Fig. 4, right). Four to six astral microtubules emanated from the SPB throughout the cell cycle, with individual microtubules splaying out in the periphery of the cytoplasm.
Figure 2.
Immunofluorescence microscopy of microtubules and dynein-GFP in a G1 cell. (A) Microtubules and (B) dynein-GFP. Cells were double labeled using a rat anti–α-tubulin antibody, YOL1/34, to label microtubules and rabbit anti-GST antibodies to label a GST epitope in the dynein-GFP fusion protein.
Figure 3.
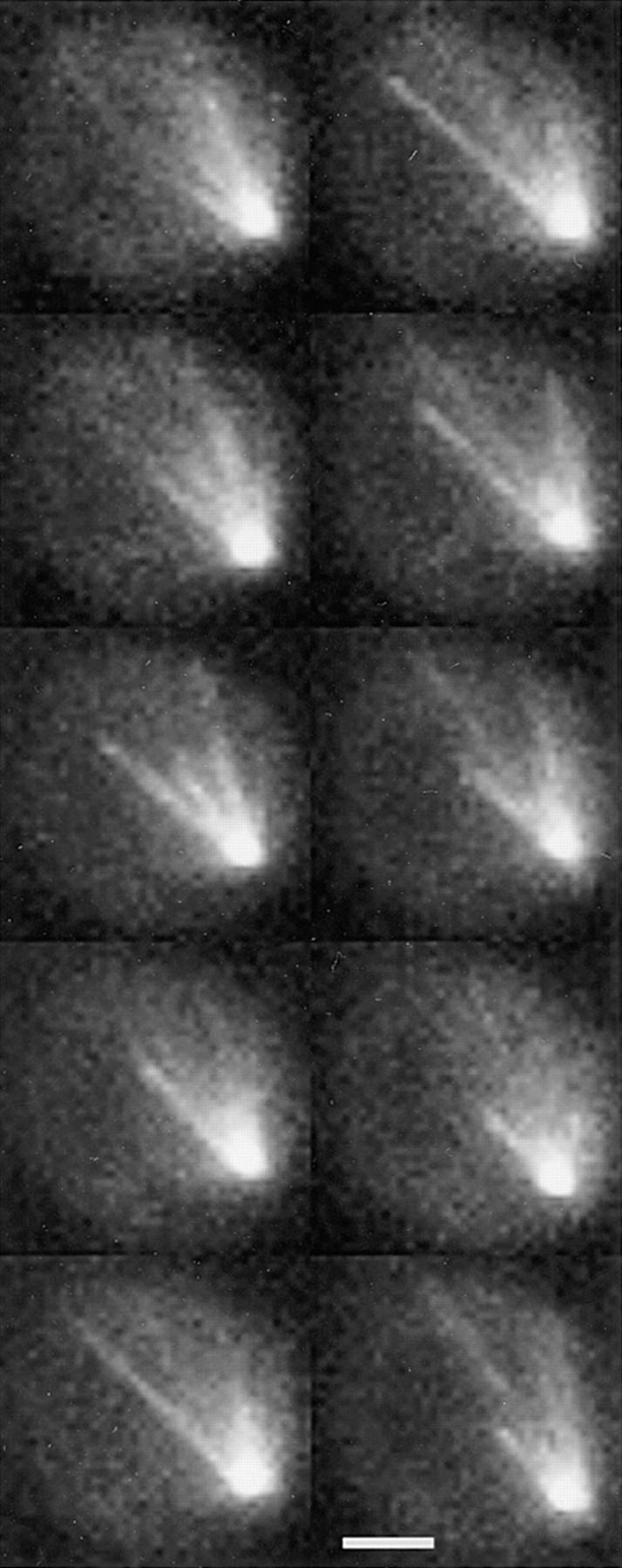
Microtubules exhibit dynamic instability. High resolution fluorescence images of unbudded G1 cells. Haploid strain 9dΔd lacks an endogenous copy of dynein and contains the GST–dynein- GFP fusion (pKBY701). A series of eight time points at 1-min intervals are displayed from top to bottom on the left and continuing on the right. Each image displayed represents a reconstruction from five exposures taken at 1-μm axial steps. A single microtubule at the bottom of each panel is shown to polymerize in the first three panels before undergoing a catastrophe and shortening in panel four. The same microtubule rescues and grows to the cell cortex before undergoing catastrophe and shortening back toward the spindle pole. Bar, 2 μm.
Figure 4.
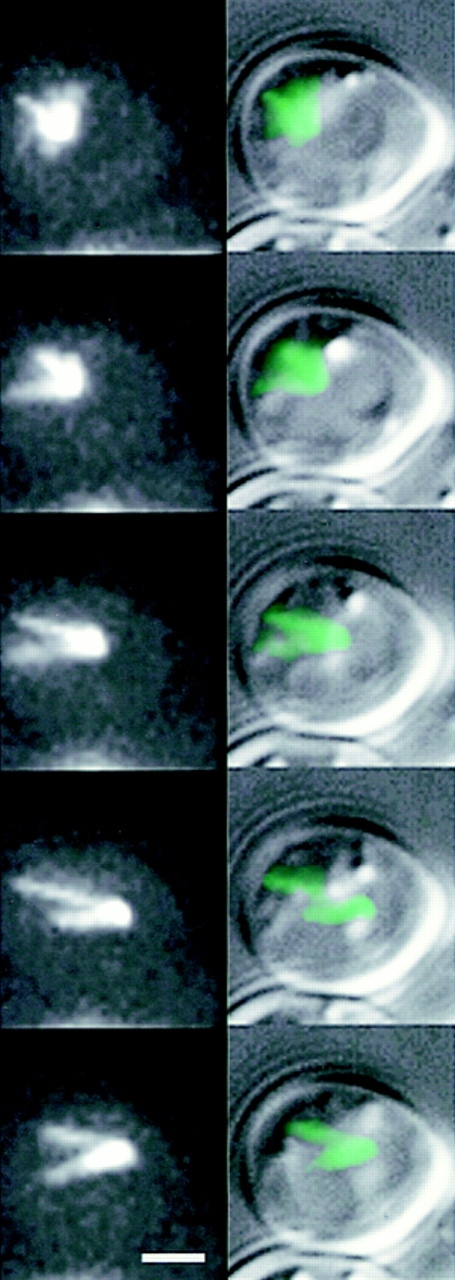
Microtubule growth in G1 is coupled to nuclear movement. Dynamic astral microtubules from a single SPB push against the cell cortex and propel the nucleus in the opposite direction. High resolution DIC with overlain fluorescence (right) and fluorescence only (left) images of unbudded G1 cells. Haploid strain 9dΔd, lack an endogenous copy of dynein and contain dynein-GFP (pKBY701). The focus of fluorescence represents the SPB, which could be seen at the edge of the nucleus in DIC (right, overlay). A series of five time points at 1-min intervals are displayed from top to bottom. Nuclear movement is from left to right over the time course. The cytoplasmic microtubules were organized into a cone-shaped array facing away from the nucleus. Note the SPB at the leading edge of the nucleus, and the cytoplasmic microtubules growing opposite to the direction of movement. As individual microtubule (or microtubule arrays) extended to the left, the nucleus was propelled rightward. Bar, 2 μm.
Dynamic instability of astral microtubules occurred throughout the cell cycle (Fig. 3). Individual microtubules exhibited growth and shortening at 0.5 μm/min (range from 0.3 to 1.5 μm/min; n = 63 microtubules). These velocities are an underestimate due to the projection of microtubules in a three-dimensional data set onto a two-dimensional plane. No statistical difference in velocities was observed from interphase to mitosis at the level of resolution of this study. The average microtubule length was 1.58 ± 0.54 μm (n = 63). Shortening microtubules were observed to switch back to growth (rescue) (Fig. 3), although determination of rescue events close to the pole was difficult to observe due to the high density of microtubules at the SPB.
In G1, Astral Microtubule Dynamics Push the SPB and Nucleus
The finding that cytoplasmic dynein decorated astral microtubules, and was concentrated at the single SPB in unbudded cells (G1 stage of the cell cycle, Fig. 4), allowed us to determine the in vivo orientation and movements of astral microtubules, the SPB, and nucleus. We examined nuclear movement by time-lapse images taken at 60-s intervals. G1 cells contained conical astral microtubule arrays (∼60° arc) undergoing constant polymerization and depolymerization (Figs. 3 and 4). These arrays apparently pushed the nucleus in the opposite direction of microtubule growth (18 cells examined), resulting in nuclear excursions through the cell. The direction of these excursions depended upon the orientation of a growing microtubule, and were reflected by the nuclear movements limited to a 60° arc in the opposite direction of the conical microtubule array. The astral microtubules were constantly growing and shortening during this time and no static interactions (interaction >3 min) with the cell cortex were observed. When the astral microtubules reached the cell cortex, continued growth was accompanied either by SPB movement away from the cortex, or in microtubule buckling (Fig. 4). In 2 cells (of 18 cells examined), the SPB could be seen to transiently move once toward the tip of a microtubule at the cell surface. These movements were short (≅0.3 μm), appeared to occur after the microtubule switched from growth to shortening, were not specific to the bud neck region, and were not significant in comparison to the movements of the SPB and nucleus associated with microtubule growth.
The nuclear movements were completely dependent upon intact microtubule arrays (Fig. 5). Nocodazole treatment produced no distinct astral microtubules, barely detectable dynein-GFP accumulation at the pole, and inhibition of SPB and nuclear migration. These results corroborate the previous report of Yeh et al. (1995) that reported that the concentration of a Dhc1p-lacZ fusion to the SPB was also dependent upon intact microtubules. Most likely, the very weak SPB pattern shown in Fig. 5 reflects short microtubule arrays at the SPB, which are not able to produce nuclear movement. SPBs marked by Nuf2p-GFP (Nuf2p, a spindle pole antigen; Kahana et al., 1995) displayed SPB movements in G1 cells typical of those observed in cells expressing the dynein-GFP fusion (data not shown). These observations confirm that movements shown in Fig. 4 are representative of wild-type nuclear motility.
Figure 5.
Nuclear movement requires cytoplasmic microtubules. Haploid strain 9dΔd (containing pKBY701) cells were treated with nocodazole (NZ) as described in the Materials and Methods. Five fluorescence images (from left to right) at 5-min intervals are shown. The cell on the lower left is small budded, and the poles have separated (note the asymmetry in intensity at the poles). The cell on the upper right has only one fluorescent SPB. No nuclear movements were detected throughout the analysis. Thus, nuclear motility is dependent upon intact cytoplasmic microtubules. Points of fluorescence, indicative of dynein-GFP, could be seen at the SPBs even upon prolonged NZ treatment. Short tufts of microtubules may be resistant to NZ treatment. Bar, 5 μm.
Nuclear Movement to the Bud Neck in S/G2 Depends on an Astral Microtubule Search and Capture Process
Time-lapse images revealed that nuclear orientation to the bud neck in S/G2 was dependent upon astral microtubules penetrating the newly formed bud. Nuclear movements as described above continued in a direction away from microtubule growth as bud emergence occurred in S/G2. There was no discernible association between the SPB or astral microtubules and the incipient site of bud emergence (Fig. 6). Growing microtubules frequently and transiently contacted the cell cortex; however, the pattern of nuclear movement was unchanged from that of G1 cells until an astral microtubule grew into the bud. Upon penetration, the SPB and nucleus moved in the direction of the polymerized microtubule, toward the bud neck. Nuclear orientations occurred in this fashion with no obvious relationship to bud size. The apparent pulling force oriented the SPB and its astral microtubule array towards the bud so that as their dynamic instability continued, more and more astral microtubules grew toward and into the bud, rather than into the mother cell. Such microtubule dependent pulling forces (sustained SPB movement in the direction of microtubule growth) occurred only with microtubules polymerized into the bud, not in the mother cell. Upon penetration of astral microtubules into the bud, nuclear movement was restricted to the mother–bud axis, and the nucleus ceased its progression around the cell.
Figure 6.
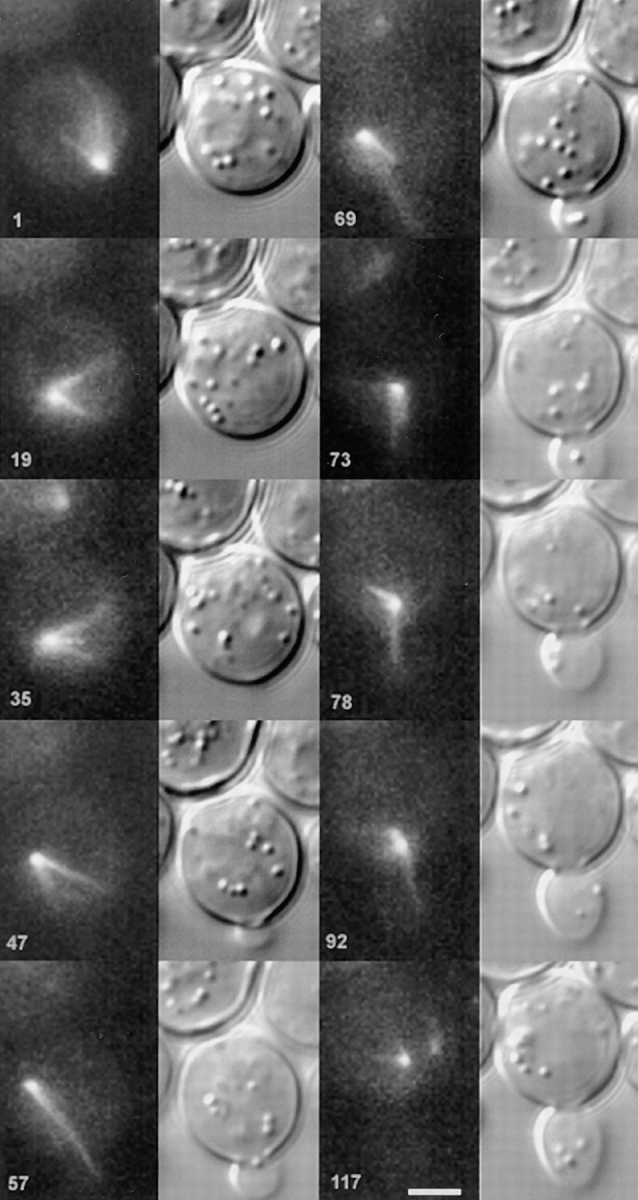
Microtubule penetration in the bud precedes nuclear movement to the neck. Montage from time-lapse experiments demonstrating that once cytoplasmic astral microtubules penetrate the bud, the nucleus migrates to the bud neck. Unbudded haploid cells (9dΔd containing pKBY701) were captured in the process of bud emergence. A series of DIC (right) and fluorescence (left) images over the time course indicated in the lower left (min) are shown from top to bottom, in consecutive series. In the upper left quadrant, the unbudded cell has a single SPB (embedded in the nuclear envelope), that is positioned at approximately five o'clock. In the next sequence, the SPB has migrated to approximately the seven o'clock position. Note that the astral microtubules trail the SPB and form a conical array. The position of bud emergence is six o'clock (see 47 min). The SPB migrates around the cell periphery until the 9–10 o'clock position (47 min), well past the emerging bud. At this time, a long microtubule (7 μm) can be seen extended toward and into the bud. The nucleus migrates in the direction of the bud after microtubule penetration of the bud (57–69 min). The nucleus continues toward the bud neck, in the direction of the microtubule over the next 23 min (69–92 min). Fluorescently labeled SPBs were apparent at 117 min. Bar, 2 μm.
In G2/M, Dynein-GFP Accumulates on the Second SPB After Spindle Pole Separation
SPB separation typically occurred after nuclear orientation to the bud neck. The spatial resolution of either dynein-GFP or Nuf2p-GFP in the fluorescence images did not allow a precise temporal determination of SPB duplication. Decoration of the second pole body with dynein-GFP could only be observed after the SPBs separated >1 μm (Fig. 7). It is important to note that in the few published electron micrographs of astral microtubules emanating from duplicated but unseparated SPBs (Byers, 1981), the microtubules originate from the bridge structure extending between the two pole bodies. Cytoplasmic dynein was clearly localized to the single array of microtubules at this time, a component of which had penetrated the bud. Once the SPBs separated during spindle formation, cytoplasmic dynein remained with the pole destined for the bud (SPBbud; Fig. 6) in three of four cells where both spindle poles could be followed discretely. There was a delay of about 10 min after visible separation of the SPBs before the spindle pole destined for the mother cell (SPBmother) accumulated cytoplasmic dynein (and astral microtubules). At this time, the SPBmother was 1–1.5 μm from the SPBbud (Fig. 7, right). We could not distinguish the old and new pole from these studies. However, the microtubule array the G1 cells were born with was most often the array of microtubules destined for the bud.
Figure 7.
Dynein-GFP accumulation at the SPB. This sequence of fluorescence stacks demonstrates the accumulation of dynein on separated SPBs in budded cells. Cell cycle progression is from left to right at 5-min intervals. The upper left cell has only one visible SPB by fluorescence. In 5 min, the second SPB can be visualized, well separated from the first (∼1.4 μm). Over time, the second SPB acquired fluorescence equal in intensity to the first. Quantitation of fluorescence accumulation over time is graphed in the panel to the right. Bar, 2 μm.
Maintenance of Nuclear Orientation to the Bud Neck Does Not Require Stable Astral Microtubule–Cortical Interactions
In G2/M, the nucleus remained near the neck of the budded cell. The SPBbud and nucleus exhibited short range movements. The SPBmother rotated about the neck site as described previously for spindles by DIC microscopy (Yeh et al., 1995). SPBs decorated with Nuf2p-GFP revealed similar short-range SPB movements (data not shown). At this time in the cell cycle, the bipolar spindle was evident within the nucleus by DIC microscopy with the spindle ∼1.5–2 μm in length. Astral microtubules from the SPBmother grew into the mother (30 out of 30 microtubules counted), whereas SPBbud initiated microtubules grew mainly into the bud (20 out of 23 microtubules counted). Stable interactions (localization >3 min) of astral microtubules with the cortex were very rare (2 out of 53 microtubules counted), indicating that the restriction in nuclear movements was not dependent upon stable attachments of astral microtubules in the mother or bud.
Spindle Elongation in Anaphase
Anaphase onset was observed as elongation of the central spindle and nuclear envelope through the bud neck by DIC (Kahana et al., 1995; Yeh et al., 1995) and rapid separation of SPBs in dynein-GFP or Nuf2-GFP fluorescence images. The average microtubule length in anaphase was 1.98 ± 1.0 (n = 12), slightly higher than the average non-anaphase microtubule length of 1.2 ± 0.2 (n = 51). As the spindle elongated (anaphase B) there was concordant decrease in the average length of the SPBbud microtubule array as the SPBbud migrated toward the tip of the bud (Fig. 8). In addition, astral microtubules in the mother were displaced during anaphase, and could be observed to trail the SPB in its movement to the cortex (astral microtubules trailing SPB in mother; Fig. 8, 15–25 min). After the SPBmother reached the distal portion of the mother cells, the astral microtubules were consistently short and difficult to resolve.
Figure 8.
Microtubule attachment in the tip of budded cells in anaphase. DIC (bottom) and fluorescence (top) images demonstrating the stabilization of a single microtubule to a bud tip and the movement of the nucleus to that site with concomitant shortening of the microtubule. This behavior was infrequent, and is not typical of most anaphases. Two SPBs of equal intensity can be seen in the upper left 10 min before anaphase onset. The SPB closest to the neck is destined for the bud. The nucleus can be seen in DIC where the positions of the spindle poles have been denoted by an asterisk. By 10 min, the fast portion of anaphase is complete and by 15 min after anaphase onset, a microtubule from SPBbud extends into the bud to the tip of the budded cell. Note that the nucleus spans the neck at this time point (15 min after anaphase onset) and that the spindle pole does not lead the nucleus. The microtubule in the bud shortens with spindle elongation (5-min intervals, 15–25 min). The cytoplasmic microtubules associated with SPBmother remain short and distant from the cortex as SPBmother moves toward the base of the mother cell. The cytoplasmic microtubules associated with SPBbud that are not in contact with the cortex remain dynamic. By the end of anaphase, the SPB migrates to the site where the microtubule in the bud is apparently “attached.” Bar, 2 μm.
Astral microtubules were capable of forming static interactions (>3 min) with the bud tip during anaphase, but they were rare (n = 2 out of 18 cells recorded) (Fig. 8). Different microtubules from SPBbud interact with the bud cortex or undergo dynamic instability (Fig. 8). Spindles undergoing anaphase without microtubules stably attached in the bud generally had several dynamic microtubules polymerizing into the bud and occasionally back into the mother cell. However by the completion of anaphase, microtubules from the SPBbud were rarely seen in the mother cell. Spindle elongation was biphasic with an initial rate of 1 μm/min followed by slower elongation into the bud at one-third that rate regardless of apparent microtubule attachment to the cell cortex (n = 9). Cells with Nuf2-GFP– labeled SPBs (containing wild-type dynein) exhibited the same rate of spindle elongation (n = 4, data not shown). These data are in accord with the rates of spindle elongation from previously published DIC (Yeh et al., 1995) and Nuf2-GFP (Kahana et al., 1995) studies.
Spindle Disassembly in Telophase
The astral microtubules and the nucleus resumed the behavior typified by G1 cells (above) before cytokinesis. The nucleus and SPB moved, in either the mother or daughter cell, in a direction opposite microtubule growth. This movement tended to push the SPB and nucleus toward and through the center of the cell. This indicates that the stiffness and pushing forces of the central spindle in anaphase may be much stronger than the pushing forces generated by the growth phases of microtubule dynamic instability.
Overexpression of Dynein-GFP Results in a Defect in Nuclear Migration
Cells expressing elevated levels of dynein-GFP were identified in the population by levels of average fluorescence equivalent to >50% of our fluorescence reference. Gross overexpression of dynein-GFP did not appear to inhibit dynamic instability, per se, but resulted in elongated and bundled microtubules preferentially in the bud during mitosis. Bundling was evidenced by occasional splaying apart of microtubule bundles. Microtubule growth in the mother cell often exhibited normal dynamic instability and bundling was much less frequent than in the bud. The hyperelongated astral microtubules exhibited sweeping motions along the inner surface of the bud, movements not characteristic of short microtubule arrays in cells expressing low levels of dynein-GFP. Mitotic cells overexpressing dynein-GFP had properly aligned spindles before anaphase (Fig. 9, row 5 column 1 and row 6 column 4). Upon anaphase onset (Fig. 9, row 6 column 5), the spindle seemed to be prevented from entering the bud by the persistent astral microtubule array extending into the bud. Further growth of this microtubule bundle pushed the nucleus and spindle towards the base of the mother cell. In a few instances, where disassembly of the stable microtubules occurred, nuclear translocation into the bud followed rapidly (Ozoy, Z., C. Yang, E. Yeh, and K. Bloom, data not shown).
Figure 9.
Overexpression of dynein-GFP. Cells with elevated levels of fluorescence (>three- to fourfold over the bulk of the population) exhibited hyperstabilized microtubules only in the bud, and only during late G2/M or anaphase. Two cells expressing different amounts of dynein-GFP are shown at 4-min intervals from left to right. The cell to the left contains low levels of dynein-GFP and completes an entire cell cycle, budding a second time within the 160-min of this time-lapse. Note in this cell that there is no apparent attachment of astral microtubules to the bud tip during anaphase. The cell on the right hand side contains three to four times more dynein-GFP fluorescence than cells normally selected for imaging. Observable in this cell are very dynamic astral microtubules from the single SPB in G1 and from both SPBs during the S/G2 stage. Note that the mitotic nucleus does not appear to remain near the bud neck, but when an astral microtubule enters the bud neck (row 4, column 3), the spindle becomes properly oriented along the mother/bud axis (row 4, column 6). The astral microtubule array in the bud remains very dynamic and undergoes numerous rescues, never depolymerizing back to the mother cell. Sweeping of the astral microtubule array laterally under the bud cortex can be observed (row 6, columns 1–3). Upon anaphase (row 6, column 4), the spindle is properly aligned but is prevented from entering the bud by the stable astral microtubule array extending into the bud. Further growth of this microtubule bundle pushes the nucleus and spindle towards the base of the mother cell. These cells often become blocked in the cell cycle at this stage.
Discussion
The ability to visualize astral microtubules with dynein-GFP has revealed robust dynamic instability in yeast. More profound, however was the ability to integrate microtubule dynamics with spindle and nuclear movements throughout an entire cell cycle. The results herein indicate: (a) a fundamental role for microtubule dynamic instability in positioning nuclei and aligning spindles; (b) an extranuclear force on the SPB arising primarily from transient interactions of astral microtubules in the bud; and (c) a possible mechanism for assuring unity of SPB transmission into the bud.
Yeast Use Dynamic Instability and Transient Microtubule Interactions to Position the Nucleus and Align the Mitotic Spindle Along the Mother/bud Axis
Live-cell imaging of interphase cells dramatically illustrates the constant polymerization and depolymerization of the conical shaped astral microtubule arrays previously documented in budding yeast (Fig. 10). These microtubule arrays pushed the nucleus in the opposite direction of microtubule growth, resulting in nuclear excursions through the cell. Stable attachments of microtubules to the cell cortex and pulling forces postulated from still images did not contribute significantly to nuclear movements. The astral microtubules, in performing this action of moving the nucleus, probed the entire cell cortex during interphase. Upon finding the emerging bud, nuclear movement was biased toward the direction of the bud. Depolymerization of astral microtubules with nocodazole completely inhibited both the slow progression of the nucleus and the directed movement to the bud neck, underscoring the requirement for astral microtubules and confirming earlier works (Huffaker et al., 1988; Jacobs et al., 1988). This stochastic process of microtubules searching for the bud via dynamic instability is reminiscent of the “search and capture” mechanisms postulated for microtubule attachment to kinetochores during prometaphase (Mitchison et al., 1986; Holy and Leibler, 1994; Hyman and Karsenti, 1996).
Figure 10.
Schematic representation of astral microtubule dynamics throughout the yeast cell cycle. The nucleus is indicated in blue, astral microtubules in green, and the intranuclear spindle microtubules in red. In unbudded cells and cells with small buds, the astral microtubules originate from the single SPB (G1, G1/S). The astral microtubules are organized most often into a conical array. Individual microtubules show asynchronous dynamic instability throughout the cell cycle with growth and shortening at ≅0.5 μm/min. As microtubules reach the cell cortex, continued growth results in nuclear movement in the opposite direction. Thus, astral microtubule growth pushes against the cell cortex to propel the nucleus around the cell interior (G1/S-S/G2). The nucleus and/or microtubules make no stable contact in unbudded cells. Once a bud has formed, the dynamically unstable microtubules penetrate the bud (S/G2). At this point, the nucleus moves toward the neck of the budded cell (G2/M). The second pole body is delayed in acquiring the dynein-GFP fusion protein (S/G2–G2/M). Only after the pole bodies have separated, is the second pole competent to nucleate astral microtubules (G2/M). Once both SPB's are labeled with dynein-GFP, and the nucleus is at the neck, biphasic spindle elongation ensues (M). Astral microtubules that contact the bud tip, shorten as the spindle elongates (Anaphase). Finally, when central spindle disassembly occurs (Telophase), the astral microtubule dynamic instability and the movements of the SPB and nucleus become typical of G1 cells.
The centrality of microtubule dynamics to the fidelity of nuclear migration was further evidenced by analysis of cells overexpressing dynein. Persistent and hyperelongated microtubules were observed in the bud in cells expressing excess dynein-GFP. Anaphase was completed in the mother cell due to the inability of the bud-resident astral microtubules to depolymerize. Spindle elongation in the mother was always accompanied by persistent microtubules in the bud, and did not result from the lack of microtubule interactions in the bud or cell cortex. Hence, eventual spindle misalignment may result from the persistence of highly stable microtubules that present a physical barrier to nuclear translocation into the bud.
The dramatic changes in microtubule dynamic instability in cells expressing no (Carminati and Stearns, 1997) or high levels of dynein reflect recent observations that Kar3p, a minus-end–directed, microtubule-based motor protein, also influences the assembly of cytoplasmic astral microtubules (Saunders et al., 1997). Loss of Kar3p results in increased number and average length of astral microtubule arrays. Whereas the mechanisms leading to the generation of hyperelongated microtubules are likely to be different in the dynein deficiency strain versus the cells overexpressing dynein-GFP, the regulation of microtubule-based motors clearly affects the distribution of astral microtubules in fundamental ways. Hence, the disruption of microtubule dynamics by microtubule-based motors, when mutant or when expression levels are modified, explains, in part, the defects observed in nuclear migration, spindle alignment, and elongation into the bud.
Orientation of the Nucleus and Mitotic Spindle Depends Primarily upon Transient Interactions of Astral Microtubules in the Bud
Alignment of the mitotic spindle along the mother–bud axis does not appear to be the consequence of the rigid interaction of cortical microtubules with the cell cortex in the mother and bud, but rather due to the net sum of less static interactions of dynamic microtubules within the bud alone. Though we were able on two occasions to document stable interactions of individual astral microtubules with the bud tip during anaphase, that interaction had no measurable effect on the rate of spindle elongation or nuclear penetration into the bud. We presume stable interactions have little to do with spindle orientation as only 2 of 11 cells imaged through anaphase exhibited any perceived stable microtubule interaction with the bud cortex. We conclude from this analysis that the majority of information for spindle alignment and the majority of force applied externally to the spindle do not come from stable interactions of astral microtubules with the cell cortex.
The movement of the nucleus in the direction of an astral microtubule that has penetrated the bud in S/G2 is indicative of pulling forces at this stage in the cell cycle, and is consistent with previous work showing that SPBs can migrate into the bud in the absence of a central spindle (Yeh et al., 1995). In the present study, seeing that the nucleus follows the microtubules, and measurements indicating that the microtubules shortened as the distance between the SPBbud and bud tip diminished, may provide additional evidence for pulling forces in the bud. Very few stable microtubules were observed in the mother cells, and microtubule shortening was not associated with migration of SPBmother during anaphase. In contrast, astral arrays associated with SPBmother were displaced upon anaphase onset and often followed the pole body as the central spindle elongated towards the distal end of the mother cell. Therefore, microtubule-dependent forces capable of pulling the SPB must be transient in nature and confined primarily to the emerging bud.
These observations provide a mechanism for understanding several classic observations of asymmetric nuclear positioning in higher animals and plants. In Caenorhabditis elegans (Hyman and White, 1987; Hyman, 1989; White and Strome, 1996) and Pelvetia fastigiata (Allen and Kropf, 1992) a nuclear rotation in early development has an essential role in determining the fate of progeny cells. Migration of spindle poles close to a site in the cell surface precedes asymmetrical cell divisions in oocytes and early embryos as well (Baker et al., 1993). These events may be related mechanistically to the orientation of the spindle in budding yeast. A microtubule-based process of searching that is mediated by dynamic instability to position the spindle with respect to the nascent bud, or other cortical sites may be a common feature of spindle and nuclear movements in a variety of cell types.
A Proposed Mechanism for the Transmission of a Single SPB into the Bud
Dynamic microtubules and transient interactions with sites in the bud could account for the mechanism that ensures unity in SPB deposition in to the bud. In G1, only one astral microtubule array is evident. Once a microtubule penetrates the bud, subsequent microtubule growth from that SPB is biased toward the bud, largely due to the SPB being partially imbedded in the nuclear envelope. One pole was usually biased toward the bud, well before the second pole was competent to acquire dynein and nucleate astral microtubule (Figs. 6 and 10). Therefore, microtubules from the second pole (SPBmother) did not accumulate in the nascent bud, avoiding potential errors that would lead to migration of both SPBs (and the accompanying spindle and nucleus) in the bud. Once the central spindle was assembled (1.5–2.0 μm) the inherent “stiffness” of the spindle oriented the SPBmother to face away from the bud site, and the restriction in astral microtubule growth to one face of the pole body further predisposed astral microtubules from this pole away from the bud and into the mother cell.
The transient nature of astral microtubule associations with the cortex may contribute to error correction mechanisms. If microtubule contacts in the bud were stable, then errors from stray microtubules would be difficult to correct. However, multiple transient interactions between microtubules and sites in the bud greatly favor the SPB first oriented to the bud to remain facing the bud. A stray microtubule from the opposite pole that does move into the bud would rapidly disassemble and not result in reorienting the distal SPB. In one case we have observed a microtubule from the SPBbud that grew into the mother well after anaphase onset. This microtubule grew quite long (>6 μm), extending well into the mother cell before catastrophe and shortening. Microtubule growth from SPBbud to the mother and vice versa were infrequent relative to the total number of events. Nevertheless, such events did not result in aberrant mitoses, and are indicative of a mechanism that does not rely on a stable microtubule–cortical interaction.
A Role for Dynein in Spindle Alignment
The interaction of dynein with dynactin, and the association of the dynactin complex with actin-related proteins (Schroer, 1994) point toward the notion that dynein– dynactin complex in the bud may be mobile and physically associated with the actin cytoskeleton. One of the most pronounced asymmetries in yeast is the polarized distribution of actin into the growing bud (Lew and Reed, 1995). A mechanism that couples dynein with the actin cytoskeleton would account for dynein's ability to transiently stabilize cytoplasmic microtubules and polarize one SPB toward the bud. As described above, there may not be a “single” site for microtubule capture in the bud. Rather, cumulative transient interactions between dynein and dynactin, the actin cytoskeleton, and dynamic astral microtubules emanating from the SPB may provide the force, as well as the mechanism that ensures spindle alignment and that only one SPB is deposited into the bud.
Dynein has been implicated in providing the directional information for nuclear migration in yeast. However, the finding that dynein is associated with microtubules in both mother and bud preclude it being the determinant in generating asymmetry. Instead, there are likely to be components in the bud and bud tip that are capable of transient interactions with dynein. The polarization of “transient stabilization” domains to the bud may result from mechanisms that contribute to actin polarization toward the bud, and the bud site selection machinery (Pringle et al., 1995). Interestingly, several proteins that mark the bud site are restricted both temporally and spatially to the tip of growing buds in yeast (Amberg et al., 1997; Evangelista et al., 1997), and in asymmetrically dividing blastomeres of the germline lineages in C. elegans (Etemad-Moghadam et al., 1995). These transient interactions may be critical in the capacity for differential positioning within the cell and mechanisms that ensure the distribution of one and only pole to the progeny.
Acknowledgments
We thank J. Kahana and P. Silver (Harvard University, Cambridge, MA) for constructing and generously supplying the bright and reliable Nuf2-GFP clone, E. Bi (University of North Carolina at Chapel Hill) for supplying the rabbit anti-GST antibodies, and S. Inoue for laboratory space at the Marine Biological Laboratory (Woods Hole, MA) at the inception of the imaging.
This work was supported by research grants from the National Institutes of Health General Medical Science (to K. Bloom and E.D. Salmon). E. Yeh was supported by the North Carolina Employment and Security Commission.
Abbreviations used in this paper
- DIC
differential interference contrast
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- SPB
spindle pole body
Footnotes
Address all correspondence to K. Bloom, Department of Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-3280. Tel.: (919) 962-1182. Fax: (919) 962-1625. E-mail: ksb.fordham@mhs.unc.edu
References
- Allen VW, Kropf DL. Nuclear rotation and lineage specification in Pelvetiaembryos. Development (Camb) 1992;115:873–883. [Google Scholar]
- Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J, Theurkauf WE, Schubiger G. Dynamic changes in microtubule configuration correlate with nuclear migration in the preblastoderm Drosophilaembryo. J Cell Biol. 1993;122:113–121. doi: 10.1083/jcb.122.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldari C, Murray JA, Ghiara P, Cesareni G, Galeotti CL. A novel leader peptide which allows efficient secretion of a fragment of human interleukin 1 beta in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1987;6:229–234. doi: 10.1002/j.1460-2075.1987.tb04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B. 1981. Cytology of the Yeast Life Cycle. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). 59–96.
- Byers B, Goetsch L. The behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerivisiae. . J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegansembryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Holy TE, Leibler S. Dynamic instability of microtubules as an efficient way to search in space. Proc Natl Acad Sci USA. 1994;91:5682–5685. doi: 10.1073/pnas.91.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker TC, Thomas JH, Botstein D. Diverse effects of β-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. . J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Karsenti E. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Jacobs CW, Adams AE, Szaniszlo PJ, Pringle JR. Functions of microtubules in the Saccharomyces cerevisiaecell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning AJ, Lum PY, Williams JM, Wright R. DiOC6 staining reveals organelle structure and dynamics in living cells. Cell Motil Cytoskeleton. 1993;25:111–128. doi: 10.1002/cm.970250202. [DOI] [PubMed] [Google Scholar]
- Kahana JA, Schnapp BJ, Silver PA. Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Evans L, Schulze E, Kirschner M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell. 1986;45:515–527. doi: 10.1016/0092-8674(86)90283-7. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin DG, Haarer B, Jones EW. Fluorescence microscopy methods in yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Bi E, Harkins HA, Zahner JE, De VC, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harb Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Salmon ED, Inoue T, Desai A, Murray AW. High resolution multimode digital imaging system for mitosis studies in vivo and in vitro. Biol Bull. 1994;187:231–232. doi: 10.1086/BBLv187n2p231. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiaekinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, W., D. Hornack, V. Lengyel, and C. Deng. 1997. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J. Cell Biol. In press. [DOI] [PMC free article] [PubMed]
- Schroer TA. New insights into the interaction of cytoplasmic dynein with the actin-related protein, Arp1. J Cell Biol. 1994;127:1–4. doi: 10.1083/jcb.127.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Bloom K, Salmon ED. Imaging green fluorescent protein fusion proteins in Saccharomyces cerevisiae. . Curr Biol. 1997;7:701–704. doi: 10.1016/s0960-9822(06)00299-5. [DOI] [PubMed] [Google Scholar]
- Snyder M, Gehrung S, Page BD. Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. . J Cell Biol. 1991;114:515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. . J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Botstein D. A gene required for the separation of chromosomes on the spindle apparatus in yeast. Cell. 1986;44:65–76. doi: 10.1016/0092-8674(86)90485-x. [DOI] [PubMed] [Google Scholar]
- White J, Strome S. Cleavage plane specification in C. elegans: how to divide the spoils. Cell. 1996;84:195–198. doi: 10.1016/s0092-8674(00)80974-5. [DOI] [PubMed] [Google Scholar]
- Winey M, Hoyt MA, Chan C, Goetsch L, Botstein D, Byers B. NDC1: A nuclear periphery component required for yeast spindle pole body duplication. J Cell Biol. 1993;122:743–751. doi: 10.1083/jcb.122.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]









