Abstract
For the analysis of microarray data, clustering techniques are frequently used. Most of such methods are based on hard clustering of data wherein one gene (or sample) is assigned to exactly one cluster. Hard clustering, however, suffers from several drawbacks such as sensitivity to noise and information loss. In contrast, soft clustering methods can assign a gene to several clusters. They can overcome shortcomings of conventional hard clustering techniques and offer further advantages. Thus, we constructed an R package termed Mfuzz implementing soft clustering tools for microarray data analysis. The additional package Mfuzzgui provides a convenient TclTk based graphical user interface.
Availability
The R package Mfuzz and Mfuzzgui are available at http://itb1.biologie.hu-berlin.de/~futschik/software/R/Mfuzz/index.html. Their distribution is subject to GPL version 2 license.
Keywords: gene expression, soft clustering, software
Background
Clustering methods are popular tools in data analysis. They can be used to reveal hidden-patterns (clusters of objects in large complex data sets). Most clustering methods assign one object to exactly one cluster. [1] While this so-called hard clustering approach is suitable for a variety of applications, it may be insufficient for microarray data analysis. Here, the detected clusters of co-expressed genes indicate co-regulation. However, genes are frequently not regulated in a simple ‘on’ ‐ ‘off’ manner, but instead their expression levels are tightly regulated by a number of fine-tuned transcriptional mechanisms. This is reflected in expression data sets generated in microarray experiments. It is a common observation that many genes show expression profiles similar to several cluster patterns. [2,3]
Ideally, clustering methods for microarray analysis should be capable of dealing with this complexity in an adequate manner. They should not only differentiate how closely a gene follows the main expression pattern of a cluster, but they should also be capable to assign genes to several clusters if their expression patterns are similar.
Soft clustering can provide these favourable capacities. Recently we have shown that applying soft clustering to microarray data analysis leads to i) more adequate clusters with information-rich structures, ii) increased noise-robustness and iii) and improved identification of regulatory sequence motifs. [4]
Methodology
Soft clustering has been implemented using the fuzzy c-means algorithm. [5] It is based on the iterative optimization of an objective function to minimize the variation of objects within clusters. Poorly clustered objects have decreased influence on the resulting clusters making the clustering process less sensitive to noise. Notably this is a valuable characteristic of fuzzy c-means method as microarray data tends to be inherently noisy. As a result, fuzzy c-means produces gradual membership values µij of a gene i between 0 and 1 indicating the degree of membership of this gene for cluster j. This strongly contrasts hard clustering e.g. the commonly used k-means clustering that generates only membership values µij of either 0 or 1. Thus, soft clustering can effectively reflect the strength of a gene's association with a cluster. Obtaining gradual membership values allows the definition of cluster cores of tightly co-expressed genes. Moreover, as soft clustering displays more noise robustness, the commonly used procedure of filtering genes to reduce noise in microarray data can be avoided and loss of the potentially important information can be prevented. [4]
Software input
Like most other clustering software, the Mfuzz package requires as input the data to be clustered and the setting of clustering parameters.
Microarray expression data can be entered either as simple table or as Bioconductor (i.e. exprSet) object. Whereas the table format is an easy and sufficient way to handle data for most experiments, Bioconductor data objects can be used for more complex experimental designs. [6] The format for tables is the same as for the standard clustering software Cluster [7], so that users can easily use both software packages without reformatting their input.
Further, the number of clusters and the so-called fuzzification parameter m have to be chosen. By variation of both parameters, users can probe the stability of obtained clusters as well as the global clustering structure [4]
Software output
As basic output, the partition matrix is supplied containing the complete set of membership values. This information can be used to define cluster cores consisting of highly correlated genes and to improve the subsequent detection of regulatory mechanism. [4] Results of the cluster analysis can be either further processed within the Bioconductor framework or stored in simple table format.
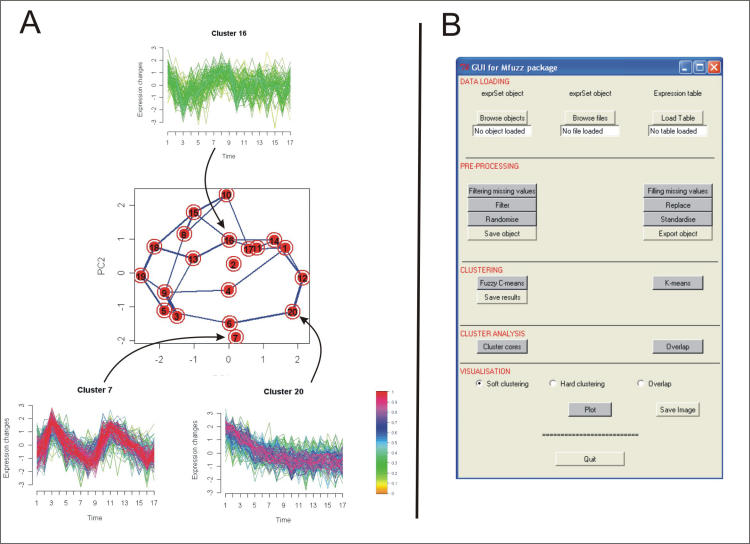
Several functions serve the visualization of the results such as internal or global cluster structures. Figure 1 shows some examples of the graphical output.
Figure 1.
A) Examples for visualization of clustering results produced by Mfuzz. Graphs show the temporal expression pattern during the yeast cell cycle (top and lower panels) and the global clustering structure (central panels). Membership values are color-encoded with red shades denoting high membership values and green shades denoting low membership values of genes. Using this color scheme, clusters with a large core of tightly co-regulated genes (e.g. cluster 7) can be easily distinguished from week or noisy clusters (e.g. cluster16). The central panel shows the principal components of the clusters. Lines between clusters indicate their overlap i.e. how many genes they share. B) Graphical user interface implemented in the Mfuzzgui package. Its outline follows the standard steps of cluster analyses of microarray data: Data loading and pre-processing, clustering, examination and visualization of results
Note that Mfuzz is not restricted to microarray data analysis, but has recently also successfully applied to examine protein phosphorrylation time series. [8]
Caveat & Future development
Mfuzz and Mfuzzgui are R packages. R is a statistical programming language and is freely available open-software. [9] Both developed packages follow conventions of the Bioconductor platform. [6] The graphical user interface implemented in Mfuzzgui demands an existing installation of Tcl/Tk. For convenience, we supply scripts for automatic installation of the software packages. Additionally, scripts are provided for a direct start of the packages enhancing their stand-alone character. Future versions will include extended export options such as automatically generated HTML pages reporting the results of the clustering analysis.
Acknowledgments
Lokesh Kumar was supported by the SFB 618 grant of the Deutsche Forschungsgemeinschaft. We would like to thank Hanspeter Herzel for his assistance of the project and B. Carlisle for critical reading of the manuscript.
References
- 1.Jain AK, et al. ACM Computing Surveys. 1999;31:264. [Google Scholar]
- 2.Cho RJ, et al. Mol Cell. 1998;2:65. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 3.Chu S, et al. Science. 1998;282:699. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 4.Futschik ME, Carlisle B. J Bioinform Comput Biol. 2005;3:965. doi: 10.1142/s0219720005001375. [DOI] [PubMed] [Google Scholar]
- 5.Hathaway R, Bezdek J. Pattern Recognition. 1985;19:477. [Google Scholar]
- 6. http://www.bioconductor.org.
- 7. http://rana.lbl.gov/EisenSoftware.htm.
- 8.Olsen JV, et al. Cell. 2006;127:635. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 9. http://www.r-project.org.