Abstract
The European Soyuz missions have been one of the main routes for conducting scientific experiments onboard the International Space Station, which is currently in the construction phase. A relatively large number of life and physical sciences experiments as well as technology demonstrations have been carried out during these missions. Included among these experiments are the Gene experiment during the Spanish “Cervantes” Soyuz mission and the ICE-1st experiment during the Dutch “Delta” mission. In both experiments, full genome microarray analyses were carried out on RNA extracted from whole animals recovered from the flight. These experiments indicated relatively large scale changes in gene expression levels in response to spaceflight for two popular model systems, Drosophila melanogaster (Gene) and Caenorabditis elegans (ICE-1st). Here we report a comparative analysis of results from these two experiments. Finding orthologous genes between the fruit fly and the nematode was far from straightforward, reducing the number of genes that we could compare to roughly 20% of the full comparative genome. Within this sub-set of the data (2286 genes), only six genes were found to display identical changes between species (decreased) while 1809 genes displayed no change in either species. Future experiments using ground simulation techniques will allow producing a better, more comprehensive picture of the putative set of genes affected in multicellular organisms by changes in gravity and getting a deeper understanding of how animals respond and adapt to spaceflight.
Keywords: Microarrays, Gene expression patterns, Microgravity, International Space Station, Drosophila melanogaster, Caenorhabditis elegans, European Soyuz missions, Spanish Cervantes mission, Dutch Delta mission
1. Introduction
The International Space Station (ISS) is the current facility designed for use in gathering the critical scientific information required for successful exploration and colonization of space. One critical scientific question is: How much are organisms affected by changes in gravity? Gravity is one of the fundamental, unchanging, physical parameters on the Earth (Marco et al., 2003). In addition to international involvement in construction and utilization of the ISS, the prevalent idea among the space scientific community was that it would be possible to perform experiments onboard the ISS under conditions similar to those normally found in laboratories on Earth. Today, the hard reality is much less encouraging. The scientific utilization of the ISS has been curtailed by delays in its completion, budgetary overruns, crew limitations, etc. The laboratory facilities are yet quite Spartan. Even when the final European and Japanese modules are installed, the number and complexity of the experiments will remain limited. Critical among these factors are the size of the permanent crew and restrictions in experiment transport and access to the station. These concerns are now amplified as NASA has stated that the Shuttle fleet is too expensive, obsolete and/or unfit for servicing the station as originally planned, namely, every three months.
One Soyuz capsule is permanently attached to the ISS, serving as a lifeboat, providing an emergency escape route for the crew. Initially conceived as an auxiliary facility, the Soyuz capsules are safe and robust, capable of providing shelter and return capability for up to three astronauts. In fact, the limited seating capacity of the Soyuz is one of the reasons why the ISS crew is limited. To assure the continued operability of capsules docked to the ISS, new capsules are launched at regular intervals. Newly launched capsules dock at the second Soyuz bay on the ISS and older capsules return to the ground. Due to the limited capacity, the Soyuz capsule does not provide much cargo space. ISS bound cargo must be shipped by the Shuttle fleet or the unmanned Russian Progress, which has a more limited cargo capacity and lacks the ability to transport large power requiring elements of the ISS (for example the European Columbus or Japanese modules).
During the past five years, the European Space Agency in collaboration with some of its member states has been using these Progress and Soyuz flights to try to implement a research program. These efforts began somewhat tentatively, but have recently become more systematic. Between 2000 and 2006, six European Soyuz missions have been flown: one French (2001), two Italian (2001 and 2005), one Belgian (2002), one Spanish (2003) and one Dutch (2004). The science run during these missions was recently reviewed at a meeting in Toledo and will be published soon (van Loon et al., in press).
During these missions, scientific experiments performed covered both physical and life sciences. During two missions, the Spanish Cervantes and the Dutch Delta missions, experiments were conducted to detect global changes in gene expression of multicellular organisms. The experiments were quite different, but they utilized two of the more important biological model systems available, namely, D. melanogaster in the Cervantes mission and the nematode C. elegans in the Delta mission. Since the genomes of these two organisms have been sequenced, it is possible to look at orthologues. The aim of this analysis is to analyze the results of both experiments in order to find genes whose expression changed in both organisms during their visits to the ISS.
2. Materials and methods
2.1. The Gene experiment in the Spanish Soyuz mission
The experiment intended to study the effects of the space environment on the gene expression pattern of D. melanogaster pupae exposed to microgravity during their development. In doing so, we could rely on previous experiments performed in our laboratory to adapt fixation methods to space conditions (Herranz et al., 2005). Because we could not use an on-board glove box to meet the level of containment imposed by safety for the use of toxic fixatives, the experiment had to be entirely executed, in a completely automatic fashion, inside a closed ESA type I container. The original Berlingot concept (Tixador et al., 1981) had to be adapted. The Berlingot is a culture chamber inside a small sealed plastic bag with two layers of polyethylene. In the Berlingot design the samples are inside this double-layered plastic bag that also holds glass ampoules containing fixatives (Fig. 1). When the ampoules are broken the fixatives are released. Dutch Space (Leiden, The Netherlands) had developed an automated system which was able to break the glass ampoules at preset times; the MAMBA hardware, “Motorized Ampoule Breaker Assembly”. A power supply had to be built and sent to the ISS in time to be able to activate the MAMBA containers during the Spanish Soyuz mission.
Fig. 1.
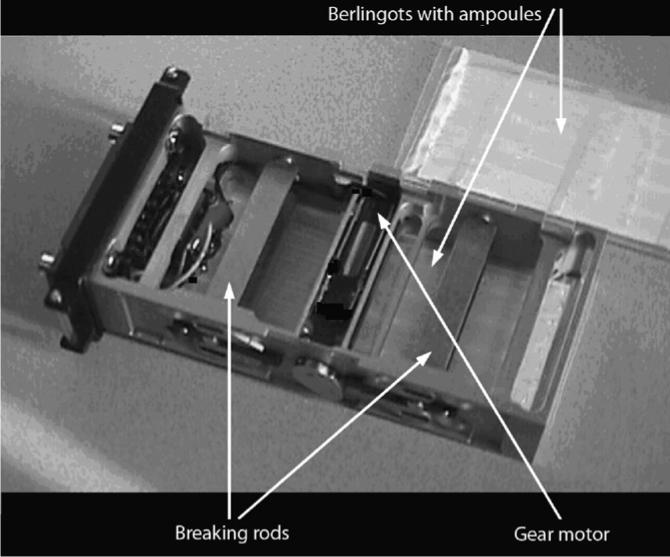
The Berlingot and the MAMBA concepts included in a Type I container are shown. The dimension of the type I container is 9 × 4 × 2 cm.
Two MAMBA containers were flown with two bags each. The same number was prepared for the near synchronous ground controls including the cold transportation step (see below) and for a posterior ground control which lacked such a cold transportation step. Due to flight constraints, we had to limit our experiment to a particular developmental process, namely pupation. The whole pupation process requires four days at normal room temperature, 22 °C. During this time, the animals remain immobile inside the pupal case. They do not require any food, only oxygen. The sample preparation process is relatively complicated and required the use of dedicated equipment available in Toulouse, which could not be easily transported to the launch site in Baikonour, Kazakhstan. We had previously shown the capability of Drosophila to survive at relatively lower temperatures than usual (Anthony et al., 1996). The animals develop normally but more slowly. This provided the possibility to prepare the experiment in Toulouse (France) and transport the samples, at 14 °C, to the launch site in Baikonour. After launch, the samples were exposed to the microgravity environment at room temperature (smoothly oscillating between 20 and 22 °C in the Soyuz capsule and fixed 22° in the aquarius incubator in the ISS) for three and half days. ESA kindly provided the Yellow Box containers for the cold transportation step.
Twenty D. melanogaster Oregon R wild type larvae/early pupae were placed onto the centre of a filter paper of the appropriate size to fit in each Berlingot bag, 3.2 × 10 cm. The larvae were already migrating out of the food and starting pupation but were still easily detached from the plastic wall of the tube and transferred to the filter paper. The glass ampoules were filled with acetone, the fixative used, under low pressure as previously described (Tixador et al., 1981). Twenty ampoules were laid inside each bag. Since each ampoule held 30 μl a total of 0.6 ml of fixative was released on the pupae when the ampoules were broken. By removing as much water as possible from the tissues using acetone we were able to preserve RNA and protein in our samples. Acetone dehydration will work through the thick cuticles of the pupae and imagoes. Once the acetone is released transferring the samples to low temperatures increases the recovery of the RNAs (Herranz et al., 2005). Once the ampoules were broken, the two containers were transferred to the ISS freezer at −22 °C in the Russian Segment. They remained there until the end of the mission, when they were brought into the passive biology transfer box and transferred to the Soyuz TMA-2 capsule that returned to the ground with Pedro Duque and the Expedition S7 crew, The containers were immediately transferred back to a Yellow Box at 3 °C after landing of the Soyuz. The samples were taken back to Moscow area where, at Star city, they were returned to the scientific team close to 12 h after landing. The containers were opened, the pupae removed and homogenized in Trizol, less than one hour later. The RNAs in the pellets were put on dry ice until further processing in our laboratory in Madrid. An almost parallel experiment, using pupae prepared a day later and exposed to 14 °C during a similar period of time as for the actual flight experiment was performed (near synchronous ground control listed above). After the successful launch, the samples were removed from the low temperature and the ampoules were broken after a similar incubation time at 22 °C as in-flight.
2.2. The ICE-1st experiment in the Dutch Soyuz mission
In the next European Soyuz flight, in April 2004, a group of International Laboratories (French, Japanese, Canadian, Americans) coordinated by Dr. Michel Viso from the French Centre National d'Etudes Spatiales (CNES), flew a set of 53 culture bags containing C. elegans (Szewczyk et al., submitted for publication). The bags were made of the same material as the Gene experiment bags and housed within the same ESA Type 1 containers. The majority of the ICE-1st Type 1 containers were vented (including the ones containing the animals utilized for the microarray analysis) as they did not contain fixative. The ICE-1st samples were prepared in Toulouse in the same laboratory as the Gene experiment and transported to Baikonour in the same Yellow Boxes. The containers were inserted in the Kubik incubator developed by ESA set at 12 °C during the day immediately preceding the launch in the Soyuz capsule when the temperature in the incubator was raised to 20 °C. After two days the Soyuz docked the International Space Station and the Kubik was transferred to the Station with the nematode samples which were kept at 20 °C for an additional period of 10 days. After landing, the samples were frozen in liquid nitrogen. Two sets of control sets of samples were prepared in the same way, but remained in either Moscow or Toulouse (1 set at each city) until landing of the space samples had taken place. They were treated in a similar way. During the 12 days at 20 °C, the animals were able to develop and produce progeny. Animal growth and development in-flight was normal (Szewczyk et al., submitted for publication), apoptosis proceeded normally in the germ line (Higashitani et al., 2005) no increase in the rate of mutation was noted (Zhao et al., 2006), and altered muscle development with a movement defect was noted upon return to Earth (Higashibata et al., 2006). RNA from three samples (4, 22 and 23) from the experimental containers ICE-08 and ICE-05 was extracted and purified from mixed stage animals as described (Selch et al., submitted for publication). The RNAs were used for microarray analysis using standard laboratory grown samples.
Two approaches were taken in analysing the microarray data. First, a biased approach of examining genes associated with biological relevant processes revealed no changes in gene expression for apoptosis related genes (Higashitani et al., 2005) and decreased expression of a number of “muscle” genes including those that encode MyoD and Myosin Heavy Chain (Higashibata et al., 2006). Second, an unbiased comparison of whole animal, whole genome transcript levels in three samples analyzed on a cDNA system and one sample on the Affymetrix system (Selch et al., submitted Advances in Space Research 2006). The unbiased analysis suggested that each population of worms had population specific changes as well as changes that were conserved between populations. In all, 21 genes displayed increased expression across all four samples and 16 displayed decreased expression. The data suggest that across populations worms display decreased expression of collagen (worm exoskeleton?), neuromuscular elements (including MyoD and Myosin Heavy Chain), and amino acid metabolism encoding genes as well as increased expression of intestinal components (worm equivalent of liver/kidney) and heat shock response genes.
2.3. Comparative gene expression profile analysis
The RNAs obtained from the pooled samples were hybridized to Affymetrix chips for both experiments (cDNA chips were also used for the C. elegans experiment). They were processed and analyzed following Affymetrix's recommendations. The equally normalized log 2 ratios of intensity of the hybridization of the space samples vs the ground control ones was used in the comparisons for the two systems, Drosophila and C. elegans. The chips detect a large number of probe-sets (14,010 for Drosophila and 22,625 for the nematode). Nevertheless, only a subset of orthologue genes can be identified in both systems. Using the Affymetrix software, it is possible to compare a relative large number of genes using the Affymetrix database. First we identified in the Affymetrix database the equivalent name of the wormbase ID gene names given in the ICE-1st analysis. Then we found the orthologue in the Drosophila genome as given by Affymetrix. As indicated below, this search did not cover the whole list of genes, probably due to the current annotation situation in both systems. Only approximately 20% of the genes in the ICE-1st chip could be identified as having Drosophila orthologues in the Affymetrix database and therefore could be compared. This limitation has to be kept in mind when evaluating the information given below. The results described in the following section correspond to this subset of genes.
3. Results
3.1. Similarities and differences in the gene expression profiles between D. melanogaster and C. elegans experiments
To establish the similarities and differences we employed two initial approaches and a third, complementary attempt. The reason for these different methods is that the only way to compare the results is to look into those genes that we can identify as orthologues.
The first approach was to take the normalized data obtained in the microarray analysis of the Gene experiment (Marco et al., in preparation) and to compare them to the normalized data of the ICE-1st experiment (Selch et al., submitted for publication). The second approach was to take the list of the genes that had been found to give a significant difference between the flight and the synchronous ground control in the C. elegans ICE-1st experiment and to look into the behaviour of their Drosophila orthologues in the Gene experiment. Finally, we tried to identify among the Caenorhabitis genes the orthologues of the genes that Beckingham's group had found to be affected in a genetic search in Drosophila identifying mutants with an altered geotropic response (Armstrong et al., 2006).
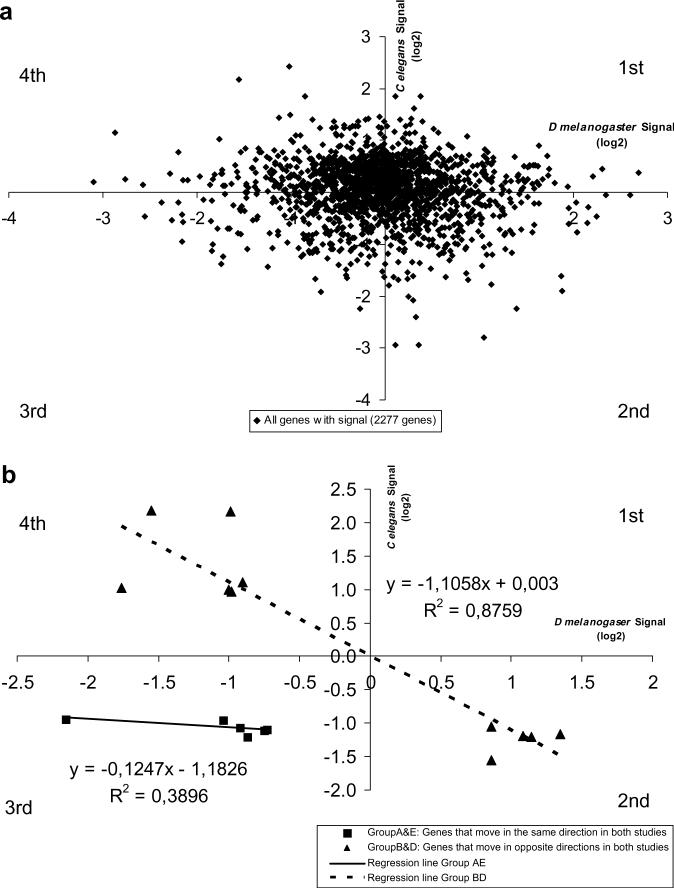
In the first approach, we started with the collection of C. elegans genes that produced reproducible, reliable data in the ICE-1st microarray data, namely 13,596 genes. For only 2277, (almost exactly 20%), we could identify the corresponding Drosophila orthologues be identified. Selch et al had used three different samples from the Delta Soyuz mission and analyzed them both independently and pooled. We decided to pool the data from the three samples as we had done with the Drosophila Gene samples and to compare the degree of variation against the controls. In Fig. 2a we can see plotted the results of such a comparison. As can also be seen in Table 1, the majority of Drosophila and Caenorhabditis orthologue genes only give relatively small changes, less than 2-fold up or down in both experiments. Moreover, they are similarly distributed among the four quadrants, indicating that there are little hints that they may be changing expression values in the same direction in both experiments. The same conclusion can be reached if we only plot the genes that give at least 2-fold changes in the ICE-1st and in the Gene experiment (Fig. 2b). Only six genes decreased both in the Gene and in the ICE-1st experiments (3d quadrant) and none increased (1st quadrant). The actual magnitude of the changes showed little correlation among the two sets. A few more (Table 1) did move oppositely in both experiments and the magnitude of the changes gave higher correlation values (2nd and 4th quadrants) but the relatively low cut-off utilized and the low number of genes made us to disregard this negative correlation.
Fig. 2.
(a) The genes for which orthologues have been found between Drosophila melanogaster and Caenorhabditis elegans are plotted indicating their relative changes (log 2 ratios) in the International Space Station. Drosophila Gene (abscissa) and Caenorhabditis ICE-1st (ordinates) against their respective controls. Being log 2 ratios positive values indicate that these genes are up-regulated in space, while negative values correspond to down-regulated ones. The number of genes in the different quadrants is as follows: 1st 590, 2nd 456, 3rd 475 and 4th 750. Twelve genes lay on the abscissa axis and three on the ordinates one. (b) The same plot limited to the genes that give relative changes of at least of ≈1.7-fold in both experiments. Most of them change in opposite directions. Only five decrease in both systems but with a weak correlation line. None increase.
Table 1.
Number of orthologue genes in the total identified 2286 genes distributed in the different categories in the Gene experiment and in the ICE-1st
| ISS D. melanogaster increase | ISS D. melanogaster decrease | ISS D. melanogaster no change | |
|---|---|---|---|
| ISS C. elegans increase | Group A (1st quadrant): 0 genes | Group B (4th quadrant): 9 genes | Group C: 80 genes |
| ISS C. elegans decrease | Group D (2nd quadrant): 5 genes | Group E (3rd quadrant): 6 genes | Group F: 125 genes |
| ISS C. elegans no change | Group G: 118 genes | Group H: 134 genes | Group I: 1809 genes |
A relatively low cut-off of ≈1.7-fold (increase or decrease) was used in reaching these numbers.
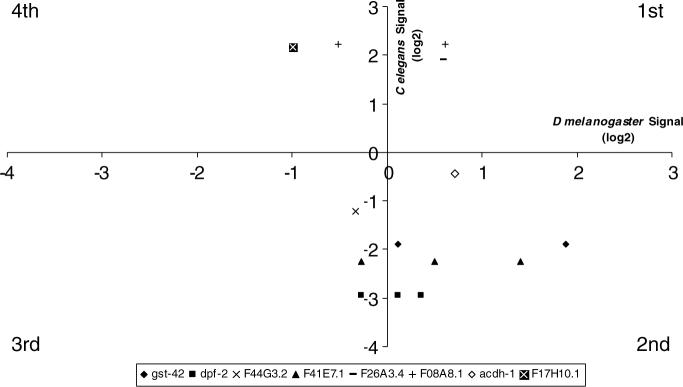
In the second approach, we started with the set of genes that had been suggested to be altered in the ICE-1st space experiment with C. elegans (Selch et al., submitted for publication). They choose a more restricting criterium, namely, those genes that differed four-fold in expression between the flight and the ground control (28 up and 23 down in sample 4, 25 up and 28 down in sample 22, 16 up and 23 down in sample 23). Only a handful of these genes were actually detected at the same time in the three replica samples (21 up and 16 down across the three populations). From this last group of genes, only 9 genes were among those that orthologues in D. melanogaster were identifiable in the Affymetrix data base, roughly again 20% of the initial list of genes. In Fig. 3, we plotted the expression changes in the Gene vs the ICE-1st experiment. Some of the genes had more than one potential orthologue form in the Drosophila Gene experiment, therefore, more than one dot appears in the case of four of the genes. The conclusion is the same, almost none of these genes changed significantly in the same direction in both of the experiments (Table 2).
Fig. 3.
The correlation among the genes identified in Caenorhabditis experiment (Selch et al., submitted for publication) as giving a large response in space, at least 4-fold increase and decrease, and the changes in the Gene experiment of their Drosophila orthologues. Only those found as orthologues are plotted. The points marked with the same type of symbols, correspond to genes that can be correlated withdifferent Drosophila ID gene numbers that show differences in relative expression.
Table 2.
Gene ontology of the 6 genes that do show at least ≈1.7-fold decreases and can be identified in both experiments (3d quadrant of Fig. 2b)
| C. elegans | D. melanogaster | Predicted function |
|---|---|---|
| T04F8.2 | CG9304 | Unknown |
| F10E7.4 (spon-1) | CG6953 (fat-spondin) | Cell adhesion//fat-spondin-like protein |
| C01B7.4 (tag-117) | CG31243 (cpo) CG32717 (sdt) CG1864 (Hr38) | Postsynaptic scaffolding-like protein |
| F35H12.4 | CG7004 (fwd) | Phosphatidyl kinase like protein |
| F31F6.6 (nac-1) | CG3979 (Indy) | Sodium sulphate membrane transporter like protein |
| F13D12.2 (ldh-1) | CG10160 (ImpL3) | Lactate dehydrogenase |
Finally, among the genes found to respond to changes in gravitaxis on the ground by the Beckingham's group (Armstrong et al., 2006) none of them was among the orthologue gene list between Caenorhabditis and Drosophila.
4. Discussion
While it is true that both experiments were able to show that multicellular organisms respond to the space environment by modifying the expression levels of a relatively large set of genes (Marco et al., in preparation; Selch et al., submitted for publication), the comparisons in this study fail to identify a common set of genes for the two main invertebrate model systems, C. elegans and D. melanogaster. It can be thought surprising that there is little overlap between the two experiments as one might suspect a common group of genes respond to the microgravity environment. On the other hand, there are many differences in the experimental design that may responsible of this apparently negative result.
First of all, it has been shown in the case of the Drosophila system that the cold exposure in the transport phase to the launching site synergistically increased the microgravity response in this system (Marco et al., in preparation). No such an effect has been reported for the C elegans system. On the one hand, C. elegans is more resistant than Drosophila to low temperatures of the magnitude used in this experiment. On the other, the C elegans animals had been maintained almost 12 days, at the normal temperature of 20 °C, much longer than the Drosophila experiment, giving more time for quenching down of any potential synergistic effects in Caenorhabditis.
Additional reasons for the difference can be found. The Drosophila experiment limited the exposure to microgravity to strictly synchronized samples during the pupal stage. The pupae are immobile inside their case and do not respond to changes in the gravity vector in ways similar to Drosophila imagoes or to C. elegans adults. The development of holometabolous organisms is logically different from the development of organisms such as the nematode. The C. elegans samples were, in this sense, a mixture of organisms at different levels of development. Almost 50% of the animals were mature adults and the other half was at different stages of development. Finally, the C. elegans samples were frozen on the ground after the experiment had been retrieved from space. The Drosophila pupae were fixed in the International Space Station, two and half days after launch and three after the cold transportation step had been completed. Finally, the set-up of the experiments is not exactly the same; in particular, there are differences between the MAMBAs and berlingots in the Drosophila experiment and the configuration of the ICE-1st experiment described above.
Thus, the idea that there may be a group of genes responding to the microgravity environment remains to be explored in more detail. More information in more strictly comparable conditions should be obtained. The Gene experiment is now being continued using ground simulation equipment, such as the random position machine (RPM) and magnetic levitation while the C. elegans results are being confirmed with another flight onboard the ISS. We already know that the Drosophila pupae respond almost identically to the neutralization of gravity achieved by the exposure to the RPM than what had been previously found in the International Space Station. Even the synergistically effect of the cold transport step has been reproduced on the ground conditions (Marco et al., in preparation). Running more experiments using these tools, complementary to the more expensive space experiments, could provide the background to make more direct proposals for future missions where the two key model systems can be compared.
A final point is the diffculty in deciding the potential orthologues of the genes responding to a similar parameter among different sequenced species. The availability of a specific bio-informatics tool capable of automatically doing so would be of great help for future comparisons and critical to the establishment of gene sets that can be connected with particular phenotypic properties of the organism(s).
Acknowledgments
The support of the all organizations and people involved in the Cervantes and Delta missions, the Spanish Space Program (Spanish Ministery of Education and Science), the Dutch Space Program, ESA, the CNES and NASA that made possible this work is gratefully acknowledged.
References
- Anthony P, Ausseil J, Bechler B, Benguría A, Blackhall N, Briarty LG, Cogoli A, Davey MR, Garesse R, Hager R, Loddenkemper R, Marchant R, Marco R, Marthy HJ, Perry M, Power JB, Schiller P, Ugalde C, Volkmann D, Wardrop J. Preservation of viable biological samples for experiments in Space laboratories. J. Biotechnol. 1996;47:377–393. doi: 10.1016/0168-1656(96)01363-6. [DOI] [PubMed] [Google Scholar]
- Armstrong JD, Texada MJ, Munjaal R, Baker DA, Beckingham KM. Gravitaxis in Drosophila melanogaster: a forward genetic screen. Genes Brain Behav. 2006;5:222–239. doi: 10.1111/j.1601-183X.2005.00154.x. [DOI] [PubMed] [Google Scholar]
- Herranz R, Husson D, Villa A, Pastor M, Medina FJ, Marco R. Modifications in basic handling techniques to study the consequences of the Drosophila melanogaster. J. Gravit. Physiol. 2005:1251–1260. [Google Scholar]
- Higashibata A, Szewczyk NJ, Conley CA, Imamizo-Sato M, Higashitani A, Ishioka N. Decreased expression of myogenic transcription factors and Myosin Heavy Chains in Caenorhabditis elegans muscles developed during spaceflight. J. Exp. Biol. 2006;209:3209–3218. doi: 10.1242/jeb.02365. [DOI] [PubMed] [Google Scholar]
- Higashitani A, Higashibata A, Sasagawa Y, Sugimoto T, Miyazawa Y, Szewczyk NJ, Viso M, Gassett G, Eche B, Fukui K, Shimazu T, Fujimoto N, Kuriyama K, Ishioka N. Checkpoint and physiological apoptosis in germ cells proceeds normally in spaceflown Caenorhabditis elegans. Apoptosis. 2005;10:949–954. doi: 10.1007/s10495-005-1323-3. [DOI] [PubMed] [Google Scholar]
- Marco R, Husson D, Herranz R, Mateos J, Medina FJ. Drosophila melanogaster and the future of ‘evo–devo’ biology in space. Challenges and problems in the path of an eventual colonization project outside the earth. Adv. Space Biol. Med. 2003;9:41–81. doi: 10.1016/s1569-2574(03)09003-8. [DOI] [PubMed] [Google Scholar]
- Selch F, Higashibata A, Imamizo-Sato M, Higashitani A, Ishioka N, Szewczyk NJ, Conley CA. Genomic Response of C. elegans to spaceflight. Adv. Space Res. doi: 10.1016/j.asr.2007.11.015. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk NJ, Tillman J, Conley CA, Granger L, Segalat L, Higashitani A, Honda S, Honda Y, Kagawa H, Higashibata A, Fujimoto N, Kuriyama K, Ishioka N, Fukui K, Baillie D, Rose A, Gasset G, Eche B, Chaput D, Viso M. Description of International Ceonorhabditis elegans experiment first flight (ICE-FIRST). Adv. Space Res. doi: 10.1016/j.asr.2008.03.017. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tixador R, Raffin J, Richoilley G, Kordium VA, Kojarinov V, Maneko G. Ampoule de verre cassable contenant un liquide sous pression, éjectable en totalité lors de la cassure de l’ampoule. Brevet B. 148 no 8007471. Innov. Tech. Biol. Med. 1981;2:12–14. [Google Scholar]
- van Loon JJ, Medina FJ, Marco R, editors. “Experiments in the European Soyuz missions to the International Space Station (2001−2005)”. Microgravity Sci. Technol. (special issue) in press. [Google Scholar]
- Zhao Y, Lai K, Cheung I, Youds J, Tarailo M, Tarailo S, Rose A. A mutational analysis of Caenorhabditis elegans in space. Mutat. Res. 2006;601:19–29. doi: 10.1016/j.mrfmmm.2006.05.001. [DOI] [PubMed] [Google Scholar]