Abstract
Although bulk chromatin is thought to have limited mobility within the interphase eukaryotic nucleus, directed long-distance chromosome movements are not unknown. Cajal bodies (CBs) are nuclear suborganelles that nonrandomly associate with small nuclear RNA (snRNA) and histone gene loci in human cells during interphase. However, the mechanism responsible for this association is uncertain. In this study, we present an experimental system to probe the dynamic interplay of CBs with a U2 snRNA target gene locus during transcriptional activation in living cells. Simultaneous four-dimensional tracking of CBs and U2 genes reveals that target loci are recruited toward relatively stably positioned CBs by long-range chromosomal motion. In the presence of a dominant-negative mutant of β-actin, the repositioning of activated U2 genes is markedly inhibited. This supports a model in which nuclear actin is required for these rapid, long-range chromosomal movements.
Introduction
The eukaryotic cell nucleus is an extremely complex, highly organized organelle. Within the nucleus, interphase chromosomes occupy discrete domains called chromosome territories (Cremer et al., 2006; Misteli, 2007). In mammalian cells, photobleaching analyses have shown that interphase chromatin is relatively immobile; the diffusion of a given locus is typically constrained within a radius of 0.4 μm (Abney et al., 1997; Chubb et al., 2002). These constraints not only reflect the boundaries of the chromosome territory but also can involve attachment of the locus to immobile nuclear substructures such as nucleoli or the nuclear periphery (Chubb et al., 2002; Gasser, 2002).
The radial positioning of genes within the nuclear volume has been linked to transcriptional activity (Thomson et al., 2004). Active genes and gene-rich chromosome regions tend to be localized within the nuclear interior, whereas transcriptionally silent loci or gene-poor regions are preferentially localized at the periphery (Chubb et al., 2002; Gasser, 2002). Importantly, the associations between various loci and subnuclear domains are dynamic and can change in response to cellular signals. Large-scale movement of a tagged chromosomal site from the periphery to the interior (over distances of 1–5 μm) was shown to depend on local chromatin decondensation (Chuang et al., 2006). The molecular mechanisms responsible for this directed chromosomal repositioning are not known but are thought to involve actin and myosin (Chuang et al., 2006; Hofmann et al., 2006).
Cajal bodies (CBs) are dynamic nuclear subdomains involved in the biogenesis of several classes of small RNPs (Matera and Shpargel, 2006; Stanek and Neugebauer, 2006; Matera et al., 2007). In human cells, CBs associate with specific genetic loci, including histone and small nuclear RNA (snRNA) genes (Frey and Matera, 1995). The association of CBs and specific gene loci has been conserved among vertebrates and invertebrates (Gall et al., 1981; Callan et al., 1991; Liu et al., 2006; Matera, 2006). Importantly, colocalization of CBs with snRNA genes is transcription dependent and requires expression of the snRNA coding region (Frey et al., 1999; Frey and Matera, 2001). These studies underline a requirement for specific factors within CBs that recognize nascent or newly transcribed RNAs to mediate interaction with their cognate genes (Frey et al., 1999; Frey and Matera, 2001). The reasons why CBs associate with histone and snRNA genes are not known, but current theories include the facilitation of transcription, RNA processing, and/or assembly of RNP complexes (Gall, 2001; Cioce and Lamond, 2005; Matera and Shpargel, 2006; Stanek and Neugebauer, 2006). Furthermore, the dynamics and mechanics of these associations are also unclear. Are CBs nucleated at sites of active snRNA and histone gene transcription in a manner similar to nucleoli and ribosomal RNA genes, or are extant CBs recruited to these chromosomal sites? If recruited, do the CBs move to the genes, or do the chromosome loci move to the CB?
To address these questions, we created an experimental system to probe the dynamic interaction of CBs with a target gene during transcriptional activation in living cells. We constructed stable cell lines containing inducible arrays of U2 snRNA genes marked with lac operator repeats that can be visualized with fluorescent protein fusions to the lac repressor. When cotransfected with a differentially fluorescent protein–tagged CB marker protein, this system allows visualization of the movements of CBs and U2 genes in response to transcriptional activation using live 4D time-lapse microscopy.
In this study, we show that before activation, U2 arrays are transcriptionally silent and do not colocalize with CBs. After induction of transcription, U2 gene arrays become closely and stably associated with CBs. Moreover, the activated U2 arrays move by long-range (2–3 μm) directed motion with apparent velocities of 0.1–0.2 μm/min. In the presence of a dominant-negative (dn) mutant of actin, the repositioning of activated U2 arrays was markedly disrupted. These findings represent the first demonstration of the repositioning of an actively transcribed locus in living mammalian cells and suggest a role for nuclear actin in this process.
Results and discussion
To investigate the mechanism whereby a specific gene locus interacts with a nuclear suborganelle, we developed an experimental system that mimics the association pattern of endogenous U2 snRNA genes with CBs. Previous studies showed that transcription of the U2 coding region from a viral RNA polymerase II promoter was sufficient for association with CBs (Frey and Matera, 2001). Therefore, we constructed a tandem array of 16 tetracycline (Tet)-inducible U2 snRNA minigenes flanked by 256 copies of the lac operator cloned into a bacterial artificial chromosome vector (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200710058/DC1; see Materials and methods for additional details of vector, U2 array, and cell line construction).
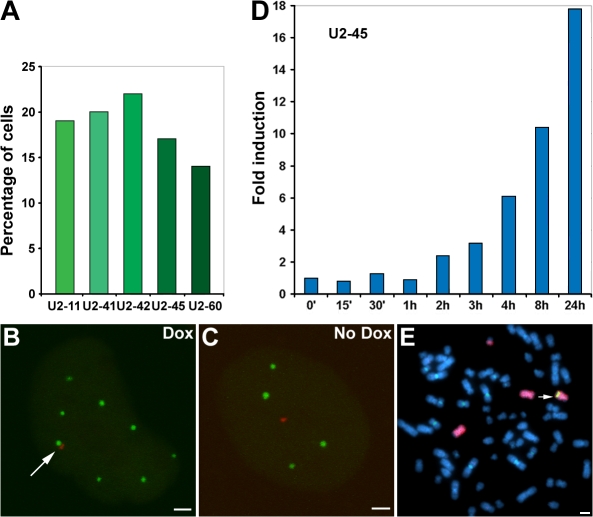
Five independent stable cell lines containing these arrays were generated and characterized. To determine whether expression of the exogenous U2 arrays was sufficient to promote CB association, we cotransfected cells with the reverse Tet-activator (pTet-ON; to activate U2 transcription), YFP-NLS-lac repressor (to mark the transgene insertion), and CFP-coilin (to mark CBs). Cells were treated with doxycycline (Dox) overnight, fixed, and imaged by fluorescence microscopy. The exogenous U2 arrays displayed CB colocalization frequencies similar to those of the individual endogenous U2 gene loci in the parental HeLa line (Fig. 1 A; Frey and Matera, 1995). Importantly, the CB–U2 array associations were Dox responsive (Fig. 1, A–C), confirming the finding that U2 transcription is required for CB association with the transgenic arrays (Frey and Matera, 2001). Note that in the absence of reverse Tet-activator protein (plus or minus Dox), the U2 arrays were essentially silent (unpublished data). Quantitative RT-PCR analysis of one of the cell lines (TetU2-45) showed that transcription of the U2 arrays was easily detectable within 2 h of Dox induction and increased by >10-fold after 8 h (Fig. 1 D).
Figure 1.
Characterization of inducible HeLa TetU2 stable cell lines. (A) Five TetU2 cell lines were generated and scored for association (as measured by overlap between the fluorescent protein–tagged coilin and lac repressor signals) with CBs after Dox induction. In the absence of Dox, no CB associations were observed in any of the cell lines (n > 50 cells per line). (B–E) TetU2-45 cells were chosen for in-depth analysis. (B) After doxycycline (Dox) induction, the U2 array (marked with YFP-NLS-lac repressor; shown in red) associated with a CB (marked with CFP-coilin; shown in green; arrow). (C) In the absence of Dox, the uninduced U2 array failed to associate with CBs. (D) Quantitative RT-PCR analysis demonstrates the inducibility of the U2 array in the TetU2-45 cell line (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200710058/DC1). (E) Mitotic chromosome spreads reveal that the U2 array (green FISH signal) in the TetU2-45 cell line integrated at a single site on distal chromosome 7q (red chromosome painting FISH signal; arrow). Bars, 2 μm.
We used live 4D cell imaging to study the dynamics of the interaction between CBs and U2 genes and found that these associations are highly photosensitive. In fact, initial attempts at live cell imaging using conventional microscopes failed to capture an association event. To reduce potential photosensitive effects of shorter wavelength light (Chuang et al., 2006), we switched the marker protein labels to mCherry-NLS-lac repressor and GFP-coilin and reduced the overall light flux through the sample by using a spinning disk confocal microscope. Notably, the spinning disk system was capable of reproducing the associations with frequencies similar to those of cells (fixed or living) that had not been exposed to light. We measured relative 3D distances between CBs and the marked U2 array in TetU2-45 cells in response to Dox induction. We analyzed videos of ∼15 cells and plotted the relative distances between CBs and U2 arrays over time. An example of such a plot is shown in Fig. 2 A, where the relative distance between the array and the nearest CB was fairly constant in the absence of Dox, indicating that the positions of these two nuclear substructures were not correlated (Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200710058/DC1).
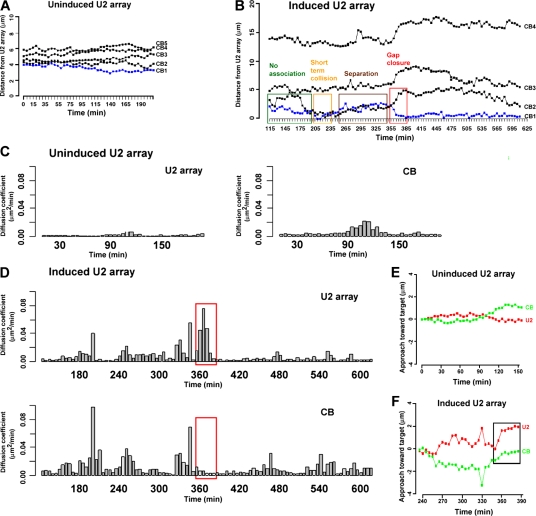
Figure 2.
3D tracking analysis of the interaction between a CB and the U2 array. (A) The relative 3D distance between the U2 array and the nearest CB is fairly constant in the absence of Dox. (B) After Dox induction, the relative 3D distance between the U2 array and the nearest CB progressively fluctuates, which is later followed by a rapid decrease in separation during the final approach (gap closure; boxed), after which time the CB and the U2 array remain stably associated. For simplicity, only plots of the three closest CBs (plus one distal CB) are shown. (C) Analysis of the movements of the U2 array and the nearest CB by calculating their diffusion coefficients over 20-min moving time windows illustrates the limited mobility of the uninduced U2 array and the CB. (D) In contrast, the induced U2 array shows transient periods of increased mobility that correlate precisely with the time frame of the final approach toward a CB (red boxes). (E and F) Targeted component tracking analysis was used to determine whether the CB moved toward the U2 array or whether the array moved toward the CB. (E) The uninduced U2 array and the closest CB exhibit restricted targeting. (F) The activated U2 array exhibits significant progression toward the CB during gap closure (boxed), whereas the CB remains relatively static.
After induction, the relative 3D distance between the U2 array and the nearest CB progressively fluctuated, with decreasing amplitude over time (Fig. 2 B). Short-lived collisions of both subdomains (as measured by the edge to edge distance between the two objects) were typically detected 3–4 h after induction and were usually followed by an abrupt increase in relative distance. At later time points (6–7 h after induction), the oscillations decreased in amplitude, and the final gaps (1.5–3 μm) were rapidly closed. In the example shown, the relative 3D distance between the U2 array and the associated CB was markedly reduced during the final approach (Fig. 2 B, boxed regions), whereas the relative distances to each of the other CBs actually increased, illustrating the specificity of the movement. Once established, the associations lasted for many hours (Fig. 2 B and not depicted). At later time points, the movements of the array and the associated CB were completely correlated, indicating that they behave as a single nuclear subdomain. As expected, there was no correlation between the movements of the array and the unassociated CBs within the same cell in the presence or absence of Dox.
We analyzed the movements of the U2 array and the nearest CB by calculating their diffusion coefficients over short (20 min) moving windows that scan the entire time course. In the absence of induction (Fig. 2 C) or after the establishment of a stable association (Fig. 2 D), CBs tend to be more mobile than U2 arrays. However, we identified transient periods of increased mobility of the U2 array that correlated well with the time frame of the final approach toward a CB (Fig. 2 D, red box). In other words, just before forming a stable association, only the U2 array displayed increased mobility.
Diffusion coefficients do not provide directional information. Therefore, to determine whether the CB moved toward the U2 array or whether the array moved toward the CB, we developed a targeted component tracking analysis to extract the directional components from the 4D datasets. At any given time point, an incremental 3D movement consists of two components with respect to a target object: a component along the direction toward (or away from) the target and a perpendicular (neutral) component that does not change the distance between the two objects. This analysis takes into account the dynamic 3D tracks of objects by recalculating the target direction at each time point. Therefore, the targeted component tracking plot shows the progression of an object toward or away from its target in a dynamic environment. When applied to the uninduced time-lapse dataset, the U2 array was relatively static throughout, and the nearest CB moved nominally toward the U2 array at the end of the time course (90–150 min; Fig. 2 E), reflecting the constrained diffusion of the CB, which was illustrated by a corresponding increase in the diffusion coefficient (Fig. 2 C). In contrast, the activated U2 array showed significant progression toward the CB during the 260–390-min time window, whereas the CB actually moved away from the U2 array until the 350-min time point and then remained in place (Fig. 2 F, box; see Fig. S2 for another example of this behavior; available at http://www.jcb.org/cgi/content/full/jcb.200710058/DC1). Thus, during the final approach, the U2 array moved rapidly toward the CB. The overall drift of the cell during the experiment was corrected at each time point by calculating the centroid (a reference point) using the 3D coordinates of the CBs and the U2 array.
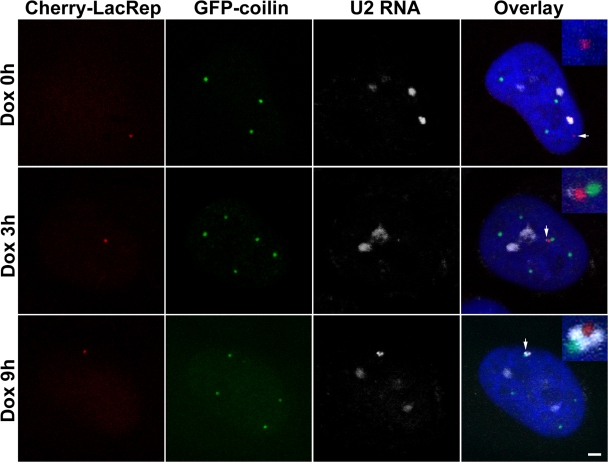
During the course of the association between the array and the CB (e.g., 385–600 min; Fig. 2 B), the topology of the two signals frequently fluctuated over time, suggesting some flexibility in their tethering elements. We investigated this aspect of the association using RNA FISH. At later time points (6–12 h after induction), small gaps were often observed between the associated CB and the U2 gene array. The gaps were consistently filled by the U2 RNA FISH signal (Fig. 3). At earlier time points (3 h), the RNA signal was considerably smaller and was randomly oriented with respect to the CB (Fig. 3). Previous experiments using GFP-coilin showed that most CBs (>80%) are relatively immobile; their movements are characterized by constrained diffusion within the interchromatin space (Platani et al., 2002). Interestingly, CB mobility was increased by the depletion of ATP or treatment with actinomycin D, suggesting that tethering to chromatin is transcription dependent (Platani et al., 2002). Our observations are consistent with these studies and a previous study (Frey and Matera, 2001) suggesting that transcripts derived from the U2 minigene arrays are responsible for tethering CBs to this locus.
Figure 3.
CBs are tethered to the U2 array by nascent transcripts. TetU2-45 cells were analyzed by RNA FISH and immunocytochemistry after 0, 3, and 9 h of Dox induction. The U2 array (marked by the mCherry-lac repressor) is shown in red, the CBs (marked by GFP-coilin) are shown in green, and the RNA FISH (using an RNU2 probe) signal is shown in white. In the absence of Dox, the U2 array (arrow) and RNU2 signals (white) do not overlap. The RNU2 signal in the uninduced cell (Dox 0 h) is presumably derived from the three endogenous RNU2 loci on chromosome 17. After 3 h of Dox activation (Dox 3 h), the U2 RNA FISH signal is detectable but not robust (arrow) and was randomly oriented with respect to the CB. After 9 h of activation (Dox 9 h), the RNA FISH signal (arrow) was invariably located in the space around and between the CB (green) and the U2 array (red). Bar, 2 μm.
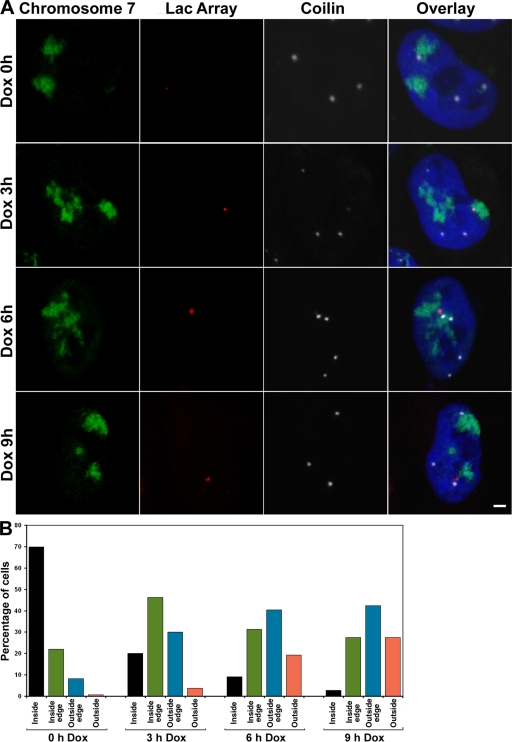
Because dynamic targeting analysis (Figs. 2 F and S2) suggested that the U2 array was primarily responsible for rapidly closing the remaining gap between the gene locus and the CB, we assayed the relative positioning of the U2 array within the chromosome territory before and after transcriptional induction. As shown in Fig. 1 E, the transgenic U2 array in TetU2-45 cells is located on the distal q arm of chromosome 7. Therefore, we performed 3D DNA FISH analysis using a chromosome 7 painting probe and a probe specific for the lac operators present in the transgenic array. In the absence of Dox, the inactive U2 array was primarily located in the interior of the chromosome territory (Fig. 4). After transcriptional induction, the U2 array was progressively repositioned toward the outer edge (and often looped outside) of the territory (Fig. 4). The net effect of these movements was to reposition the U2 array toward the nuclear interior (Fig. 4 and not depicted). These data are consistent with previous findings that transcriptionally inactive sequences localize to the nuclear periphery, whereas actively transcribed loci localize within the interior (Chambeyron and Bickmore, 2004; Chuang et al., 2006). Furthermore, our in vivo analysis suggests the existence of an active mechanism for repositioning genes after transcriptional induction.
Figure 4.
Intranuclear repositioning of the U2 array after transcriptional induction. The relative positioning of the U2 array within the chromosome 7 territory was analyzed before and after Dox induction. TetU2-45 cells were analyzed by DNA FISH and immunocytochemistry using a chromosome 7 painting probe (green), a lac operator probe (red), and antibodies targeting coilin (white). (A) The inactive U2 array (Dox 0 h) is primarily located in the interior of the chromosome 7 territory. After induction, the U2 array is progressively repositioned toward the inner edge of the chromosome 7 territory (Dox 3 h) and then toward the outer edge of the territory (Dox 6 h) and is often looped entirely outside of the territory (Dox 9 h). Note that the CB-associated U2 arrays were predominantly located either near the outer edge or outside of the chromosome territory. (B) The corresponding quantification of the relative repositioning of the U2 array with respect to the chromosome 7 territory was scored and graphed. Note that this analysis was performed using 3D z-axis projections, as 2D maximum intensity projections can lead to inaccurate scoring. n > 85 cells per time point. Bar, 2 μm.
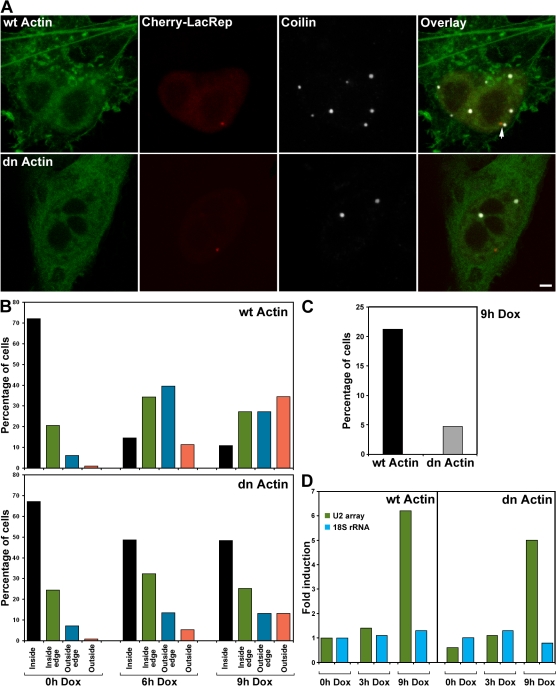
To investigate this mechanism, we tested whether the association between U2 genes and CBs is dependent on nuclear actin. Chuang et al. (2006) recently showed that intranuclear repositioning of a chromosomal locus after targeting a transcriptional activator to a promoterless cassette was perturbed by overexpression of a dn actin mutant. Therefore, we transiently overexpressed wild-type (wt) YFP-actin or a nonpolymerizable mutant, YFP-actin(G13R), along with the mCherry-NLS-lac repressor in TetU2-45 cells. The dn actin mutant not only abolished the association between CBs and the U2 array (Fig. 5 C) but also inhibited the repositioning of the locus within the chromosome 7 territory (Fig. 5 B). In the presence of wt YFP-actin, the U2 array was repositioned normally (compare Figs. 4 B with 5 B); however, in the presence of the mutant actin, the intranuclear repositioning of the U2 array was significantly reduced. To test the effect of actin overexpression on transcription of the U2 array, we transfected with wt or mutant YFP-actin constructs and sorted cells by FACS (gating on YFP) such that all cells analyzed were expressing the desired constructs. After sorting, total RNA was isolated, and array-specific U2 RNA transcription was analyzed by quantitative PCR. No appreciable difference was observed (Fig. 5 D). Thus, the overexpression of dn actin has little overall effect on transcription of the U2 array but has a significant effect on intranuclear repositioning and CB association.
Figure 5.
The association between U2 genes and CBs was disrupted by the expression of a β-actin mutant. TetU2-45 cells were transfected with mCherry-lac repressor, reverse Tet-activator, and wild-type (wt) or dominant-negative (dn) YFP-actin constructs. (A) In the presence of wt YFP-actin, association of CBs and U2 arrays was not disrupted (arrow). However, in the presence of the nonpolymerizable dn YFP-actin, the association between CBs and the U2 array was significantly diminished. (B) Quantification of the relative repositioning of the U2 array within the chromosome 7 territory. Cells were scored (n > 85 cells per time point) on 3D z-axis projections as in Fig. 4 B in the presence of wt actin or dn actin. (C) Quantification of the effect of wt and dn YFP-actin on association of the U2 array with CBs. n > 250 cells. (D) The effect of dn YFP-actin on transcription of the U2 array as analyzed by quantitative PCR. The levels of 18S ribosomal RNA were monitored as a control. Bar, 2 μm.
In summary, we have shown that preexisting CBs are recruited to sites of active snRNA gene transcription. Although diffusion of CBs to within striking distance of the gene locus may be important, our 4D imaging analyses reveal that stable interphase associations between CBs and U2 genes are the result of rapid and directed long-range chromosomal movements. Importantly, the interaction of CBs and U2 genes was inhibited by the overexpression of a nonpolymerizable actin mutant. Together with previous results (Chuang et al., 2006), our data support a model wherein nuclear actin plays an important role in repositioning chromosomal loci in response to the chromatin remodeling that is associated with transcriptional activation.
Materials and methods
Vector construction
To stabilize its expression in bacteria, a 10-kb insert containing a 256-copy lac operator array was transferred from pSV2-8.32 (gift of A. Belmont, University of Illinois, Champaign, IL) into pBeloBAC11 (New England Biolabs, Inc.) by BamHI and SphI restriction followed by ligation to create pJO1. A neomycinr/dihydrofolate reductase cassette was excised from pE1 (gift of A. Weiner, University of Washington, Seattle, WA) by restriction with BamHI and BglII and ligated into the BamHI site of pJO1 to generate pJO3. An alternative version of pJO3 (pJO3-MCS) containing an expanded eight-cutter multiple cloning site is also available.
To construct a Tet-responsive system, primers were used to introduce BglII and SgfI sites flanking the Tet-responsive elements and minimal cytomegalovirus (CMV) promoter of pTRE2 (Clontech Laboratories, Inc.). PCR was conducted using these primers, and the resulting fragment was cloned into pCR-Blunt II-TOPO with the Zero Blunt TOPO Cloning kit (Invitrogen). The Tet-CMV fragment was then subcloned into pRc/RSV-U2 (Frey and Matera, 2001) at BglII and SgfI. The use of SgfI places the U2 snRNA gene precisely at the +1 start site of transcription; the insert was confirmed by sequencing. The Tet-CMV U2 cassette was then excised with BamHI and BglII and ligated into the BamHI and BglII sites of pUC18 Bgl (gift of A. Weiner) to create pJO8.1. Multimerization of the Tet-CMV U2 cassette was then conducted by serial digestion and ligation reactions using BamHI, BglII, and HindIII. An eight-copy Tet-CMV U2 cassette was excised with BamHI and BsrBI and ligated into pVJ104 (gift of H. Willard, Duke University, Durham, NC) at BamHI and SwaI. One round of multimerization yielded a cassette containing 16 copies of Tet-inducible U2 genes. This 16-copy array was then excised with BamHI and HpaI and ligated into pJO3, yielding pJO11.
An ampicillinr/blasticidinr cassette was added to the vector as follows: QuikChange mutagenesis (Stratagene) was used to introduce BamHI into pGEX-3X (Invitrogen) at nucleotide 2,356, yielding pGEX-3X- Bam. A blasticidinr cassette was excised from pPAC4 (gift of H. Willard) with EcoRI and ligated into pGEX-3X-Bam, yielding pGEX-3X-Bam-blast. The ampicillinr/blasticidinr cassette from pGEX-3X-Bam-Blast was then ligated into pJO11 at BamHI, yielding pJO11B (Fig. S1).
Establishment of stable cell lines
pJO11B was digested with SpeI, purified by phenol/chloroform extraction, and concentrated by ethanol precipitation. Self-ligation using 25 μg of recovered linear DNA was performed using T4 DNA ligase (New England Biolabs, Inc.) for 15 min at 25°C. The resulting arrays were ethanol precipitated and transfected into HeLa cells (American Type Culture Collection) at 50% confluency using Calcium Phosphate Transfection reagent (Invitrogen). After 48 h, cells were seeded to 150-mm dishes (Thermo Fisher Scientific) and selected with 4 μg/ml blasticidin (Invitrogen). After 10 d of selection, isolated colonies were picked to 24-well dishes (Thermo Fisher Scientific). Cells were selected for an additional 7 d, replica plated, and frozen in DME containing 10% DMSO. Cell lines were then seeded to eight-well chamber slides (Thermo Fisher Scientific) and transiently transfected (Superfect) with YFP-NLS-lac repressor. After fixation, cell lines were examined by fluorescence microscopy to confirm the presence of the lac operator array. Positive insertion was scored by the presence of one or more nuclear YFP foci in addition to diffuse nuclear YFP fluorescence.
Plasmid construction
The GFP-coilin, YFP–β-actin, and YFP–dn mutant actin (G13R; provided by P. de Lanerolle, University of Illinois, Chicago, IL) constructs were described previously (Hebert and Matera, 2000; Chuang et al., 2006). The untagged nuclear β-actin and G13R mutant actin cDNAs were PCR amplified and resubcloned into pEGFP-N3 as EcoRI–BamHI fragments. The mCherry tag was added to NLS-lac repressor as an NheI–BsrGI fragment.
Cell culture and transfection
HeLa cells stably expressing the U2 array were grown in DME (Invitrogen) supplemented with 10% FCS (Invitrogen), 1% glutamine, penicillin, and streptomycin at 37°C in 5% CO2. For live cell imaging, cells were plated in Lab-Tek II chambers (Nalgene) overnight and transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Before imaging, the medium was changed to DME with 25 mM Hepes without phenol red (Invitrogen).
Live cell imaging
Cells were visualized using a fast-spinning wheel confocal system (UltraVIEW ERS; PerkinElmer) equipped with a progressive interline CCD camera (Orca ERII; Hamamatsu) and filters suitable for the visualization of both EGFP and mCherry. Samples were illuminated with either the 488- or 568-nm laser lines from a krypton/argon laser. Live images were collected as vertical z-stacks of 40 focal planes that were projected in three dimensions every 5 min for up to 8.5 h. All images were collected using a 100× plan-Apochromat 1.45 NA objective (Carl Zeiss, Inc.). The temperature of the sample was maintained at 37°C using a hot air blower (Nevtek) or a temperature-controlled sample chamber for live cell imaging (Carl Zeiss, Inc.) and an objective heater (Bioptechs).
Image processing and 3D distance position measurements
Each time point in the time-lapse video was projected as a combined maximum intensity projection for the GFP and mCherry channels. For 3D tracking analysis, time-lapse videos were exported from UltraVIEW imaging software (PerkinElmer) as 16-bit TIFF files and imported into Imaris 4.5 (Bitplane), and 3D surface rendering was performed for both the GFP and mCherry channels. The x, y, and z coordinates of the centroid of the U2 array and the CBs in each cell for each time point were individually calculated.
3D mobility analysis of the U2 array and CBs
Distances (edge to edge) between objects were calculated as the distance between the two centers of mass minus the radii of the objects. Therefore, this distance measurement is positive for two objects with separated boundaries, zero when they touch the boundaries, and negative when they overlap partially or completely. In a few cases in which the U2 array was ellipsoidal rather than spherical in 3D, the radius from the axis in the direction of the nearest CB was used. To adjust the 3D coordinates for the global drift of the cell, the 3D coordinates of the centroid (calculated as described in the previous paragraph) were subtracted from the coordinates of each object. These drift-adjusted 3D coordinates were used for the diffusion and tracking analyses. The diffusion coefficient was calculated for a 20-min time window centered at each time point because the object mobility could change during our long time-lapse experiments. For a given time window, the diffusion coefficient was obtained by estimating the slope of the mean square displacement curve over time.
Targeted component tracking analysis
We developed a targeted component tracking analysis method to isolate the directional movement from the total object movement over time. Each object displacement vector at any given time point can be decomposed into two vector components: one along the direction toward the target object (the targeted component) and the other in the plane perpendicular to this direction (the neutral component). Separation of the targeted component from the total movement makes it easier to assess whether or not a given displacement of a mobile object resulted in a productive approach toward the target. For two mobile objects associating with each other (as shown by the decreasing distance between them), we identify the object whose targeted component tracking shows greater movement than the one that approached the other.
Hybridization probes
The U2 hybridization probe was generated using a plasmid containing a 1.7-kb fragment of the human RNU2 repeat (Frey et al., 1999) by nick translation with biotin-16-dUTP. The lac operon hybridization probe was generated using the plasmid pSV2-8.32 containing an array of 256 lac operon repeats by nick translation with biotin-16-dUTP.
RNA FISH
Cells grown on glass coverslips were washed with PBS and fixed with 4% PFA in PBS for 15 min at room temperature. After rinsing in PBS, cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min on ice and were washed with PBS and finally with 2× SSC. The hybridization mixture was prepared using 100 ng of biotinylated probe and 20 μg of yeast tRNA per coverslip and was dried under vacuum. 6 μl of deionized formamide was added per coverslip, and the mixture was denatured for 10 min at 80°C. The probe was immediately chilled on ice/water, and the hybridization mixture was made to a final concentration of 2× SSC, 10 mM Tris-HCl, pH 7.2, 1 mM EDTA, and 10% dextran sulfate. A hybridization mixture (20 μl) was placed onto each coverslip, and coverslips were sealed to microscopic slides by rubber cement and allowed to incubate in a chamber moistened with 2× SSC overnight at 37°C. The coverslips were rinsed twice for 15 min with 2× SSC/0.05% Triton X-100, for 15 min with 2× SSC, and for 15 min with 4× SSC at room temperature. The cells were incubated with streptavidin-conjugated Cy5 in 4× SSC/0.25% BSA for 2 h and rinsed four times for 15 min with 4× SSC and PBS. Coverslips were mounted in Vectashield with DAPI (Invitrogen) and sealed by nail polish. Cells were observed on a microscope (LSM510 Meta; Carl Zeiss, Inc.) using a 100× 1.45 NA objective.
3D DNA chromosome painting
Cells were grown on coverslips and transfected with pTet-On and plasmids encoding the untagged wt actin or the dn actin mutant by Lipofectamine 2000 overnight. Cells were washed with PBS, fixed in 4% PFA in PBS for 10 min, washed in PBS, and permeabilized in 0.5% saponin/0.5% Triton X-100 in PBS for 20 min. After washing in PBS, cells were incubated in 20% glycerol in PBS for 30 min, freezed/thawed five times by dipping into liquid nitrogen, and thawed at room temperature. Subsequently, cells were incubated in PBS for at least 30 min to remove glycerol, incubated in 0.1 N HCl for 15 min, washed in PBS several times, and washed in 2× SSC for 10 min and in 50% formamide/2× SSC for 30 min. For one 12-mm round coverslip, ∼150 ng of biotinylated DNA probe generated from a plasmid containing 256 lac operator repeats were precipitated with C0t1 DNA (Roche) and tRNA in ice-cold absolute ethanol by sodium acetate, spun down, and resuspended in 7 μl of hybridization solution (50% formamide and 2× SSC) containing an FITC-labeled human whole chromosome 7–specific painting probe (Open Biosystems). The cocktail was denatured at 95°C for 5 min, put briefly on ice, incubated with cells at 75°C for 5 min, sealed with rubber cement, and hybridized in a moisture chamber for three nights at 37°C.
After hybridization, cells were washed three times in 50% formamide and 2× SSC for 5 min at 45°C, washed three times in 1× SSC for 5 min at 60°C, and washed in 0.05% Tween 20 and 4× SSC for 5 min. Then, cells were blocked in 3% BSA, 0.05% Tween 20, and 4× SSC for 20 min. After blocking, cells were incubated with the monoclonal antibody against coilin (Sigma-Aldrich) and avidin conjugated with Texas red for 1 h and were washed three times in 4× SSC/0.05% Tween 20 for 5 min. Cells were then incubated with donkey anti–mouse secondary antibody conjugated with Cy5 (Jackson ImmunoResearch Laboratories) for 1 h, washed three times in 0.05% Tween 20/4× SSC for 5 min, and mounted in Vectashield with DAPI (Invitrogen). Cells were observed on an LSM510 Meta microscope using a 100× 1.45 NA objective.
Online supplemental material
Fig. S1 shows the construction and characterization of inducible U2 arrays. Fig. S2 shows 3D tracking analysis of the interaction between a CB and the U2 array. Video 1 shows the uninduced TetU2-45 HeLa stable cell line coexpressing Cherry-LacRep and GFP-coilin monitored by live time-lapse microscopy. Video 2 shows the TetU2-45 HeLa stable cell line coexpressing Cherry-LacRep and GFP-coilin induced for 2 h by Dox and monitored by live time-lapse microscopy. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200710058/DC1.
Supplementary Material
Acknowledgments
We thank P. de Lanerolle, A. Weiner, and H. Willard for reagents. We are particularly grateful to K. Meaburn for assistance with 3D DNA chromosome painting and to V. Barr for help with the fast-spinning disc confocal microscope system. We also thank T. Misteli for helpful suggestions and critical reading of the manuscript.
This work was supported, in part, by the National Institutes of Health (NIH; grants R01-GM53034 and R01-NS41617 to A.G. Matera), the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (grants to G.L. Hager and T. Ried), and by startup funds from the Rosalind Franklin University of Medicine and Science (to M. Dundr). J.K. Ospina was supported, in part, by NIH predoctoral traineeship T32-GM08613.
M. Dundr and J.K. Ospina contributed equally to this paper.
Abbreviations used in this paper: CB, Cajal body; CMV, cytomegalovirus; dn, dominant negative; Dox, doxycycline; snRNA, small nuclear RNA; Tet, tetracycline; wt, wild type.
References
- Abney, J.R., B. Cutler, M.L. Fillbach, D. Axelrod, and B.A. Scalettar. 1997. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J. Cell Biol. 137:1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan, H.G., J.G. Gall, and C. Murphy. 1991. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma. 101:245–251. [DOI] [PubMed] [Google Scholar]
- Chambeyron, S., and W.A. Bickmore. 2004. Does looping and clustering in the nucleus regulate gene expression? Curr. Opin. Cell Biol. 16:256–262. [DOI] [PubMed] [Google Scholar]
- Chuang, C.H., A.E. Carpenter, B. Fuchsova, T. Johnson, P. de Lanerolle, and A.S. Belmont. 2006. Long-range directional movement of an interphase chromosome site. Curr. Biol. 16:825–831. [DOI] [PubMed] [Google Scholar]
- Chubb, J.R., S. Boyle, P. Perry, and W.A. Bickmore. 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12:439–445. [DOI] [PubMed] [Google Scholar]
- Cioce, M., and A.I. Lamond. 2005. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 21:105–131. [DOI] [PubMed] [Google Scholar]
- Cremer, T., M. Cremer, S. Dietzel, S. Muller, I. Solovei, and S. Fakan. 2006. Chromosome territories–a functional nuclear landscape. Curr. Opin. Cell Biol. 18:307–316. [DOI] [PubMed] [Google Scholar]
- Frey, M.R., and A.G. Matera. 1995. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA. 92:5915–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, M.R., and A.G. Matera. 2001. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J. Cell Biol. 154:499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, M.R., A.D. Bailey, A.M. Weiner, and A.G. Matera. 1999. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol. 9:126–135. [DOI] [PubMed] [Google Scholar]
- Gall, J.G. 2001. A role for Cajal bodies in assembly of the nuclear transcription machinery. FEBS Lett. 498:164–167. [DOI] [PubMed] [Google Scholar]
- Gall, J.G., E.C. Stephenson, H.P. Erba, M.O. Diaz, and G. Barsacchi-Pilone. 1981. Histone genes are located at the sphere loci of newt lampbrush chromosomes. Chromosoma. 84:159–171. [DOI] [PubMed] [Google Scholar]
- Gasser, S.M. 2002. Visualizing chromatin dynamics in interphase nuclei. Science. 296:1412–1416. [DOI] [PubMed] [Google Scholar]
- Hebert, M.D., and A.G. Matera. 2000. Self-association of coilin reveals a common theme in nuclear body localization. Mol. Biol. Cell. 11:4159–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, W.A., T. Johnson, M. Klapczynski, J.L. Fan, and P. de Lanerolle. 2006. From transcription to transport: emerging roles for nuclear myosin I. Biochem. Cell Biol. 84:418–426. [DOI] [PubMed] [Google Scholar]
- Liu, J.-L., C. Murphy, M. Buszczak, S. Clatterbuck, R. Goodman, and J.G. Gall. 2006. The Drosophila melanogaster Cajal body. J. Cell Biol. 172:875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera, A.G. 2006. Drosophila Cajal bodies: accessories not included. J. Cell Biol. 172:791–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera, A.G., and K.B. Shpargel. 2006. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell Biol. 18:317–324. [DOI] [PubMed] [Google Scholar]
- Matera, A.G., R.M. Terns, and M.P. Terns. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8:209–220. [DOI] [PubMed] [Google Scholar]
- Misteli, T. 2007. Beyond the sequence: cellular organization of genome function. Cell. 128:787–800. [DOI] [PubMed] [Google Scholar]
- Platani, M., I. Goldberg, A.I. Lamond, and J.R. Swedlow. 2002. Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 4:502–508. [DOI] [PubMed] [Google Scholar]
- Stanek, D., and K.M. Neugebauer. 2006. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 115:343–354. [DOI] [PubMed] [Google Scholar]
- Thomson, I., S. Gilchrist, W.A. Bickmore, and J.R. Chubb. 2004. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr. Biol. 14:166–172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.