Abstract
Par3 is a PDZ protein important for the formation of junctional complexes in epithelial cells. We have identified an additional role for Par3 in membrane biogenesis. Although Par3 was not required for maintaining polarized apical or basolateral membrane domains, at the apical surface, Par3 was absolutely essential for the growth and elongation of the primary cilium. The activity reflected its ability to interact with kinesin-2, the microtubule motor responsible for anterograde transport of intraflagellar transport particles to the tip of the growing cilium. The Par3 binding partners Par6 and atypical protein kinase C interacted with the ciliary membrane component Crumbs3 and we show that the PDZ binding motif of Crumbs3 was necessary for its targeting to the ciliary membrane. Thus, the Par complex likely serves as an adaptor that couples the vectorial movement of at least a subset of membrane proteins to microtubule-dependent transport during ciliogenesis.
Introduction
Par3 is a PDZ protein first described as being required for asymmetric cell division during embryogenesis in Caenorhabditis elegans (Kemphues et al., 1988). Genetic analysis in Drosophila melanogaster demonstrated that the Par3 homologue Baz played a different but no less crucial role later in embryogenesis, being concentrated between the apical and basolateral domains of epithelial cells and needed for the morphogenesis of tissues (Müller and Wieschaus, 1996; Zallen and Wieschaus, 2004). Biochemically, Par3 is part of a complex consisting of a second PDZ protein, Par6, the Rho GTPase Cdc42, and an atypical PKC (aPKC; Joberty et al., 2000; Lin et al., 2000). In mammalian epithelial cells, the complex has been found to regulate tight junction formation. MDCK cells deficient in Par3 expression by RNAi were slow to reestablish transepithelial resistance (Chen and Macara, 2005).
Although the aPKC component is known to regulate Cdc42 and Par3 is recruited with Tiam1 to initiate junction assembly (Mizuno et al., 2003; Chen and Macara, 2005; Mertens et al., 2005), the actual functions of the Par complex remain unclear. A possible link to polarized membrane traffic was suggested by observations that the junctional complex is a preferred site for insertion of basolateral membrane proteins (Grindstaff et al., 1998; Nejsum and Nelson, 2007). Expression of Cdc42 mutants disrupted traffic to the basolateral domain (Kroschewski et al., 1999; Musch et al., 2001). The Par complex may also be involved in the biogenesis of the apical domain, having recently been localized to the primary cilia (Fan et al., 2004). Pharmacological inhibition of aPKC prevented cilium regrowth after microtubule depolymerization, although it is unclear if Par3-associated aPKC or some other isoform was responsible.
Whatever its precise functions, the complex does play an essential role because disruption of the Par3 gene in mice is embryonically lethal, causing aberrant development of epithelia (Hirose et al., 2006). We therefore sought to define the roles of Par3 in controlling epithelial cell biogenesis.
Results and discussion
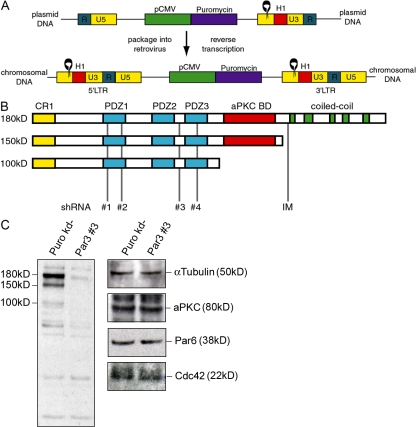
Although several groups have used RNAi to investigate the role of Par3 in tight junction assembly in MDCK cells, its role in polarized membrane traffic has not been explored. Because such studies would be facilitated by stable, Par3 knockdown (Par3-kd) cell lines, we modified a retrovirus for this purpose (Barton and Medzhitov, 2002; Schuck et al., 2004). The pac gene was inserted into the LTRH1 vector with short hairpin RNA (shRNA) ligated at the U3 region (Fig. 1 A), and upon infection, the U3 and 5 regions were duplicated, yielding two copies of the H1 cassette. This enabled stable knockdown even if the provirus recombined and deleted one of the long terminal repeat sequences. Of the sequences targeting all Par3 splice variants (Fig. 1 B), two (shRNAs nos. 3 and 4) resulted in >95% knockdown (Fig. 1 C, left, Par3 no. 3 vs. Puro kd). Knockdown was stable, lasting >2 mo in culture. Reduced Par3 expression did not affect expression of Par6, Cdc42, or aPKC (Fig. 1 C, right).
Figure 1.
Stable reduction of Par3 expression in MDCK cells. (A) Schematic representation of a retroviral vector designed to express shRNAs. Black rings represent shRNA sequences under control of the H1 promoter (H1). Puromycin resistance under control of the CMV promoter (pCMV) is in the body of the provirus (plasmid DNA). Duplication of the U3 and 5 regions upon reverse transcription and integration into chromosomal DNA results in expression of two identical shRNAs. The direct repeat is designated R. (B) Par3 is expressed in MDCK cells as three major splice variants. A dimerization (CR1) and PDZ domains are common to all variants. The 150-kD variant truncates the C-terminal coiled-coil domains and the 100-kD variant further truncates the aPKC binding domain. shRNAs were targeted to sequences common to all Par3 mRNAs. Previously published sequences (IM) only targeted the largest variant of Par3 (Chen and Macara, 2005). (C) Each of the three splice variants of Par3 were specifically reduced in MDCK cells expressing shRNA no. 3. Expression of the other components of the Par complex was unaltered by the reduction of Par3. Cells transduced with a viral vector that does not express an shRNA are indicated as Puro kd−. Lysates were subjected to SDS-PAGE and then probed with the indicated antibodies.
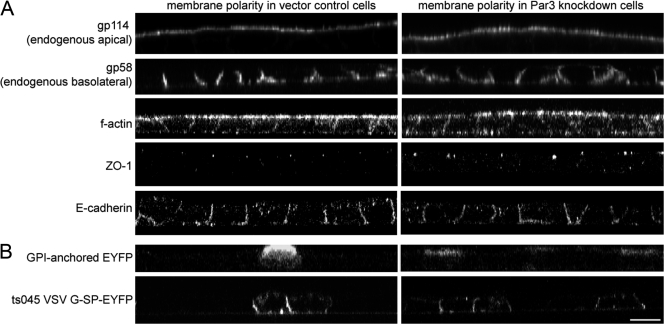
Par3-kd cells exhibited normal distributions of the endogenous membrane proteins gp114 and 58 (Fig. 2 A). F-actin was also unchanged, as were markers for junctional complexes (Fig. 2 A). Nor was there any alteration in the polarized appearance of newly synthesized membrane proteins (Fig. 2 B). Transient Par3-kd cells delayed the reassembly of tight junctions (Chen and Macara, 2005), which we confirmed in the stable lines (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200709111/DC1).
Figure 2.
The plasma membrane of MDCK cells grown on transwell filters remains polarized in the absence of Par3. (A) Endogenous membrane proteins are sorted to the appropriate domains of the plasma membrane in Par3-kd cells. The apical and basolateral markers gp114 and 58, respectively, were properly localized in control and Par3-kd cells as visualized by confocal microscopy (z axis reconstructions) using specific monoclonal antibodies on fixed, permeabilized cells. The cortical actin network (F-actin), the tight junction marker ZO-1, and the adherence junction marker E-cadherin were also properly localized in the Par3-kd cells. (B) Reduction of Par3 does not affect the sorting of proteins along the biosynthetic pathway. Newly synthesized apical glycophosphatidylinositol-anchored EYFP and basolateral vesicular stomatitis virus G–EYFP expressed using recombinant adenoviruses were sorted to appropriate membrane domains in control and Par3-kd cells. Bar, 10 μm.
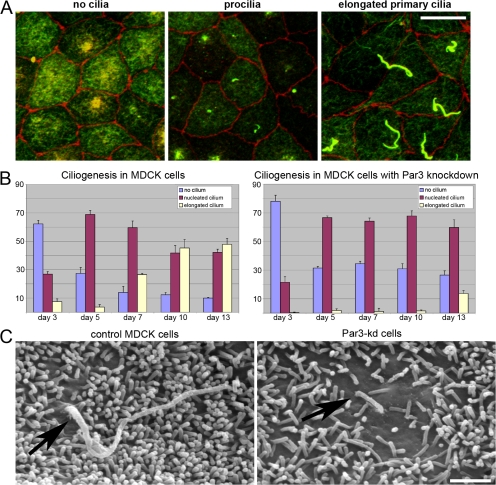
An additional role for Par3 was suggested by its recent localization to the primary cilium (Fan et al., 2004). Whether Par3 is required for ciliogenesis, however, is unknown. After confirming that cilia in MDCK cells contained Par3 (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200709111/DC1), we assayed the time course of ciliogenesis. In young monolayers of control cells (3 d), cilia were rarely observed, although basal bodies were located in the subapical cytoplasm. Subsequently, acetylated tubulin–positive cilia were detected more frequently, first appearing (5 d) as short (<3 μm) structures ("procilia") and later (7–10 d) as "elongated cilia" (>3 μm; Fig. 3, A and B).
Figure 3.
Reduction of Par3 causes defects in ciliogenesis. (A) Definition of ciliary phenotype by immunofluorescence. MDCK cells were seeded on polycarbonate filters, fixed, and stained for acetylated tubulin (green) and ZO-1 (red) 3, 5, and 10 d later. At the earliest time points, cilia were rarely seen, although acetylated tubulin was always found in the apical cytoplasm (left). At later times, short (<3 μm) acetylated tubulin–positive structures were observed, defined as a "procilium" (center). Later, cells exhibiting an "elongated primary cilium" typically >3 μm in length were observed at the apical membrane (right). (B) Par3-kd cells are capable of nucleating but not elongating cilia. More than 300 cells per time point were scored based on the presence and length of the primary cilium using the criteria defined in A. The results indicate that Par3-kd cells failed to elongate the procilia within the time course of the experiment. Nearly 50% of control MDCK cells exhibited an elongated primary cilium. Error bars indicate the standard error of categorized cells in nine independent fields. (C) Scanning EM of the apical membrane on MDCK cells. Images of control cells verified the presence of a primary cilium emerging from the apical membrane (left, arrow). In Par3-kd cells, it was occasionally possible to identify apical protrusions that appeared thicker than the typical microvilli in regions of the apical membrane where microvilli were less dense or absent (right, arrow); these could represent procilia. Bars: (A) 10 μm; (B) 1 μm.
These events were markedly altered in Par3-kd cells, which generated procilia but failed to form elongated cilia even after 10 d (Fig. 3 B). Similar results were obtained for independently derived Par3-kd cell lines. Differences in cilia length were confirmed by scanning EM, although it was difficult to identify the procilia in Par3-kd cells (Fig. 3 C, arrows). By thin section EM, it was possible to capture occasional images of basal bodies or cilia in control cells and procilia in the Par3-kd cells. Despite variation in the morphology of cilia even in control cells, no consistent differences were observed between the cell lines (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200709111/DC1). Furthermore, the basal bodies in the Par3-kd cells appeared properly positioned (Fig. S2 B). The procilia and basal bodies in Par3-kd cells also did not contain obvious inclusions that might suggest the accumulation of intraflagellar transport (IFT) particles, which have previously been observed when retrograde transport is blocked (Blacque et al., 2004). Collectively, these observations suggested that Par3 is required for the elongation of primary cilia, possibly by facilitating anterograde transport of cargo.
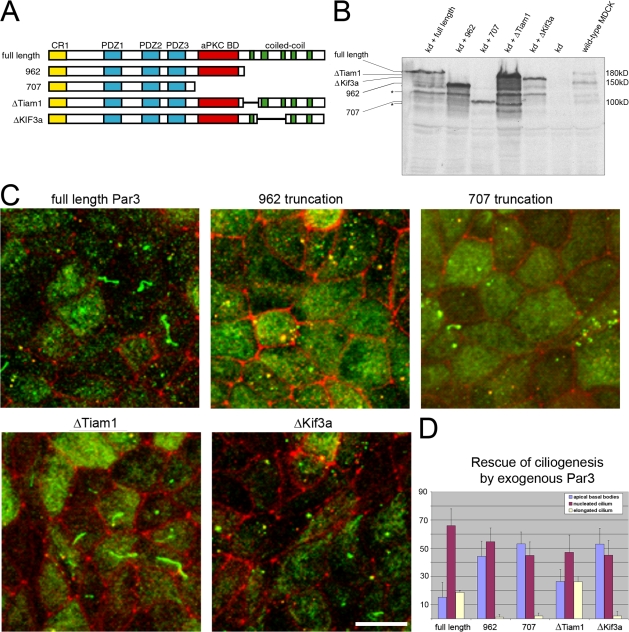
To confirm that the cilium phenotype was caused by Par3-kd, we generated rescue cell lines. The human cDNA was rendered resistant to Par3 shRNA no. 3, and stop codons were inserted after amino acids 962 and 707 to mimic the splice variants (Fig. 4 A; Lin et al., 2000). Using simultaneous transduction, endogenous Par3 was reduced and reexpressed (Fig. 4 B). Each cell line was seeded on filters for 7 d and scored for cilium length after staining with acetylated tubulin. Par3-kd cells reexpressing only the full-length Par3 displayed both procilia and elongated cilia in proportions similar to control cells (Fig. 4, C and D; compare to Fig. 3 B, day 7). However, neither of the truncated forms was able to rescue ciliogenesis (Fig. 4, C and D), despite being expressed at levels comparable to or higher than full-length Par3 (Fig. 4 B). These results indicate that the C terminus of Par3 is required for ciliogenesis.
Figure 4.
Rescue of ciliogenesis defects in Par3-kd cells. (A) Schematic diagram of mutants generated in the Par3 cDNA. Truncations that reproduce splice variants were generated by inserting stop codons at amino acids 962 and 707. These mutants were designated full length, 962, and 707. Mutants were also generated by deleting the coding region of coiled-coil domains 1 and 2–3, which correspond to the binding domains of Tiam1 and Kif3a, which have been defined previously (Nishimura et al., 2004). (B) Expression of exogenous Par3 proteins in Par3-kd cells. Simultaneous transduction of MDCK cells with vectors expressing Par3 shRNA and Par3 mutants resistant to knockdown results in replacement of the endogenous Par3 protein. Lines on the side of the image indicate the position of Par3-related proteins (full length is 180 kD, 962 is 150 kD, and 707 is 100 kD). Asterisks show degradation products. (C) Rescue of defective ciliary phenotype requires the Par3 kinesin binding domain. Rescued cell lines were seeded on polycarbonate filters for 7 d and stained for acetylated tubulin (green) and ZO-1 (red). Elongated cilia were only observed in cells expressing full-length Par3 and the ΔTiam-1 deletion mutant. Cells expressing Par3 constructs that lacked the Kif3a binding site did not rescue the defective ciliary phenotype. Bar, 10 μm. (D) Quantitation of monolayers imaged in C indicate that only the Par3 proteins containing the Kif3a binding site (full length and ΔTiam1) were able to rescue the defect in ciliogenesis caused by reduction of Par3. Individual cells of monolayers lacking endogenous Par3 and expressing one of the indicated exogenous Par3 cDNAs were categorized as having no primary cilium, a procilium, or an elongated primary cilium. Error bars indicate the standard error of categorized cells in five independent fields.
Recently, Par3 was found to interact with the microtubule motor of cilia and flagella Kif3a, and the precise regions of Par3 involved were mapped to the second and third coiled-coil domains of Par3 (Fig. 4 A; Fan et al., 2004; Nishimura et al., 2004). The first of the coiled-coil domains interacts with the nucleotide exchange factor Tiam1 (Chen and Macara, 2005; Zhang and Macara, 2006). Because both the Tiam1 and Kif3a binding sites were deleted in the truncations that failed to complement the ciliogenesis defect (Fig. 4 A), we removed each individually and used these mutants to replace endogenous Par3. Interestingly, the Tiam1 mutant was as effective as full-length human Par3 at correcting the ciliary defect (Fig. 4, C and D). Thus, although the Tiam1 binding region is required for tight junction formation (Chen and Macara, 2005), it is dispensable for Par3 function at the primary cilium.
In contrast, Par3 lacking the region required for interaction with Kif3a did not facilitate ciliary growth (Fig. 4, C and D). Although expression of this construct permitted the assembly of procilia, it did not allow the growth of primary cilia found in cells expressing wild-type Par3 or the Tiam1 mutant. Thus, the Par3 coiled-coil domain that specifies interaction with Kif3a is required for ciliogenesis.
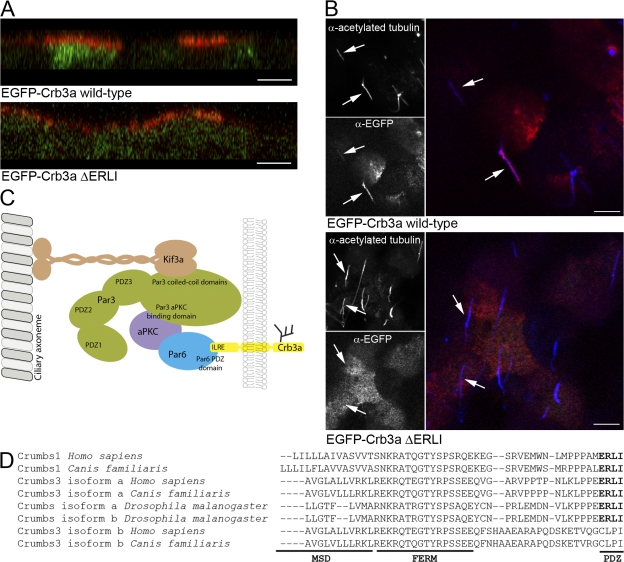
The ciliary defect caused by Par3-kd is phenotypically similar to defects caused by mutations that impair IFT. Crumbs3 (Crb3a) has been shown to interact directly with Par6 via PDZ binding. Interestingly, knockdown of Crb3 also inhibits ciliogenesis (Fan et al., 2004; Lemmers et al., 2004). Consequently, we investigated whether Crb transport to the primary cilium is dependent on its ability to interact with PDZs and is therefore dependent on the Par complex. For this purpose, EGFP was fused to Crb3a or a mutant Crb3a lacking its PDZ binding motif (EGFP-Crb3ΔERLI). Both constructs were expressed in MDCK cells and the apical and basolateral surface of the cells was exposed to an anti-EGFP antibody. The EGFP-Crb3a and the EGFP-Crb3ΔERLI both accumulated at the apical domain (Fig. 4 A) as in the endogenous Crb3a (Makarova et al., 2003). Further, the EGFP-Crb3a construct could be identified along the length of the primary cilium in 82% of the cells (Fig. 4 B). However, despite being delivered to the apical plasma membrane, EGFP-Crb3ΔERLI was mostly absent from the primary cilium. Only 7% of the cells expressing the ΔERLI construct showed colocalization of the acetylated tubulin and EGFP stains (Fig. 4 B). Thus, the PDZ binding motif of Crb3a is required for transport along the axoneme, strongly suggesting that Par3 (via Par6) acts as a linker between at least one integral membrane protein essential for ciliogenesis and the ciliary axoneme via kinesin-2 (Fig. 5 C).
Figure 5.
The PDZ binding motif of Crb3a is necessary for targeting to the primary cilium in MDCK cells. (A) Surface EGFP-Crb3a proteins are restricted to the apical domain. MDCK cells expressing EGFP-Crb3a or a form lacking the PDZ binding motif were stained with an anti-EGFP antibody (red) applied to the apical and basolateral surface. The images show that surface EGFP was restricted to the apical plasma membrane domain. Bars, 5 μM. (B) The PDZ binding motif of Crb3a is necessary for targeting to the primary cilium. MDCK cells expressing the EGFP-Crb3a constructs were surface stained with an anti-EGFP antibody (red) to enhance the signal of the proteins and then fixed and permeabilized before staining with anti-acetylated tubulin (blue). EGFP-Crb3a was identified in 82% of cilia. Only 7% of cilia were positive for the Crb3a construct lacking the PDZ binding motif. Arrows indicate identical locations for each panel. Bars, 5 μM. (C) Model of the role of Par3 in ciliogenesis. The data presented here supports a model in which Par3 functions as a tether for the transmembrane protein Crb3a to the axoneme. The Kif3a and aPKC proteins bind to Par3 through previously characterized interactions involving the coiled-coil domains and an aPKC binding domain, respectively. The tethering mechanism is also facilitated through direct interactions between the PDZ binding motif of Crb3a and the PDZ Par6, which also binds to aPKC. (D) Protein sequence alignment of Crb C terminus. The membrane spanning domain (MSD), FERM binding domain (FERM), and PDZ binding motif (PDZ) are aligned for various Crb isoforms from Homo sapiens, Canis familiaris, and D. melanogaster.
In neurons, Kif3a localizes Par3 to the tip of the growing axon, where it serves to recruit Tiam1 (Shi et al., 2003; Nishimura et al., 2004, 2005). In epithelial cells, however, Par3 is used differently at junctions and at the cilium. In early steps of establishing polarity, Tiam1 and Par3 are recruited to points of cell–cell contact and to initiate junction assembly (Mizuno et al., 2003; Chen and Macara, 2005; Mertens et al., 2005). Later, at the cilium, the ability of Par3 to interact with Kif3a is essential for ciliogenesis, whereas its ability to interact with Tiam1 is dispensable. Because Par6 binds to the Crb3a cytoplasmic domain (Lemmers et al., 2004), Par3 seems likely to link at least one critical membrane protein component to the axoneme via Kif3a (Fig. 5 C). The PDZ binding domain of Crb3a is identical to C-terminal sequences of other Crb family members across different species (Fig. 5 D). However, alternative splicing of the Crb gene generates heterogeneity in its cytoplasmic tail (Fan et al., 2007). The splice variant Crb3b is also targeted to the cilia and is required for ciliogenesis (Fig. 5 D; Fan et al., 2007) but lacks a PDZ binding motif. Thus, both Crb3 variants seem essential for ciliogenesis in MDCK cells, possibly for nonredundant reasons.
Interestingly, the Crb3-kd phenotype (no cilia) appears different from the one caused by Par3-kd (shortened cilia). The latter phenotype was similar to that resulting from antibody inhibition of kinesin-2 function in sea urchins, which demonstrated that the formation of a procilium was kinesin-2 independent, although the appearance of elongated cilia was prevented (Morris and Scholey, 1997). Unfortunately, knockdown of Kif3a has thus far proved cell lethal in MDCK cells (unpublished data). It is also possible that another kinesin-2 family member couples with Par3 to mediate membrane protein transport into the growing cilium in light of recent findings that Kif17 may be involved in the transport of odorant receptors in specialized ciliated olfactory cells (Jenkins et al., 2006).
Insights into the mechanism of ciliary growth have been provided by the analysis of mutations in proteins that cause Bardet-Biedl syndrome (Blacque et al., 2004; Nachury et al., 2007). Bardet-Biedl syndrome proteins (e.g., BBS-7 and -8) allow IFT particles to target to the base of the cilium, but the particles are too inefficiently loaded onto the axoneme to facilitate normal elongation. The accumulation of procilia in the Par3 knockdown suggests that the Par complex may play an analogous role. It remains possible, however, that the short cilium in Par3-kd cells reflects residual (<5%) Par3; a complete deficiency might cause a complete defect in ciliogenesis. In any event, our results clearly indicate that Par3 presumably together with other members of its well-characterized, multifunctional complex plays an essential role in the formation of cilia. Whether the loss of this essential role, as opposed to Par3's role in junction assembly, was responsible for the multiorgan defects that lead to embryonic lethality in mice bearing a deletion of the Par3 gene (Hirose et al., 2006) is an intriguing question. The identification of a requirement for the interaction of Par3 with kinesin during ciliogenesis, however, will greatly facilitate dissection of the manifold roles attributed to the Par complex.
Materials and methods
Antibodies and reagents
Hybridomas expressing mouse anti-gp114 and anti-gp58 IgGs, were obtained from the American Type Culture Collection. Monoclonal anti-acetylated tubulin clone 6-11B-1 and anti–γ-tubulin clone GTU-88 mouse ascities fluid was obtained from Sigma-Aldrich. Mouse anti–E-cadherin was provided by W.J. Nelson (University of California, San Francisco, San Francisco, CA). Rabbit polyclonal IgG1 anti-Par3 was purchased from Millipore and provided by T. Pawson (University of Toronto, Toronto, ON, Canada). Mouse anti–ZO-1 was obtained from Invitrogen. HRP-conjugated secondary antibodies were purchased from Thermo Fisher Scientific. Alexa Fluor 633 phalloidin, Alexa Fluor 647–conjugated streptavidin, Alexa Fluor 488–conjugated goat anti–mouse, Alexa Fluor 647–conjugated goat anti–rat, and Alexa Fluor 568–conjugated goat anti–rabbit antibodies were obtained from Invitrogen. Vitrogen was purchased from Cohesion Laboratories. All other reagents were purchased from Sigma-Aldrich unless otherwise indicated.
Cell culture
MDCK and GP2-293 cells (Clontech Laboratories, Inc.) were maintained in Dulbecco's minimum essential medium (Invitrogen) supplemented with 10% FBS (vol/vol; Invitrogen) and 2 mM l-glutamine. All cells were maintained at 37°C in a 5% CO2 incubator. Cells were seeded at a density of 4.5 × 106 onto 10-mm polycarbonate filters (Corning) and the medium changed every day for the entire analysis. For surface biotinylation of membrane proteins, 1 mg/ml EZ-link sulfo NHS-LC-LC biotin reagent (Thermo Fisher Scientific) in PBS++ was added to the upper chamber of the filter apparatus for 5 min and the conjugation reaction was quenched with PBS++ containing 100 mM glycine. Expression of ts045 vesicular stomatitis virus G sp-EYFP and ECFP-GL-glycophosphatidylinositol using adenovirus vectors was described previously (Keller et al., 2001; Ang et al., 2004). MDCK cells were grown in a collagen matrix as described previously (O'Brien et al., 2006).
Immunofluorescence microscopy
Cells grown on coverslips were fixed with either 4% PFA for 15 min at room temperature or methanol for 20 min at −20°C and blocked for 1 h with PBS containing 1 mM MgCl2, 2.5 mM CaCl2, 0.2% saponin, and 0.5% bovine serum albumin (IF buffer). Both primary and secondary antibodies were diluted in IF buffer, used to probe membranes for 1 h, and washed four times with IF buffer. The filters were then mounted with glass coverslips using Prolong Gold (Invitrogen). Images of the cells were acquired at room temperature on a confocal microscope (LSM-510 META) with associated 3.0 software (both from Carl Zeiss, Inc.) using a Plan-Apochromat 100× 1.40 NA oil differential interference contrast objective (Carl Zeiss, Inc.) to detect Alexa 488-, 568-, and 633-nm fluorochromes. Image sizes were adjusted using Photoshop CS3 (Adobe).
EM
MDCK cells grown on polycarbonate filters were fixed in PBS containing 100 mM sodium cacodylate, 4% PFA, 2% gluteraldehyde, 1 mM MgCl2, and 2.5 mM CaCl2, pH 7.4, for 20 min at room temperature. For transmission EM, the cells were then rinsed with 100 mM cacodylate buffer, postfixed for 1 h in 1% osmium tetroxide, en bloc uranyl acetate stained, dehydrated through a graded ethanol series, and embedded using EMBed 812 (Electron Microscopy Sciences). To image the primary cilium and basal bodies, the embedded cells were sliced at 60° angles so that each section contained apical membrane and subapical cytoplasm. Digital images were captured using a transmission EM (Tecnai 20; FEI Company) and a charge-coupled device camera (Morada) using iTEM acquisition software (Olympus Soft Imaging Solutions). For scanning EM, the fixed cells were rinsed with 100 mM of cacodylate buffer, dehydrated through a graded ethanol series, washed with hexamethyldisilazane (Electron Microscopy Sciences), dried for 5 min at 60°C, coated with platinum, and analyzed on a scanning EM (FEI ESEM; Philips).
Retroviral transduction
The LTRH1 vector was a provided by R. Medzhitov (Howard Hughes Medical Institute, Yale University, New Haven, CT; Barton and Medzhitov, 2002). The CD4 gene of LTRH1 was replaced with the puromycin resistance gene as described previously to create LTRH1-puro (Schuck et al., 2004). Oligos encoding short hairpin DNA sequences targeting Par3 were annealed, phosphorylated, and ligated to the pSUPER vector as described previously (Schuck et al., 2004). The oligo sequences are as follows: Par3 oligo no. 1 (5′- GATCCCCGCCTCTGGGAATCCATGTAGTGCCTTCGAAAAGGCACTACATGGATTCCCAGAGGTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAACCTCTGGGAATCCATGTAGTGCCTTTTCGAAGGCACTACATGGATTCCCAGAGGCGGG-3′), Par3 oligo no. 2 (5′-GATCCCCGCCAAGCCATGCGTACACCCATCATTCGAAAATGATGGGTGTACGCATGGCTTGGTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAACCAAGCCATGCGTACACCCATCATTTTCGAATGATGGGTGTACGCATGGCTTGGCGGG-3′), Par3 oligo no. 3 (5′-GATCCCCGCCAAGGGAACTGAATGCAGAGCCAACGAATTGGCTCTGCATTCAGTTCCCTTGGTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAACCAAGGGAACTGAATGCAGAGCCAATTCGTTGGCTCTGCATTCAGTTCCCTTGGCGGG-3′), and Par3 oligo no. 4 (5′-GATCCCCGGCTTCGGGTGAATGATCAACTGATACGAATATCAGTTGATCATTCACCCGAAGCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGCTTCGGGTGAATGATCAACTGATATTCGATCAGTTGATCATTCACCCGAAGCCGGG-3′). The H1 cassette containing the ligated shRNA sequence was then excised using EcoRI and XhoI and ligated to the U3 region of LTRH1-puro using SalI. Human Par3 cDNA was provided by I. Macara (University of Virginia, Charlottesville, VA) and Crb3a cDNA was provided by B. Margolis (University of Michigan, Ann Arbor, MI). The codon of leucine 555 was mutated from CTG to TTA in the human Par3 shRNA to render it resistant to oligo no. 3. This mutation was carried through with all subsequent mutations. The Par3 cDNA was amplified and inserted into pQCXIH (Clontech Laboratories, Inc.). Truncations in the Par3 cDNA were designed by inserting stop sequences after the codons for amino acids 976 and 707. Deletions within the Par3 open reading frame were generated by PCR amplification of the entire plasmid using primers designed directly outside the sequence to be deleted. The resulting amplified DNA was then phosphorylated with T4 DNA kinase and ligated to itself using the T4 DNA ligase (both from New England Biolabs, Inc.). All constructs were sequenced through the Par3 gene after mutagenesis. GP2-293 cells (Clontech Laboratories, Inc.) were seeded at 90% confluence and transfected with pMD.G and either the pLTRH1-puro or pQCXIH Par3 according to the instructions from the manufacturer of Lipofectamine 2000 (Invitrogen). Virus-like particles (VLPs) were collected in 24-h periods beginning 16 h after transfection. VLP-containing supernatants were immediately cleared of cell debris by centrifugation at 2,000 g for 5 min and stored at −80°C or used directly for transduction. Subconfluent MDCK cells were infected by adding VLP-containing supernatants into the well with 4 μg/ml polybrene (Sigma-Aldrich) and centrifuged at 1,500 g for 45 min. Selection with either 4 μg/ml puromycin and/or 800 μg/ml hygromycin B was initiated 24 h after infection.
Online supplemental material
Fig. S1 shows that knockdown of Par3 delays generation of transepithelial resistance. Fig. S2 shows Par3 localization. Fig. S3 shows transmission EM of Par3-kd and control cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200709111/DC1.
Supplementary Material
Acknowledgments
We thank Cecile Chalouni for outstanding assistance with confocal microscopy, Fernando Bazan for helpful discussion, and the Mellarren laboratory.
This work was supported by grants from the National Institutes of Health (RO1-GM29765) and the Ludwig Institute for Cancer Research. J. Sfakianos is the recipient of a Ruth L. Kirschstein Fellowship (National Institutes of Health grant 5F32GM072162-03).
Abbreviations used in this paper: aPKC, atypical PKC; Crumbs3, Crb3a; IFT, intraflagellar transport; shRNA, short hairpin RNA.
References
- Ang, A.L., T. Taguchi, S. Francis, H. Folsch, L.J. Murrells, M. Pypaert, G. Warren, and I. Mellman. 2004. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 167:531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, G.M., and R. Medzhitov. 2002. Retroviral delivery of small interfering RNA into primary cells. Proc. Natl. Acad. Sci. USA. 99:14943–14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque, O.E., M.J. Reardon, C. Li, J. McCarthy, M.R. Mahjoub, S.J. Ansley, J.L. Badano, A.K. Mah, P.L. Beales, W.S. Davidson, et al. 2004. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 18:1630–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., and I.G. Macara. 2005. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat. Cell Biol. 7:262–269. [DOI] [PubMed] [Google Scholar]
- Fan, S., T.W. Hurd, C.J. Liu, S.W. Straight, T. Weimbs, E.A. Hurd, S.E. Domino, and B. Margolis. 2004. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr. Biol. 14:1451–1461. [DOI] [PubMed] [Google Scholar]
- Fan, S., V. Fogg, Q. Wang, X.W. Chen, C.J. Liu, and B. Margolis. 2007. A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin β interactions. J. Cell Biol. 178:387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindstaff, K.K., C. Yeaman, N. Anandasabapathy, S.C. Hsu, E. Rodriguez-Boulan, R.H. Scheller, and W.J. Nelson. 1998. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell. 93:731–740. [DOI] [PubMed] [Google Scholar]
- Hirose, T., M. Karasawa, Y. Sugitani, M. Fujisawa, K. Akimoto, S. Ohno, and T. Noda. 2006. PAR3 is essential for cyst-mediated epicardial development by establishing apical cortical domains. Development. 133:1389–1398. [DOI] [PubMed] [Google Scholar]
- Jenkins, P.M., T.W. Hurd, L. Zhang, D.P. McEwen, R.L. Brown, B. Margolis, K.J. Verhey, and J.R. Martens. 2006. Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr. Biol. 16:1211–1216. [DOI] [PubMed] [Google Scholar]
- Joberty, G., C. Petersen, L. Gao, and I.G. Macara. 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2:531–539. [DOI] [PubMed] [Google Scholar]
- Keller, P., D. Toomre, E. Diaz, J. White, and K. Simons. 2001. Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat. Cell Biol. 3:140–149. [DOI] [PubMed] [Google Scholar]
- Kemphues, K.J., J.R. Priess, D.G. Morton, and N.S. Cheng. 1988. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 52:311–320. [DOI] [PubMed] [Google Scholar]
- Kroschewski, R., A. Hall, and I. Mellman. 1999. Cdc42 controls secretory and endocytic transport to the basolateral plasma membrane of MDCK cells. Nat. Cell Biol. 1:8–13. [DOI] [PubMed] [Google Scholar]
- Lemmers, C., D. Michel, L. Lane-Guermonprez, M.H. Delgrossi, E. Medina, J.P. Arsanto, and A. Le Bivic. 2004. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol. Biol. Cell. 15:1324–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D., A.S. Edwards, J.P. Fawcett, G. Mbamalu, J.D. Scott, and T. Pawson. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2:540–547. [DOI] [PubMed] [Google Scholar]
- Makarova, O., M.H. Roh, C.J. Liu, S. Laurinec, and B. Margolis. 2003. Mammalian Crumbs3 is a small transmembrane protein linked to protein associated with Lin-7 (Pals1). Gene. 302:21–29. [DOI] [PubMed] [Google Scholar]
- Mertens, A.E., T.P. Rygiel, C. Olivo, R. van der Kammen, and J.G. Collard. 2005. The Rac activator Tiam1 controls tight junction biogenesis in keratinocytes through binding to and activation of the Par polarity complex. J. Cell Biol. 170:1029–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, K., A. Suzuki, T. Hirose, K. Kitamura, K. Kutsuzawa, M. Futaki, Y. Amano, and S. Ohno. 2003. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J. Biol. Chem. 278:31240–31250. [DOI] [PubMed] [Google Scholar]
- Morris, R.L., and J.M. Scholey. 1997. Heterotrimeric kinesin-II is required for the assembly of motile 9 + 2 ciliary axonemes on sea urchin embryos. J. Cell Biol. 138:1009–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, H.A., and E. Wieschaus. 1996. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J. Cell Biol. 134:149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musch, A., D. Cohen, G. Kreitzer, and E. Rodriguez-Boulan. 2001. cdc42 regulates the exit of apical and basolateral proteins from the trans-Golgi network. EMBO J. 20:2171–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury, M.V., A.V. Loktev, Q. Zhang, C.J. Westlake, J. Peranen, A. Merdes, D.C. Slusarski, R.H. Scheller, J.F. Bazan, V.C. Sheffield, and P.K. Jackson. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213. [DOI] [PubMed] [Google Scholar]
- Nejsum, L.N., and W.J. Nelson. 2007. A molecular mechanism directly linking E-cadherin adhesion to initiation of epithelial cell surface polarity. J. Cell Biol. 178:323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, T., K. Kato, T. Yamaguchi, Y. Fukata, S. Ohno, and K. Kaibuchi. 2004. Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat. Cell Biol. 6:328–334. [DOI] [PubMed] [Google Scholar]
- Nishimura, T., T. Yamaguchi, K. Kato, M. Yoshizawa, Y. Nabeshima, S. Ohno, M. Hoshino, and K. Kaibuchi. 2005. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat. Cell Biol. 7:270–277. [DOI] [PubMed] [Google Scholar]
- O'Brien, L.E., W. Yu, K. Tang, T.S. Jou, M.M. Zegers, and K.E. Mostov. 2006. Morphological and biochemical analysis of Rac1 in three-dimensional epithelial cell cultures. Methods Enzymol. 406:676–691. [DOI] [PubMed] [Google Scholar]
- Schuck, S., A. Manninen, M. Honsho, J. Fullekrug, and K. Simons. 2004. Generation of single and double knockdowns in polarized epithelial cells by retrovirus-mediated RNA interference. Proc. Natl. Acad. Sci. USA. 101:4912–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, S.H., L.Y. Jan, and Y.N. Jan. 2003. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 112:63–75. [DOI] [PubMed] [Google Scholar]
- Zallen, J.A., and E. Wieschaus. 2004. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell. 6:343–355. [DOI] [PubMed] [Google Scholar]
- Zhang, H., and I.G. Macara. 2006. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat. Cell Biol. 8:227–237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.