Abstract
The calcium-activated phosphatase calcineurin (Cn) transduces physiological signals through intracellular pathways to influence the expression of specific genes. Here, we characterize a naturally occurring splicing variant of the CnAβ catalytic subunit (CnAβ1) in which the autoinhibitory domain that controls enzyme activation is replaced with a unique C-terminal region. The CnAβ1 enzyme is constitutively active and dephosphorylates its NFAT target in a cyclosporine-resistant manner. CnAβ1 is highly expressed in proliferating myoblasts and regenerating skeletal muscle fibers. In myoblasts, CnAβ1 knockdown activates FoxO-regulated genes, reduces proliferation, and induces myoblast differentiation. Conversely, CnAβ1 overexpression inhibits FoxO and prevents myotube atrophy. Supplemental CnAβ1 transgene expression in skeletal muscle leads to enhanced regeneration, reduced scar formation, and accelerated resolution of inflammation. This unique mode of action distinguishes the CnAβ1 isoform as a candidate for interventional strategies in muscle wasting treatment.
Introduction
The serine/threonine phosphatase calcineurin (Cn) plays a main role in a wide variety of physiological and pathological processes, including the immune response, neuronal plasticity, and cardiac development and hypertrophy (Schulz and Yutzey, 2004). In response to calcium increase, Cn induces dephosphorylation, nuclear translocation, and activation of the NFAT transcription factors, a process sensitive to the action of the immunosuppressive drug cyclosporine-A (CsA). Cn enzymatic activity requires a catalytic (CnA) and a regulatory (CnB) subunit, variants of which are encoded by multiple genes (Hogan et al., 2003). The CnA subunit includes protein domains conferring catalytic activity, CnB interaction, calmodulin-binding and a C-terminal autoinhibitory domain, which blocks the catalytic site and is removed in response to calcium increase. Three CnA genes have been described: CnAα and CnAβ are ubiquitously expressed, whereas CnAγ is restricted to brain and testis. Two CnAβ isoforms, CnAβ1 and CnAβ2, which differ in their C-terminal domain, are encoded by alternatively spliced transcripts (Guerini and Klee, 1989). The typical autoinhibitory domain present in CnAβ2 and other CnA isoforms is absent from CnAβ1, in which an unrelated C-terminal domain is generated by the translation of intronic sequences (Fig. 1 A; and Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200704179/DC1). This novel domain is preserved in the CnAβ1 orthologues from different species (Fig. S1 B), especially in higher vertebrates, suggesting an evolutionarily conserved role for this Cn variant.
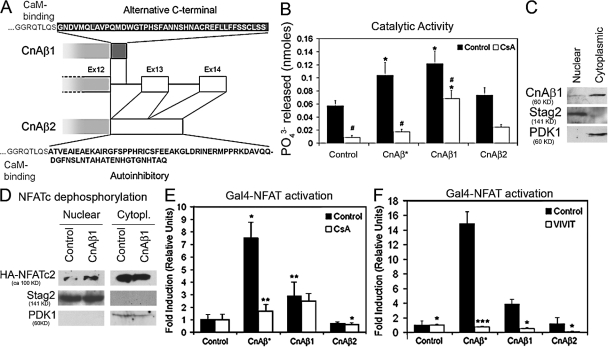
Figure 1.
CnAβ1 is a constitutively active Cn isoform. (A) Schematic diagram of CnAβ1 and CnAβ2 isoforms, alternative splicing variants of the CnAβ gene. CnAβ1 encodes an alternate C-terminal domain encoded by intronic sequences (top) whereas CnAβ2 includes a canonical autoinhibitory domain encoded by exons 13–14 (bottom). (B) HEK293 cells were transfected with CnA expression vectors or empty pcDNA3.1 (control), grown for 48 h in the absence (black bars) or presence (white bars) of 1 μg/ml CsA and Cn phosphatase activity was assayed. (C) Nuclear and cytoplasmic extracts were analyzed for the presence of CnAβ1. Anti-Stag2 and anti-PDK1, respectively show equal nuclear and cytoplasmic protein loading. (D) C2C12 myoblasts were transiently cotransfected with a HA-NFATc2 expression vector and pcDNA3.1-CnAβ1 or empty pcDNA3.1. After 2 d in DM, nuclear and cytoplasmic extracts were analyzed by Western blot using an anti-HA antibody. Arrow indicates increased dephosphorylated NFAT. (E) C2C12 myoblasts were transiently transfected with CnA expression vectors (or empty pcDNA3.1 as a control), the pGal4-Luc reporter and pGal4-NFAT-1-415 and grown in DM for 2 d. Where indicated, 1 μg/ml cyclosporine A (white bars) or EtOH as a vehicle (black bars) was added to the culture after transfection. (F) C2C12 myoblasts were transfected as in D together with a VIVIT expression plasmid or control vector and luciferase activity was analyzed. Results show fold induction over the control value ±SD and represent the average of at least three independent experiments. *, P < 0.05; **, P < 0.005.
In skeletal muscle, the Cn/NFAT pathway mediates myotube differentiation, enhances myoblast recruitment, controls muscle fiber type specification, and ameliorates injury to dystrophic muscles (Friday et al., 2000; Naya et al., 2000; Horsley et al., 2001, 2003; Parsons et al., 2003; Stupka et al., 2006). The function of the CnAβ1 isoform has not been explored because no obvious phenotype was reported in a germline knockout of the CnAβ gene (Bueno et al., 2002). Notably, in that study the knockout scheme involved deletion of catalytic domain encoded by exon 2, which would still allow transcription of in-frame transcripts encoding phosphatase-dead CnAβ2 or CnAβ1 protein. Interestingly, increased CnAβ1 expression was noted in response to the muscle-restricted overexpression of a local IGF-1 isoform (mIGF-1), which preserved muscle integrity and enhanced the response to chronic degeneration in a mouse model of amyotrophic lateral sclerosis (ALS) (Dobrowolny et al., 2005), suggesting a role for CnAβ1 in satellite cell activation.
To establish a function for CnAβ1, we overexpressed the protein or blocked its production in muscle cell culture and found that, unlike other CnA variants, CnAβ1 inhibits FoxO transcriptional activity independently of its phosphatase function, regulates myoblast proliferation, and prevents myotube atrophy under starvation conditions. In addition, CnAβ1 overexpression in post-mitotic muscle enhances regeneration, reducing scar formation and accelerating the resolution of inflammation. The unique properties of the CnAβ1 isoform expand the spectrum of biological functions for calcineurins and provide new avenues for prevention of muscle atrophy.
Results
CnAβ1 is a constitutively active Cn isoform
To determine whether the absence of an autoinhibitory domain rendered the CnAβ1 isoform constitutively active without high intracellular calcium, we analyzed Cn phosphatase activity in HEK293 cells transfected with expression plasmids encoding CnAβ1, CnAβ2, or CnAβ* in which the autoinhibitory domain has been artificially deleted. Phosphatase activity in CnAβ1-transfected cells was as high as those expressing constitutively active CnAβ*, compared with cells transfected with CnAβ2 (Fig. 1 B). Cell fractionation and subsequent analysis by Western blot showed that CnAβ1 is localized both in the cytoplasm and the nucleus in proliferating C2C12 cells, with most of the protein mainly localized in the cytoplasm (Fig. 1 C). Co-transfection of C2C12 muscle cells with CnAβ1 and an HA-NFAT expression vectors resulted in increased nuclear translocation of NFAT (Fig. 1 D). Accordingly, cotransfection of C2C12 myoblasts with different Cn constructs together with a Gal4-NFAT chimera and a Gal4-Luc reporter plasmid showed that in the absence of further stimulus, CnAβ1 activated NFAT in C2C12 muscle cells (Fig. 1 E; Fig. S1, C and D), but not in Jurkat T cells (Fig. S1 E), albeit to a lower degree than the truncated, constitutively active CnAβ* form. Interestingly, both the increased phosphatase activity and the activation of NFAT induced by CnAβ1 were partially resistant to the action of CsA, whereas truncated Cn forms were inhibited by the immunosuppressant drug (Fig. 1, B and E). Conversely, overexpression of the VIVIT peptide, which blocks the interaction between Cn and NFAT, prevented the activation of NFAT by all Cn forms (Fig. 1 F). Together, these results show that CnAβ1 is a physiological Cn form that is constitutively active and is partially insensitive to the action of CsA.
CnAβ1 expression is increased during skeletal muscle regeneration
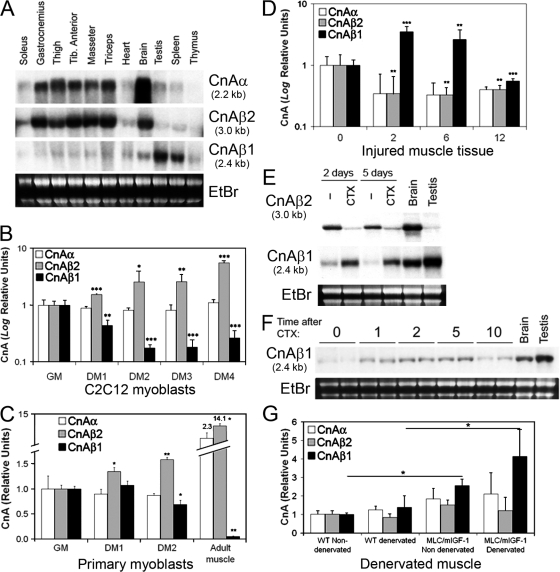
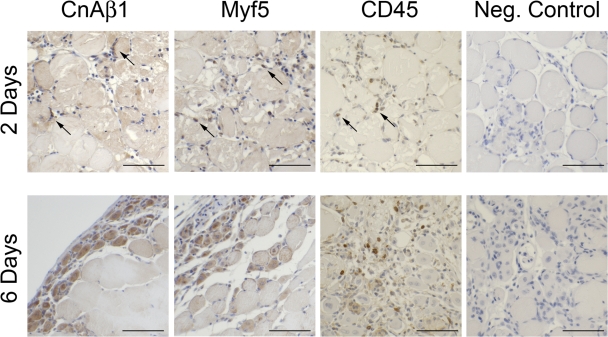
We analyzed the CnA expression patterns in differentiating myoblasts as well as different healthy and cardiotoxin-injured muscles by Northern blot and quantitative RT-PCR (qRT-PCR). All uninjured muscles showed high expression levels of CnAα and CnAβ2 and weaker expression of CnAβ1 (Fig. 2 A). Differentiation of primary and C2C12 myoblasts, both of which expressed high CnAβ1 levels, was accompanied by a progressive increase in CnAβ2 and a decline in CnAβ1 levels (Fig. 2, B and C). In vivo, skeletal muscle regeneration in response to injury induced a transient rise in CnAβ1 transcripts and a concomitant decrease in CnAβ2 transcripts (Fig. 2, D–F). IGF-1 was able to induce CnAβ1 expression both in vivo, in MLC/mIGF-1 transgenic mice especially during recovery from muscle atrophy induced by sciatic denervation (Fig. 2 G), and in vitro in starving myotubes (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200704179/DC1). To determine whether CnAβ1 was expressed by muscle precursors and regenerating fibers or by the immune infiltrate, we analyzed its tissue distribution during skeletal muscle regeneration in the tibialis anterior of wild-type mice 2 and 6 d after cardiotoxin injection. CnAβ1 was strongly expressed by muscle precursors and regenerating muscle fibers, showing a pattern similar to that of Myf5, which labels satellite cells and early regenerating fibers (Cooper et al., 1999), whereas uninjured fibers or the immune infiltrate (marked by anti-CD45) expressed little or no CnAβ1 (Fig. 3). These results implicate the predominant CnAβ2 isoform in skeletal muscle differentiation and maturation, whereas CnAβ1 might play a role during myoblast proliferation and muscle regeneration.
Figure 2.
Profile of CnA isoforms expression in skeletal muscle. (A) Expression patterns of CnAα, CnAβ2, and CnAβ1 isoforms were analyzed in different striated muscles by Northern blot. (B) CnA isoform expression was determined by qRT-PCR in C2C12 myoblasts incubated in growth medium (GM) or differentiation medium (DM) for days indicated. Results are expressed as fold induction over the value in GM (note Log relative units ±SD). (C) Comparison of CnA isoform expression in proliferating primary myoblasts and adult skeletal muscle. Satellite cells were isolated using the single fiber technique from rat EDL muscle, induced to proliferate in GM, and then shifted to DM for days indicated. RNA was extracted and CnAα, CnAβ2, and CnAβ1 expression analyzed by qRT-PCR. Results are expressed as fold induction over myoblasts in GM. (D) 2-mo-old wild-type mice were injured in the quadriceps muscle by cardiotoxin injection and allowed to recover for 2, 6, or 12 d (0, uninjured). Total muscle RNA was extracted and CnAα, CnAβ2, and CnAβ1 expression was determined by qRT-PCR. Results are expressed as fold induction over uninjured muscle (note Log relative units ±SD). (E and F) The switch between CnAβ1 and CnAβ2 expression during skeletal muscle regeneration and the transient increase in CnAβ1 mRNA expression were confirmed by Northern blot. (G) CnA isoform expression in denervated muscle from wild-type and MLC/mIGF-1 transgenic mice was analyzed by qRT-PCR 28 d after denervation and expressed as fold induction ±SD over nondenervated wild type. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Figure 3.
CnAβ1 is expressed in myoblasts and regenerating skeletal muscle fibers. 2-mo-old wild-type mice were injured in the tibialis anterior (TA) muscle by cardiotoxin injection and allowed to recover for 2 or 6 d. The TA muscle was extracted, fixed, sectioned and immunostained with anti-CnAβ1, anti-Myf5, anti-CD45, or secondary antibody alone (Control). Arrows indicate positive cells. Bar, 100 μm.
CnAβ1 regulates myoblast proliferation and differentiation
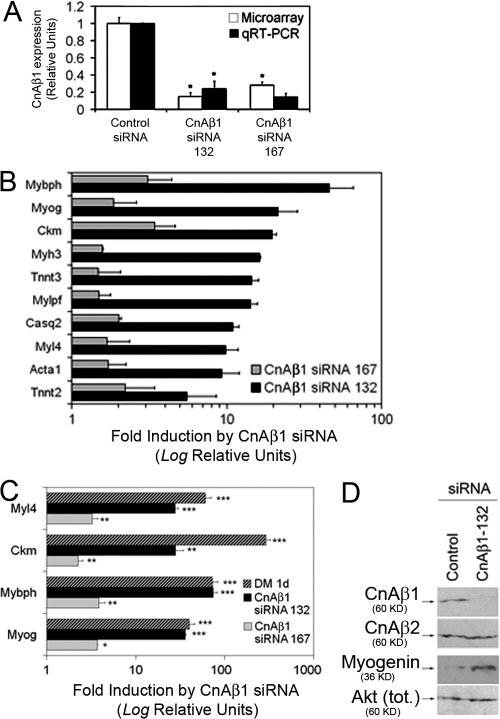
To characterize the function of CnAβ1, we knocked down CnAβ1 expression by transfecting proliferating C2C12 myoblasts with two different siRNAs specific for mouse CnAβ1 (termed 132 and 167) or a luciferase siRNA as a control (Fig. 4 A). After 48 h in growth medium (GM), oligonucleotide-based microarray analysis showed an induction of muscle differentiation-specific gene expression in cells transfected with the CnAβ1 siRNAs (Fig. 4 B). The induction of the muscle differentiation program was confirmed by qRT-PCR and was found to be similar to that obtained with cells in differentiation medium (DM) for 1 d (Fig. 4 C). Although the response obtained with the siRNA 167 was weaker than that obtained with the 132 siRNA, similarities in the profiles of genes up-regulated by each of the CnAβ1 siRNAs (P < 4.63 × 10−138; Table S1 and S2, available at http://www.jcb.org/cgi/content/full/jcb.200704179/DC1) and down-regulated genes (P < 4.39 × 10−60; Table S3 and S4) indicated that the observed effects were due to specific knockdown of CnAβ1 rather than to off-target effects. Correspondingly, an increase in myogenin expression was detected by Western blot after transfection of C2C12 cells with the CnAβ1 siRNA (Fig. 4 D).
Figure 4.
CnAβ1 knock-down results in myoblast differentiation. (A) C2C12 myoblasts were transfected with CnAβ1-specific siRNAs 132 (black bars) and 167 (gray bars) or a control siRNA, grown for 48 h in growth medium (GM), and the expression of CnAβ1 was assayed by microarray and qRT-PCR. (B and C) RNA was extracted from myoblasts transfected with control or CnAβ1 siRNAs and subjected to microarray (B) or qRT-PCR (C) analysis. Comparison with untransfected cells differentiated for 24 h (DM 1d) is shown. Gene expression is represented as fold induction ±SD (note Log. relative units) of values from cells transfected with each CnAβ1 siRNA over those transfected with control siRNA. (D) C2C12 myoblasts were transfected with control or CnAβ1 siRNA and the expression of CnAβ1, CnAβ2, and myogenin was analyzed by Western blot. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Given that CnAβ1 knockdown results in decreased myoblast proliferation and increased differentiation and because its expression declines during myotube formation, we studied whether CnAβ1 could actually interfere with myoblast differentiation. We generated C2C12 stable transfectants in which the expression of the different CnAβ isoforms is driven by the CMV promoter (pcDNA3.1 vector) and thus is not affected by differentiation. When induced to differentiate, C2C12 cells stably overexpressing CnAβ1 showed a mild significant increase in differentiation markers only at early time points (Fig. S2, B and C) compared with the mock transfectants. These data suggest that CnAβ1 may slightly accelerate the initial differentiation steps when artificially overexpressed. It is noteworthy that this effect was only detected after switching C2C12 cells to differentiation medium and was not observed under growth conditions. A similar dual action has been previously reported for IGF-1, which is able to both enhance myoblast proliferation and up-regulate differentiation markers once the cells have been induced to differentiate (Engert et al., 1996; Musarò and Rosenthal, 1999). On the contrary, continuous overexpression of CnAβ1 had no impact on differentiation markers 5 d after shifting the cells to differentiation medium. In addition, 8 d after differentiation, myotubes overexpressing CnAβ1 showed a similar diameter to that of myotubes bearing the expression vectors for CnAβ*, CnAβ2, or the parental vector alone (see Fig. 6 H; Fig. S3 C, available at http://www.jcb.org/cgi/content/full/jcb.200704179/DC1), indicating that although CnAβ1 expression is required to prevent myoblast differentiation, overexpression of this Cn isoform does not interfere with myotube differentiation.
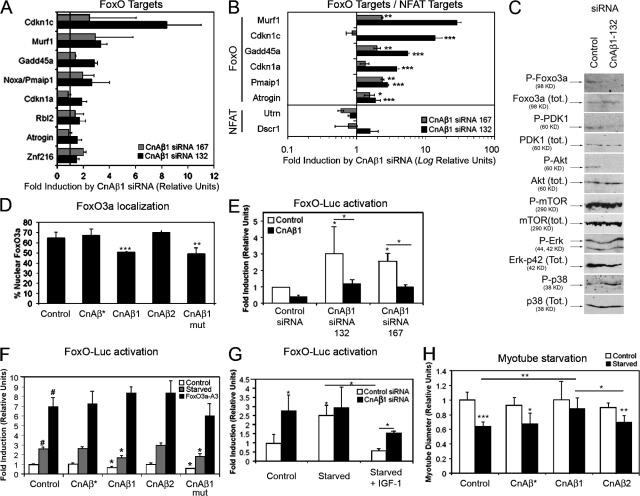
Figure 6.
CnAβ1 inhibits FoxO. (A and B) C2C12 myoblasts were transfected with CnAβ1 siRNAs 132 (black bars) and 167 (gray bars) or a control siRNA and grown for 48 h in GM. RNA was analyzed by microarray (A) and qRT-PCR (B) for induction of FoxO (A and B) or NFAT (B) targets and expressed as fold induction ±SD of values from cells transfected with each CnAβ1 siRNA over those transfected with control siRNA. (C) C2C12 cells were transfected with CnAβ1 siRNA 132 or a control siRNA and the phosphorylation status of Foxo3a, PDK1, Akt, mTOR, Erk, and p38 was analyzed by Western blot. (D) C2C12 myoblasts were transfected with pGFP-FoxO3a and CnA expression vectors and the number of cells with preferentially nuclear GFP-Foxo3a localization was quantified. (E–G) C2C12 myoblasts were cotransfected with the p6xDBE-Luc reporter and the following vectors and/or siRNAs: (E) control or CnAβ1 siRNAs 132 and 167, together with CnAβ1 (black bars) or control (white bars) expression plasmids; (F) control or different CnA expression vectors; (G) control or CnAβ1 siRNA 132. (F and G) Cells were incubated for 48 h in GM (Control) or starving conditions. FoxO3a-A3 vector (F) or 25 ng/ml IGF-1 (G) were added where indicated. Luciferase activity is expressed as fold induction ±SD over the value of empty pcDNA3.1 in GM (F) or control siRNA in GM (E and G). (H) C2C12 myoblasts stably transfected with the different CnAβ or empty (control) expression vectors were differentiated for 8 d and incubated overnight in starving conditions. The myotube diameter was determined and expressed as relative units ±SD. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
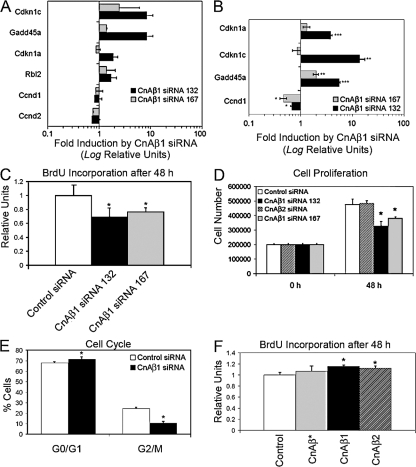
Interestingly, CnAβ1 knockdown induced an increase in negative cell cycle regulators and a decrease in genes involved in cell cycle progression (Fig. 5, A and B). A corresponding decrease in BrdU incorporation (Fig. 5 C) and cell proliferation (Fig. 5 D) was observed after CnAβ1, but not CnAβ2, knock-down, accompanied by a decrease in the percentage of cells in the G2/M phase of the cell cycle and an increase in G1 (Fig. 5 E), indicating that the CnAβ1 isoform is necessary for cell cycle progression. C2C12 stable transfectants overexpressing CnAβ1 showed only a modest increase in BrdU incorporation (Fig. 5 F). Together, these results suggest that CnAβ1 is required for myoblast proliferation and that CnAβ1 knockdown results in exit from the cell cycle and subsequent myoblast differentiation.
Figure 5.
CnAβ1 knock-down reduces myoblast proliferation. (A and B) C2C12 myoblasts were transfected with control or CnAβ1 siRNAs 132 and 167, grown for 48 h, and mRNA expression was analyzed by microarray (A) and qRT-PCR (B). Gene expression is shown as fold induction ±SD (note Log. relative units) of values from cells transfected with each CnAβ1 siRNA over those transfected with control siRNA. (C, D, and F) C2C12 cells transfected with the different siRNAs (C and D) or stably transfected with CnAβ expression plasmids (F) were allowed to grow for 48 h and their proliferative capacity was determined by BrdU incorporation (C and F) and MTT (D) assays. (E) C2C12 cells were transfected as in A, with control or CnAβ1 siRNA, grown for 48 h, and the percentage of cells in the G0/G1 and G2/M phases of the cell cycle was determined by flow cytometry after staining the cells with propidium iodide. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
CnAβ1 inhibits FoxO transcription factors
Many of the negative cell cycle regulators induced by CnAβ1 inhibition, such as p21, Rbl2, and Gadd45a, are known to be regulated by the FoxO family of transcription factors (Furukawa-Hibi et al., 2002; Kops et al., 2002; Tran et al., 2002; Seoane et al., 2004). We observed that additional FoxO target genes were induced after CnAβ1 knockdown as observed by microarray (Fig. 6 A) and qRT-PCR analysis (Fig. 6 B), including increased expression of atrogenes Murf1 and MAFbx/Atrogin that are known to mediate FoxO-induced muscle atrophy (Sandri et al., 2004; Stitt et al., 2004). Inhibition of CnAβ1 by siRNA abrogated FoxO3a phosphorylation at Ser253, (Fig. 6 C) and also resulted in defective PDK1 and Akt phosphorylation, without affecting mTOR or Erk, suggesting that CnAβ1 is required for the activation of the PDK1-Akt-FoxO phosphorylation cascade that leads to FoxO inhibition. An increase in p38 phosphorylation was observed that is likely to be a direct consequence of C2C12 differentiation (Perdiguero et al., 2007), rather than a direct target of CnAβ1 knockdown. Moreover, overexpression of CnAβ1, but not CnAβ2 or the constitutively active CnAβ*, in C2C12 cells inhibited FoxO nuclear localization (Fig. 6 D; Fig. S3 A) and the activity of a FoxO-dependent luciferase reporter (Fig. 6 F), whereas transfection of C2C12 myoblasts with the CnAβ1 siRNAs 132 or 167 activated the reporter (Fig. 6 E). This activation was prevented by cotransfection with an expression plasmid for human CnAβ1, confirming the specificity of the response.
To determine whether FoxO inhibition by CnAβ1 can prevent myotube atrophy, C2C12 stable transfectants overexpressing CnAβ*, CnAβ1, or CnAβ2 were differentiated into myotubes for 8 d and then starved for 16 h. As shown in Fig. 6 H and Fig. S3 C, starvation caused a 35–40% decrease in myotube diameter that was prevented by CnAβ1, but not by CnAβ* or CnAβ2. Accordingly, CnAβ1 overexpression repressed FoxO in starving cells (Fig. 6 F) and CnAβ1 knockdown blocked repression of FoxO by IGF-1 (Fig. 6 G). However, CnAβ1 had no effect on a constitutively active FoxO3a-A3 mutant lacking the three Akt/SGK phosphorylation sites (Nakae et al., 2000) (Fig. 6 F), which also renders the transcription factor insensitive to IGF-1 (Zheng et al., 2000; Brunet et al., 2001). Because IGF-1 inhibits FoxO-regulated atrogenes (Sandri et al., 2004; Stitt et al., 2004; and Fig. S3 B) and CnAβ1 expression is induced by IGF-1 in atrophied and regenerating muscle (Dobrowolny et al., 2005; Fig. 2 G, Fig. S2 A), IGF-1 and CnAβ1 may share common pathways for the inhibition of FoxO and FoxO-regulated processes. Interestingly, inhibition of FoxO activity and nuclear localization by CnAβ1 did not require a functional phosphatase domain because a phosphatase-dead CnAβ1 mutant (CnAβ1mut) also reduced FoxO nuclear localization and activation (Fig. 6, D and F). In addition, NFAT-regulated genes were only slightly affected by CnAβ1 knockdown in myoblasts (Fig. 6 B). These results suggest that the main role of CnAβ1 in proliferating myoblasts is to inhibit FoxO, allowing cell proliferation and preventing differentiation, rather than to activate NFAT, which is involved in myoblast differentiation (Delling et al., 2000) after CnAβ1 expression has been shut down.
CnAβ1 enhances skeletal muscle regeneration
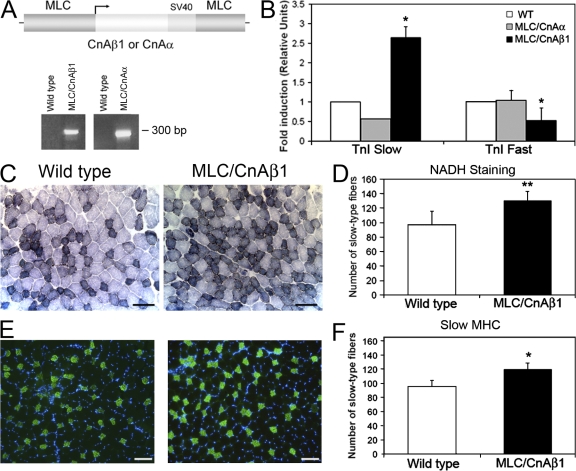
Given that CnAβ1 expression is induced by mIGF-1 during skeletal muscle regeneration (Fig. 2 G; Dobrowolny et al., 2005) and is necessary for the activation of the PDK1/Akt signaling pathway that leads to FoxO inhibition, we explored whether CnAβ1 shared some of the regenerative abilities of mIGF-1. We generated MLC/CnAβ1 transgenic mice, expressing CnAβ1 in a skeletal muscle-specific, post-mitotic restricted fashion using a myosin light chain (MLC)1/3 expression cassette (Fig. 7 A), and MLC/CnAα transgenic mice expressing a full-length CnAα construct, including the autoinhibitory domain, as control. DNA microarray-based transcript expression profiling of the quadriceps muscles of wild-type, MLC/CnAα, and MLC/CnAβ1 uninjured mice at 2 mo of age revealed an induction of slow fiber-associated genes including β-myosin heavy chain (Myh7), myosin light chains (Myl3, Myl2), troponins (Tnni1, Tnnt1), tropomyosins (Tpm3), and Serca2a in MLC/CnAβ1 mice compared with wild- type and MLC/CnAα mice (Table S4). Quantitative RT-PCR corroborated the up-regulation of TnI1 (slow) and slight decrease of TnI2 (fast) expression in MLC/CnAβ1 mice compared with wild-type and MLC/CnAα (Fig. 7 B). Moreover, NADH-tetrazolium reductase staining of EDL sections from wild-type and MLC/CnAβ1 mice showed a significant increase in the number of slow fibers in mice overexpressing CnAβ1 (Fig. 7, C and D). and similar results were obtained when the amount of slow MHC-expressing fibers was determined by immunohistochemistry (Fig. 7, E and F).
Figure 7.
CnAβ1 induces an increase in slow-type muscle fibers. (A) Transgenic construct used in the generation of MLC/CnAβ1 and MLC/CnAα mice. The CnAβ1 or CnAα cDNA was placed under the control of the myosin light chain promoter and enhancer, which allows skeletal muscle-specific, post- mitotic expression. Expression of CnAβ1 and CnAα transgenes in the quadriceps muscle of the transgenic mice was confirmed by RT-PCR using specific primers for the transgene. (B) Total RNA was extracted from the quadriceps of 2-mo-old MLC/CnAβ1, MLC/CnAα, and wild-type mice and the expression of TnI-slow and TnI-fast was determined by qRT-PCR. Results are given as relative units referred to the values obtained in the wild-type mouse quadriceps ±SD. (C and E) The EDL muscle from wild-type and MLC/CnAβ1 mice was stained for NADH activity (C and D) or slow MyHC expression (E and F) and the number of slow fibers per field was determined (D and F). At least seven fields per mouse were counted. **, P < 0.01. Bar, 100 μm.
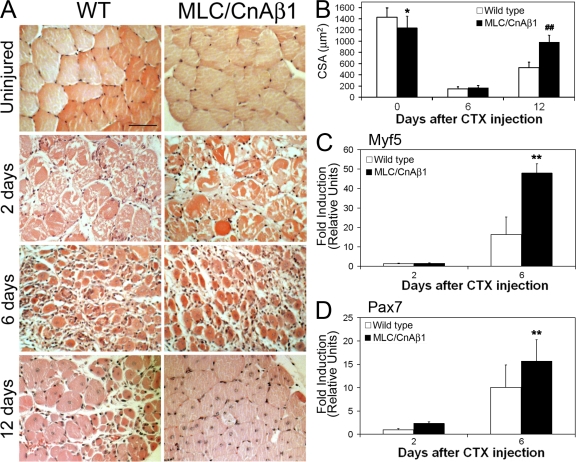
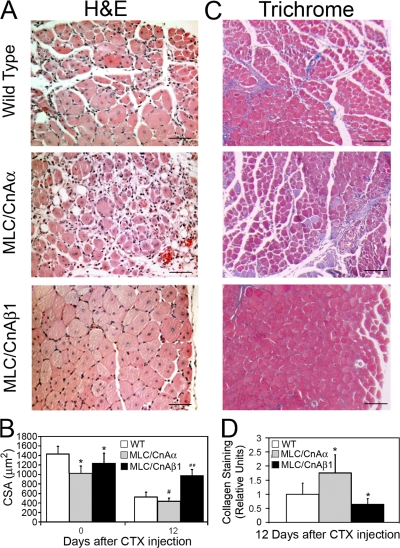
To determine whether supplemental CnAβ1 transgene expression contributed to muscle regeneration, we injected cardiotoxin into the tibialis anterior (TA) and quadriceps muscles of wild-type (WT), MLC/CnAα, and MLC/CnAβ1 mice and allowed regeneration to proceed for 2, 6, or 12 d (Musarò et al., 2001). Both wild-type and MLC/CnAβ1 mice presented similar muscle damage 2 d after the injury, suggesting analogous susceptibility to the toxin (Fig. 8 A). In contrast, regenerating TA muscle from MLC/CnAβ1 mice 12 d after CTX injury contained larger myotubes with centralized nuclei and less extracellular matrix accumulation when compared with regenerating wild-type muscle (Fig. 8 A). The enhanced regenerative capacity induced by CnAβ1 was also reflected in the increased mean cross-sectional area (CSA) of the MLC/CnAβ1 myofibers 12 d after CTX injection, compared with those of wild-typeT mice (2.1-fold increase; Fig. 8 B). Interestingly, an increase in the satellite cell markers Myf5 and Pax7 was observed 6 d after the injury in MLC/CnAβ1 mice compared with wild type (Fig. 8, C and D), paralleling the response obtained in response to IGF-1 in a murine model of ALS (Dobrowolny et al., 2005). Unlike mIGF-1 (Musarò et al., 2001), CnAβ1 overexpression did not result in muscle hypertrophy, indicating that the regenerative and hypertrophic responses induced by mIGF-1 in skeletal muscle may be driven by different mechanisms.
Figure 8.
CnAβ1 enhances skeletal muscle regeneration. Wild-type and MLC/CnAβ1 mice were given five cardiotoxin (CTX) injections in the tibialis anterior (TA) and allowed to recover for 6 or 12 d (0, uninjured). The TA was analyzed by hematoxylin and eosin (H&E) staining (A) and the average cross- sectional area (CSA) of the regenerating myofibers was measured (B). Results are expressed as μm2 ±SD. ##, P < 10−12 (t test; n = 2,500 fibers (three mice)/mouse line). Bar, 40 μm.(C and D) RNA from the quadriceps of wild-type and MLC/CnAβ1 mice 2 and 6 d after cardiotoxin injection was analyzed for Myf5 (C) and Pax7 (D) expression by qRT-PCR. **, P < 0.005 compared with wild type.
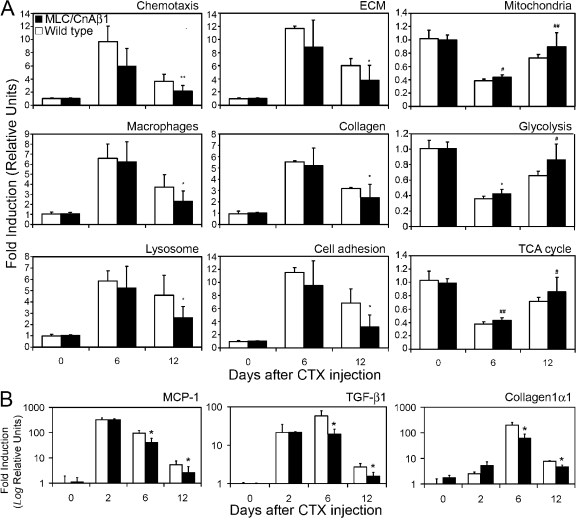
To elucidate the mechanisms underlying the enhanced skeletal muscle regeneration observed in MLC/CnAβ1 mice, we analyzed the microarray transcription profiles of wild-type, MLC/CnAβ1, and MLC/CnAα quadriceps during the regeneration process. In CTX-injured wild-type muscle, induction of genes involved in the immune response included chemotaxis and macrophage-specific genes, and extracellular matrix production, especially collagen, paralleled by a strong decrease in the expression of genes associated with the mitochondria and energy production, likely reflecting myocyte necrosis (Fig. 9 A; Table S5, available at http://www.jcb.org/cgi/content/full/jcb.200704179/DC1). In contrast, MLC/CnAβ1 muscle significantly decreased the expression of genes associated with both immune response and scar formation 12 d after the muscle injury compared with wild-type mice and showed a faster recovery of mitochondrial gene expression (Fig. 9 A). Quantitative RT-PCR analysis of the chemokine macrophage chemoattractant protein (MCP-1), the profibrotic factor TGF-β1, and collagen I confirmed the results seen with the microarrays, with a significant decrease of these factors in MLC/CnAβ1 mice at both 6 and 12 d after the injury, and showed no differences between the two genotypes 2 d after CTX injection (Fig. 9 B), confirming that wild-type and MLC/CnAβ1 mice were equally susceptible to the action of cardiotoxin. We quantified the presence of different leukocyte populations during muscle regeneration by qRT-PCR and found decreased expression of the monocyte/macrophage marker CD14 in MLC/CnAβ1 mice compared with wild type 6 and 12 d after the injury (Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200704179/DC1). This effect was paralleled by a lower degree of endothelial activation (Selectin P expression) and decreased expression of chemokines Ccl2/MCP-1 and Ccl5/Rantes in MLC/CnAβ1 mice (Fig. S4 A and Fig. 9), together with increased expression of the anti-inflammatory cytokine IL-10, an immunosuppressive mediator involved in the resolution of inflammation and the regulation of extracellular matrix. No significant differences were observed in CD4 and CD8 lymphocyte subpopulations between wild-type and transgenic mice, and we did not detect neutrophil markers by microarray or qRT-PCR at the analyzed time points. Together, our results suggest that the negative regulation of macrophage recruitment through Ccl2/MCP-1 and Ccl5/Rantes among others, combined with the secretion of anti-inflammatory mediators such as IL-10, may be responsible for the accelerated resolution of inflammation observed in MLC/CnAβ1 mice.
Figure 9.
MLC/CnAβ1 mice show decreased inflammation and scar formation 12 d after muscle injury. (A) Total RNA was extracted from the quadriceps muscle of injured (6 and 12 d after CTX injection) and uninjured mice and subjected to microarray analysis. The average expression ±SD of the genes in the indicated gene ontology lists is shown in wild-type (white bars) and MLC/CnAβ1 mice (black bars), referred to the uninjured mice. (B) The expression of MCP-1, Collagen1α1 and TGF-β1 mRNA was analyzed in the quadriceps of wild-type (white bars) and MLC/CnAβ1 mice (black bars) at different days after CTX injection (0, uninjured). Results are expressed as fold induction ±SD over the values of uninjured mice. *, P < 0.05; **, P < 0.005; #, P < 10−5; ##, P < 10−13.
Notably, regenerating MLC/CnAα muscle showed increased matrix deposition, with small, separated myotubes and persistent presence of infiltrating inflammatory cells (Fig. 10 A) that was reflected in the smaller cross-sectional area presented by the regenerating myofibers (Fig. 10 B) and the increased collagen staining (Fig. 10, C and D). Microarray analysis of MLC/CnAα mice 12 d after CTX injection revealed increased expression levels of genes associated with macrophage presence and extracellular matrix production (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200704179/DC1), indicating enhanced inflammation and fibrosis and supporting the impaired muscle regeneration displayed by these mice at this stage. In summary, the improved muscle regeneration associated with CnAβ1 transgene expression was characterized by enhanced muscle precursor expansion, decreased fibrosis and inflammation, and faster muscle recovery, whereas supplemental expression of CnAα exacerbated rather than improved the regenerative capacity of skeletal muscle.
Figure 10.
Overexpression of full-length CnAα isoform inhibits skeletal muscle regeneration. (A and C) Wild-type, MLC/CnAα and MLC/CnAβ1 mice were given five CTX injections into the tibialis anterior and the muscle analyzed after 12 d by hematoxylin and eosin (H&E) or Masson trichrome (C) staining. Bar, 40 μm (A) or 100 μm (C). (B) The average cross-sectional area (CSA) of uninjured (0 d) and regenerating myofibers 12 d after CTX injection was measured and expressed as μm2 ±SD. #, P < 10−5; ##, P < 10−12 (t test; n = 2,500 fibers (three mice)/mouse line). (D) Collagen staining from C was quantified and expressed as fold induction ±SD over the value of wild-type mice. *, P < 0.05.
Discussion
This study establishes CnAβ1 as an evolutionarily conserved, partially CsA-insensitive Cn splicing variant that regulates cell proliferation and improves muscle regeneration, inhibiting FoxO independently of its phosphatase activity. The unexpected connection between FoxO and calcineurin pathways, specific for the CnAβ1 isoform, may have important implications for the regulation of cell proliferation and for treatment of muscle wasting.
Cn activity is tightly regulated by calcium through CnB and calmodulin. In response to changes in intracellular calcium levels, the autoinhibitory domain of the enzyme is removed and the catalytic site is exposed (Ke and Huai, 2003). The lack of an autoinhibitory domain confers constitutive phosphatase activity and NFAT activation on the CnAβ1 variant in the absence of calcium increase. Although constitutive enzymatic activity has been previously described for artificially truncated CnA forms (Molkentin et al., 1998), CnAβ1 is, to our knowledge, the first naturally occurring CnA isoform with this capacity. Contrary to artificially truncated CnA isoforms, CnAβ1 is partially insensitive to the action of the immunosuppressant CsA, suggesting that the alternative C-terminal domain in CnAβ1 may interfere with the binding of the CsA–cyclophilin complex to the enzyme and thus prevents its inactivation. Together, these findings indicate that the Cn/NFAT pathway can be activated without the need for a rise in intracellular calcium levels by alternative splicing of the CnAβ gene, and that this activation is partially resistant to the action of CsA.
The inhibition of FoxO by CnAβ1 has important implications. The family of FoxO transcription factors control key physiological processes, including cell proliferation, growth, atrophy, metabolism, and longevity (Greer and Brunet, 2005). In myoblasts, FoxO is sufficient to induce Gadd45 expression and prevent cell cycle progression (Furukawa-Hibi et al., 2002). The abundant expression of CnAβ1 in proliferating myoblasts is necessary to prevent FoxO activation. Indeed, CnAβ1 knockdown results in the induction of negative cell cycle regulators, blockade of cell proliferation, and subsequent myoblast differentiation. It is noteworthy that C2C12 cells overexpressing CnAβ1 fuse normally into myotubes (Fig. 6 H and Fig. S4 B), suggesting that CnAβ1 expression is necessary for myoblast proliferation rather than for inhibition of differentiation. Interestingly, when overexpressed in transgenic mice, CnAβ1 induces an increase in slow muscle fibers, an effect compatible with both FoxO inhibition and NFAT activation (Naya et al., 2000; Kamei et al., 2004; McCullagh et al., 2004).
FoxO factors play a crucial role in skeletal muscle atrophy through the induction of MAFbx/Atrogin and Murf1 (Sandri et al., 2004; Stitt et al., 2004). In response to IGF-1, Akt phosphorylates and inhibits FoxO, preventing Atrogin expression and muscle atrophy (Stitt et al., 2004; Schulze et al., 2005). Interestingly, CnAβ1 is induced by IGF-1 in regenerating muscle and is necessary for the inhibition of FoxO activity by IGF-1 (Fig. 2 G, Fig. 6 G, Fig. S3 A; Dobrowolny et al., 2005). In addition, CnAβ1 itself is able to inhibit FoxO and prevent myotube atrophy induced by starvation. FoxO induction of MAFbx/Atrogin has recently been shown to inhibit Cn activity (Li et al., 2004; Ni et al., 2006); thus, the inhibition of FoxO by CnAβ1 might indirectly contribute to the increased Cn activity in CnAβ1-expressing cells. Like IGF-1, CnAβ1 needs the three Akt target Ser/Thr residues in FoxO in order to inhibit its activity and furthermore, CnAβ1 expression is necessary for PDK1 and Akt activation in both myoblasts and tumor cells (Fig. 4 C and Fig. 6 D), suggesting that IGF-1 and CnAβ1 share a common signaling pathway.
Supplemental expression of CnAβ1 in post-mitotic muscle leads to enhanced muscle regeneration. Our results suggest that the quicker expansion of muscle precursors combined with a faster resolution of both the inflammatory response and subsequent extracellular matrix deposition allows the regenerating fibers to grow more rapidly in response to CnAβ1, whereas mice overexpressing the full-length CnAα isoform show delayed regeneration, with persistent inflammation and increased deposition of extracellular matrix. Interestingly, mIGF-1 enhances muscle regeneration in various models of muscle injury and disease by reducing scar formation and resolving inflammation faster, facilitating the recruitment of stem and progenitor cells (Musarò et al., 2001, 2004; Barton et al., 2002; Dobrowolny et al., 2005). Thus, although the role of Cn in skeletal muscle regeneration remains controversial and both pro- and anti-regenerative effects have been reported depending on the mouse model used (Stupka et al., 2004; Parsons et al., 2007), the present study shows that CnAβ1 is able to recapitulate the skeletal muscle regenerative response induced by mIGF-1, probably sharing a common mechanism, suggesting that this naturally occurring Cn isoform could prove a valuable tool in therapies aiming to counteract muscle atrophy and enhance regeneration.
Materials and methods
Cells, plasmids, and transfection
Cells.
The murine myoblast cell line C2C12 was propagated in growth medium (GM) (Dulbecco's modified Eagle's medium [DMEM] 4.5 g/liter glucose, 10% FCS, 2 mM l-glutamine, and 5 mM penicillin/streptomycin) and induced to differentiate in differentiation medium (DM) (growth medium, 0% FCS, and 2% horse serum). C2C12 cells were starved by incubating in PBS, 1 mM CaCl2, and 1 mM MgCl2 for 16 h as described (Sandri et al., 2004). Muscle satellite cells were isolated from rat EDL muscles using the single fiber technique as previously described (Suzuki et al., 2001). HEK293 cells were grown in DMEM, 1 g/liter glucose, 10% FCS, 2 mM l-glutamine, and 5 mM penicillin/streptomycin. Jurkat T cells were grown in RPMI, 10% FCS, 2 mM l-glutamine, and 5 mM penicillin/streptomycin.
Plasmids.
The expression vectors pcDNA3-CnAβ1, pcDNA3-CnAβ1mut, pcDNA3-CnAβ2, pcDNA3-CnAα, pcDNA3.1-CnAα*, and pcDNA3.1-CnAβ* carry cDNAs of the respective CnA isoforms under the control of a CMV promoter (CnAα* and CnAβ* represent the artificially truncated forms of CnAα and CnAβ2; CnAβ1mut carries a D130N mutation and lacks phosphatase activity). pGal4-Luc and p6xDBE-Luc express the luciferase reporter gene under the control of three tandem binding sites for the yeast transcription factor Gal4 or six tandem binding sites for the Caenorhabditis. elegans FoxO homologue Daf16, respectively. pGal4-NFAT (1–415) expresses a chimerical protein containing the Gal4 DNA-binding domain linked to NFAT transcription activation and regulatory domains (amino acids 1–415) (Luo et al., 1996). pGFP-VIVIT carries the NFAT-specific inhibitory peptide VIVIT linked to GFP (15). pHA-NFAT expresses NFATc1 tagged with the hemagglutinin peptide. pGFP-FoxO3a and pFoxO3a-A3 express, respectively, a GFP-FoxO3a fusion protein and a FoxO3a mutant in which the three Thr/Ser known to be phosphorylated by Akt and SGK are mutated into Ala, rendering FoxO3a insensitive to inhibition by Akt or IGF-1 (Dijkers et al., 2000; Potente et al., 2005).
Transfection.
For NFAT activity analysis, C2C12 myoblasts were transfected for 6 h with 0.5 μg of Gal4-Luc, 4 ng of the different pGal4-NFAT vectors along with 3 μg of pcDNA3-CnA, or empty vector, by using 10 μg of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Where indicated, 1 μg of pGFP-VIVIT or the control vector pGFP was added. After 18 h in GM, cells were induced to differentiate in DM for 48 h and luciferase activity was measured. Where indicated, 10 ng/ml of cyclosporin A (or equivalent volume of EtOH) was added with DM. Jurkat T cells were grown and transfected as described (Lara-Pezzi et al., 2004), following the same protocols as for C2C12 cells. For FoxO activity analysis, C2C12 cells were transfected with 0.25 μg of p6xDBE-Luc and either 2.5 μg Cn expression vectors and 0.8 μg of pFoxO3a-A3 (or pcDNA3.1 as negative control), or 100 pmol of CnAβ2 or CnAβ1 siRNA. Where indicated, 25 ng/ml IGF-1 was added to the culture for 16 h. IGF-1 and cyclosporin a (CsA) were purchased from Sigma-Aldrich. Similar expression levels of all CnAβ isoforms after transfection were detected by qRT-PCR (Fig. S3). Stable C2C12 transfectants bearing the pcDNA3.1-CnAβ1 construct or the empty pcDNA3.1 vector were generated by selecting the cells with 0.5 mg/ml G418 after transfection. Two different polyclonal populations of transfectants were used in this study. All stable transfectants expressed similar levels of the transfected CnAβ isoform, as quantified by qRT-PCR (Fig. S3 B).
NADH staining, immunohistochemistry, muscle injury, and denervation
Nicotinamide adenine dinucleotide nitro-blue tetrazolium (NADH)-TR staining was used to differentiate fast, intermediate, and slow fibers. Mice were killed by cervical dislocation and muscles were removed as fast as possible, embedded in Tragacanth gum (Sigma-Aldrich), and frozen in isopentane-submerged liquid nitrogen. Frozen muscle sections (8 μm) were incubated in NADH-TR solution (NADH solution (Sigma-Aldrich)/nitro blue tetrazolium (NBT) solution (Sigma-Aldrich) [vol/vol]) for 30 min at 37°C, followed by three washes in ddH2O and three changes of increasing (30, 60, 90%) and decreasing acetone solutions (30 s each) to remove unbound NBT. Slides were rinsed with ddH2O and mounted with Aqua-Mount (Lerner Laboratories). For MyHC staining, frozen sections were cut at 16 μm, air-dried, and fixed in 4% PFA for 10 min on ice. Sections were then incubated in PBT buffer for 15 min (0.3% Tween 20 in PBS) and blocked in 5% goat serum in PBT for 1 h. Primary antibodies were applied over night at 4°C at a 1:400 dilution (Anti-Myosin Skeletal, fast, Clone MY-32; Anti-Myosin Skeletal, slow, Clone NOQ7.5.4D; both from Sigma-Aldrich). Samples were washed three times for 15 min in PBT buffer and incubated with goat anti rabbit-Alexa488 secondary antibody for 45 min at RT, washed three times for 15 min in PBT and once in PBS containing 1× Hoechst. Images were analyzed using a microscope (Axiovert 200; Carl Zeiss, Inc.) with an Achroplan 10×/0.25 Ph1 ∞/− objective at room temperature, using 70% glycerol/ Tris-HCl as imaging medium. Pictures were captured using a digital camera (DXM 1200; Nikon) and acquisition software from Nikon (ACT-1, ver. 2). Brightness/contrast enhancement was applied to the whole picture at the same time using Adobe Photoshop.
For skeletal muscle regeneration experiments, tibialis anterior (TA) and quadriceps muscles from 2-mo MLC/CnAβ1, MLC/CnAα or wild-type mice were given five (TA) or eight (quadriceps) 5-μl injections of 10 μM cardiotoxin in PBS (Musarò et al., 2001). Quadriceps muscles were collected 2, 6, and 12 d (2, 5, and 10 d for the Northern blot) after cardiotoxin injection and snap-frozen in liquid nitrogen, and RNA was extracted. MLC/mIGF-1 and denervation studies have been previously described (Musarò et al., 2001; Mourkioti et al., 2006). For histology, tibialis anterior was fixed in 4% paraformaldehyde, dehydrated, and included in paraffin. Sections were incubated with anti-CnAβ1 (developed by the EMBL Monoclonal Core Facility), anti-Myf5 (Santa Cruz Biotechnology, Inc.), or anti-CD45 (BD Biosciences) followed by the appropriate HRP-conjugated secondary antibody and counterstained with hematoxylin and eosin. For collagen staining, TA sections were stained using the Trichrome Stain (Masson) kit from Sigma-Aldrich, following the manufacturer's recommendations. All images were acquired using a microscope (Axioskop; Carl Zeiss, Inc.) and a digital camera (DXM 1200F; Nikon) and analyzed using Nikon software (ACT-1, ver. 2.62). Brightness/contrast enhancement was applied to the whole picture at the same time using Adobe Photoshop. All experiments involving animals were approved by the EMBL Animal Ethics Committee and were performed according to local and institutional guidelines.
siRNA, RNA isolation, cDNAs, Northern blot, and real-time PCR
The following stealth siRNAs (Invitrogen) were used: mouse CnAβ1 132 (AGUUCCUGUCUUAGCAGCUGACAUA), mouse CnAβ1 167 (GGGUAUUAUGUGAUAGGCAUCUGAU), human CnAβ1 136 (GGUAGUGUAUUAGCUAGUGUCUCAU), human CnAβ1 247 (GAUAUGGGAGCAGCUCAUAUCAUAA), mouse and human CnAβ2 (GACUGGCAACCAUAGUGCCCAGUGA), and Luciferase, used as a negative control (GCCCGCGAACGAGAUUUAUAAUGAA). Total RNA was extracted from snap-frozen tissue or cell cultures by using Trizol (Sigma-Aldrich). 20 μg total RNA were resolved by agarose gel, blotted and hybridized with CnAα-, CnAβ2-, and CnAβ1-specific probes. For real-time PCR analysis, cDNA was generated using a Roche kit from 2 μg of total RNA in a 40-μl volume. Human tumor cDNA (matching tumor and nontumor samples from the same patient) from five lung tumor patients and five colon tumor patients was purchased from Clontech Laboratories, Inc.
A total of 2 μl of the cDNA reaction were amplified by PCR in duplicate by using either Taqman probes (Applied Biosystems) or a SYBR green kit (Finnzymes), and the following annealing temperatures and primers: CnAβ1, 57° (fwd-AGAAGGTGAAGACCAGT, rev-AGCAAGTTGCATAACATCATT); CnAβ2 58° (fwd-AGGCTATTGAGGCTGAAA, rev-CGGATCTCAGAAAGCAC); CnAα, 57° (fwd-CAAGGCGATTGATCCCA, rev-TCGAAGCACCCTCTGTTA); CnB I (fwd 5′-ATGCAGATAAGGATGGA-3′, rev 5′-GAAAGCAAAAGTGTTGGG-3′); ubiquitin B, 58° (fwd 5′-TGGCTATTAATTATTCGGTCTGCAT, rev GCAAGTGGCTAGAGTGCAGAGTAA); TnI-slow, 55° (fwd 5′-TGCTGAAGAGCCTGATGCTA-3′, rev 5′-GAACATCTTCTTGCGACCTTC-3′); TnI-fast, 55° (fwd 5′-GAAGGAGAACTACCTGTCAGA-3′, rev 5′-TGGGCAGTTAGGACTCAGACTC-3′). MAFbx/Atrogin primers have been previously described (Mourkioti et al., 2006). Data were normalized with ubiquitin B values and expressed as fold induction over a control sample.
Microarray analysis and sequence comparison
Total RNA from the different samples was hybridized in triplicate on Affymetrix mouse MG_430A2.0 or the human HG-U133A2.0 oligonucleotide-based arrays. Raw data were normalized and analyzed using GeneSpring-7.2 (Silicon Genetics). After chip normalization to the 50th percentile, samples were normalized to cells transfected with control siRNA and ANOVA statistical analysis was performed. For the analysis of cardiotoxin-injured mice, statistically significant genes were subjected to gene tree hierarchical clustering and lists of up-regulated and down-regulated genes were compared with annotated gene ontology lists, choosing those lists with at least 10 genes in common and a P value < 10−11 (wild type) or P < 10−7 (MLC-CnAβ1). The average of the relative gene expression variation of the genes in these lists (mitochondria, 661 genes; immune response, 200 genes; macrophage-associated, 6 genes; extracellular matrix, 164 genes; collagen, 26 genes; complete gene lists available upon request) in the different experimental conditions was calculated and expressed as fold induction over the value of the uninjured wild-type mice.
Intronic sequences obtained from Ensemble (www.ensemble.org) were translated using the Expasy translation engine (www.expasy.org) and compared using ClustalW (www.ebi.ac.uk/clustalw).
Proliferation analysis
Cell proliferation was quantified using the MTT cell proliferation assay (Sigma-Aldrich) 0 and 48 h after cell transfection. BrdU incorporation was measured 0 and 48 h after transfection with the different siRNAs by using the BrdU ELISA Assay Kit (Roche) following the manufacturer's instructions. Cell cycle analysis was performed 48 h after transfection of C2C12 myoblasts with the different siRNAs by fixing the cells in 70% ethanol for 2 h, staining the nuclei with 20 μg/ml propidium iodide and analyzing them in a flow cytometer (Beckman Coulter).
Western blots and ELISA
Western blots were performed as previously described (Lara-Pezzi et al., 2002). Antibodies against CnAβ1 were generated using the CGMDWGTPHSFANNSHNA peptide as an immunogen. Antibodies against CnAβ2, myogenin, and p53 were obtained from Santa Cruz Biotechnology, Inc. and used at 1 μg/ml. Anti-phospho-Ser253-FoxO3a, anti-FoxO3a, anti-phospho-PDK1, anti-PDK1, anti-phospho-Thr308-Akt, anti-Akt anti-phospho-mTOR, anti-mTOR, anti-phospho-Erk, anti-Erk(p42), anti-phospho-p38, and anti-p38 were purchased from Cell Signaling Technology. Anti-Stag2 was a gift of Dr. José Luis Barbero (CNB, Madrid, Spain; Prieto et al., 2002). Anti-HA was purchased from Roche. Secondary HRP-conjugated anti–rabbit and anti–mouse antibodies were obtained from GE Healthcare and used at a 1:3,000 dilution. Peroxydase activity was visualized using a SuperSignal kit (Thermo Fisher Scientific). Films were scanned and brightness/contrast was enhanced for the whole picture following a dust and scratches filter using Adobe Photoshop. The Cn activity was assayed by ELISA (BIOMOL International, L.P.) in extracts of 293 cells transfected with 2.5 μg of different Cn-expression vectors or empty pcDNA3.1 as a control. Cn activity was determined in the presence or absence of EGTA and okadaic acid following the manufacturer's instructions and the results expressed as the phosphatase activity obtained in the presence of okadaic acid minus the activity obtained in the presence of EGTA.
Online supplemental material
Fig. S1 includes a schematic representation of the functional domains of the different Cn isoforms and shows that the C-terminal region of CnAβ1 is conserved among vertebrates. It also illustrates how the Gal4 luciferase system works and shows that both CnAβ1 and the artificially truncated CnAβ* form are constitutively able to activate NFAT-dependent transcription in C2C12 myoblasts, but not in Jurkat. Fig. S2 shows that IGF-1 induces CnAβ1 expression in starving myotubes and shares some targets with CnAβ1. It also demonstrates that C2C12 stable transfectants overexpress the different CnAβ isoforms at similar levels and that CnAβ1 does not interfere with myotube differentiation. Fig. S3 shows GFP-FoxO3a localization in the presence of the different CnAβ isoforms and includes phase-contrast pictures corresponding to Fig. 6 H. Fig. S4 analyzes the presence of different leukocyte subpopulations and the expression of FoxO and NFATc targets in regenerating muscle in both wild-type and MLC/CnAβ1 mice. Fig. S5 shows microarray analysis of regenerating muscle in wild-type, MLC/CnAβ1, and MLC/CnAα mice. The supplementary tables include lists of genes generated during the different microarray analyses. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200704179/DC1.
Supplementary Material
Acknowledgments
We are grateful to Jose González, Arianna Nenci, Emerald Perlas, and Annabel Varela-Carter for technical assistance and to Michael Potente, Stefanie Dimmeler, Frank McKeon, Claude Klee, Manuel López-Cabrera, José Luis Barbero, and Nigel Brand for providing valuable reagents and unpublished information. We thank the EMBL Monoclonal Core Facility for the development of the anti-CnAβ1 antibody.
This work was supported by grants to N. Rosenthal from the Leducq Foundation (04-CVD-03) and EU (MYORES: LSHG-CT-2004-511978). E. Lara-Pezzi was supported by a 3+3 Award from the Spanish Centro Nacional de Investigaciones Cardiovasculares (CNIC) and a Marie Curie Fellowship.
The authors declare there is no conflict of interest related to this work.
A. Paul's present address is Novartis Institutes for BioMedical Research, Musculoskeletal Diseases, 100 Technology Square, Cambridge, MA 02139.
Abbreviations used in this paper: Cn, calcineurin; CnA, calcineurin A; CnAα* and CnAβ*, artificially truncated forms of CnAα and CnAβ2 lacking the autoinhibitory domain; CnB, calcineurin B; CsA, cyclosporine A; FoxO, Forkhead box O; IGF-1, insulin-like growth factor 1; NFAT, nuclear factor of activated T cells.
References
- Barton, E.R., L. Morris, A. Musarò, N. Rosenthal, and H.L. Sweeney. 2002. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J. Cell Biol. 157:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet, A., J. Park, H. Tran, L.S. Hu, B.A. Hemmings, and M.E. Greenberg. 2001. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Mol. Cell. Biol. 21:952–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno, O.F., E.B. Brandt, M.E. Rothenberg, and J.D. Molkentin. 2002. Defective T cell development and function in calcineurin A beta-deficient mice. Proc. Natl. Acad. Sci. USA. 99:9398–9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, R.N., S. Tajbakhsh, V. Mouly, G. Cossu, M. Buckingham, and G.S. Butler-Browne. 1999. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J. Cell Sci. 112:2895–2901. [DOI] [PubMed] [Google Scholar]
- Delling, U., J. Tureckova, H.W. Lim, L.J.D. Windt, P. Rotwein, and J.D. Molkentin. 2000. A calcineurin-NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Mol. Cell. Biol. 20:6600–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkers, P.F., R.H. Medema, C. Pals, L. Banerji, N.S. Thomas, E.W. Lam, B.M. Burgering, J.A. Raaijmakers, J.W. Lammers, L. Koenderman, and P.J. Coffer. 2000. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol. Cell. Biol. 20:9138–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolny, G., C. Giacinti, L. Pelosi, C. Nicoletti, N. Winn, L. Barberi, M. Molinaro, N. Rosenthal, and A. Musarò. 2005. Muscle expression of a local Igf-1 isoform protects motor neurons in an ALS mouse model. J. Cell Biol. 168:193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert, J.C., E.B. Berglund, and N. Rosenthal. 1996. Proliferation precedes differentiation in IGF-I-stimulated myogenesis. J. Cell Biol. 135:431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friday, B.B., V. Horsley, and G.K. Pavlath. 2000. Calcineurin activity is required for the initiation of skeletal muscle differentiation. J. Cell Biol. 149:657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa-Hibi, Y., K. Yoshida-Araki, T. Ohta, K. Ikeda, and N. Motoyama. 2002. FOXO forkhead transcription factors induce G(2)-M checkpoint in response to oxidative stress. J. Biol. Chem. 277:26729–26732. [DOI] [PubMed] [Google Scholar]
- Greer, E.L., and A. Brunet. 2005. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 24:7410–7425. [DOI] [PubMed] [Google Scholar]
- Guerini, D., and C.B. Klee. 1989. Cloning of human calcineurin A: evidence for two isozymes and identification of a polyproline structural domain. Proc. Natl. Acad. Sci. USA. 86:9183–9187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, P., L. Chen, J. Nardone, and A. Rao. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205–2232. [DOI] [PubMed] [Google Scholar]
- Horsley, V., B.B. Friday, S. Matteson, K.M. Kegley, J. Gephart, and G.K. Pavlath. 2001. Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J. Cell Biol. 153:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley, V., K.M. Jansen, S.T. Mills, and G.K. Pavlath. 2003. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 113:483–494. [DOI] [PubMed] [Google Scholar]
- Kamei, Y., S. Miura, M. Suzuki, Y. Kai, J. Mizukami, T. Taniguchi, K. Mochida, T. Hata, J. Matsuda, H. Aburatani, et al. 2004. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J. Biol. Chem. 279:41114–41123. [DOI] [PubMed] [Google Scholar]
- Ke, H., and Q. Huai. 2003. Structures of calcineurin and its complexes with immunophilins–immunosuppressants. Biochem. Biophys. Res. Commun. 311:1095–1102. [DOI] [PubMed] [Google Scholar]
- Kops, G.J., R.H. Medema, J. Glassford, M.A. Essers, P.F. Dijkers, P.J. Coffer, E.W. Lam, and B.M. Burgering. 2002. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol. Cell. Biol. 22:2025–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Pezzi, E., M.V. Gomez-Gaviro, B.G. Galvez, E. Mira, M.A. Iniguez, M. Fresno, C. Martinez-A, A.G. Arroyo, and M. Lopez-Cabrera. 2002. The hepatitis B virus X protein promotes tumor cell invasion by inducing membrane-type matrix metalloproteinase-1 and cyclooxygenase-2 expression. J. Clin. Invest. 110:1831–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Pezzi, E., N. Pezzi, I. Prieto, I. Barthelemy, C. Carreiro, A. Martínez, A. Maldonado-Rodríguez, M. López-Cabrera, and J.L. Barbero. 2004. Evidence of a transcriptional co-activator function of cohesin STAG/SA/Scc3. J. Biol. Chem. 279:6553–6559. [DOI] [PubMed] [Google Scholar]
- Li, H.H., V. Kedar, C. Zhang, H. McDonough, R. Arya, D.Z. Wang, and C. Patterson. 2004. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Invest. 114:1058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, C., E. Burgeon, and A. Rao. 1996. Mechanisms of transactivation by nuclear factor of activated T cells-1. J. Exp. Med. 184:141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh, K.J., E. Calabria, G. Pallafacchina, S. Ciciliot, A.L. Serrano, C. Argentini, J.M. Kalhovde, T. Lomo, and S. Schiaffino. 2004. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proc. Natl. Acad. Sci. USA. 101:10590–10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin, J.D., J.R. Lu, C.L. Antos, B. Markham, J. Richardson, J. Robbins, S.R. Grant, and E.N. Olson. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 93:215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourkioti, F., P. Kratsios, T. Luedde, Y.-H. Song, P. Delafontaine, R. Adami, V. Parente, R. Bottinelli, M. Pasparakis, and N. Rosenthal. 2006. Targeted ablation of IKK2 improves skeletal muscle strength, maintains mass and promotes regeneration. J. Clin. Invest. 116:2945–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarò, A., and N. Rosenthal. 1999. Maturation of the myogenic program in induced by post-mitotic expression of IGF-1. Mol. Cell. Biol. 19:3115–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarò, A., K. McCullagh, A. Paul, L. Houghton, G. Dobrowolny, M. Molinaro, E.R. Barton, H.L. Sweeney, and N. Rosenthal. 2001. Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat. Genet. 27:195–200. [DOI] [PubMed] [Google Scholar]
- Musarò, A., C. Giacinti, G. Borsellino, G. Dobrowolny, L. Pelosi, L. Cairns, S. Ottolenghi, G. Cossu, G. Bernardi, L. Battistini, et al. 2004. Stem cell-mediated muscle regeneration is enhanced by mIGF-1. Proc. Natl. Acad. Sci. USA. 101:1206–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae, J., V. Barr, and D. Accili. 2000. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J. 19:989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya, F.J., B. Mercer, J. Shelton, J.A. Richardson, R.S. Williams, and E.N. Olson. 2000. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J. Biol. Chem. 275:4545–4548. [DOI] [PubMed] [Google Scholar]
- Ni, Y.G., K. Berenji, N. Wang, M. Oh, N. Sachan, A. Dey, J. Cheng, G. Lu, D.J. Morris, D.H. Castrillon, et al. 2006. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation. 114:1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, S.A., B.J. Wilkins, O.F. Bueno, and J.D. Molkentin. 2003. Altered skeletal muscle phenotypes in calcineurin Aalpha and Abeta gene-targeted mice. Mol. Cell. Biol. 23:4331–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, S.A., D.P. Millay, M.A. Sargent, F.J. Naya, E.M. McNally, H.L. Sweeney, and J.D. Molkentin. 2007. Genetic disruption of calcineurin improves skeletal muscle pathology and cardiac disease in a mouse model of limb-girdle muscular dystrophy. J. Biol. Chem. 282:10068–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero, E., V. Ruiz-Bonilla, L. Gresh, L. Hui, E. Ballestar, P. Sousa-Victor, B. Baeza-Raja, M. Jardi, A. Bosch-Comas, M. Esteller, et al. 2007. Genetic analysis of p38 MAP kinases in myogenesis: fundamental role of p38alpha in abrogating myoblast proliferation. EMBO J. 26:1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potente, M., C. Urbich, K. Sasaki, W.K. Hofmann, C. Heeschen, A. Aicher, R. Kollipara, R.A. DePinho, A.M. Zeiher, and S. Dimmeler. 2005. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J. Clin. Invest. 115:2382–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto, I., N. Pezzi, J.M. Buesa, L. Kremer, I. Barthelemy, C. Carreiro, F. Roncal, A. Martínez, L. Gómez, R. Fernández, et al. 2002. STAG2 and Rad21 mammalian mitotic cohesins are implicated in meiosis. EMBO Rep. 3:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri, M., C. Sandri, A. Gilbert, C. Skurk, E. Calabria, A. Picard, K. Walsh, S. Schiaffino, S.H. Lecker, and A.L. Goldberg. 2004. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 117:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, R.A., and K.E. Yutzey. 2004. Calcineurin signaling and NFAT activation in cardiovascular and skeletal muscle development. Dev. Biol. 266:1–16. [DOI] [PubMed] [Google Scholar]
- Schulze, P.C., J. Fang, K.A. Kassik, J. Gannon, M. Cupesi, C. MacGillivray, R.T. Lee, and N. Rosenthal. 2005. Transgenic overexpression of locally acting insulin-like growth factor-1 inhibits ubiquitin-mediated muscle atrophy in chronic left-ventricular disfunction. Circ. Res. 97:418–426. [DOI] [PubMed] [Google Scholar]
- Seoane, J., H.V. Le, L. Shen, S.A. Anderson, and J. Massague. 2004. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 117:211–223. [DOI] [PubMed] [Google Scholar]
- Stitt, T.N., D. Drujan, B.A. Clarke, F. Panaro, Y. Timofeyva, W.O. Kline, M. Gonzalez, G.D. Yancopoulos, and D.J. Glass. 2004. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell. 14:395–403. [DOI] [PubMed] [Google Scholar]
- Stupka, N., P. Gregorevic, D.R. Plant, and G.S. Lynch. 2004. The calcineurin signal transduction pathway is essential for successful muscle regeneration in mdx dystrophic mice. Acta Neuropathol. 107:299–310. [DOI] [PubMed] [Google Scholar]
- Stupka, N., D.R. Plant, J.D. Schertzer, T.M. Emerson, R. Bassel-Duby, E.N. Olson, and G.S. Lynch. 2006. Activated calcineurin ameliorates contraction-induced injury to skeletal muscles of mdx dystrophic mice. J. Physiol. 575:645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., B. Murtuza, R.T. Smolenski, I.A. Sammut, N. Suzuki, Y. Kaneda, and M.H. Yacoub. 2001. Cell transplantation for the treatment of acute myocardial infarction using vascular endothelial growth factor-expressing skeletal myoblasts. Circulation. 104:I207–I212. [DOI] [PubMed] [Google Scholar]
- Tran, H., A. Brunet, J.M. Grenier, S.R. Datta, A.J. Fornace, P.S. DiStefano, L.W. Chiang, and M.E. Greenberg. 2002. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 296:530–534. [DOI] [PubMed] [Google Scholar]
- Zheng, W.H., S. Kar, and R. Quirion. 2000. Insulin-like growth factor-1-induced phosphorylation of the forkhead family transcription factor FKHRL1 is mediated by Akt kinase in PC12 cells. J. Biol. Chem. 275:39152–39158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.