Abstract
In the RNA interference (RNAi) pathway, small interfering RNAs (siRNAs) play important roles as intermediates. Primary siRNAs are produced from trigger dsRNAs by an RNaseIII-related enzyme called Dicer; in some organisms, secondary siRNAs are also produced by processes involving RNA-dependent RNA polymerases (RdRPs), which act on target mRNAs. Using a cell-free assay system prepared from Caenorhabditis elegans, we analyzed the production and activity of secondary siRNAs. In this cell-free system, RdRP activity acts on mRNA-derived templates to produce small RNAs. The RRF-1 complex is predominantly responsible for the RdRP activity, and synthesizes secondary-type siRNA molecules in a Dicer-independent manner. Notably, secondary-type siRNAs induce a prominent Slicer activity to cleave target mRNAs far more effectively than primary-type siRNAs. An Argonaute protein, CSR-1, is responsible for the Slicer activity induced by secondary-type siRNAs. Secondary rather than primary siRNAs may play a major role in the destabilization of target transcripts during RNAi in C. elegans.
Keywords: Argonaute, RdRP, RNAi, siRNA
Introduction
RNA interference (RNAi) is a form of sequence-specific gene silencing induced by double-stranded RNA (dsRNA) (Fire et al, 1998). RNAi and related gene silencing mechanisms are known to be involved in silencing transposons (Ketting et al, 1999; Tabara et al, 1999) and/or viruses. In addition, RNAi shares features with gene regulatory mechanisms mediated by some classes of endogenous small RNA, such as micro-RNAs (reviewed in Mello and Conte, 2004). RNAi and related gene silencing phenomena are termed ‘RNA silencing' in a broad sense and have been studied by various approaches.
RNA silencing accompanies an accumulation of small-RNA species homologous to a target gene, as first detected by a study in plants (Hamilton and Baulcombe, 1999). The small-RNA species generated during RNAi and the closely related RNA silencing are generally termed small interfering RNAs (siRNAs). The results of genetic studies in fungi and plants indicate that the mechanisms of RNA silencing require various factors, including cell-encoded RNA-dependent RNA polymerases (RdRPs) (Cogoni and Macino, 1999; Dalmay et al, 2000; Mourrain et al, 2000). Studies in Caenorhabditis elegans have isolated various RNAi-deficient mutants and have shown that specific genetic factors, such as rde-1 and rde-4, are required for RNAi (Tabara et al, 1999, 2002).
In Drosophila and mammals, cell-free systems for analyzing RNAi have been constructed and used in biochemical studies (Tuschl et al, 1999; Martinez et al, 2002). These studies have shown that RNAi involves two major enzymatic activities. First, dsRNA is processed into 21- to 22-nucleotide (nt) siRNAs by an RNase III activity (Zamore et al, 2000; Elbashir et al, 2001). Dicer is the core enzyme responsible for this activity (Bernstein et al, 2001), producing duplex siRNAs bearing monophosphorylated 5′ ends and protruding 3′ ends (Elbashir et al, 2001). Then, siRNAs are incorporated into a complex termed the RNA-induced silencing complex (RISC), which has a so-called Slicer activity that cleaves the target mRNA in a sequence-specific manner (Hammond et al, 2000; Zamore et al, 2000; Elbashir et al, 2001). Studies in several organisms have shown that the Slicer activity of RISC is attributed to some Argonaute proteins (Hammond et al, 2001; Martinez et al, 2002; Liu et al, 2004). Cell-free analyses of RNA silencing have been performed, even in plant systems (Tang et al, 2003; Qi et al, 2005). Unfortunately, studies in C. elegans have lacked a cell-free system suitable for the in vitro analysis of RNAi.
RdRP-encoding genes have not been found in the genomes of insects and vertebrates, but they are present in the genomes of many other eukaryotes. In addition to Dicer and Slicer activities, the third major enzymatic activity for RNAi may be provided by RdRPs. The mechanism of RNAi in C. elegans also involves RdRPs (Smardon et al, 2000; Sijen et al, 2001). C. elegans exhibiting RNAi accumulate secondary siRNAs, which are produced by RdRPs using target mRNAs as their templates (Sijen et al, 2001). These secondary siRNAs bear 5′ triphosphorylated ends, which makes them a new type of small RNA (Pak and Fire, 2007; Sijen et al, 2007).
In this study, using a cell-free system prepared from C. elegans, we analyzed the mechanism of production of secondary siRNAs and the enzymatic activities induced by several types of siRNA.
Results
Construction of a cell-free system for in vitro analysis of RNAi in C. elegans
Using cell lysates from C. elegans, we constructed an in vitro system that allows the analysis of enzymatic activities related to RNAi (Figure 1A). First, a cytoplasmic fraction was obtained from worm homogenates and was passed through a weak cation-exchange (C-type) chromatography column to diminish nonspecific nuclease activity often detected in the cytoplasmic fraction. Factors such as RDE-1, RDE-4 and ribosomes did not bind to the column (data not shown); however, most of the nonspecific nucleases appeared to be retained on the column. The flow-through fraction was buffer-exchanged and served as the standard lysate for in vitro analyses.
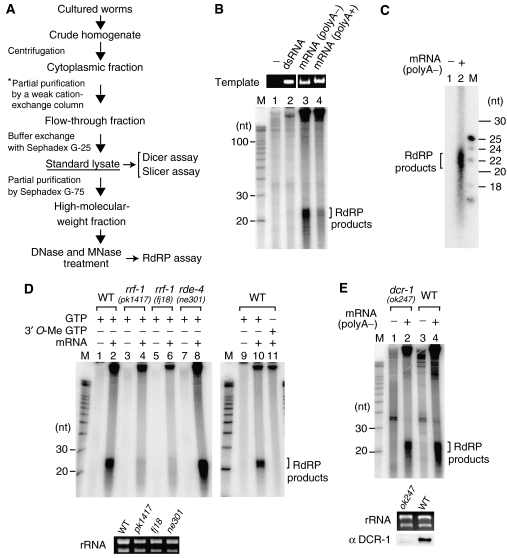
Figure 1.
RdRP activity in a fraction of the cell lysate and its impairment by rrf-1 mutations. (A) Scheme for the preparation of cell lysates. Lysates from wild-type animals and viable mutants were prepared from large-scale cultures of mixed-stage populations. Lysates from sterile mutants were made from homozygotes at L4 or adult stages on a small scale, omitting the step indicated by the asterisk. (B) Aberrant mRNAs effectively induced RdRP activity in the cell-free system. In the presence of [α-32P]UTP and other ribonucleotides, high-MW extracts pretreated with both MNase and DNase were reacted with the following templates (final 12.5 ng/μl): a dsRNA (542 bp), aberrant mRNA (1299 nt RNA lacking poly-A), or a normal mRNA (1359 nt). The RNA products were analyzed on a 15% sequencing gel. The electrophoretic data of template RNAs served as quality and loading controls. (C) The length of the small RNAs produced via the RdRP activity. (D) RdRP activity to produce the small RNAs was impaired in rrf-1(pk1417; fj18) mutants but was observed in the rde-4 mutant and wild-type (WT) animals. For the reactions to distinguish between RdRP activity and terminal transferase activity, 3′-O-methyl GTP was used instead of GTP. (E) RdRP activity to produce small RNAs was present in the dcr-1(ok247) mutant. High-MW extracts were prepared on a small scale from the dcr-1 mutant and wild-type animals. The template used for the RdRP assay in panels C, D and E was the aberrant mRNA. Ribosomal RNAs prepared from extracts served as loading controls.
To check whether the lysate prepared by our protocol was appropriate for the in vitro analysis of RNAi, we first tested the Dicer activity in the lysate. DCR-1 (Dicer) and RDE-4 are known to form a complex (Tabara et al, 2002) and act to process dsRNA into siRNAs during RNAi (Parker et al, 2006). Dicer activity to process radiolabeled dsRNAs into siRNAs was detected in lysates from rde-1(ne300), rde-3(ne298) and rrf-1(pk1417) mutants as well as in those from wild-type animals, whereas this activity was absent in the lysate from the rde-4(ne301) mutant (data not shown). This result is consistent with previous studies; therefore, lysates prepared as shown in Figure 1A continued to be used for the analyses of other enzymatic activities.
RdRP activity in a fraction of the cell lysate
RNAi in C. elegans involves two types (primary and secondary) of siRNAs. To investigate the biochemical mechanism of secondary siRNA production, we tried to detect RdRP activity in worm lysates. First, standard lysate treated with DNase I was reacted with an RNA template and radiolabeled ribonucleotides. RdRP activities were difficult to detect by this experiment, because of strong background signals.
To overcome this problem, we used a partially fractionated lysate. RdRP-related proteins involved in RNA silencing are generally larger than 100 kDa. Considering this feature, we applied standard lysate onto a Sephadex G-75 gel-filtration column and recovered the void fraction, representing a high-molecular-weight (MW) fraction roughly greater in size than 50 kDa. We then incubated a long single-stranded RNA, [α-32P]UTP and other ribonucleotides in the high-MW fraction treated with DNase I. We detected the production of small RNAs, possibly reflecting RdRP activity (Supplementary Figure 1). Weak production of small RNAs was detected even in the absence of exogenous RNA and was thought to originate from reactions probably occurring on endogenous RNAs. However, the introduction of an exogenous RNA template resulted in a significant increase in small-RNA production.
We next investigated the type of RNA structure that was preferred as the template for small-RNA production via RdRP-like activity. Because the RdRP reaction on endogenous RNAs is obstructive to the analysis of substrate specificity, we treated high-MW extracts with micrococcal nuclease (MNase) and DNase I to remove endogenous nucleic acids. We then set up an RdRP assay with EGTA to inactivate the MNase. Pretreatment of extracts with MNase successfully suppressed the signals from endogenous RNAs (Figure 1B, lane 1). High-MW extracts were incubated with a long dsRNA, a normal mRNA or an aberrant mRNA lacking poly-A. The dsRNA was inefficient at producing small RNAs. By contrast, both types of mRNA induced small-RNA production, whereas the aberrant mRNA was several fold more efficient at producing small RNAs than normal mRNA (Figure 1B, lanes 3 and 4). The small RNAs produced with the aberrant mRNA template were 21–23 nt in length (Figure 1C). Small-RNA production was inhibited by 3′-O-methyl GTP, which acts as an RNA chain elongation terminator (Figure 1D, lane 11); therefore, we believe the small RNAs to be the products of RdRP reactions.
Impairment of RdRP activity by rrf-1 and not dcr-1 mutations
To determine the genetic factors responsible for the RdRP activity toward mRNA-derived templates, we tested the RdRP activity in high-MW extracts of several viable mutants (rrf-1 and rde-4) with impaired RNAi responses. rrf-1 is one of four RdRP-encoding genes in C. elegans (Sijen et al, 2001). The RdRP activity to produce small RNAs in rrf-1 mutant strains (pk1417; fj18) turned out to be less than 10% of that in wild-type animals (Figure 1D). This result suggests that RRF-1 is predominantly responsible for producing small RNAs on mRNA-derived templates. The somatic cells of rrf-1 mutants are known to be deficient in RNAi; therefore, residual RdRP activity might result from other RdRPs, which could be redundantly expressed in the germ line. By contrast, RdRP activity to produce small RNAs was detected in extracts of rde-4(ne301) mutants (Figure 1D).
Moreover, we examined whether Dicer is required for small-RNA production via RdRP activity. DCR-1 is the sole Dicer protein in C. elegans, and DCR-1-null mutant animals are sterile (Grishok et al, 2001). We prepared high-MW extracts of dcr-1 sterile homozygotes collected from a balanced dcr-1(ok247) mutant strain. Small-RNA production via RdRP activity was detected even in extracts of dcr-1 homozygotes (Figure 1E), which strongly suggests that the small-RNA production we observed is Dicer independent.
DRH-3 is present in the RRF-1 complex and is also required for RdRP activity
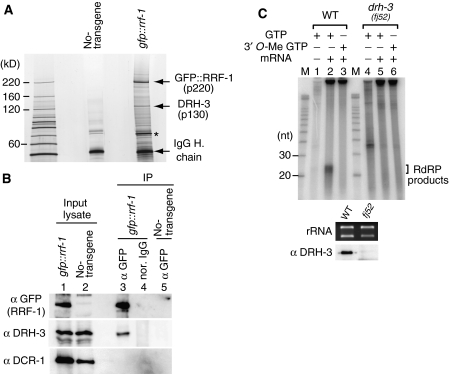
Because RRF-1 turned out to be involved in small-RNA production via RdRP activity in lysates, we further analyzed RRF-1. First, we introduced a transgene expressing GFP-tagged RRF-1 into the rrf-1(pk1417) mutant and established a stable transgenic line. This transgene rescued the RNAi-deficient phenotype of the rrf-1 mutant, because transgenic animals responded appropriately to a dsRNA against the muscle gene unc-22 (data not shown). Therefore, the GFP∷RRF-1 fusion protein is thought to be functional. Analysis of transgenic animals showed the GFP signal to be present predominantly in the cytoplasm and weakly in the nucleus (Supplementary Figure 2A).
Using transgenic animals, we investigated whether RRF-1 interacts with any other protein. GFP∷RRF-1 and its associated proteins were immunopurified from transgenic animals using an anti-GFP monoclonal antibody (Figure 2A). A 130-kDa protein was detected in the GFP∷RRF-1 immune complex; this was identified as DRH-3 by mass spectrometry analyses. DRH-3 is a DExH-box helicase that was previously identified as a factor interacting with DCR-1, and the drh-3 mutant was shown to have defects in oogenesis and in RNAi in germ cells (Duchaine et al, 2006). Our immunoblotting experiments with antibodies raised against DRH-3 and DCR-1 showed that the GFP∷RRF-1 complex contained DRH-3 but not DCR-1 (Figure 2B).
Figure 2.
RRF-1 interacts with DRH-3. (A) The GFP∷RRF-1 complex was immunopurified from lysates of transgenic animals. The proteins in the complex were resolved by SDS–PAGE and visualized by silver staining. Prominent protein bands were subjected to peptide mass fingerprinting analyses by MALDI-TOF mass spectrometry. The 130-kDa protein (p130) corresponded to DRH-3. The 75-kDa protein (asterisk) was Hsc70. (B) GFP∷RRF-1 immunoprecipitates did not contain a detectable amount of DCR-1. The immunoprecipitates and a portion of the input lysate were analyzed by immunoblotting using antibodies against GFP, DRH-3 and DCR-1. (C) RdRP activity to produce small RNAs was impaired in the drh-3(fj52) mutant. High-MW extracts were prepared on a small scale, and their RdRP activities towards an aberrant mRNA template were assayed. The 35-nt signal in lane 4 is background noise that was often detected in experiments with lysates prepared on a small scale.
To determine whether DRH-3 is also required for RdRP activity, we prepared high-MW extracts of drh-3 sterile homozygotes on a small scale. Extracts of drh-3 mutants were deficient in small-RNA production via RdRP activity (Figure 2C).
Furthermore, we investigated what cellular components colocalize with RRF-1. Cytoplasmic lysates were prepared from transgenic animals and further fractionated by sucrose gradient centrifugation. Immunoblotting experiments showed that a considerable amount of the GFP∷RRF-1 molecules co-fractionated with polysomes (Supplementary Figure 2B).
RdRP activity of RRF-1 immunoprecipitates
We next examined whether the RRF-1 complex has any RdRP activity. GFP∷RRF-1 immunoprecipitates were reacted with an mRNA template, [α-32P]CTP and other ribonucleotides, and the reaction products were analyzed. We detected two types of signal: one corresponding to small RNAs and the other matching the full-length template RNA (Figure 3A). The signal for small RNAs was not generated in the absence of UTP; therefore, we concluded that the small-RNA species were products of an RdRP reaction. The length of the small RNAs was approximately 22 to 23 nt (Figure 3B, lane 3). By contrast, we concluded that the full-length signal was caused mainly by a terminal transferase activity, because the signal was generated even in the absence of UTP (Figure 3A, lane 3). Thus, the RRF-1 complex appears to have both RdRP and terminal transferase activities.
Figure 3.
The GFP∷RRF-1 complex has an RdRP activity to generate small RNAs. (A) Enzymatic activity of the GFP∷RRF-1 complex. Immunoprecipitations from the lysates of transgenic animals expressing GFP∷RRF-1 or wild-type animals were performed using the anti-GFP monoclonal antibody. Immunoprecipitates were treated with MNase to avoid contamination from endogenous RNAs and then washed with a buffer containing EGTA. Finally, the immunoprecipitates were reacted with an RNA template (393-nt mRNA), [α-32P]CTP and other ribonucleotides. The RNA products were analyzed on a 15% sequencing gel. (B) Small RNAs generated by the RRF-1 complex were complementary to the template RNA. The GFP∷RRF-1 immunoprecipitates were reacted with an RNA template (1299-nt mRNA lacking poly-A) and radiolabeled ribonucleotides. Half of the RNA products were treated with a mixture of RNase A and T1 in the presence of 0.5 M NaCl. As a control, primary-type siRNAs produced from dsRNA by the Dicer activity in the RDE-4 complex were also analyzed. The RNA products were resolved on sequencing gels. (C) The RNA products described in B were resolved by 15% native PAGE. A chemically-synthesized 23-bp duplex siRNA (m) served as a size marker. (D) Small RNAs generated by the RdRP activity of GFP∷RRF-1 immunoprecipitates were found to be sensitive to treatment with vaccinia virus capping enzyme. M represents single-stranded RNA size markers.
In addition, we checked whether small RNAs produced by the RRF-1 complex are complementary to RNA templates. GFP∷RRF-1 immunoprecipitates were reacted with an RNA template and radiolabeled ribonucleotides, and then the reaction products and RNA template were annealed and treated with a mixture of RNase A and T1, which degrades single-stranded RNA but not duplex RNA. Denaturing gel analysis showed that RNase treatment did not erase the signal for small RNAs produced by the RdRP reaction and did not alter the length of small RNAs; however, it did erase the signal corresponding to the full-length template RNA (Figure 3B, lane 4). On native gels, reaction products not treated with RNase migrated as large molecules, whereas those treated with RNase migrated at a size similar to duplex siRNA (Figure 3C). As control, primary-type siRNAs were prepared by the Dicer activity in the RDE-4 complex and were also treated with RNase. RNase treatment slightly shortened the length of primary-type siRNAs, which are known to be small duplex RNAs bearing protruding 3′ ends (Figure 3B and C). Thus, the small RNAs produced by the RRF-1 complex were perfectly complementary to the RNA template and were structurally different from the siRNAs produced by Dicer activity.
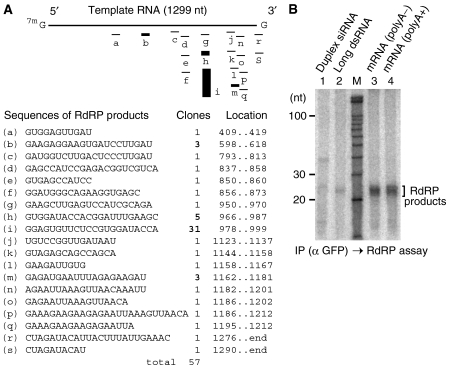
We amplified the RdRP products by RT–PCR and subjected them to cloning and sequencing analyses (Figure 4A): 93% of RdRP products possessed guanine at their 5′ ends and 96% of RdRP products matched at internal sites rather than at the ends of the RNA template, and several internal sites were hot spots corresponding to multiple clones with the same sequences.
Figure 4.
Products and templates of the RdRP reactions. (A) Sequences of RdRP products. An RdRP reaction corresponding to lane 4 in Figure 3B was performed with non-radioactive nucleotides. The RdRP products were amplified by RT–PCR and cloned. (B) Long single-stranded RNAs are suitable templates for small-RNA synthesis by the RRF-1 complex. The GFP∷RRF-1 immunoprecipitates were reacted with the following RNA templates: primary-type siRNA (23-nt duplex), long dsRNA (542 bp), mRNA lacking poly-A (1299 nt) and normal mRNA (1359 nt).
Our data suggested that Dicer activity is not involved in small-RNA production via RdRP activity on mRNA-derived templates. To confirm this interpretation, we examined whether the GFP∷RRF-1 complex has Dicer activity to process radiolabeled dsRNA. DCR-1 immunoprecipitates processed dsRNA into siRNAs, but GFP∷RRF-1 immunoprecipitates were unable to process dsRNA (Supplementary Figure 2C).
Moreover, we examined whether RNA synthesis by the RRF-1 complex requires a primer for its initiation. First, we incubated an mRNA template, a 5′ end-labeled single-stranded siRNA complementary to the template and ribonucleotides with GFP∷RRF-1 immunoprecipitates or a high-MW fraction of cell lysate. We detected no significant primer-extension activity in either case (data not shown). Next, we incubated an RNA template, [γ-32P]GTP and other ribonucleotides with the GFP∷RRF-1 immunoprecipitates and looked for a signal corresponding to small-RNA synthesis. If gamma-labeled GTP is used for RNA synthesis, the synthesized RNA is not labeled internally, but a radiolabeled triphosphate is attached to the 5′ end of the synthesized RNA only in cases in which RNA synthesis is initiated de novo. We observed a small-RNA signal (Supplementary Figure 2D) and concluded that at least some of the small RNAs were synthesized in a primer-independent fashion.
Furthermore, we reacted an RNA template, [α-32P]CTP and other ribonucleotides with the GFP∷RRF-1 immunoprecipitates and then tested whether the small RNAs produced by the GFP∷RRF-1 complex were sensitive to a capping enzyme. Almost all small RNAs were lengthened by treatment with the capping enzyme (Figure 3D), which attaches a methylated guanosine cap to the 5′ triphosphorylated RNA. This result also suggests that the RRF-1 complex supports de novo RNA synthesis and produces small RNAs, predominantly in a triphosphorylated form.
We examined the type of RNA structure that is the preferred template for the synthesis of small RNAs by the GFP∷RRF-1 complex. In the presence of radiolabeled ribonucleotides, the GFP∷RRF-1 immunoprecipitates were challenged with a duplex siRNA, a long dsRNA, a normal mRNA and an aberrant mRNA lacking poly-A. Both types of mRNA were equally effective in inducing small-RNA synthesis, whereas the siRNA and the long dsRNA were inefficient (Figure 4B). C. elegans exhibiting RNAi are known to accumulate secondary siRNAs complementary to target mRNAs (Pak and Fire, 2007; Sijen et al, 2007). Given the product similarity and template preference, we interpreted these findings to suggest that the RdRP activity detected in this study reflects the reaction that produces secondary siRNAs.
Secondary-type siRNAs strongly induce target mRNA destruction in vitro
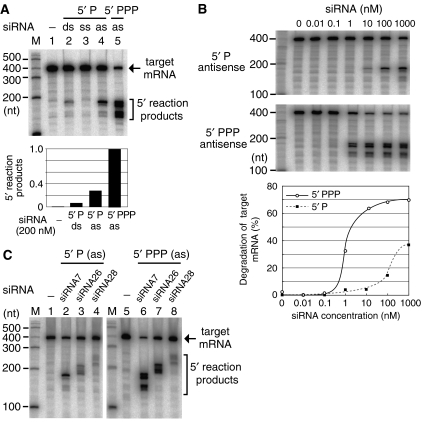
In a typical RNAi pathway, siRNAs and certain Argonaute proteins form a complex termed RISC, which has a Slicer activity that recognizes and slices target mRNAs. To examine the Slicer activity in C. elegans, we incubated cell lysates with a radiolabeled mRNA and 23-nt synthetic siRNAs. Cleavages of the target mRNA were induced weakly by a monophosphorylated duplex siRNA (primary type), moderately by a monophosphorylated single-stranded siRNA and strongly by a triphosphorylated single-stranded siRNA (secondary type) (Figure 5A). We paid attention to the effectiveness of single-stranded siRNAs in inducing cleavage and next examined the effect of siRNA concentration on mRNA cleavage. The secondary-type siRNA was one order of magnitude more effective at inducing mRNA cleavage than the monophosphorylated single-stranded siRNA (Figure 5B).
Figure 5.
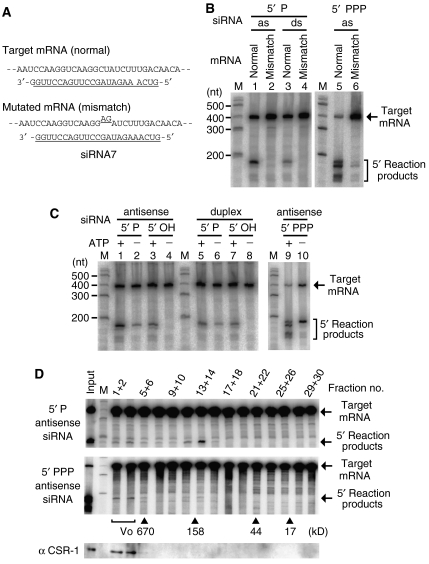
Detection of mRNA cleavage (Slicer) activities induced by siRNAs. (A) Sequence-specific mRNA cleavage activities were induced by several types of siRNA in cell lysates from C. elegans. The cell lysates were reacted with a cap-radiolabeled mRNA and any of the following siRNAs: a monophosphorylated (5′ P) duplex (ds) siRNA, monophosphorylated single-stranded siRNAs and triphosphorylated (5′ PPP) single-stranded siRNA. The reaction products were analyzed on a 6% sequencing gel. ss, sense-oriented single strand; as, antisense-oriented single strand. (B) In the cell lysates, the triphosphorylated single-stranded siRNA mimicking secondary siRNAs was one order of magnitude more effective at inducing mRNA cleavage than the monophosphorylated single-stranded siRNA. (C) Target mRNA cleavages induced by several single-stranded siRNAs with different sequences. The siRNA no. 7 was used for the assays in panels A and B.
Meanwhile, we designed multiple siRNAs corresponding to three different regions in the target mRNA. Regarding mono- and triphosphorylated single-stranded siRNAs, all three siRNAs from different regions induced Slicer activities in cell lysates (Figure 5C). The introduction of triphosphorylated single-stranded siRNAs led to the generation of multiple bands of 5′ reaction products from the target mRNA, but the banding patterns of the 5′ reaction products varied depending on the sequence of each siRNA. Regarding duplex siRNAs mimicking Dicer products, one of the three siRNAs induced a Slicer activity with detectable intensity (data not shown). In later experiments, we used the siRNA sequence (no. 7) that was most effective at inducing mRNA cleavage.
Similarities between the Slicer activity in lysates and that of the typical RISC
We next examined whether the Slicer activity induced in cell lysates from C. elegans shares features with RISC in other organisms. Previous studies have shown that RISC requires Mg2+ ions for its endonuclease activity (Schwarz et al, 2004). The Slicer activity detected in our cell-free system also required Mg2+ (Supplementary Figure 3A).
mRNA cleavage by RISC is known to generally occur near the center of the region covered by the siRNAs (Elbashir et al, 2001); therefore, we checked whether the Slicer activities in our system show this feature. mRNA cleavages were induced by single-stranded siRNAs, and then the 5′ reaction products from the target mRNA were recovered and subjected to RT–PCR and cloning analyses. The ends of some RT–PCR products matched an mRNA site at the center of the region covered by the siRNA with the following frequency: seven out of 15 clones in a monophosphorylated siRNA-mediated reaction and four out of 27 clones in a triphosphorylated siRNA-mediated reaction (Supplementary Figure 3B). Based on this result, we made a mutated mRNA bearing a 2-nt mismatch at the probable cleavage target site (Figure 6A). None of the three siRNAs with different structures efficiently induced cleavage of the mutated mRNA (Figure 6B).
Figure 6.
Similarities and differences in the Slicer activities induced by mono- and triphosphorylated siRNAs. (A) mRNA substrates to confirm the site of mRNA cleavage by Slicer activity. A 2-nt mismatch mutation was introduced into the mRNA site where cleavage was suggested by RT–PCR analysis. (B) mRNA carrying the 2-nt mismatch was not cleaved efficiently by the activities induced by any of the three types of siRNA. The abbreviations are the same as those described in the legend for Figure 5A. (C) Effect of ATP on the target mRNA destruction induced by several types of siRNA. 5′ phosphorylated siRNAs (5′ P or 5′ PPP) or 5′ non-phosphorylated siRNAs (5′ OH) were introduced into standard lysates containing the ATP regeneration system (lanes 1, 3, 5, 7 and 9) and ATP-depleted lysates (lanes 2, 4, 6, 8 and 10). (D) Gel-filtration analysis of Slicer factors that respond to single-stranded siRNAs. The standard lysate was fractionated into 30 fractions using a Superdex 200 (GE) column, and adjacent pairs were pooled into 15 samples. siRNAs and labeled mRNAs were reacted in each sample. The fractionation pattern of molecular weight markers is indicated by the arrowheads. Fractions 1, 2, 3 and 4 correspond to the void fraction (Vo).
Mono- and triphosphorylated siRNAs induce distinct Slicer activities
We examined the effect of ATP on the sequence-specific mRNA destruction induced by siRNAs (Figure 6C). In the presence of ATP, non-phosphorylated as well as monophosphorylated siRNAs, in both duplex and single-stranded forms, induced Slicer activity in cell lysates. By contrast, non-phosphorylated siRNAs were unable to induce Slicer activity in the absence of ATP, as reported previously (Nykanen et al, 2001). In the presence of ATP, triphosphorylated single-stranded siRNAs induced production of not only 5′ reaction products matching mRNA cleavage at the siRNA target site but also shorter forms of 5′ reaction products. These shorter 5′ reaction products were not generated from the target mRNA in the absence of ATP (Figure 6C, lane 10). We interpreted this finding to suggest that primary products generated by Slicer activity were attacked at their 3′ regions by ATP-dependent nucleases, and subsequently became the shorter forms of reaction products. The generation of 5′ reaction products shorter than primary Slicer products was prominent in reactions in lysates from mixed-stage animals, but was not prominent in lysates from animals at L4 and adult stages, which were used for the analyses of sterile mutants.
Next, we fractionated standard lysates by gel-filtration chromatography and incubated several types of siRNA in each fraction (Figure 6D; Supplementary Figure 4A). Monophosphorylated single-stranded siRNA induced Slicer activity in fractions of approximately 150 kDa and weakly in void fractions. Triphosphorylated single-stranded siRNA induced Slicer activity predominantly in void fractions. These results suggest that the Slicer activities induced by mono- and triphosphorylated siRNAs may not be mediated by the same set of proteins.
CSR-1 is responsible for the Slicer activity in conjunction with secondary-type siRNAs
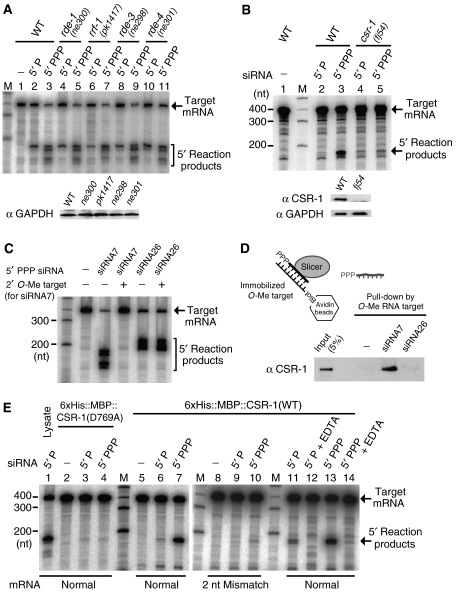
Finally, to identify the genetic factors responsible for Slicer activities in C. elegans, we tested some RNAi-deficient mutants. Cell lysates prepared from rde-1, rrf-1, rde-3 and rde-4 mutants were reacted with mono- and triphosphorylated single-stranded siRNAs. In this study, we used a viable hypomorphic mutant of rde-3 (also termed mut-2), an essential gene that encodes a member of the nucleotidyltransferase superfamily (Chen et al, 2005). The lysates of all four mutants and those of wild-type animals showed Slicer activities with almost equal intensities (Figure 7A). More than 25 Argonaute-related genes are present in C. elegans, and RDE-1 is an Argonaute protein known to interact with primary siRNAs (Yigit et al, 2006). Therefore, the Slicer activity induced by primary-type duplex siRNA was also checked in the rde-1 mutant, but this turned out to be almost equal to the activity in the lysates of wild-type animals (Supplementary Figure 4B).
Figure 7.
CSR-1 is predominantly responsible for the Slicer activity induced by secondary-type siRNAs. (A) Single-stranded siRNAs properly induced target mRNA cleavages in cell lysates from rde-1, rrf-1, rde-3 and rde-4 mutants and those from wild-type animals. To confirm equality of the lysate concentration, the amount of GAPDH was monitored by immunoblotting. (B) The Slicer activity induced by a secondary-type siRNA bearing a 5′ triphosphorylated end was impaired in the csr-1(fj54) mutant. Cell lysates were prepared on a small scale from the csr-1 mutant and wild-type animals, and served for the Slicer assay. (C) Inhibition of Slicer activities by a 2′-O-methyl oligonucleotide. Reactions were performed with 100 nM secondary-type siRNAs and 400 nM 2′-O-methyl oligonucleotide complementary to siRNA no. 7. (D) CSR-1 interacted in vitro with the secondary-type siRNA. The siRNA/Slicer complex was captured by the 2′-O-methyl oligonucleotide mimicking the target mRNA, and was examined by immunoblotting. (E) Enzymatic activities of CSR-1 recombinant proteins. 6 × His∷MBP∷CSR-1 fusion proteins were expressed in bacteria and purified. The catalytic domain of 6 × His∷MBP∷CSR-1(D769A) was designed to have a substitution of a conserved aspartic acid by an alanine. Recombinant proteins (final 5.3 ng/μl) were incubated with mono- or triphosphorylated siRNAs and then reacted with a radiolabeled target mRNA.
Meanwhile, we supposed that mutants lacking Slicer proteins to work in conjunction with secondary siRNAs might have phenotypes similar to that of another mutant that lacks any component of the RdRP complex producing the secondary siRNAs. We then focused on csr-1, encoding an Argonaute protein, because csr-1 mutants are phenotypically similar to drh-3 mutants in that both mutants have defects in oogenesis and RNAi (Yigit et al, 2006). We prepared cell lysates from csr-1 sterile homozygotes collected from a balanced csr-1(fj54) mutant strain and examined the Slicer activity in these lysates using single-stranded siRNAs. mRNA cleavage activity induced by secondary-type siRNAs in the csr-1 mutant was approximately only 10% the level of that observed in wild-type animals (Figure 7B).
Using antibodies raised against CSR-1, we examined which gel-filtration fractions contain CSR-1. CSR-1 was detected predominantly in void fractions, in which Slicer activity induced by the secondary-type siRNA was also detected (Figure 6D). We next examined the interaction between secondary-type siRNA and CSR-1 with the following experiments. We designed a nuclease-resistant 2′-O-methyl oligonucletotide, which mimics target mRNA and blocks Slicer activity (Figure 7C). A secondary-type siRNA/Slicer complex formed in vitro was captured by the 2′-O-methyl oligonucletotide immobilized on beads. As a result, CSR-1 was detected in the captured complex (Figure 7D).
We then prepared a CSR-1 recombinant protein fused with a His tag and MBP tag (Supplementary Figure 4C). We mixed the CSR-1 recombinant protein and single-stranded siRNAs and tested for the presence of Slicer activities. Interestingly, the CSR-1 recombinant protein showed a strong Slicer activity with secondary-type siRNAs, whereas it showed a weak Slicer activity with monophosphorylated single-stranded siRNAs (Figure 7E).
Discussion
Biochemical features of RdRP in C. elegans
In some organisms, the mechanisms of RNAi and related gene silencing are thought to involve signal amplification steps using RdRPs, which share homology with a cell-encoded enzyme originally found in tomato (Schiebel et al, 1998). In cell lysates from C. elegans, we detected an RdRP activity able to produce small RNAs using mRNA-derived templates. The polymerase RRF-1, which is involved in RNAi, was responsible for the RdRP activity in cell lysates. The RRF-1 complex showed an RdRP activity to synthesize small RNAs with 5′ triphosphorylated ends that are complementary to template RNAs. Previous studies in C. elegans have shown animals exhibiting RNAi to accumulate secondary siRNAs, which have 5′ triphosphorylated ends and are complementary to target mRNAs (Sijen et al, 2001, 2007; Pak and Fire, 2007). The biochemical features of the RdRP activity detected by our in vitro analyses were consistent with the known features of the in vivo production of secondary siRNAs; therefore, we believe that the RdRP activity detected by our in vitro analyses reflects secondary siRNA production.
We observed that aberrant mRNA lacking poly-A was several fold more efficient at generating RdRP products in worm extracts than normal mRNA. This observation might imply the possibility that normal mRNA coated with proteins could be occasionally cleaved by a weak reaction with primary siRNA, and then become aberrant mRNA accessible to RdRPs. However, the several-fold difference is not enough to deny the possibility that RdRPs act on normal mRNA activated without cleavage. Thus, an important question to address in the future is ‘what is the mechanism by which primary siRNAs aid the selective recruitment of RdRPs onto the target mRNA'.
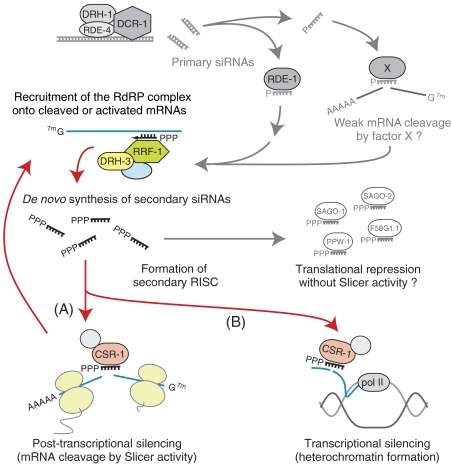
Based on the analyses in this study, we suggest that the RdRP complex for secondary siRNA production consists of RRF-1, DRH-3 and possibly a few other factors in C. elegans (Figure 8). DRH-3 is a DExH-box helicase involved in RNAi (Duchaine et al, 2006). We detected an interaction between DRH-3 and RRF-1 and showed that small RNA production via RdRP activity was impaired in drh-3 as well as rrf-1 mutants. An RdRP involved in RNA silencing in fission yeast is also known to form a complex. The complex in fission yeast consists of Rdp1 (RdRP core enzyme), Hrr1 (Upf1-like helicase) and Cid12 (nucleotidyltransferase-related protein) (Motamedi et al, 2004). In some organisms, appropriate functions of RdRPs in RNA silencing may be achieved by the formation of multi-subunit complexes.
Figure 8.
Model for the RNAi pathway in C. elegans (see text).
The triphosphorylated structure of secondary siRNAs indicates that the 5′ ends of secondary siRNAs are defined due to the de novo initiation of RNA syntheses by RdRPs (Pak and Fire, 2007; Sijen et al, 2007), although it is not yet clear whether the 3′-end formation of secondary siRNAs is also a Dicer-independent reaction. The data from this study indicate that secondary siRNA production is a Dicer-independent reaction, mainly because siRNA production via RdRP activity was detected even in lysates from the dcr-1 mutant.
qde-1 in Neurospora crassa is known to encode an RdRP required for a type of RNA silencing (Cogoni and Macino, 1999). The RdRP domain in QDE-1 was shown to be structurally similar to the active domains in DNA-dependent RNA polymerases (Salgado et al, 2006), and it was also shown that QDE-1 recombinant proteins were able to initiate RNA synthesis de novo (Makeyev and Bamford, 2002) just as DNA-dependent RNA polymerases do. Because RRF-1 in C. elegans has an RdRP domain homologous to the domain in QDE-1, it is easy to analogize that RRF-1 can initiate RNA synthesis de novo. However, information regarding the structure of the RdRP domain may be insufficient to explain how the RRF-1 complex synthesizes small RNAs whose major populations resemble Dicer products in length. It is tempting to speculate that the length of small RNAs synthesized by the RRF-1 complex could be modulated by non-core subunits.
On the other hand, recent studies have reported that RdRP and Dicer in Tetrahymena and fission yeast interact physically and may accomplish the production of small RNAs via coupled reactions (Colmenares et al, 2007; Lee and Collins, 2007). We did not detect a direct physical interaction between RRF-1 and DCR-1 in C. elegans. Thus, different organisms appear to use different strategies for small-RNA production via RdRP activities.
Slicer activity induced by a new class of small RNAs
In several organisms, primary-type siRNAs produced by Dicer were shown to have 5′ monophosphorylated ends and an ability to induce Slicer activities to cleave target mRNAs. However, it has not been well elucidated what kind of biochemical reactions are induced by secondary siRNAs that belong to a new class of small RNAs. We showed that secondary-type siRNAs bearing triphosphorylated ends strongly induce cleavage of target mRNAs in cell lysates from C. elegans. By contrast, both duplex and single-stranded forms of monophosphorylated siRNAs were far less effective at inducing Slicer activity than secondary-type siRNAs. These data imply the possibility that secondary rather than primary siRNAs play a major role in the destabilization of target transcripts during RNAi in C. elegans.
In C. elegans, it is difficult to conclude that primary siRNAs produced by the DCR-1 complex are directly involved in mRNA cleavage during the in vivo RNAi response, because the Slicer activity induced by primary-type duplex siRNAs was weak in our in vitro experiments. Meanwhile, we showed that the introduction of monophosphorylated single-stranded siRNAs also induced Slicer activity in cell lysates. These monophosphorylated single-stranded siRNAs may mimic an activated state of primary duplex siRNAs. Alternatively, the Slicer activity induced by the monophosphorylated single-stranded siRNAs may reflect a reaction in the endogenous RNAi-related silencing pathway, but not in the RNAi pathway induced by exogenous dsRNA.
We also showed that CSR-1 is predominantly responsible for the Slicer activity induced by secondary-type siRNAs in cell lysates. CSR-1 appears to be an Argonaute protein suited for Slicer activity in conjunction with secondary siRNAs, because a CSR-1 recombinant protein showed strong Slicer activity in conjunction with secondary-type siRNAs and weak activity in conjunction with monophosphorylated single-stranded siRNAs. The germline cells of csr-1 mutants are known to be partially deficient in RNAi (Yigit et al, 2006); we detected the expression of CSR-1 in somatic cells and observed that the somatic cells of csr-1 mutants were also partially deficient in RNAi (Supplementary Figure 5A and B). CSR-1 and other factors interacting with secondary siRNAs could take on partially overlapping functions. In fact, some part of the secondary siRNA molecules is known to interact with SAGO factors, which are Argonaute proteins in which the key amino-acid residues for endonuclease activity are not well conserved (Yigit et al, 2006).
Several models can be presumed to explain the roles of CSR-1. The first model is that CSR-1, along with secondary siRNAs, forms a secondary RISC and causes the destruction of target mRNAs (Figure 8, route A). Alternatively, it is also possible that CSR-1 is involved in silencing via a different mechanism, because it was reported that a csr-1 mutant has defects not only in RNAi and fertility but also in chromosomal segregation (Yigit et al, 2006). The chromosomal segregation defect in the csr-1 mutant implies that CSR-1 is involved in an endogenous pathway for transcriptional silencing. For this reason, we assume in the second model that CSR-1 and secondary siRNAs are involved in transcriptional silencing (Figure 8, route B). Indeed, CSR-1 may be involved in both post-transcriptional and transcriptional silencing processes in the exogenous RNAi pathway, or the involvement of CSR-1 in transcriptional silencing could be the case only in the endogenous pathway. Different approaches will be required in future to verify the possible involvement of CSR-1 in transcriptional silencing, because cytoplasmic fractions were used for the cell-free analyses in this study.
Materials and methods
Strains and transgenic animals
N2 was used as a wild-type C. elegans strain. Some experiments were performed using the following RNAi-deficient mutants: rde-1(ne300), rde-3(ne298), rde-4(ne301), rrf-1(pk1417), rrf-1(fj18), dcr-1(ok247)/hT2[qIs48], drh-3(fj52)/hT2[qIs48] and csr-1(fj54)/nT1[qIs51]. fj18 was obtained by a forward genetic screen. fj52 and fj54 were obtained by screening deletion mutants. Transgenic animals expressing RRF-1 tagged with GFP at its N-terminus were produced by microinjection.
Antibodies
Polyclonal antibodies against RDE-4, DCR-1, DRH-3 and CSR-1 were raised in rabbits or rats. Details are described in the Supplementary data.
Preparation of cell lysates for cell-free analyses
Animals were generally cultured on a large scale, purified using the sucrose flotation method and agitated in M9 buffer for 10 min. To weaken the cuticles, animals were agitated in 10 mM DTT and 25 mM borate-NaOH (pH 9) for 20 min. Subsequently, animals were washed three times with TBS and once with 1 × acetate buffer (100 mM potassium acetate, 10 mM HEPES-KOH (pH 7.6) and 4 mM magnesium acetate). One volume of animals was mixed with four volumes of homogenization buffer (150 mM KCl, 25 mM Hepes-KOH (pH 7.6), 5 mM magnesium acetate, 100 mM sucrose, 1 mM DTT and 1 mM CaCl2) containing 1 × Pefabloc Plus (Roche) and homogenized using a stainless steel homogenizer. The homogenate was centrifuged five times at 13 000 g for 1.5 min each, and then the supernatant was filtrated once using a 1.2-μm syringe filter. The cleared lysate was loaded onto a spin column bearing a weak cation exchanger (Vivapure C, Vivascience), and the flow-through from the column was recovered after centrifugation at 600 g. The flow-through fraction was loaded onto the same column, and again, the flow-through was recovered. The second flow-through from the C-type column was loaded onto a gel-filtration spin column of Sephadex G-25 (GE) equilibrated with freezing buffer (1 × acetate buffer containing 10% ethylene glycol, 0.2 mM DTT, and 0.1 × Pefabloc Plus) and centrifuged at 600 g for 2 min. The eluate from the G-25 column was supplemented with Pefabloc to 1 × concentration and centrifuged at 13 000 g for 2 min to remove insoluble material. The supernatant was dispensed into small aliquots and quickly frozen in liquid nitrogen. This type of lysate served as the standard lysate.
A high-MW fraction was used to detect RdRP activity. Standard lysate was loaded onto a Sephadex G-75 (GE) spin column equilibrated with freezing buffer. The eluate from the spin column served as the high-MW fraction.
RNAs for enzymatic assays
RNA substrates were mainly made from a cDNA clone (yk719g6 obtained from Y Kohara) corresponding to F46H5.3. An internally labeled dsRNA (542 bp) for the Dicer assay and cap-labeled mRNAs (393 nt) for the Slicer assay were prepared using methods based on those described by Zamore et al (2000). Monophosphorylated siRNAs were synthesized by an outside manufacturer. Triphosphorylated siRNAs were made by in vitro transcription and purified by electrophoresis. For the RdRP assay, normal mRNAs, aberrant mRNAs lacking poly-A and long dsRNAs were made by in vitro transcription and used as RNA templates. Details are described in the Supplementary data.
Basic conditions for enzymatic assays with the cell-free system
Each in vitro assay was performed at 25°C for 2 h in a total reaction volume of 20 μl, comprising 10 μl of lysate, 1 μl of HMD solution (0.2 M HEPES-KOH (pH 7.6), 40 mM magnesium acetate and 2 mM DTT) and 9 μl of RNA substrates and supplements. The final concentration of potassium was 50 mM because of carry-over from the lysate. Typical final concentrations of supplements were 1 mM ATP, 0.2 mM GTP, 0.2 mM CTP, 0.2 mM UTP, 20 mM creatine phosphate, 0.2 mg/ml creatine kinase and 0.3 U/μl RNasin (Promega). The supplement content was modified for each assay.
To stop the reaction, nine volumes of 1 mg/ml Proteinase K in PK buffer (200 mM Tris–HCl (pH 6.8), 0.3 M NaCl, 25 mM EDTA and 1% lauroylsarcosine) were added to the reaction mixture, which was then incubated at 50°C for 30 min. RNAs extracted from the reaction mixture were resuspended in formamide/EDTA gel-loading buffer and electrophoresed on sequencing gels.
RdRP assay
To remove endogenous nucleic acids, high-MW extracts were treated with 2 mM CaCl2, 0.02 U/μl DNase I and 0.8 U/μl MNase at 25°C for 30 min. EGTA was later added to the fraction to a final concentration of 4.8 mM. Then, a 35-μl reaction mixture was prepared from 20 μl of the high-MW fraction treated with DNase I and MNase, 15 μl of RNA templates (final 12.5 ng/μl) and supplements (omitting UTP and CTP) and then preincubated for 10 min. Next, 3 μl of [α-32P]UTP (3000 Ci/mmol) and 2 μl of non-radioactive UTP and CTP were added to the reaction, and the mixture was further incubated for 2 h. The final concentration of UTP was 10.3 μM and that of the other supplements was the same as in the basic conditions for the in vitro assay. The procedure for RdRP assays on immunoprecipitates of GFP∷RRF-1 is described in the Supplementary data.
Slicer assay
A 19-μl reaction mixture was prepared from 10 μl of the standard lysate and 9 μl of synthetic siRNAs and supplements, which were preincubated for 10 min. Next, 1 μl of cap-labeled mRNA was added to the reaction, and the mixture was further incubated for 2 h. The final siRNA concentration was typically 200 nM and the supplement content was the same as in the basic conditions for the in vitro assay.
Recombinant proteins
The cDNA for csr-1 was obtained by RT–PCR and cloned into a pET plasmid carrying a His tag (6 × His) and maltose-binding protein (MBP). The plasmid was transformed into ArcticExpress (DE3) bacterial cells (Stratagene), and then 6 × His∷MBP∷CSR-1 fusion protein was expressed at 12°C. The CSR-1 fusion protein was purified first using a nickel chelate resin and then with an amylose resin.
Supplementary Material
Supplementary Information
Supplementary Figures and Legends
Acknowledgments
We thank Y Kaziro, Y Kohara, A Fire and CC Mello for encouragement; Y Ogawa and T Asanuma for technical assistance; K Nishiwaki, R Kuroki and H Kimura for their guidance; T Ohta for useful suggestions; Y Kohara for the cDNA clones and the C. elegans Genetics Center (supported by NIH) for the strains. This work was supported by Special Coordination Funds for Promoting Science and Technology from MEXT in Japan, MEXT Grants-in-aid for Scientific Research on Priority Areas (Decode; RNA; Genome) and for Young Scientists Type B, an Encouragement Research Grant from the Asahi Glass Foundation, and PRESTO from JST.
References
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Chen CC, Simard MJ, Tabara H, Brownell DR, McCollough JA, Mello CC (2005) A member of the polymerase β nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr Biol 15: 378–383 [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G (1999) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166–169 [DOI] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D (2007) Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 [DOI] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, Yates JR III, Mello CC (2006) Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH (1999) Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99: 133–141 [DOI] [PubMed] [Google Scholar]
- Lee SR, Collins K (2007) Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol 14: 604–610 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Bamford DH (2002) Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol Cell 10: 1417–1427 [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574 [DOI] [PubMed] [Google Scholar]
- Mello CC, Conte D Jr (2004) Revealing the world of RNA interference. Nature 431: 338–342 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Remoue K, Sanial M, Vo TA, Vaucheret H (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321 [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A (2007) Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Parker GS, Eckert DM, Bass BL (2006) RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA 12: 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing in plants. Mol Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Salgado PS, Koivunen MR, Makeyev EV, Bamford DH, Stuart DI, Grimes JM (2006) The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol 4: e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel W, Pelissier T, Riedel L, Thalmeir S, Schiebel R, Kempe D, Lottspeich F, Sanger HL, Wassenegger M (1998) Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell 10: 2087–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Tomari Y, Zamore PD (2004) The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol 14: 787–791 [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476 [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH (2007) Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315: 244–247 [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol 10: 169–178 [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC (1999) The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132 [DOI] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC (2002) The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-Box helicase to direct RNAi in C. elegans. Cell 109: 861–871 [DOI] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA (1999) Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev 13: 3191–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC (2006) Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figures and Legends