Abstract
Intracellular membrane fusion requires SNARE proteins in a trans-complex, anchored to apposed membranes. Proteoliposome studies have suggested that SNAREs drive fusion by stressing the lipid bilayer via their transmembrane domains (TMDs), and that SNARE complexes require a TMD in each docked membrane to promote fusion. Yeast vacuole fusion is believed to require three Q-SNAREs from one vacuole and the R-SNARE Nyv1p from its fusion partner. In accord with this model, we find that fusion is abolished when the TMD of Nyv1p is replaced by lipid anchors, even though lipid-anchored Nyv1p assembles into trans-SNARE complexes. However, normal fusion is restored by the addition of both Sec18p and the soluble SNARE Vam7p. In restoring fusion, Sec18p promotes the disassembly of trans-SNARE complexes, and Vam7p enhances their assembly. Thus, either the TMD of this R-SNARE is not essential for fusion, and TMD-mediated membrane stress is not the only mode of trans-SNARE complex action, or these SNAREs have more flexibility than heretofore appreciated to form alternate functional complexes that violate the 3Q:1R rule.
Keywords: membrane fusion, Nyv1p, R-SNARE, trans-SNARE complex, yeast vacuole
Introduction
Intracellular membrane fusion requires conserved lipids and proteins, including SNARE proteins. SNAREs contain a variable N-terminal domain, a central SNARE domain with conserved heptad repeats that can assemble into four helical complexes, and (in most cases) a single transmembrane domain (TMD) at the C terminus (Jahn et al, 2003). SNARE proteins are classified as Q- or R-SNAREs based on the glutamine (Q) or arginine (R) at the center of their SNARE domain (Fasshauer et al, 1998). Though most SNAREs are membrane-anchored through a single C-terminal TMD, several SNAREs which lack a TMD are still required for fusion: SNAP-25 and SNAP-23 have a palmitate anchor, while Vam7p, a Q-SNARE for yeast vacuole fusion, lacks a proteinaceous TMD but has an N-terminal PX domain with affinity for PI(3)P (Cheever et al, 2001). A four-helical coiled-coils complex of SNARE proteins is required for fusion. Each of four SNARE proteins, three Q-SNAREs and one R-SNARE, can contribute a single helix to generate a four-helix bundle in cis (four SNAREs bound to the same membrane) or in trans (three Q-SNAREs from one membrane and one R-SNARE from the other). Studies of the fusion of proteoliposomes bearing recombinant SNAREs have led to a model (Hanson et al, 1997; Parlati et al, 2000) in which a trans-SNARE complex clamps two apposed membranes together, then triggers fusion. The trans-SNARE complexes are thought to promote fusion by applying force to the membranes as the energy of SNARE complex assembly is transmitted to the apposed bilayers through TMDs in each bilayer. SNARE-mediated membrane fusion is only thought to occur if at least one SNARE in the trans-SNARE complex is anchored to each membrane by a TMD (McNew et al, 2000b). The replacement of the TMD of the yeast vacuolar Q-SNARE Vam3p with a lipid-anchor allows trans-SNARE complex formation with its cognate R-SNARE Nyv1p but completely abolishes fusion (Rohde et al, 2003); since the three Q-SNARE vacuolar complex would still be expected to have a TMD from Vti1p, the loss of fusion potency may not simply reflect the absence of a trans-SNARE complex TMD in each bilayer. The TMD of Pep12p, a Q-SNARE for yeast endosomal fusion, can be removed without affecting its function in membrane fusion (Gerrard et al, 2000); presumably, it is associated with another Q-SNARE which has a TM domain. Overexpression of geranylgeranylated versions of Snc1p or Sso1p, which are normally required for Golgi-to-plasma membrane trafficking, dominantly inhibits exocytosis at a stage after vesicle docking and SNARE complex assembly (Grote et al, 2000). Since the wild-type Snc1p and Sso1p still enter SNARE complexes, the mode of dominant inhibition by their lipid-anchored mutant forms is not clear. Ykt6p, an R-SNARE in yeast, is naturally lipid-anchored (McNew et al, 1997). Though this SNARE does not support liposome fusion, an artificially protein-anchored version of Ykt6p triggers fusion in reconstituted liposome studies (McNew et al, 2000a). Each of these results is consistent with the model that at least one SNARE on each bilayer must be anchored by a TMD to mediate fusion, but none of these studies establishes this mechanism.
Homotypic yeast vacuole fusion is a technically accessible model for studying membrane fusion. Purified vacuoles fuse when incubated with ATP. Vacuole fusion requires three Q-SNAREs (Vti1p, Vam3p, and Vam7p) and one R-SNARE (Nyv1p) to form a four-helix complex. Studies using reconstituted proteoliposomes were the first to suggest that trans-SNARE complexes mediating vacuolar fusion exhibit the 3Q:1R topology, with three Q-SNAREs provided by one membrane and the R-SNARE Nyv1p from the other (Fukuda et al, 2000). This idea is also supported by studies of the fusion of purified vacuoles (Jun et al, 2006). In these studies, fusion was blocked by addition of the recombinant soluble domains of all three Q-SNAREs, but not by combinations of any two soluble Q-SNARE domains, suggesting that these three Q-SNAREs function together to engage a lone R-SNARE and thereby prevent its pairing with membrane-anchored Q-SNARE triads. Overexpression of only the R-SNARE increased the amount of the three soluble Q-SNARE domains needed for inhibition, and inhibition was relieved by pre-incubation of the three soluble Q-SNAREs with the soluble domain of the R-SNARE. The agreement between these studies of fusion on the intact organelle and studies with model liposomes indicates that vacuole fusion normally conforms to a rule of three Q-SNAREs engaging a lone R-SNARE in trans. If trans-SNARE complex function relies on a TMD in each bilayer, the TMD of the vacuolar R-SNARE Nyv1p would be essential for fusion.
We have now examined this model by replacing the TMD of Nyv1p with lipid anchors. In accord with previous studies, the lipid-anchored Nyv1p readily assembled into trans-SNARE complexes but did not support vacuole fusion. Strikingly, however, the fusion of vacuoles bearing the lipid-anchored Nyv1p was completely restored by the addition of both Sec18p (yeast NSF) and Vam7p (the soluble Q-SNARE for vacuole fusion) to remodel trans-SNARE complexes. This restored fusion still relies on Nyv1p, excluding the possibility that another R-SNARE might substitute for Nyv1p in the fusion of lipid-anchored Nyv1p vacuoles. Thus, either vacuole fusion can be promoted efficiently by trans-SNARE pairing which is not 3Q:1R, or the vacuolar trans-SNARE complex can lead to lipid bilayer merger without TMD-mediated membrane stress.
Results
The R-SNARE Nyv1p mediates vacuole fusion through forming trans-SNARE complexes with its cognate Q-SNAREs (Ungermann et al, 1998b). While nyv1Δ vacuoles do not fuse in vitro (Nichols et al, 1997) and antibody to Nyv1p blocks in vitro vacuole fusion (Ungermann et al, 1998a), yeast cells lacking NYV1 have morphologically normal vacuoles, an indication of normal fusion in vivo (Nichols et al, 1997), which may be supported by the lipid-anchored R-SNARE Ykt6p (Thorngren et al, 2004). We have now re-examined the requirement for Nyv1p during vacuole fusion in vitro: is the inactivation of fusion by antibody to Nyv1p due to nonspecific crossreactivity with other proteins on the vacuole, or is it direct? Is the loss of fusion of nyv1Δ vacuoles caused by an indirect effect, such as a defect in the trafficking to the vacuole of other proteins that are required for fusion?
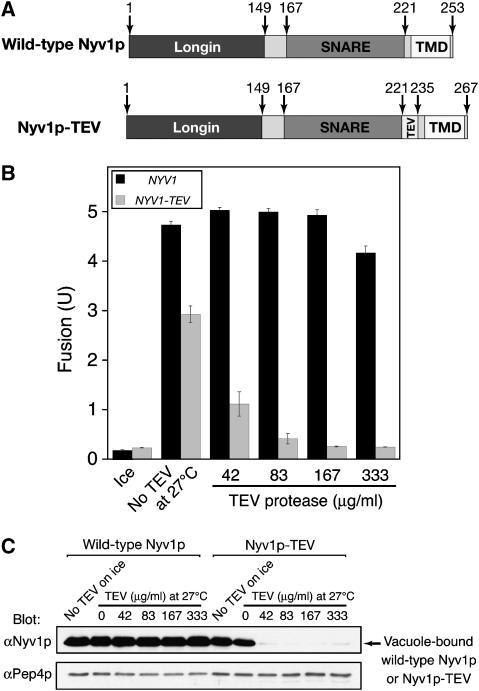
To measure fusion, vacuoles are isolated from two strains. One contains normal proteases but lacks the major vacuolar phosphatase encoded by the PHO8 gene. The other strain has a wild-type PHO8 gene but is deleted for vacuolar protease genes and thus accumulates catalytically inactive pro-Pho8p. Though neither vacuole population bears active Pho8p phosphatase, fusion allows proteases to gain access to pro-Pho8p and cleave it to active Pho8p, which is assayed colorimetrically. To selectively inactivate Nyv1p during in vitro vacuole fusion, we generated a mutant form of Nyv1p that contains a TEV protease cleavage sequence between its SNARE motif and TMD (named Nyv1p-TEV hereafter; Figure 1A) and expressed it in either the protease-deficient BJ3505 background or in the Pho8p-deficient DKY6281 background as a sole source of Nyv1p.
Figure 1.
TEV protease-mediated Nyv1p inactivation prevents homotypic yeast vacuole fusion in vitro. (A) Schematic representation of wild-type Nyv1p and Nyv1p-TEV. (B) TEV protease specifically inactivates Nyv1p-TEV vacuoles for fusion. BJ3505 (NYV1) and DKY6281 (NYV1) vacuoles or BJ3505 NYV1-TEV and DKY6281 NYV1-TEV vacuoles were incubated at 27°C in fusion reactions (see Materials and methods) in the presence of indicated concentrations of TEV protease. After 90 min, a portion was assayed for fusion (B), and the rest centrifuged to sediment vacuoles, which were resuspended in SDS sample buffer and analyzed by SDS–PAGE and immunoblotting (C). (C) Fusion inactivation by TEV protease correlates with the removal of Nyv1p-TEV from vacuoles. The anti-Pep4p blot serves as a loading control. Data are mean±s.e.m. (n=3).
Vacuoles isolated from cells expressing Nyv1p-TEV fuse almost as well as wild-type vacuoles (Figure 1B), indicating that Nyv1p-TEV is functional. The modest reduction in the fusion of Nyv1p-TEV vacuoles (Figure 1B; gray bars versus black bars) could be due to the insertion of 14 amino acids (the TEV protease cleavage and linker sequences) between the SNARE domain and the TMD of Nyv1p, as the length of linker between SNARE domain and TMD is critical for fusion (McNew et al, 1999; Wang et al, 2001). While the fusion of wild-type vacuoles is hardly affected by increasing concentrations of TEV, the fusion of Nyv1p-TEV vacuoles is fully inhibited by TEV protease (Figure 1B). The reduction in fusion by TEV protease is accompanied by reduced levels of Nyv1p-TEV on vacuoles (Figure 1C), demonstrating that fusion was inhibited by TEV protease-mediated removal of Nyv1p-TEV from vacuoles. These data provide an independent and definitive confirmation of the essential role of Nyv1p in yeast vacuole fusion, as none of the proteins encoded in the Saccharomyces cerevisiae genome contain a TEV protease cleavage sequence. The lipid-anchored SNARE Ykt6p, which can function with high levels of Vam7p to restore fusion to nyv1Δ vacuoles (Thorngren et al, 2004), may fulfill the same role in vivo and thereby allow normal vacuole morphology.
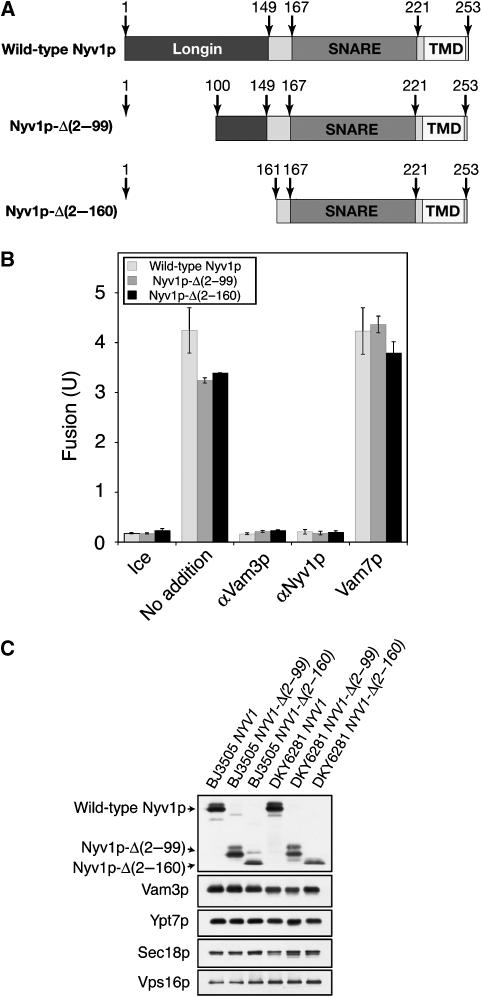
Nyv1p consists of three domains: an N-terminal longin domain (Wen et al, 2006), a SNARE motif, and a single TMD. While replacing the zero-layer arginine of the SNARE domain with glutamine inhibits fusion (Wang et al, 2001; Fratti et al, 2007), the other domains are less characterized. We have therefore examined whether Nyv1p function requires the longin domain and TMD of Nyv1p.
The genome of S. cerevisiae encodes only five R-SNAREs. Three R-SNAREs (Ykt6p, Sec22p, and Nyv1p) have a longin domain at their N termini, whereas the other two (Snc1p and Snc2p) have a short N-terminal sequence. The longin domain of Ykt6p, another R-SNARE found on the vacuole that shares sequence homology with Nyv1p, inhibits SNARE complex assembly by engaging its own SNARE domain in an ‘autoinhibited' conformation (Tochio et al, 2001). Ykt6p also has acyltransferase activity, contributing to palmitoylation of the fusion factor Vac8p (Dietrich et al, 2004). The roles of the longin domains of Nyv1p and Sec22p in membrane fusion are unknown. To test whether the longin domain of Nyv1p participates in vacuole fusion, we generated Nyv1p mutants that are deleted for some or all of their longin domain (Figure 2A). Partial truncation or full deletion of the longin domain only caused modest reduction in fusion (Figure 2B). This partial fusion defect could be due to a reduced level of mutant Nyv1p on the vacuole (Figure 2C), consistent with the recent finding that efficient sorting of Nyv1p to the limiting membrane of the vacuole relies on the Y31GTI34 motif in its longin domain (Wen et al, 2006), but the apparent reduction in levels of truncated Nyv1p could be due to loss of epitopes. These results show that the N-terminal domain of Nyv1p is not essential for vacuole fusion.
Figure 2.
The Nyv1p N-terminal longin domain is not essential for vacuole fusion. (A) Nyv1p and its derivatives. (B) Nyv1p longin domain deletions support fusion. BJ3505 NYV1 and DKY6281 NYV1 vacuoles, BJ3505 NYV1-Δ(2–99) and DKY6281 NYV1-Δ(2–99) vacuoles, or BJ3505 NYV1-Δ(2–160) and DKY6281 NYV1-Δ(2–160) vacuoles were incubated at 27°C in the absence or presence of antibodies to Vam3p, antibodies to Nyv1p, or recombinant Vam7p and, after 90 min, assayed for fusion. Data are mean±s.e.m. (n=3). (C) Vacuoles from BJ3505 NYV1, BJ3505 NYV1-Δ(2–99), BJ3505 NYV1-Δ(2–160), DKY6281 NYV1, DKY6281 NYV1-Δ(2–99), and DKY6281 NYV1-Δ(2–160) were analyzed by SDS–PAGE and immunoblotting.
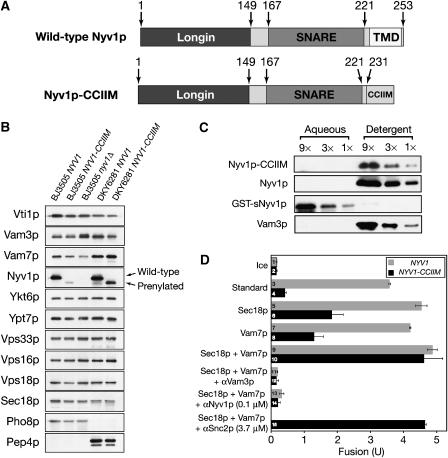
Most SNAREs are membrane-anchored through their single TMD, and truncation or deletion of the TMD of any SNARE tested abolishes or reduces fusion (Nonet et al, 1998; Saifee et al, 1998; Grote et al, 2000; McNew et al, 2000b; Rohde et al, 2003; Xu et al, 2005), suggesting an essential role of TMD in SNARE-mediated membrane fusion. To determine whether the TMD of Nyv1p is required for fusion, we replaced it with the Ykt6p prenylation motif (Figure 3A) and expressed it in either the protease-deficient BJ3505 background or in the Pho8p-deficient DKY6281 background as a sole source of Nyv1p. Levels of most vacuolar proteins tested were comparable between wild-type NYV1 and NYV1-CCIIM vacuoles (Figure 3B), and this mutation had no effect on vacuole morphology or cell growth rate (data not shown). Less Nyv1p-CCIIM was found on the vacuole than wild-type Nyv1p, though their levels in total cell lysates are similar (data not shown), suggesting that the TMD of Nyv1p contributes to its targeting. After vacuoles have undergone a Sec18p and ATP-dependent priming reaction, which releases Nyv1p from its association with other SNAREs, Nyv1p-CCIIM remains bound to the vacuole (Dietrich et al, 2005), indicating that it (like the R-SNARE Ykt6p, which also has CCIIM at its C terminus) is derivatized by a combination of prenyl and acyl groups. After vacuole priming, Nyv1p-CCIIM is released from other SNAREs (data not shown) and the detergent-solubilized Nyv1p-CCIIM partitions into Triton X-114 micelles as efficiently as wild-type Nyv1p (Figure 3C), and in contrast to purified recombinant soluble domain of Nyv1p, which lacks any membrane anchor but retains its activity of combining with other SNAREs (Jun et al, 2006). This is in accord with findings of Dietrich et al (2005) that Nyv1p-CCIIM remained vacuole bound after priming.
Figure 3.
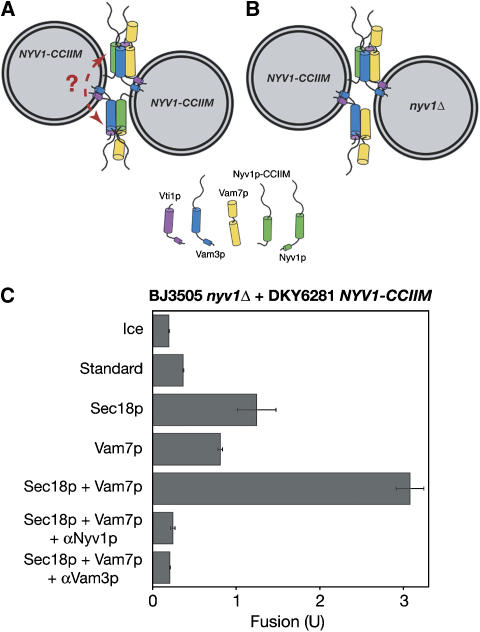
Fusion is restored to vacuoles that lack the TMD of Nyv1p by added Sec18p and Vam7p. (A) Wild-type Nyv1p and Nyv1p-CCIIM. (B) Protein profiles of NYV1, nyv1Δ, and NYV1-CCIIM vacuoles. Vacuoles purified from BJ3505 NYV1, BJ3505 NYV1-CCIIM, BJ3505 nyv1Δ, DKY6281 NYV1, and DKY6281 NYV1-CCIIM were analyzed by SDS–PAGE and immunoblotting. (C) Triton X-114 phase partitioning analysis of the hydrophobicity of wild-type Nyv1p with its TMD, Nyv1p-CCIIM, and the cytoplasmic domain of Nyv1p without an apolar anchor was done with primed BJ3505 NYV1 vacuoles, BJ3505 NYV1-CCIIM vacuoles, and BJ3505 nyv1Δ vacuoles supplemented with recombinant GST-sNyv1p, as described (Bordier, 1981). (D) The addition of both Sec18p and Vam7p enables Nyv1p-CCIIM vacuoles to fuse. Wild-type or Nyv1p-CCIIM vacuoles were incubated on ice or at 27°C in fusion reactions with indicated proteins, added from the start of the incubation. After 90 min, reactions were assayed for fusion. Data are mean±s.e.m. (n=3).
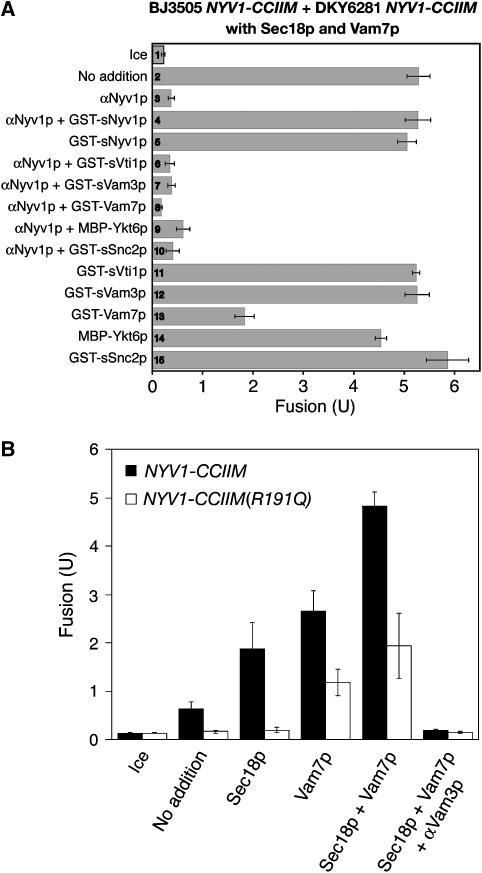
In agreement with previous studies of SNAREs with TMDs replaced by lipid anchors (Grote et al, 2000; Rohde et al, 2003; Giraudo et al, 2005), vacuoles isolated from yeast strains expressing the lipid-anchored Nyv1p (named Nyv1p-CCIIM) did not fuse (Figure 3D, bar 4). The addition of either recombinant Sec18p or recombinant Vam7p restored some fusion (bars 6 and 8); the levels of partial fusion restoration by either Sec18p or Vam7p showed some variation among vacuole preparations but were not significantly different (Figures 3D, 4 and 5). Strikingly, nearly full fusion was achieved upon addition of both Sec18p and Vam7p (Figure 3D, compare bars 9 and 10). This fusion is blocked by antibodies to Vam3p or Nyv1p, indicating SNARE dependence (bars 12 and 14). This fusion is not sensitive to even 40-fold higher concentrations of antibody to Snc2p (bar 15), an R-SNARE which is found on the plasma membrane, endosomes and vacuoles (Robinson et al, 2006). Vacuoles from NYV1-CCIIM snc2Δ strains also show full fusion, which depends on the addition of both Vam7p and Sec18p (data not shown). The sensitivity of fusion to anti-Nyv1p (Figure 3D, bar 14) suggests that this fusion is mediated by the lipid-anchored Nyv1p as a functional R-SNARE rather than by another R-SNARE, which might have replaced Nyv1p. Antibody to Nyv1p does not block the disassembly of cis-SNARE complexes by Sec18p (Ungermann et al, 1998a). To confirm the specificity of inhibition by antibodies to Nyv1p, fusion reactions containing NYV1-CCIIM vacuoles, Sec18p and Vam7p were incubated with anti-Nyv1p, GST-tagged cytosolic domain of Nyv1p (GST-sNyv1p), or both. Though GST-sNyv1p had little effect on fusion, it completely relieved anti-Nyv1p-mediated fusion inhibition (Figure 4A, bars 2–5). In contrast, none of the other SNARE protein soluble domains could relieve the inhibitory effect of anti-Nyv1p upon fusion (bars 6–10). These results strongly suggest that anti-Nyv1p inhibits fusion through inactivation of Nyv1p, though another non-SNARE target that shares an epitope with Nyv1p is not formally excluded. As an independent means of testing whether Nyv1p-CCIIM remains involved in vacuole fusion, we compared the fusion capacity of vacuoles bearing Nyv1p-CCIIM with its normal arginyl residue at the central zero-layer of the SNARE domain to the fusion capacity of vacuoles with Nyv1p-CCIIM bearing an R to Q mutation at the zero layer (Nyv1p-CCIIM R191Q). As reported for wild-type Nyv1p (Fratti et al, 2007), exchange of the Nyv1p-CCIIM zero-layer arginyl residue for a glutamyl residue strongly depresses fusion (Figure 4B), though high levels of Vam7p at least partially restore fusion as they do for vacuoles with a normal transmembrane-anchored Nyv1p with the R191Q mutation (ibid). Taken together, these data on antibody inhibition and zero-layer involvement establish that the Nyv1p-CCIIM is required for the fusion of NYV1-CCIIM vacuoles.
Figure 4.
Nyv1p-CCIIM engages in vacuole fusion. (A) The Sec18p/Vam7p-mediated fusion of Nyv1p-CCIIM vacuoles is inhibited by antibodies to Nyv1p, relieved only by recombinant sNyv1p. Vacuoles from BJ3505 NYV1-CCIIM and DKY6281 NYV1-CCIIM were incubated in fusion reactions containing both Sec18p and Vam7p at 27°C in the presence of indicated proteins. After 90 min, reactions were assayed for fusion. Data represent mean±s.e.m. (n=3). To optimize the chance of seeing relief from αNyv1p inhibition, we employed 3.3 μM GST-Vam7p (lanes 8 and 13), a level which itself often causes some fusion inhibition (lane 13). (B) The zero-layer arginine of Nyv1p-CCIIM is important for Nyv1p-CCIIM vacuole fusion. Standard fusion assays (27°C, 90 min) bore BJ3505 nyv1Δ vacuoles and either DKY6281 NYV1-CCIIM or DKY6281 NYV1-CCIIM R192Q vacuoles. Vam7p (638 nM), his6-Sec18p (63.8 nM), and anti-Vam3p (444 nM) were added where indicated.
Figure 5.
BJ3505 nyv1Δ vacuoles can fuse with DKY6281 NYV1-CCIIM vacuoles upon addition of Sec18p and Vam7p. Schematic representation of fusion between BJ3505 NYV1-CCIIM vacuoles and DKY6281 NYV1-CCIIM vacuoles (A) or BJ3505 nyv1Δ vacuoles and DKY6281 NYV1-CCIIM vacuoles (B). The dotted line indicates a hypothetical interaction between trans-SNARE complexes. (C) Vacuoles from BJ3505 nyv1Δ and DKY6281 NYV1-CCIIM were incubated for 90 min in fusion reactions on ice or at 27°C with indicated proteins and assayed for fusion. Data are mean±s.e.m. (n=3).
Might multiple trans-SNARE complexes be bound together, directly or through other proteins such as HOPS, providing TMD anchors on apposed membranes and thereby allowing the TMD anchored in the two apposed bilayers to transmit strain (Figure 5A, dotted arrow)? Vacuoles from BJ3505 nyv1Δ and DKY6281 NYV1-CCIIM (Figure 5B) fuse upon addition of Vam7p and Sec18p (Figure 5C). These data indicate that oligomerization of conventional four-SNARE trans-complexes is not required to provide TMDs in each bilayer for fusion, unless a novel functional complex is formed between three Q-SNAREs and a conventional four-SNARE trans-complex.
Remodeling trans-SNARE complexes
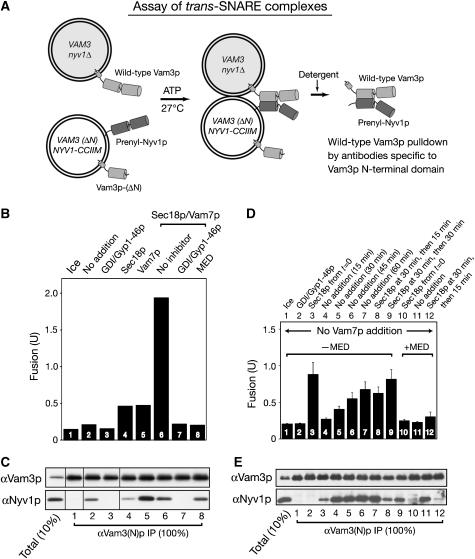
To study how Sec18p and Vam7p support the fusion of vacuoles bearing Nyv1p-CCIIM, we assayed the assembly of Nyv1p-CCIIM into trans-SNARE complexes. Trans-SNARE complexes were analyzed in fusion reactions with vacuoles from BJ3505 VAM3 nyv1Δ and DKY6281 VAM3(ΔN) NYV1-CCIIM, bearing N-terminally deleted Vam3p and Nyv1p-CCIIM (Figure 6A). The Vam3p N-domain is dispensable for fusion (Wang et al, 2001). With these vacuoles, the only four-SNARE trans-complex that can form will include the full-length Vam3p from the BJ3505-derived strain and the Nyv1p-CCIIM from the DKY6281-derived strain. When supplemented with Sec18p and Vam7p, these vacuoles fuse (Figure 6B, bar 6) at a rate that is comparable to the fusion rate of reactions with BJ3505 nyv1Δ vacuoles with DKY6281 vacuoles bearing wild-type Vam3p and Nyv1p (Nichols et al, 1997). Detergent extracts were prepared from these same incubations for analysis of trans-SNARE associations (Figure 6C).
Figure 6.
Nyv1p-CCIIM-mediated vacuole fusion requires Sec18p remodeling of trans-SNARE complexes. (A) Assay of trans-SNARE complexes. (B–E) Remodeling of trans-SNARE complexes is required for Nyv1p-CCIIM to support fusion. BJ3505 nyv1Δ and DKY6281 VAM3(ΔN) NYV1-CCIIM vacuoles were incubated in fusion reactions on ice or at 27°C with indicated reagents. After 45 min, aliquots were assayed for fusion (B) and trans-SNARE complexes (C). (D, E) The kinetics of trans-SNARE complex assembly and disassembly. BJ3505 nyv1Δ and DKY6281 VAM3(ΔN) NYV1-CCIIM vacuoles (396 μg each) were mixed with standard reaction buffer and ATP in 3.96 ml. At indicated times, portions (330 μl) were transferred to tubes with Sec18p, Gyp1-46p/Gdi1p, or control buffer before continuing incubation, either on ice or at 27°C. Reactions were stopped by transfer to ice. Aliquots (30 μl) were removed to measure fusion. The remaining 300 μl was mixed with 6 μl of 0.5 mM EDTA, then assayed for trans-SNARE complexes as described in Materials and methods, with proportionate reduction in solubilization buffer from 600 to 400 μl. Incubations were either on ice (lane 1) or at 27°C, with Sec18p added from the start of the incubation (lanes 3 and 10) or after 30 min at 27°C (lanes 8, 9 and 12), and with Gyp1-46p and Gdi1p added from the start of incubation (lane 2). Incubations at 27°C were for 15, 30, 45, or 60 min (lanes 4–7, respectively). After Sec18p addition, the samples in lanes 8 and 9 were incubated for an additional 15 or 30 min at 27°C, respectively. Samples in lanes 10–12 had MED from the start of the incubation; the sample in lane 12 received Sec18p after 30 min of incubation and was then incubated for a further 15 min before analysis.
Using DKY6281 VAM3(ΔN) NYV1-CCIIM vacuoles, the immunoprecipitation of wild-type Vam3p by means of antibodies specific to the N-terminal domain of Vam3p co-precipitates Nyv1p-CCIIM (Figure 6C, lane 2) in a temperature- and Rab GTPase Ypt7p-dependent manner (lanes 1 and 3). Thus the normal Rab-dependent pathway allows Nyv1p-CCIIM to assemble into trans-SNARE complexes. This result, along with the anti-Nyv1p sensitivity of the fusion of vacuoles bearing this lipid-anchored Nyv1p, shows that Nyv1p-CCIIM is a functional version of Nyv1p that associates with vacuolar Q-SNAREs, or at least the Q-SNARE Vam3p, in trans to mediate fusion.
How do Vam7p and Sec18p cooperate with the Nyv1p-CCIIM SNARE to promote fusion? Unsupplemented fusion reactions that include lipid-anchored Nyv1p-CCIIM form trans-SNARE complexes (Figure 6C, lane 2) but do not go on to fuse (Figure 6B, bar 2). The addition of Sec18p diminishes the amount of trans-SNARE complex (Figure 6C, lane 4), while Vam7p promotes the formation of substantially more trans-SNARE complex (lane 5; see also Collins and Wickner, 2007), yet the addition of Vam7p or Sec18p alone only gives a minor additional fusion signal (Figure 6B, bars 4 and 5). Both Vam7p and Sec18p must be added for vigorous fusion (Figure 6B, bar 6), yet together they yield less trans-SNARE complex than is seen with the addition of Vam7p alone (compare Nyv1p in lanes 5 and 6 of Figure 6C). Sec18p disassembly of cis-SNARE complexes would make more SNAREs, not less, available to form trans-SNARE complexes; the diminution of trans-SNARE complex levels by added Sec18p (Figure 6C) must therefore reflect Sec18p action on trans-SNARE complexes, whose re-formation can be driven by Vam7p. The level of trans-SNARE complex increases for 15–30 min during our fusion incubations, then reaches a steady state (Figure 6E, lanes 4–7; also see Collins and Wickner, 2007). The addition of Sec18p at 30 min, to vacuoles which had already formed the steady-state level of trans-SNARE complex (Figure 6E, lane 5), results in dramatic complex disassembly (Figure 6E, lanes 8 and 9). We also employed the MARCKS effector domain (MED) peptide, a high-affinity ligand to the phosphoinositides which are required for vacuole fusion (Fratti et al, 2004). In accord with studies of NYV1 vacuoles (Collins and Wickner, 2007), MED permits full assembly of trans-SNARE complex with Nyv1p-CCIIM without accompanying fusion (Figure 6B and C, lane 8 and Figure 6D and E, lane 11), and this trans-SNARE complex can be disassembled by added Sec18p (Figure 6E, lane 12).
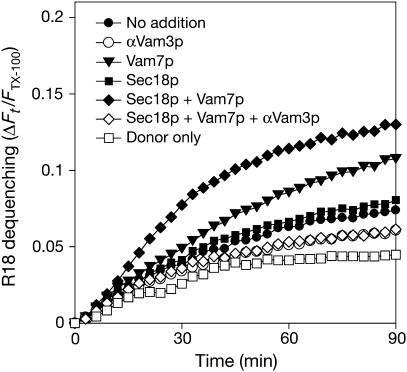
Though vacuoles with Nyv1p-CCIIM undergo little lumenal compartment mixing when neither Vam7p nor Sec18p is added (Figure 3D, lane 4; Figure 5C, lane 2; or Figure 6B, lane 2), it remained possible that they had undergone hemifusion, allowing the lipidic anchor of Nyv1p-CCIIM to move laterally in the fused cytoplasmic leaflet and thereby enter an essentially cis-SNARE complex with full-length Vam3p. Such hemifusion would be accompanied by lipid mixing. We therefore labeled a portion of vacuoles bearing Nyv1p-CCIIM with the self-quenching lipidic fluorophore octadecyl rhodamine B (R18), incubated them with unlabeled vacuoles bearing Nyv1p-CCIIM, and assayed for hemifusion- or fusion-induced lipid mixing, as reported for vacuoles with wild-type Nyv1p (Jun and Wickner, 2007). The initial rate of dequenching (Figure 7, filled circles) was hardly above the background rates seen in presence of antibody to Vam3p (open circles) or in the absence of unlabeled vacuoles (open squares), and there was only modest stimulation by adding Sec18p (filled squares) or Vam7p (triangles) alone. Maximal dequenching was seen when both Sec18p and Vam7p were added (filled diamonds), and this dequenching signal was fully blocked by antibody to Vam3p (open diamonds). This absence of SNARE-dependent lipid mixing seen when there was no added Sec18p or Vam7p indicates that the Nyv1p-CCIIM association with full-length Vam3p (Figure 6) was truly in trans.
Figure 7.
Vacuoles bearing Nyv1p-CCIIM require additional Sec18p and Vam7p for lipid mixing. A portion of BJ3505 NYV1-CCIIM vacuoles were labeled with self-quenching levels of octadecyl rhodamine B (R18), reisolated, and incubated with an unlabeled portion of these vacuoles under standard fusion conditions to assay for lipid mixing-induced dequenching, as described (Jun and Wickner, 2007). Measurements were taken every 2 min for 90 min, yielding fluorescence values at the onset (F0) and during the reaction (Ft). The final 10 measurements of a sample containing 0.33% (v/v) Triton X-100 were averaged and used as a value for the fluorescence after infinite dilution (FTX100). The relative total fluorescence change ΔFt/FTX100=(Ft−F0)/FTX100 was calculated.
Fusion of vacuoles with lipid-anchored Vam3p
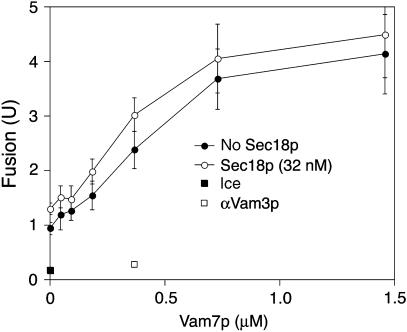
The above results could be explained if the remodeling of trans-SNARE complex, by cycles of Sec18p-mediated disassembly and Vam7p-promoted reassembly, allows vacuoles to fuse without a SNARE proteinaceous TMD in each bilayer. This model rests on the consensus view that vacuole SNAREs pair 3Q∷1R, yet this is not completely established and is very hard to establish. However, it has been clearly shown that the SNAREs Vam3p and Nyv1p enter trans-SNARE complexes from opposing vacuoles (Nichols et al, 1997; Collins and Wickner, 2007). Since Vam7p has no TMD, the question then becomes whether Vti1p enters trans-SNARE complexes from the same side as Vam3p or Nyv1p. We therefore examined the fusion of VAM3-CCIIM vacuoles. Like their Nyv1p-CCIIM counterparts, the poor fusion of VAM3-CCIIM vacuoles (Rohde et al, 2003) can be restored to near normal levels by the addition of Vam7p and Sec18p (Figure 8), though in this case the major dependence is on the Vam7p SNARE and there is less need for additional Sec18p. Thus, fusion is blocked when the number of TMDs that anchor the vacuolar trans-SNARE complex is reduced from three to two, whether the TMD that is lost was on Nyv1p or Vam3p, and fusion is restored by Sec18p and Vam7p-mediated remodeling. This indicates that either productive trans-SNARE pairing is not limited to 3Q:1R, or TMDs anchoring trans-SNARE complexes to each docked membrane are not required for fusion.
Figure 8.
Vam7p restores fusion to vacuoles from BJ3505 VAM3-CCIIM and DKY6281 VAM3-CCIIM. Standard fusion assays (90 min, 27°C) were supplemented with Vam7p and/or 32 nM Sec18p. Anti-Vam3p IgG (888 nM) was added with 365 nM Vam7p and 32 nM Sec18p where indicated. The assay value for a sample held on ice was subtracted from all points except the anti-Vam3p and ice points. Error bars represent standard deviations from three independent experiments. The error bars for the anti-Vam3p and the ice points are covered by their symbols.
Discussion
SNARE proteins are comprised of a heptad-repeat SNARE domain, flanked by an N-terminal domain and a C-terminal TMD. N-terminal domains are not conserved, and are not always required for membrane fusion (Wang et al, 2001), in accord with our finding that the N-domain of Nyv1p only confers a modest enhancement to fusion rates (Figure 2). The well-studied SNARE domains are crucial for fusion (Rizo and Sudhof, 2002). Yeast vacuole fusion requires the presence of the Nyv1p SNARE domain (Figure 1) and depends upon its contribution of a zero-layer arginyl residue to the four-helical bundle (Fratti et al, 2007). It has been proposed that the TMDs of SNAREs not only maintain membrane attachment but are also the major effectors whereby SNARE complexes contribute to bilayer fusion. We find that the replacement of the TMD of Nyv1p or Vam3p by lipid anchors creates a conditional defect in fusion, which can be overcome by adding the SNARE chaperone Sec18p and the soluble Q-SNARE Vam7p.
Sec18p diminishes the steady-state level of trans-SNARE complexes while promoting fusion (Figure 6). We have previously suggested that Sec18p can disassemble the SNARE pairs that form in trans between Vam3p on nyv1Δ vacuoles and Nyv1p on vam3Δ vacuoles (Ungermann et al, 1998b). However, vam3Δ vacuoles fuse poorly and are highly fragmented, and it was not clear whether the complex between Vam3p and Nyv1p represented an authentic trans-SNARE complex or post-fusion cis-SNARE complex. We have recently documented an assay of trans-SNARE complexes between vacuoles of normal size, composition, and fusion potency (Collins and Wickner, 2007), and shown that stable trans-SNARE complexes can form even when fusion is blocked by MED. We now show that Sec18p depresses the steady-state level of trans-SNARE pairs while strongly enhancing fusion for vacuoles with lipid-anchored Nyv1p (Figure 6). Earlier studies also reported that added Sec18p or NSF had little or no effect on the level of vacuole fusion (Ungermann et al, 1998b) or of SNARE proteoliposome lipid mixing (Weber et al, 2000); it nonetheless regulates the steady-state level of SNARE pairs (Figure 6). Our current studies of vacuoles with Nyv1p-CCIIM reveal a condition where Sec18p remodeling of trans-SNARE complexes is actually needed for normal rates of fusion. This remodeling may entail disassembly and reassembly with different partners, such as Sec17p or HOPS, in a different microdomain, such as boundary membrane versus vertex ring (Wang et al, 2003), in different state of oligomerization with other four-helical SNARE complexes (Peters et al, 2004; Roy et al, 2006), or through trans-SNARE complexes other than 3Q:1R. Further experiments will be needed to directly test these possibilities.
While SNAREs can pair promiscuously in solution (Yang et al, 1999), studies of lipid mixing with SNARE-bearing liposomes have suggested that fusion at each organelle can only be supported by the association of one combination of SNAREs from each apposed bilayer (Parlati et al, 2000). For yeast vacuoles, this combination is Nyv1p on one membrane and the three Q-SNAREs (Vam3p, Vti1p, and Vam7p) from the other (Fukuda et al, 2000). In accord with this model, Nyv1p has been shown to normally associate with Vam3p in trans (Collins and Wickner, 2007). It has also been proposed that a fusogenic trans-SNARE complex requires at least one TMD in each apposed membrane (McNew et al, 2000b). This would predict that the TMD of Nyv1p is critical for vacuole fusion. In this study, we replaced the TMD of Nyv1p with lipid anchors and found that fusion was blocked. Surprisingly, fusion can be restored by Sec18p/Vam7p-mediated trans-complex remodeling (Figures 3, 4, 5, 6 and 7). One simple model to explain this observation is that trans-SNARE complex does not require TMDs in each bilayer to support fusion. Remodeling may either spatially redistribute the trans-SNARE complex or alter its associations with Sec17p (which facilitates Sec18p-mediated disassembly), HOPS (which does not facilitate Sec18p-mediated disassembly but which supports fusion), or other factors. However, if TMDs are in fact required in each apposed membrane for fusion, different combinations of SNAREs which are not anchored in the apposed membranes according to 3Q:1R might provide functional complexes. For example, on Nyv1p-CCIIM mutant vacuoles, a functional trans-SNARE complex might be formed by Vam3p/Vam7p from one membrane and Vti1p/Nyv1p-CCIIM from the other, thus maintaining one TMD on each membrane. Similarly, on Vam3p-CCIIM mutant vacuoles (Figure 8), a functional trans-SNARE complex could be formed by Vam3p-CCIIM/Vti1p from one membrane and Vam7p/Nyv1p from the other. These ‘abnormal' combinations may not necessarily be the dominant forms, but under remodeling conditions may become more abundant and fusogenic via a yet-to-be-identified mechanism. This model would violate the 3Q∷1R rule but there is precedent for this, at least for endosomal SNARE-mediated liposome fusion (Zwilling et al, 2007). Further experiments are clearly required to test each of these concepts.
Previous studies with vacuoles bearing wild-type SNAREs have also suggested a remodeling of trans-SNARE complexes. We observed that the Ca2+ efflux from vacuole stores that accompanies membrane fusion (Peters and Mayer, 1998) occurs at trans-SNARE pairing events (Merz and Wickner, 2004). The addition of Vam7p and Sec18p caused a synergistic increase in Ca2+ release (ibid, Figure 5A), possibly due to the additional trans-SNARE pairing events, which are inherent in multiple cycles of Sec18p-mediated disassembly of trans-SNARE complexes and Vam7p-driven reassembly. Our current studies have the advantage of uncovering a condition (lipid-anchored Nyv1p) in which trans-SNARE complexes can be assayed directly and for which remodeling is needed for fusion; the earlier studies (Merz and Wickner, 2004) employed wild-type vacuoles and an entirely independent assay of trans-SNARE complex dynamics. Taken together, they provide complementary support for trans-SNARE complex remodeling.
It has been proposed (McNew et al, 1999, 2000b) that SNAREs mediate membrane fusion through applying stress to the bilayer. The energy for this stress would derive from the assembly of the SNAREs into a four-helical trans-complex, and be transmitted to their TMDs. In accord with this idea, the fusion of liposomes bearing SNAREs is inhibited by inserting amino acyl residues between the SNARE and TMDs of SNAREs (McNew et al, 1999; Wang et al, 2001), though the introduction of helix-disrupting prolyl or glycyl residues had little effect on fusion. While these studies are consistent with the torsional stress model, there is no direct assay of force transmission. The replacement of other SNARE TMDs with a lipidic anchor which occupies half of the bilayer blocks fusion (McNew et al, 2000b; Rohde et al, 2003). This has been presumed to be due to an inability of this lipid anchor to exert stress on the distal bilayer leaflet, since longer lipid anchors will restore fusion (McNew et al, 2000b). Lipid-anchored Nyv1p also fails to support vacuole fusion (Figure 3D). However, fusion is readily restored by additional Sec18p and Vam7p. The restored fusion reaction remains fully dependent on the lipid-anchored Nyv1p (Figure 4), even when that Nyv1p is only present on one fusion partner (Figure 5). This finding does not demonstrate that an intact TMD does not provide torsional bilayer stress, or that this stress does not contribute to fusion reactions. Rather, it suggests that either fusion is not driven exclusively by canonical 3Q:1R trans-SNARE complexes or that TMDs are not required to induce stress for the Rab- and SNARE-dependent fusion of yeast vacuoles. These findings re-open these questions for other membrane fusion reactions as well.
What else besides torsional stress might cause apposed bilayers to rearrange their lipids to yield fusion? SNAREs accumulate in membrane microdomains (Lang et al, 2001), and vacuole fusion occurs at a membrane microdomain termed the vertex ring. The proteins and lipids that are required for fusion become highly enriched in this fusion microdomain. SNAREs are required for the vertex ring enrichment of other proteins (Wang et al, 2003) and of fusion-essential ‘regulatory' lipids (Fratti et al, 2004). Trans-SNARE pairing may contribute to fusion by aligning the vertex rings on each pair of docked vacuoles and allowing the enrichment of inherently fusogenic lipids such as diacylglycerol (Allan et al, 1978; Fratti et al, 2004; Jun et al, 2004) and of other proteins such as HOPS (Wang et al, 2003).
Materials and methods
Yeast strains
S. cerevisiae strains BJ3505 (Matα ura3–52 trp1-Δ101 his3-Δ200 lys2-801 gal2 (gal3) can1 prb1-Δ1. 6R pep4∷HIS3) (Jones, 2002) and DKY6281 (Matα ura3-52 leu2-3,112 trp1-Δ901 his3-Δ200 lys2-801 suc2-Δ9 pho8∷TRP1) (Haas et al, 1994), or their derivatives described below were used to purify vacuoles for in vitro fusion assays. BJ3505 nyv1Δ∷TRP1 and DKY6281 nyv1Δ∷HIS3 were transformed with EcoNI-linearized pRS406-NYV1-TEV, pRS406-NYV1-(Δ2–99), pRS406-NYV1-(Δ2–160), or pRS406-NYV1-CCIIM to generate BJ3505 or DKY6281 expressing Nyv1p-TEV, Nyv1p-(Δ2–99), Nyv1p-(Δ2–160), or Nyv1p-CCIIM, respectively. DKY6281 vam3Δ∷HIS3 was transformed with NruI-digested pRS406-VAM3(ΔN), generating DKY6281-VAM3(ΔN). To generate DKY6281-VAM3(ΔN) NYV1-CCIIM, the NYV1 gene was deleted first in the DKY6281-VAM3(ΔN) strain by PCR product-mediated gene deletion, generating DKY6281-VAM3(ΔN) nyv1Δ∷KanR. This strain was then transformed with EcoNI-linearized pRS408-NYV1-CCIIM, creating DKY6281-VAM3(ΔN) NYV1-CCIIM. BJ3505 VAM3-CCIIM and DKY6281 VAM3-CCIIM (Rohde et al, 2003) were generous gifts from Dr Christian Ungermann (University of Osnabrück, Germany).
Reagents
Antibodies were prepared as described and dialyzed into PS buffer (10 mM PIPES/KOH, pH 6.8, 200 mM sorbitol) with 125 mM KCl. Concentrations used (unless otherwise noted) were 200 nM affinity-purified anti-Nyv1p antibody (Thorngren et al, 2004); 3.7 μM affinity-purified anti-Snc2p antibody (described below) and 444 nM anti-Vam3p IgG (Wang et al, 2003). Purified recombinant proteins were dialyzed into PS buffer with 125 mM KCl and used at the following concentrations: 5 μM his6-Gyp1-46p (Wang et al, 2003); 1.2 μM Gdi1p (Starai et al, 2007); 10 μM MED (Wang et al, 2001); 66 nM his6-Sec18p (Thorngren et al, 2004), unless indicated otherwise. Recombinant Vam7p, purified via chitin affinity chromatography and intein cleavage as described (Starai et al, 2007), was stored in 20 mM HEPES-NaOH, pH 8.0, 300 mM NaCl and added to fusion reactions at 0.67 μM. GST-tagged soluble domains of Nyv1p, Vti1p, Vam7p, and Vam3p were produced from Escherichia coli as described (Thorngren et al, 2004; Jun et al, 2006). MBP fusion of Ykt6p cytosolic domain was produced as described (Thorngren et al, 2004). The cytosolic domain (1–93) of Snc2p was produced as a GST fusion from E. coli bearing pGST-sSnc2p, and the purified protein was injected into a rabbit to generate polyclonal antibodies. The N-terminal domain of Vam3p was produced as a GST fusion (Dulubova et al, 2001) from E. coli containing the plasmid pGEX-KT-Vam3 (5–135), a generous gift of Dr Josep Rizo (University of Texas Southwestern Medical Center) and immobilized on SulfoLink Coupling Resin (Pierce) according to the manufacturer's instruction. By using these Vam3p N-terminal domain-immobilized agarose beads, antibodies specific to the N-terminal domain of Vam3p were affinity-purified from anti-Vam3p rabbit serum. The purified antibodies were then immobilized on AminoLink Coupling Resin (Pierce).
MBP-TEV(S219V)-Arg5 was produced as described (Kapust et al, 2001) with modifications, using E. coli containing the plasmid pRK1043 (Addgene plasmid 8835), a generous gift of Dr David Waugh (National Institute of Cancer). Briefly, E. coli Rosetta-2 (Novagen) cells bearing pRK1043 were grown in TB plus ampicillin (100 μg/ml) and chloramphenicol (37 μg/ml) to an OD600 of 0.8–1.0 at 37°C. Fusion protein expression was induced by 0.5 mM IPTG at 20°C for 14 h. After harvesting cells by centrifugation, cells were resuspended in 200 ml of MBP buffer (50 mM HEPES (pH 7.4), 200 mM NaCl, 1 mM EDTA, 1 mg/ml benzamidine, and one protease inhibitor cocktail tablet (Roche)) and lysed by French press. After centrifugation (15 000 g, 20 min, 4°C), the supernatant was loaded onto an amylose (NEB) column equilibrated with MBP buffer. After the column was washed five times with 20 ml MBP buffer, the MBP fusion protein was eluted with this buffer containing 10 mM maltose. The eluate was dialyzed against PS buffer with 125 mM KCl and stored at −80°C.
Vacuole isolation and in vitro vacuole fusion assay
Standard 30 μl in vitro fusion reactions at 27°C contained 20 mM PIPES-KOH, pH 6.8, 200 mM sorbitol, 125 mM KCl, 6 mM MgCl2, 1 mM ATP, 1 mg/ml creatine kinase, 29 mM creatine phosphate, 10 μM coenzyme A, 264 nM purified Pbi2p (IB2), 3 μg pep4Δ vacuoles (from BJ3505 or its derivatives), and 3 μg pho8Δ vacuoles (from DKY6281 or its derivatives). Pho8p phosphatase activity was assayed as a measure of vacuole fusion. Fusion units (U) are micromole p-nitrophenolate formed per minute per microgram pep4Δ vacuole.
Assay of trans-SNARE complexes
Standard trans-SNARE and fusion assays (16 × ) contained 48 μg of vacuoles from BJ3505 nyv1Δ and 48 μg of vacuoles from DKY6281 VAM3-(ΔN) NYV1-CCIIM. After 45 min, reactions were placed on ice (5 min). From each 16 × reaction, 30 μl was withdrawn to assay Pho8p maturation, and the rest (450 μl) received 10 mM EDTA, was incubated on ice for 10 min to stop the ATPase activity of Sec18p, and was then centrifuged (11 000 g, 5 min, 4°C). The supernatant was removed, and the sedimented vacuoles were overlaid with ice-cold solubilization buffer (200μl; 20 mM TrisCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, 10% glycerol, 1 × protease inhibitor cocktail (0.46 μg/ml leupeptin, 3.5 μg/ml pepstatin, 2.4 μg/ml pefabloc-SC, 1 mM PMSF), resuspended on ice, and solubilization buffer was added to a final volume of 600 μl. The extracts were mixed on a nutator at 4°C for 20 min, and the detergent-insoluble material was removed by centrifugation (16 000 g, 20 min, 4°C). A portion (80 μl) of the resulting supernatant was removed for a ‘total' sample, and the remainder (520 μl) received anti-Vam3p N-terminal domain antibody-conjugated agarose beads and was incubated on a nutator at 4°C overnight. Beads were collected by brief centrifugation (4000 g, 2 min, 4°C) and resuspended five times with the solubilization buffer followed by bead sedimentation. Bound proteins were eluted by boiling beads in SDS sample buffer (94°C, 5 min), separated by SDS–PAGE, and analyzed by immunoblotting.
Supplementary methods are available at The EMBO Journal Online (http://embojournal.org).
Supplementary Material
Supplementary Methods
Acknowledgments
We thank Drs David Waugh and Josep Rizo for plasmids and Dr Christian Ungermann for strains. This work was supported by NIH grant GM23377.
References
- Allan D, Thomas P, Michell RH (1978) Rapid transbilayer diffusion of 1,2-diacylglycerol and its relevance to control of membrane curvature. Nature 276: 289–290 [DOI] [PubMed] [Google Scholar]
- Bordier C (1981) Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem 256: 1604–1607 [PubMed] [Google Scholar]
- Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M (2001) Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol 3: 613–618 [DOI] [PubMed] [Google Scholar]
- Collins KM, Wickner WT (2007) Trans-SNARE complex assembly and yeast vacuole membrane fusion. Proc Natl Acad Sci USA 104: 8755–8760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LE, Gurezka R, Veit M, Ungermann C (2004) The SNARE Ykt6 mediates protein palmitoylation during an early stage of homotypic vacuole fusion. EMBO J 23: 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LE, Peplowska K, LaGrassa TJ, Hou H, Rohde J, Ungermann C (2005) The SNARE Ykt6 is released from yeast vacuoles during an early stage of fusion. EMBO Rep 6: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Yamaguchi T, Wang Y, Sudhof TC, Rizo J (2001) Vam3p structure reveals conserved and divergent properties of syntaxins. Nat Struct Biol 8: 258–264 [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R (1998) Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA 95: 15781–15786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti RA, Collins KM, Hickey CM, Wickner W (2007) Stringent 3Q{middle dot}1R composition of the SNARE 0-layer can be bypassed for fusion by compensatory SNARE mutation or by lipid bilayer modification. J Biol Chem 282: 14861–14867 [DOI] [PubMed] [Google Scholar]
- Fratti RA, Jun Y, Merz AJ, Margolis N, Wickner W (2004) Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J Cell Biol 167: 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R, McNew JA, Weber T, Parlati F, Engel T, Nickel W, Rothman JE, Sollner TH (2000) Functional architecture of an intracellular membrane t-SNARE. Nature 407: 198–202 [DOI] [PubMed] [Google Scholar]
- Gerrard SR, Mecklem AB, Stevens TH (2000) The yeast endosomal t-SNARE, Pep12p, functions in the absence of its transmembrane domain. Traffic 1: 45–55 [DOI] [PubMed] [Google Scholar]
- Giraudo CG, Hu C, You D, Slovic AM, Mosharov EV, Sulzer D, Melia TJ, Rothman JE (2005) SNAREs can promote complete fusion and hemifusion as alternative outcomes. J Cell Biol 170: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote E, Baba M, Ohsumi Y, Novick PJ (2000) Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion. J Cell Biol 151: 453–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A, Conradt B, Wickner W (1994) G-protein ligands inhibit in vitro reactions of vacuole inheritance. J Cell Biol 126: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE (1997) Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90: 523–535 [DOI] [PubMed] [Google Scholar]
- Jahn R, Lang T, Sudhof TC (2003) Membrane fusion. Cell 112: 519–533 [DOI] [PubMed] [Google Scholar]
- Jones EW (2002) Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae. Methods Enzymol 351: 127–150 [DOI] [PubMed] [Google Scholar]
- Jun Y, Fratti RA, Wickner W (2004) Diacylglycerol and its formation by phospholipase C regulate Rab- and SNARE-dependent yeast vacuole fusion. J Biol Chem 279: 53186–53195 [DOI] [PubMed] [Google Scholar]
- Jun Y, Thorngren N, Starai VJ, Fratti RA, Collins K, Wickner W (2006) Reversible, cooperative reactions of yeast vacuole docking. EMBO J 25: 5260–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun Y, Wickner W (2007) Assays of vacuole fusion resolve the stages of docking, lipid mixing, and content mixing. Proc Natl Acad Sci USA 104: 13010–13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng 14: 993–1000 [DOI] [PubMed] [Google Scholar]
- Lang T, Bruns D, Wenzel D, Riedel D, Holroyd P, Thiele C, Jahn R (2001) SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J 20: 2202–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew JA, Parlati F, Fukuda R, Johnston RJ, Paz K, Paumet F, Sollner TH, Rothman JE (2000a) Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature 407: 153–159 [DOI] [PubMed] [Google Scholar]
- McNew JA, Sogaard M, Lampen NM, Machida S, Ye RR, Lacomis L, Tempst P, Rothman JE, Sollner TH (1997) Ykt6p, a prenylated SNARE essential for endoplasmic reticulum–Golgi transport. J Biol Chem 272: 17776–17783 [DOI] [PubMed] [Google Scholar]
- McNew JA, Weber T, Engelman DM, Sollner TH, Rothman JE (1999) The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol Cell 4: 415–421 [DOI] [PubMed] [Google Scholar]
- McNew JA, Weber T, Parlati F, Johnston RJ, Melia TJ, Sollner TH, Rothman JE (2000b) Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol 150: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz AJ, Wickner WT (2004) Trans-SNARE interactions elicit Ca2+ efflux from the yeast vacuole lumen. J Cell Biol 164: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols BJ, Ungermann C, Pelham HR, Wickner WT, Haas A (1997) Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature 387: 199–202 [DOI] [PubMed] [Google Scholar]
- Nonet ML, Saifee O, Zhao H, Rand JB, Wei L (1998) Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J Neurosci 18: 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F, McNew JA, Fukuda R, Miller R, Sollner TH, Rothman JE (2000) Topological restriction of SNARE-dependent membrane fusion. Nature 407: 194–198 [DOI] [PubMed] [Google Scholar]
- Peters C, Baars TL, Buhler S, Mayer A (2004) Mutual control of membrane fission and fusion proteins. Cell 119: 667–678 [DOI] [PubMed] [Google Scholar]
- Peters C, Mayer A (1998) Ca2+/calmodulin signals the completion of docking and triggers a late step of vacuole fusion. Nature 396: 575–580 [DOI] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC (2002) Snares and Munc18 in synaptic vesicle fusion. Nat Rev Neurosci 3: 641–653 [DOI] [PubMed] [Google Scholar]
- Robinson M, Poon PP, Schindler C, Murray LE, Kama R, Gabriely G, Singer RA, Spang A, Johnston GC, Gerst JE (2006) The Gcs1 Arf-GAP mediates Snc1,2 v-SNARE retrieval to the Golgi in yeast. Mol Biol Cell 17: 1845–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde J, Dietrich L, Langosch D, Ungermann C (2003) The transmembrane domain of Vam3 affects the composition of cis- and trans-SNARE complexes to promote homotypic vacuole fusion. J Biol Chem 278: 1656–1662 [DOI] [PubMed] [Google Scholar]
- Roy R, Peplowska K, Rohde J, Ungermann C, Langosch D (2006) Role of the Vam3p transmembrane segment in homodimerization and SNARE complex formation. Biochemistry 45: 7654–7660 [DOI] [PubMed] [Google Scholar]
- Saifee O, Wei L, Nonet ML (1998) The Caenorhabditis elegans unc-64 locus encodes a syntaxin that interacts genetically with synaptobrevin. Mol Biol Cell 9: 1235–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Jun Y, Wickner W (2007) From the cover: feature article: excess vacuolar SNAREs drive lysis and Rab bypass fusion. Proc Natl Acad Sci USA 104: 13551–13558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorngren N, Collins KM, Fratti RA, Wickner W, Merz AJ (2004) A soluble SNARE drives rapid docking, bypassing ATP and Sec17/18p for vacuole fusion. EMBO J 23: 2765–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochio H, Tsui MM, Banfield DK, Zhang M (2001) An autoinhibitory mechanism for nonsyntaxin SNARE proteins revealed by the structure of Ykt6p. Science 293: 698–702 [DOI] [PubMed] [Google Scholar]
- Ungermann C, Nichols BJ, Pelham HR, Wickner W (1998a) A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J Cell Biol 140: 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C, Sato K, Wickner W (1998b) Defining the functions of trans-SNARE pairs. Nature 396: 543–548 [DOI] [PubMed] [Google Scholar]
- Wang L, Merz AJ, Collins KM, Wickner W (2003) Hierarchy of protein assembly at the vertex ring domain for yeast vacuole docking and fusion. J Cell Biol 160: 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dulubova I, Rizo J, Sudhof TC (2001) Functional analysis of conserved structural elements in yeast syntaxin Vam3p. J Biol Chem 276: 28598–28605 [DOI] [PubMed] [Google Scholar]
- Weber T, Parlati F, McNew JA, Johnston RJ, Westermann B, Sollner TH, Rothman JE (2000) SNAREpins are functionally resistant to disruption by NSF and alphaSNAP. J Cell Biol 149: 1063–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Chen L, Wu H, Sun X, Zhang M, Banfield DK (2006) Identification of the yeast R-SNARE Nyv1p as a novel longin domain-containing protein. Mol Biol Cell 17: 4282–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang F, Su Z, McNew JA, Shin YK (2005) Hemifusion in SNARE-mediated membrane fusion. Nat Struct Mol Biol 12: 417–422 [DOI] [PubMed] [Google Scholar]
- Yang B, Gonzalez L Jr, Prekeris R, Steegmaier M, Advani RJ, Scheller RH (1999) SNARE interactions are not selective. Implications for membrane fusion specificity. J Biol Chem 274: 5649–5653 [DOI] [PubMed] [Google Scholar]
- Zwilling D, Cypionka A, Pohl WH, Fasshauer D, Walla PJ, Wahl MC, Jahn R (2007) Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J 26: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods