Abstract
The second messenger cyclic diguanylate (c-di-GMP) controls the transition between motile and sessile growth in eubacteria, but little is known about the proteins that sense its concentration. Bioinformatics analyses suggested that PilZ domains bind c-di-GMP and allosterically modulate effector pathways. We have determined a 1.9 Å crystal structure of c-di-GMP bound to VCA0042/PlzD, a PilZ domain-containing protein from Vibrio cholerae. Either this protein or another specific PilZ domain-containing protein is required for V. cholerae to efficiently infect mice. VCA0042/PlzD comprises a C-terminal PilZ domain plus an N-terminal domain with a similar β-barrel fold. C-di-GMP contacts seven of the nine strongly conserved residues in the PilZ domain, including three in a seven-residue long N-terminal loop that undergoes a conformational switch as it wraps around c-di-GMP. This switch brings the PilZ domain into close apposition with the N-terminal domain, forming a new allosteric interaction surface that spans these domains and the c-di-GMP at their interface. The very small size of the N-terminal conformational switch is likely to explain the facile evolutionary diversification of the PilZ domain.
Keywords: allostery, bacterial pathogenesis, cyclic diguanylate (cyclic-di-GMP), fluorescence resonance energy transfer, X-ray crystallography
Introduction
The cyclic mono-nucleotides cAMP and cGMP have long been recognized as major intracellular second messengers in eukaryotes and prokaryotes. Recently, bis-(3′-5′)-cyclic dimeric guanosine monophosphate (cyclic diguanylate or c-di-GMP—Figure 6 and Supplementary Figure S6) has emerged as a novel second messenger species controlling the transition between motile and sessile growth modes in many eubacterial species (Galperin et al, 2001; D'Argenio and Miller, 2004; Galperin, 2004; Jenal, 2004; Romling et al, 2005; Jenal and Malone, 2006; Romling and Amikam, 2006; Cotter and Stibitz, 2007). Similar to cyclic mono-nucleotides, the intracellular level of c-di-GMP is controlled via the offsetting action of specific cyclase and phosphodiesterase enzymes that are regulated in response to changing physiological conditions. The so-called GGDEF domains function as c-di-GMP cyclases (Paul et al, 2004; Ryjenkov et al, 2005). The so-called HD-GYP and EAL domains function as c-di-GMP phosphodiesterases that degrade c-di-GMP into two GMP molecules or pGpG, respectively (Ausmees et al, 2001; Christen et al, 2005; Schmidt et al, 2005; Tamayo et al, 2005); pGpG is then further degraded into two GMP molecules by an uncharacterized phosphodiesterase (Ross et al, 1985; Romling et al, 2005). These two domains were found to be among the most abundant domains encoded in eubacterial genomes, suggesting that c-di-GMP-dependent regulation is widespread in this kingdom (Galperin et al, 2001; Galperin, 2005).
C-di-GMP concentration has been shown to control a variety of cellular processes, including motility, pilus expression, biofilm formation, and photosynthesis (Huang et al, 2003; Kirillina et al, 2004; Simm et al, 2004; Thomas et al, 2004). While different biochemical processes seem to be controlled by c-di-GMP in response to different extracellular signals in different eubacterial species depending on their physiology and environmental niche, this second messenger generally controls the transition from motile to sessile growth mode (Romling et al, 2005). Low concentrations of c-di-GMP promote motile growth, while high concentrations promote sessile growth (e.g. biofilm formation).
Although clear evidence links changes in cytosolic c-di-GMP level to specific phenotypic alterations, there is little information on the proteins that bind c-di-GMP to effect these changes. It is generally assumed that such effector domains come from a variety of different sequence–structure families including the ‘PilZ' domain (Amikam and Galperin, 2006). However, the identity of these domains is just beginning to be elucidated, and no protein structure has yet been determined for any candidate effector domain bound to c-di-GMP.
An insightful bioinformatics analysis has suggested that PilZ domains (Figure 1 and Supplementary Figure S1) generally function as c-di-GMP effectors (Amikam and Galperin, 2006). This domain is found at the C terminus of cellulose synthetase, the product of the bcsA gene. C-di-GMP was first discovered as an allosteric regulator of this enzyme from the fruit-degrading eubacterium Gluconacetobacter xylinus in seminal studies conducted by Benziman and coworkers (Ross et al, 1985). The PilZ protein from Pseudomonas aeruginosa comprises a single 118-residue domain with significant sequence similarity to the C terminus of BcsA (Amikam and Galperin, 2006). PilZ is one of the few proteins of unassigned function in the pil operon that is responsible for pilus biosynthesis and function, a process known to be regulated as part of the transition from motile to sessile growth modes even though the mechanistic details of this regulation have not been elucidated (Mattick, 2002). PilZ deletion mutants (ΔpilZ) are able to generate normal amounts of pilin protomer but are not capable of assembling functional pili (Alm et al, 1996). Other PilZ-domain proteins recently have been shown to participate in the control of motility in several organisms, including the YcgR protein in Salmonella enterica (Ryjenkov et al, 2006) and the DgrA and DgrB proteins in Caulobacter crescentus (Christen et al, 2007). These latter proteins have been shown to be required for the inhibition of motility at high cytosolic c-di-GMP concentration. There are two examples of PilZ domains being located at the C terminus of proteins participating in processes known to be sensitive to c-di-GMP level (Amikam and Galperin, 2006) (right side of Figure 1A). In addition to BcsA, another example is Alg44 from P. aeruginosa, an essential protein in the c-di-GMP-regulated process of alginate biosynthesis (Merighi et al, 2007). All of these observations are consistent with the hypothesis that PilZ domains function as effectors binding c-di-GMP.
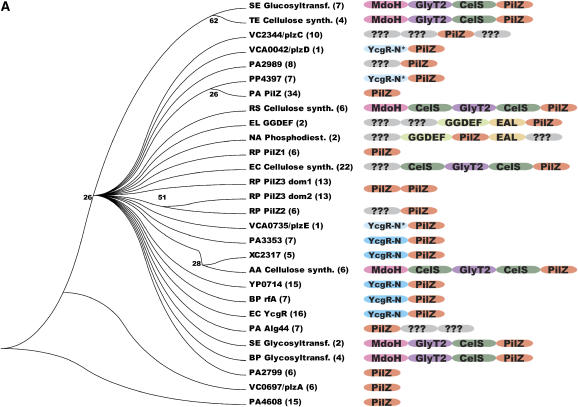
Figure 1a.
Phylogenetic analysis of eubacterial PilZ domains. These analyses were modeled on those of Amikam and Galperin (2006). (A) Cladogram showing evolutionary relationships between PilZ domains inferred from maximum parsimony analysis of a multiple sequence alignment (containing only the PilZ domain from each protein). The name of a representative member of each functional protein family is given, with the first two letters representing the SwissProt species code and the open-reading-frame number used for proteins lacking functional annotation. The number of likely orthologs identified in 295 fully sequenced eubacterial genomes is indicated in parentheses, while the diagrams schematize the overall domain organization inferred using RPS-BLAST analysis of the Conserved Domain Database (CDD) (Marchler-Bauer et al, 2005). Numbers at internal nodes in the tree represent the support for a split at the indicated evolutionary distance (but are not shown for 100% support). Protein domains are labeled as follows: CelS, cellulose synthetase domain; EAL, c-di-GMP phosphodiesterase domain; GGDEF, c-di-GMP cyclase domain; GlyT2, glycosyl transferase domain; MdoH, domain of membrane glycosyltransferases in COG2943; YcgR-N, strong sequence homology to the N-terminal domain of the E. coli YcgR protein at a level equivalent to that found in proteins in COG5581; YcgR-N*, weaker sequence homology to the YcgR-N domain; ???, uncharacterized conserved domain. The single PilZ-domain proteins DgrA and DgrB from C. crescentus are not represented here because likely orthologous proteins were not identified in other organisms.
The interaction of c-di-GMP with PilZ domains is supported by recent binding and mutagenesis studies of several proteins, including the YcgR protein from Escherichia coli (Ryjenkov et al, 2006), the DgrA and DgrB proteins from C. crescentus (Christen et al, 2007), and the Alg44 protein from P. aeruginosa (Merighi et al, 2007). These studies demonstrate c-di-GMP binding with sub-micromolar affinity dependent on residues in the RxxxR and D/NxSxxG sequence motifs conserved in PilZ domains (Figure 1B). Furthermore, NMR studies of PA4608, a PilZ domain protein from P. aeruginosa show large chemical shift perturbations during c-di-GMP titration, consistent with high-affinity binding of this ligand (Christen et al, 2007; Ramelot et al, 2007).
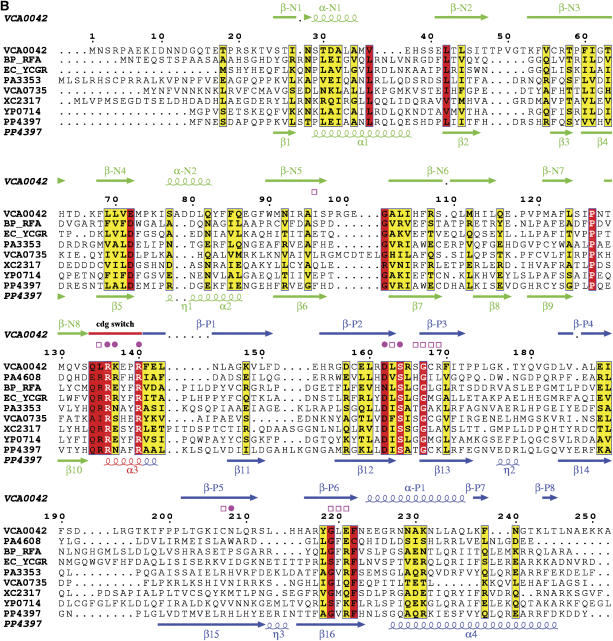
Figure 1b.
(B) Sequence–structure alignment generated by ESPRIPT (Gouet et al, 1999). Arrows represent β-strands and coils represent α-helices. Secondary structural elements are numbered independently in the two domains and colored according to domain of origin, with the YcgR-N* domain green, the c-di-GMP switch red, and the remainder of the PilZ domain blue. Magenta symbols represent van der Waals contacts to c-di-GMP, with closed circles and open squares indicating the presence or absence of H-bonds, respectively. The sequence alignment, which has the most strongly conserved sites highlighted in red, includes the other PilZ-domain-containing proteins of known structure plus one representative from each functional protein family in (B) whose domain organization is equivalent to that of VCA0042/PlzD. These were aligned initially using a position-specific score matrix (Altschul et al, 1997) derived from the alignment of Amikam and Galperin (2006) but manually adjusted to reflect the structural alignment of the individual domains.
C-di-GMP signaling plays an important role in the life cycle of Vibrio cholerae. This natural inhabitant of aquatic environments is the causative agent of the human disease cholera (Islam et al, 1993), which involves potentially life-threatening diarrhea caused by the secretion of cholera toxin. C-di-GMP has been shown to regulate the expression of virulence genes in V. cholerae (Tischler and Camilli, 2005). VieA is a c-di-GMP phosphodiesterase that regulates cholera toxin production through its effect on cytosolic c-di-GMP level. The VCA0042 protein is one of five PilZ domain-containing proteins identified in the genome of V. cholerae. There proteins diverged in sequence prior to the root of the Vibrio lineage (Figure 1A) and presumably function as regulators of various cellular processes controlled by c-di-GMP. VCA0042 has recently been named PlzD and shown to bind c-di-GMP when immobilized on nitrocellulose in the native conformation (Pratt et al, 2007). This study also shows that either VCA0042/PlzD or VC2344/PlzC, another PilZ domain-containing protein, in V. cholerae is required for the El Tor biotype of V. cholerae to efficiently colonize the intestines of mice in a virulence assay.
We herein present a high-resolution crystal structure of VCA0042 bound to c-di-GMP. The crystal structure of apo VCA0042 was previously determined by the Midwest Center for Structural Genomics (PBD ID 1YLN; R Zhang, M Zhou, S Moy, F Collart, and A Joachimiak). Our results provide the first information on the stereochemical basis of c-di-GMP recognition by PilZ domains as well as a model for the structural basis of c-di-GMP-dependent signal transduction by these domains.
Results
PilZ domain phylogeny
A phylogenetic analysis of eubacterial PilZ domains supports an early divergence into different functional protein families (i.e. ortholog groups) characterized by diverse overall domain organizations (Figure 1A). Even organisms in the same phylogenetic family (e.g. E. coli, V. cholerae, and Yersinia pestis in the enterobacteriaceae) typically encode a different set of PilZ domain-containing proteins from one another as judged by divergence in their sequences and domain organizations. These differences presumably reflect the diversity of the molecular processes subject to c-di-GMP regulation in different environmental niches. Sequence analysis suggests that there are five PilZ domain-containing proteins in V. cholerae—VC0697, VC1885, VC2344, VCA0042, and VCA0735 (Amikam and Galperin, 2006). These have recently been named PlzA through PlzE (Pratt et al, 2007), and they share equivalently low levels of sequence identity in their PilZ domains to each other and to the bulk of the PilZ domain-containing functional families found in other eubacteria (Figure 1B and Supplementary Figure S1). The PilZ domain from VCA0042/PlzD is found in the largest sub-tree within the PilZ superfamily, which itself contains at least 23 different protein families that are likely to have diverged in function at an ancient point in the eubacterial lineage (Figure 1A). The position of the PilZ domain from VCA0042/PlzD within this sub-tree indicates that it should be a representative model for the structure and function of PilZ domains.
Combined sequence profiling and structural analyses (described at the top of page 2 in the Supplementary data) indicate that the overall domain organization of VCA0042/PlzD is equivalent to that of seven other functional protein families (i.e. distinct ortholog groups) found in eubacteria (Figure 1A). These proteins comprise a C-terminal PilZ domain approximately 100 residues in length plus an N-terminal domain of slightly larger size. Their common domain organization suggests that proteins in all seven families share a common mechanism of c-di-GMP-dependent signal transduction. One of these families contains the YcgR protein from E. coli, which was recently demonstrated to bind c-di-GMP and regulate motility in vivo (Ryjenkov et al, 2006). Because of the more advanced functional characterization of this protein compared to the paralogous families, we have labeled their N-terminal domains either ‘YcgR-N' or ‘YcgR-N*' (in Figure 1A and elsewhere). The designation YcgR-N is used for domains whose homology to E. coli YcgR is strong enough to be detected in a single BLAST search, while YcgR-N* is used for domains like that in VCA0042/PlzD for which sequence profiling techniques must be used to detect this homology.
Intriguingly, the N-terminal YcgR-N* domain in VCA0042/PlzD has essentially the same fold as its C-terminal PilZ domain (see below) even though they do not share detectable sequence homology. This observation suggests that this protein and the others sharing a common domain organization with YcgR may have evolved from a primordial protein containing a tandem pair of PilZ domains, an organization still found in some proteins such as PilZ3 from Rhodobacter sphaeroides (Figure 1A).
C-di-GMP binds to VCA0042 with high affinity but unfavorable entropy
Isothermal titration calorimetry (ITC) was used to demonstrate that c-di-GMP binds tightly to VCA0042/PlzD in solution at 25°C (Figure 2 and Supplementary Table S1). The experiment shows a significant release of heat upon binding, indicating that the reaction is exothermic. As explained in the methods section of the Supplementary data (on page 9), analyses associated with the ITC experiment suggest that UV-absorbance measurements overestimate c-di-GMP stock concentration by a factor of ∼2, creating minor uncertainty in net binding affinity but more significant uncertainty in the enthalpy and entropy of the reaction. (For this reason, the c-di-GMP concentration is empirically adjusted by a factor of 1/2 in all graphs presented in this paper, while thermodynamic parameters calculated using either adjusted or nominal concentration are both presented in Supplementary Table S1.) The affinity of VCA0042/PlzD for c-di-GMP is estimated to be ∼100–300 nM, depending on the details of the fitting procedure (Supplementary Table S2). This range is consistent with the results of additional binding experiments reported below (Figure 5) as well as previously reported measurements on this protein (Pratt et al, 2007) and E. coli YcgR (Ryjenkov et al, 2006).
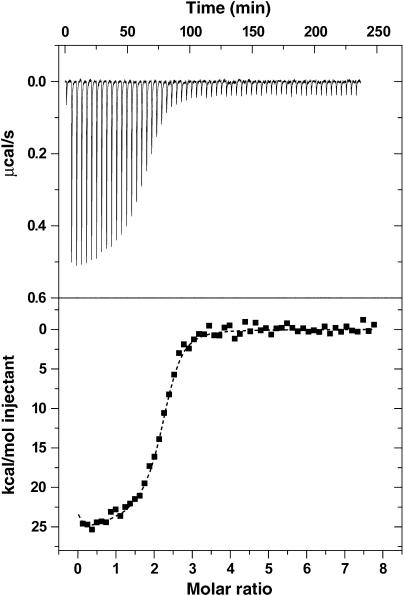
Figure 2.
Isothermal titration calorimetry shows that c-di-GMP binds to VCA0042/PlzD with sub-micromolar affinity. The top trace shows baseline-corrected data collected at 25°C in binding buffer (5 mM MgCl2, 10 mM KCl, 300 mM NaCl, 10% glycerol, 8 mM β-mercaptoethanol, and 10 mM Tris-Cl, pH 8.0), while the bottom trace shows the integrated heat released during each injection as a function of the molar ratio of c-di-GMP to VCA0042/PlzD dimer. The dotted line shows the results of curve fitting using a two-site sequential binding model with c-di-GMP concentration adjusted to give a binding stoichiometry of ∼1. (See page 9 in the Supplementary data for a detailed explanation.) Supplementary Table S1 gives the thermodynamic parameters estimated from these data using a variety of curve-fitting procedures, showing that c-di-GMP binds with affinity better than 350 nM (probably ∼100 nM) in an enthalpically favorable (ΔH<−12 kcal/mol) but entropically unfavorable (TΔS<−3 kcal/mol) reaction.
The ITC data indicate that the binding reaction is strongly exothermic, with an enthalpy estimated at −12 kcal/mol using the nominal c-di-GMP concentration or −25 kcal/mol using the adjusted concentration (Supplementary Table S2). The reaction is entropically unfavorable, with TΔS at 25°C of at least −3 kcal/mol (calculated using nominal ligand concentration) and probably closer to −16 kcal/mol (calculated using adjusted concentration). The latter number is high enough to imply either loss of conformational freedom or increased exposure of hydrophobic surfaces to solvent upon ligand binding (i.e. contributions beyond the loss of translational entropy by the ligand).
Thermal shift assays, which are faster and can be performed at a lower protein concentration, were used to further characterize the interaction of c-di-GMP with VCA0042/PlzD in solution (Supplementary Figure S2 and bottom of page 9 in the Supplementary data). The melting temperature (Tm) of the protein increases from 52°C in the absence of ligand to 64°C in the presence of saturating c-di-GMP (Supplementary Figure S2A). Measurements at progressively lower c-di-GMP concentrations suggest an affinity constant of ∼10–40 μM at the effective temperature of these experiments around 60°C (Supplementary Figure S2B). The reduction in affinity at elevated temperature confirms the inference that the binding reaction is entropically unfavorable (i.e. ΔSbinding must be negative for increasing temperature to reduce the magnitude of ΔGbinding=ΔHbinding−TΔSbinding because ΔGbinding is negative). The steep reduction in affinity at elevated temperature is consistent with a large loss in entropy on the order of that calculated from the ITC experiments using the adjusted c-di-GMP concentration.
Thermal shift experiments conducted in the presence or absence of Mg++ (5 mM MgCl2 versus 10 mM Na-EDTA and 0.1 mM MgCl2) and at high or low K+ concentrations (10 mM versus 0.22 mM) show no difference in affinity or stability of the ligand complex (data not shown). These results indicate that Mg++ is not required and that K+ may not be required for c-di-GMP binding to VCA0042/PlzD, consistent with the inability to find a well-ordered cation associated with c-di-GMP in the crystal structure reported in this paper (Table I) but contrary to results previously reported for E. coli YcgR (Ryjenkov et al, 2006).
Table 1.
Crystallographic statistics for the VCA0042/PlzD/c-di-GMP complex structurea
| Crystal parameters | |||
| Space group | P1 | ||
| Unit cell at 100 K | 47.0, 55.4, 58.0 Å | 99.9°, 97.1°, 106.8° | |
| Matthews number (Å3/Da) | 2.5 | ||
| Data quality | |||
| Resolution (Å) | 50–1.9 (2.0–1.9) | ||
| No. of measured reflections | 160 324 (15 779) | ||
| No. of unique reflections | 40 956 (4033) | ||
| Rsym (%) | 6.2 (39.1) | (I⩾–3σI for observations) | |
| Mean redundancy | 3.9 (3.9) | ||
| Completeness (%) | 97.9 (96.6) | (All measured reflections) | |
| 88.6 (65.1) | (I⩾2σI) | ||
| Mean I/σI | 19.7 (11.1) | (I⩾σI after merging) | |
| Refinement residuals (F⩾0F) | |||
| Rfree (%) | 20.9 | ||
| Rwork (%) | 19.5 | ||
| Model quality | |||
| RMSD bond lengths (Å) | 0.005 | ||
| RMSD bond angles (%) | 1.3° | ||
| Ramachandran plot (%) | 90.3% | Core | |
| 8.7 | Allowed | ||
| 1.0 | Generously allowed | ||
| Average B-factors (Å2) | |||
| All | 32.1 | ||
| Main chain | 27.6 | ||
| Side chain | 36.6 | ||
| Waters | 40.9 | ||
| RMSD non-bonded B-factors | |||
| Main chain | 2.9 | ||
| Side chain | 3.5 | ||
| Model contents | |||
| Protein residues | A23-A247, B24-B248 | ||
| Ligands | 2 c-di-GMP | ||
| Water molecules | 541 | ||
| PDB accession codeb | 2RDE | ||
| aStandard definitions were used for all parameters. Data reduction and refinement statistics come from SCALEPACK (Otwinowski and Minor, 1997) and CNS (Brünger et al, 1998), respectively. | |||
| bBoth coordinates and structure factors have been deposited in the Protein Data Bank. The numbers in parentheses represent data in the highest resolution shell. | |||
VCA0042/PlzD protein structure
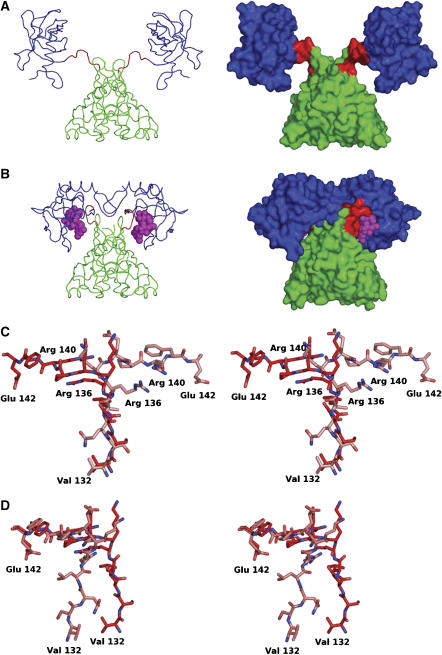
The crystal structure of the c-di-GMP complex with VCA0042/PlzD (Figure 3) was solved by molecular replacement from the apo structure (PBD ID 1YLN; R Zhang, M Zhou, S Moy, F Collart, and A Joachimiak) and refined at 1.9-Å resolution to working and free R-factors of 19.5 and 20.9%, respectively (Table I). The YcgR-N* domain in VCA0042/PlzD shares a similar six-stranded β-barrel topology with the C-terminal PilZ domain (Figures 3A and C). These domains are connected by a short hinge comprising a well-ordered seven-residue loop at the extreme N terminus of the PilZ domain sequence identified by Amikam and Galperin (red in Figures 1B, 3A and B, and elsewhere). This loop contains one of the two most strongly conserved motifs in PilZ sequences (Figure 1B) and directly contacts the c-di-GMP ligand (Figures 1B, 3B, 4B, 6A and 6B, and Supplementary Figure S4) while playing a pivotal role in the conformational change induced by its binding (Figure 4 and Supplementary Figure S4). We therefore designate this loop at residues 134–140 as the ‘c-di-GMP switch'.
Figure 3.
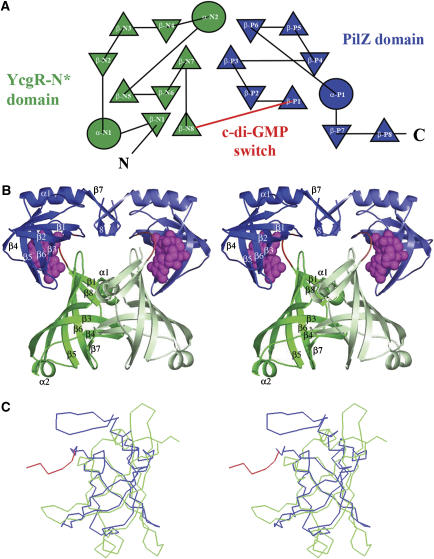
Crystal structure of the VCA0042/PlzD complex with c-di-GMP. In all panels in this paper, the YcgR-N* domain is colored green, the c-di-GMP switch red, and the remainder of the PilZ domain blue. (A) Topology diagram generated by TOPS (Michalopoulos et al, 2004), with circles and triangles respectively representing α-helices and β-strands running roughly perpendicular to the plane of the page. Connecting segments penetrating into the symbols pass above the plane while those stopping at the boundary of the symbol pass below. The β-strands make exclusively antiparallel H-bonding interactions. (B) Stereo ribbon diagram of the protein dimer with c-di-GMP shown in magenta space-filling representation. Darker or lighter colors are used to distinguish the two subunits in the dimer. Secondary structural elements are numbered separately in the two domains like in Figure 1B. (C) Cα traces showing structural superposition of the YcgR-N* domain and the PilZ domain performed using DALI (Holm and Sander, 1993).
Figure 4.
Comparison of apo and c-di-GMP complex structures of VCA0042/PlzD. (A) Cα trace (left) and molecular surface (right) of the apo structure (PBD ID 1YLN; R Zhang, M Zhou, S Moy, F Collart, and A Joachimiak) colored as in Figure 3. (B) Equivalent representations of the c-di-GMP complex structure. (C, D) The alternative conformations of the c-di-GMP switch (residues 134–140) are shown in ball-and-stick representation following least-squares alignment of the Cα atoms in either the N-terminal YcgR-N* domain (C) or the C-terminal PilZ domain (D). Carbon atoms in the apo and complex structures are colored lighter and darker shade of red, respectively. Nitrogen atoms are colored blue and oxygen atoms are colored red.
Both β-barrel domains in VCA0042/PlzD are formed by the interaction of a pair of three-stranded antiparallel β-sheets (Figures 3A and B), which are connected by a short α-helix in the N-terminal YcgR-N* domain (residues 1–133) but a loop without regular secondary structure in the C-terminal PilZ domain (residues 134–242). In the YcgR-N* domain, the β-barrel is preceded by a single β-strand and short α-helix and followed by a single β-strand that makes antiparallel hydrogen-bonding (H-bonding) interactions bridging the N-terminal β-strand and the β-barrel core. In contrast, in the PilZ domain, the β-barrel is followed by an α-helix and an antiparallel β-hairpin that does not interact with the β-strands in the β-barrel core. Structural superposition of the N- and C-terminal domains in VCA0042/PlzD using the program DALI yields a Z-score of 6.6 and a root mean square deviation of 2.7 Å for alignment of 76 Cα atoms from residues sharing only 5% sequence identity (Figure 3C).
Detailed analyses of the relationship between these domains and others of known structure are presented in the Supplementary data (on pages 2 and 3). In brief, the C-terminal PilZ domain in VCA0042/PlzD shows the strongest structural similarity to two known PilZ domain-containing proteins. One of these is the single-domain protein PA4608 (PDB ID 1YWU) (Ramelot et al, 2007), which shares 15% sequence identity with the PilZ domain in VCA0042. Yet stronger structural similarity is observed to the C-terminal domain in protein PP4397 from Pseudomonas putida KT2440 (PDB ID 2GJG; IA Wilson, the Joint Center for Structural Genomics), which shares only 11% sequence identity with the PilZ domain in VCA0042. The N-terminal domain in PP4397 also shows the strongest similarity of any known structure to the N-terminal YcgR-N* domain in VCA0042/PlzD, even though these domains again share only 11% sequence identity. Therefore, VCA0042/PlzD and PP4397 share a common domain organization with each other and with YcgR. The critical residues in the c-di-GMP switch are also conserved in sequence in PP4397 and YcgR (Figure 1B and Supplementary Figure S1), indicating that these proteins probably bind c-di-GMP in a similar manner to VCA0042/PlzD.
Hydrodynamic analyses demonstrate that VCA0042/PlzD forms a stable and monodisperse dimer either in the presence or absence of c-di-GMP (Supplementary Table S2, Figure S3, and pages 3 and 10 in the Supplementary data). The apo structure shows an extensive intersubunit dimer interface burying 1590 Å2 of solvent-accessible surface area per subunit in contacts exclusively between the N-terminal YcgR-N* domains (Figure 3A). This interface involves primarily homotypic contacts between α-helix 1, β-strands 3 and 4, and the antiparallel β-hairpin connecting β-strands 6 and 7 (Supplementary Figure S4D).
Binding of c-di-GMP causes a major change in interdomain interactions in crystal structures of VCA0042/PlzD
Comparison of the c-di-GMP-bound structure of VCA0042/PlzD to the earlier apo structure reveals that a major change in interdomain interactions occurs upon ligand binding (Figure 4 and Supplementary Figure S4). In the apo structure, the C-terminal PilZ domain is essentially detached from the YcgR-N* domain. This domain is located far from the two-fold axis in the apo structure and makes no contact either with the YcgR-N* domain in the same protomer or either domain in the other protomer. In contrast, in the c-di-GMP bound structure, the two domains in a single protomer are found in close apposition with the c-di-GMP molecule packed tightly in their mutual interface. The inferred conformational change involves a 123° rotation of the PilZ domain towards the YcgR-N* domain forming the core of the VCA0042/PlzD dimer. This change converts an extended apo structure (Figure 4A) into a much more compact ligand-bound structure in which the PilZ domains make new contacts with each other across the dimer interface (Figures 3B and 4B). These contacts almost double the solvent-accessible surface area buried in the inter-protomer interface (to 2760 Å2 per subunit) based primarily on new contacts made by the β-hairpin at the C terminus of the PilZ domain to itself and to two protein segments near the interface between the PilZ domain and the YcgR-N* domain (Figures 3B and 4B, and Supplementary Figure S4C). However, the dimer interface between the N-terminal YcgR-N* domains shows minimal perturbation upon binding c-di-GMP (Figures 4A and B and Supplementary Figure S4C), with their relative packing angle changing by only 0.6° (Supplementary Figure S5D).
A substantial portion of the c-di-GMP-binding site is formed by the seven-residue c-di-GMP switch at the N terminus of the PilZ domain (red in Figures 1B and 3, and elsewhere) that attaches it to the YcgR-N* domain. In the apo structure, this protein segment adopts an extended conformation that is well ordered in the crystal lattice (as evidenced by backbone B-factors of 22–40 Å2 compared to an overall mean of 32 Å2). However, the lack of any structural interactions between the flanking domains suggests that the c-di-GMP switch could be flexible when the apo protein is not constrained in a crystal lattice, as observed in solution NMR experiments on the PilZ domain of PA4608 in the absence of c-di-GMP (Ramelot et al, 2007). In the ligand-bound structure of VCA0042/PlzD, the c-di-GMP switch has undergone a large conformational change involving alterations in backbone dihedral angles at residues ser-133, leu-135, arg-136, and glu-138 (Supplementary Figure S4A). This backbone conformational change results in rotation of the C terminus of the switch back towards its N terminus (Figures 4C and D), thereby bringing the C-terminal PilZ domain in close proximity to the YcgR-N* domain to form the c-di-GMP-binding cavity lined by residues from both domains and the c-di-GMP switch itself (Figures 3B, 4B and 6A and B). Moreover, the c-di-GMP switch makes extensive contacts to both flanking domains as well as to the c-di-GMP ligand, indicating that its conformation is tightly constrained by local intra-protomer interactions in the ligand-bound state (Figure 4B). Therefore, this protein segment will lose conformational entropy upon ligand binding if it is flexible in the apo state. Given that no new hydrophobic surfaces are exposed to solvent in the ligand-bound structure (Figures 4A and B), the large change in entropy inferred to accompany c-di-GMP binding (Supplementary Table S1) is likely to derive from conformational restriction of the c-di-GMP switch.
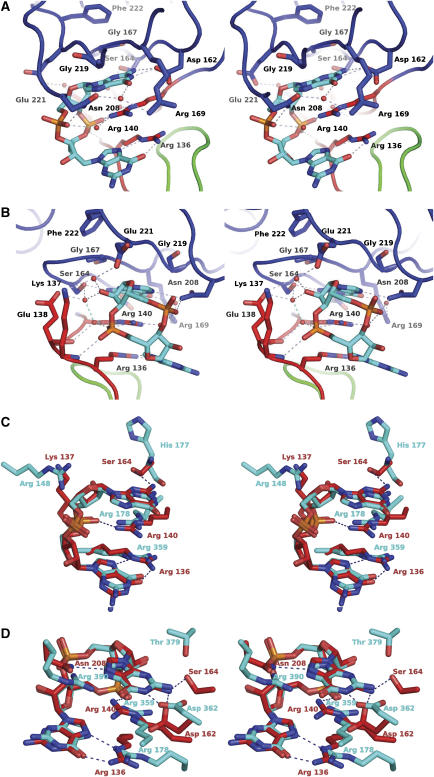
Figure 6.
Stereochemistry of the c-di-GMP-binding site in VCA0042/PlzD.The stereo pairs have nitrogen, oxygen, and phosphorus atoms colored blue, red, and orange, respectively. Dotted lines indicate H-bonds. (A, B) Two views of the c-di-GMP-binding site in the complex structure. Carbon atoms and backbone worms are colored according to domain/region of origin (green for the YcgR-N* domain, red for the c-di-GMP switch, and blue for the C-terminal PilZ domain). A subset of the ordered water molecules in the cooperative H-bonding network is shown. See also Supplementary Figure S6. (C, D) Least-squares superposition of the c-di-GMP molecule in VCA0042/PlzD with each of the two different c-di-GMP molecules bound in the allosteric regulatory site of the PleD guanylate cyclase (PDB ID 1W25) (Chan et al, 2004). Carbon atoms and residue numbers from PleD or VCA0042 are colored cyan or red, respectively. The c-di-GMP molecules from PleD shown in these two panels interact with each other to form a base-stacked dimer (illustrated in context in the PleD structure in Supplementary Figure S7). Therefore, many of the PleD side chains shown in these two panels are the same, including the two arginine residues forming the dual guanidino motif.
Outside of the c-di-GMP switch, few conformational differences are observed in comparing the individual domains in the apo and c-di-GMP complex structures of VCA0042/PlzD. The minor conformational differences that are observed are documented in detail in the Supplementary data (on pages 3 and 4).
One of these differences is a modest conformational change in the β-hairpin at the C terminus of the PilZ domain (formed by β-strands 7 and 8) that is likely to be functionally related to ligand binding. Cα atoms in this structure move by up to 3.2 Å relative to the core of the PilZ domain between the apo and c-di-GMP complex structures (Supplementary Figure S5B), enabling it to make several new contacts across the intersubunit interface (Figures 4A and B). This β-hairpin contacts itself, forming a π–π stacking interactions between the side chains of residue phe-239 in the two subunits (not shown), and also contacts β-strand 5 in the YcgR-N* domain and the junction between the c-di-GMP switch and β-strand 1 in the PilZ domain (Figure 3B and Supplementary Figures S4C and D). These additional intersubunit interactions observed in the complex structure could produce a degree of positive cooperativity in the c-di-GMP-binding reaction, as suggested by curve fitting of some of the binding isotherms reported in this paper (Figure 5, Supplementary Figure S2, and Supplementary Table S1). Furthermore, these inter-protomer structural interactions are likely to contribute to stabilizing the new relative orientation of the two PilZ domains observed in the complex structure, which could play a role in allosteric activation of downstream effectors (see Discussion).
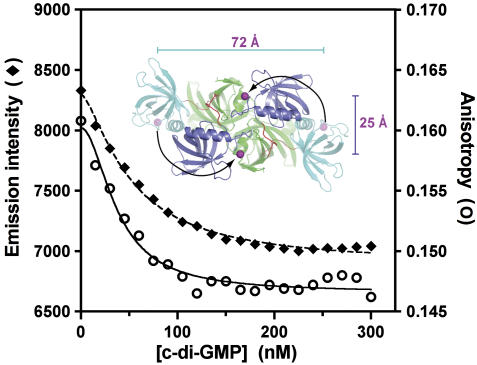
Figure 5.
Fluorescence resonance energy transfer experiments support a change in relative orientation of the C-terminal PilZ domains in VCA0042/PlzD upon c-di-GMP binding. Total fluorescence and anisotropy (measured as described on page 11 in the Supplementary data) are shown for an Alexa-Fluor 488 dye covalently bound to residue 247 in VCA0042/PlzD (in a C207A/N247C double mutant protein). The magenta spheres in the inset show the separations of the Cα atoms of residue 247 in the protein dimers observed in the apo (72 Å) and c-di-GMP-bound (25 Å) crystal structures of the wild-type protein. The experiment was conducted in binding buffer at 25°C (like the ITC experiment in Figure 2).
Fluorescence-resonance energy transfer experiments demonstrate reorientation of the PilZ domains in VCA0042/PlzD upon binding c-di-GMP in solution
Fluorescence-resonance energy transfer (FRET) experiments were designed to evaluate whether the ligand-dependent change in interdomain interactions inferred from the crystal structures also occurs in solution (Figure 5). Alexa-Fluor 488 was covalently bound at position 247 in an N247C mutant of VCA0042/PlzD that was engineered to have just this single solvent-exposed cys residue. (See pages 4, 5, and 10 in the Supplementary data for details.) Asn-247 in the wild-type protein is the final or penultimate C-terminal residue observed crystallographically. Its Cα atom is separated by ∼72 Å in the two protomers of the dimer in the conformation observed in the apo structure but only by ∼25 Å in the conformation observed in the c-di-GMP complex structure (Figure 4A, B and inset in Figure 5). Fluorophores bound near this site in both protomers forming a dimer should move substantially closer to one another if a similar change in interdomain interactions occurs upon c-di-GMP binding in solution. Such a movement would result in a reduction of fluorescence emission intensity for a self-quenching dye like Alexa-Fluor 488 (which has a Förster radius of ∼44 Å). Indeed, titration of c-di-GMP onto the labeled protein shows a progressive quenching of its fluorescence emission intensity (Figure 5), consistent with the mean separation between the dyes being reduced as the ligand binds to the protein. Curve-fitting of these data shows that the affinity of the binding interaction is consistent with that measured by ITC (Supplementary Table S1). Moreover, the quenching is accompanied by a significant reduction in fluorescence anisotropy (Figure 5), which indicates that the dye molecules reorient more rapidly in the c-di-GMP-bound conformation. This observation is most easily explained by formation of a more compact and therefore more rapidly tumbling structure upon ligand binding. Therefore, the FRET data support the occurrence of a c-di-GMP-dependent conformational change that brings the C termini of the PilZ domains into closer apposition in a more compact dimer structure, in agreement with the crystallographic observations.
Stereochemistry of the c-di-GMP-binding site
A single molecule of c-di-GMP is bound in an equivalent manner to each protomer in the VCA0042/PlzD dimer (Figures 6A and B and Supplementary Figure S6). It binds in a cis conformation with the planes of both guanine bases oriented parallel to one another. One of the two bases makes more extensive contacts than the other to a binding pocket formed primarily by the c-di-GMP switch (at residues 134–140), the antiparallel β-hairpin formed by strands β-P2/β-P3 (at residues 162–170), and strand β-P6 (at residues 219–221) in the PilZ domain. These protein segments contain all nine sequence positions that are strongly conserved in the PilZ domain (highlighted in red in the sequence alignment in Figure 1B), with four residing in the c-di-GMP switch, three in the β-hairpin formed by strands β-P2/β-P3, and two in strand β-P6.
A key c-di-GMP recognition motif involves residues arg-136 and arg-140 in the c-di-GMP switch (Figures 6A and B and Supplementary Figure S6), which are almost invariant in the PilZ domain superfamily (Figure 1B and Supplementary Figure S1). Their side chains cross at an oblique angle, making close van der Waals interactions between the atoms of the Cδ-Nɛ bond linking their terminal guanidino groups to the hydrocarbon moieties of the side chains. Both of these guanidino groups directly contact the c-di-GMP ligand with their planes parallel to the planes of its guanine bases. The positively charged guanidino group of arg-140 makes π–π stacking interactions with the lower face of the upper guanine base (as shown in Figures 6A and B), while simultaneously forming a direct H-bond to one of the phosphate groups on the backbone of c-di-GMP and a water-mediated H-bond to the other. Two of the nitrogen atoms in the guanidino group on the side chain of arg-136 make a pair of edge-to-edge H-bonds to the N7 and O6 atoms of the other guanine base, as frequently observed in DNA-binding proteins (Seeman et al, 1976; Ades, 1995). In addition, the backbone nitrogen of the next residue (lys-137) makes an H-bond to the same phosphate group salt-bridging to arg-140. Therefore, this phosphate group is clamped between the guanidino group of one arginine and the backbone amide group on the C-terminal side of the other arginine. In summary, the c-di-GMP switch has intimate structural complementarity to a significant portion of the molecular surface of c-di-GMP, with the most important side-chain interactions coming from its two highly conserved arginine residues that form a dual guanidino motif in the ligand-bound conformation (Figure 6A).
Additional contacts to the c-di-GMP come from several regions of VCA0042/PlzD (Figures 6A and B and Supplementary Figure S6). After the c-di-GMP switch, the most strongly conserved region in the PilZ domain sequence alignment (Figures 1B and Supplementary Figure S1) is located at residues 162–170 in the β-hairpin formed by β-strands 2 and 3 in VCA0042/PlzD (Figure 3B). This protein segment forms most of a binding pocket with intimate complementarity to the upper surface of the upper guanine base (as shown in Figures 6A and B). The side chains of invariant residues asp-162 and ser-164 and partially conserved residue arg-169 H-bond to the Watson–Crick base-pairing atoms of the guanine, while the backbone atoms of residues 167 and 168 in this protein segment and residues 219 and 220 in β-strand 6 make van der Waals contacts to its planar surface (Figure 6A and Supplementary Figure S6). The Cα atoms of the glycine residues at positions 167 and 219 both make direct contacts to the surface of the base, and introduction of a Cβ atom at either position would create a steric clash. This interaction explains the very strong sequence conservation at both of these sites in proteins with equivalent domain organization (Figure 1B). An additional H-bond to the upper guanine base is made by the side-chain amide of asn-208, which also H-bonds to one of the backbone phosphate groups (Figure 6B and Supplementary Figure S6). This residue is not conserved in either the narrow (Figure 1B) or broad (Supplementary Figure S1) PilZ domain alignments. However, the level of sequence conservation in this region is so weak that inaccuracies in these alignments are possible at this site.
Van der Waals contacts to the planar surface of the lower guanine base come from residues leu-135 in the c-di-GMP switch and ile-96 in β-strand 5 in the YcgR-N* domain. These contacts along with the intersubunit contacts between the C-terminal β-hairpins in the PilZ domain stabilize the new orientation of the PilZ domain relative to the YcgR-N* domain observed in the c-di-GMP complex. Finally, the phosphoribosyl backbone of c-di-GMP participates in a cooperative H-bonding network, including side-chain and backbone atoms from more than a dozen protein residues that interact with an extensive set of ordered water molecules (partially shown in Figures 6A and B and Supplementary Figure S6). This network is described in more detail in the Supplementary data (on pages 5 and 6).
The observed binding interactions of VCA0042/PlzD with c-di-GMP are consistent with existing results on PilZ domains. Mutation of the RxxxR and D/NxSxxG sequence motifs conserved in PilZ domains (Figure 1B) abrogate low micromolar to nanomolar affinity binding of c-di-GMP to E. coli YcgR (Ryjenkov et al, 2006), C. crescentus DgrA (Christen et al, 2007), or P. aeruginosa Alg44 (Merighi et al, 2007). NMR studies of P. aeruginosa protein PA4608 show that the binding of c-di-GMP causes significant chemical shift perturbations in residues clustered near the equivalent region and suggest that the c-di-GMP switch undergoes a disorder-to-order transition upon ligand binding to this PilZ domain protein (Christen et al, 2007). Finally, alanine substitutions at residues arg-136, arg-140, asp-162, ser-164, or gly-169 in VCA0042/PlzD have been demonstrated to block c-di-GMP binding to purified native VCA0042/PlzD transferred to a nitrocellulose membrane (Pratt et al, 2007).
An equivalent dual guanidino motif is observed in an unrelated protein binding c-di-GMP
The only protein structure previously deposited in the PDB containing c-di-GMP has two arginine residues that form a dual guanidino motif with close stereochemical similarity to that formed by arg-136 and arg-140 from the c-di-GMP switch in VCA0042/PlzD (Figures 6C and D and Supplementary Figure S7). The catalytic domain of the PleD diguanylate cyclase (PDB ID 1W25) has an allosteric binding site sensing c-di-GMP concentration in addition to the catalytic site mediating its synthesis (Chan et al, 2004). The allosteric site binds two molecules of c-di-GMP forming a base-stacked dimer of c-di-GMP (as shown in Supplementary Figure S7) that has been observed in crystals of c-di-GMP itself and may be present in solution (Egli et al, 1990; Liaw et al, 1990). Although PleD has no sequence similarity or structural homology to the PilZ domain, it contains a pair of arginine residues (arg-178 and arg-359) in its allosteric binding site that form a dual guanidino motif with equivalent stereochemistry to that present in VCA0042/PlzD. This motif simultaneously makes similar structural interactions with both c-di-GMP molecules bound to PleD (Figures 6C and D and Supplementary Figure S7). In both cases, one guanidino group stacks with one of the guanine bases and also salt-bridges to one of the backbone phosphate groups, while the other guanidino group makes a pair of edge-on H-bonds to the other guanine base. For one of the c-di-GMP molecules, the interaction geometry is substantially different from that observed in VCA0042/PlzD even though it preserves these chemical interactions (Figure 6D and Supplementary Figures S6 and S7). However, for the other c-di-GMP molecule, the interaction geometry is essentially the same as that observed in VCA0042/PlzD (Figure 6C), including formation of a phosphate clamp by one of the guanidino groups together with the nitrogen atom from the backbone amide group on the C-terminal side of the other (not shown).
Discussion
Evolutionary selection of PilZ domains as c-di-GMP sensors
The fact that the c-di-GMP binding pocket is formed primarily by two local protein segments (Figures 1B, 3B, and 6A, B and Supplementary Figure S6) explains the low level of overall sequence conservation observed in the PilZ domain superfamily (Figure 1B). Outside of these sites, the sequence is constrained only by the relatively loose requirement of maintaining compatibility with the overall domain fold. Broader sequence conservation covering more extensive regions of the surface is likely to be observed in individual functional families to maintain the structure of specific inter-protein interaction sites involved in individual signal-transduction systems. Given the diverse physiological functions that are controlled by c-di-GMP in eubacteria, the protein architecture used to sense this ligand must be readily adapted to function in different biochemical pathways. The small number of residues required to preserve high-affinity binding of c-di-GMP combined with the very compact size (seven residues) and N-terminal location of the c-di-GMP switch provide great evolutionary flexibility in PilZ domain architecture. This flexibility allows facile functional diversification and is likely to explain the evolutionary success of the PilZ domain as a c-di-GMP signal transducer.
Thermodynamic analyses presented above suggest that the c-di-GMP switch is disordered in VCA0042/PlzD in the absence of ligand, as previously observed in NMR studies of the PA4608 PilZ domain (Ramelot et al, 2007). The necessity of ordering this segment during c-di-GMP binding creates an entropic penalty (negative ΔSbinding) that leads to a steep reduction in binding affinity as temperature is increased (Supplementary Table S1). Sequence variations that reduce the flexibility of the c-di-GMP switch in the apo state would reduce the steepness of this temperature dependence by reducing the net entropy loss incurred upon ligand binding. Tuning the net entropy loss upon ordering the c-di-GMP switch could also be used to adjust ligand-binding affinity at any single temperature and thereby contribute to the wide range of c-di-GMP-binding affinities reported for different PilZ domains (Ryjenkov et al, 2006; Christen et al, 2007; Merighi et al, 2007). While fine-tuning protein–ligand interactions in the bound state could also contribute to these observed differences, the disorder–order transition in the c-di-GMP switch provides a facile mechanism for thermodynamic optimization of c-di-GMP signal transduction processes in response to physiological needs, and it therefore represents an additional evolutionary advantage of PilZ domain architecture.
Implications of the VCA0042/PlzD complex with c-di-GMP for the mechanism of signal transduction
The conformational change observed in the c-di-GMP switch furthermore suggests a simple mechanism for signal transduction by VCA0042/PlzD and other PilZ domain-containing proteins. The c-di-GMP-bound conformation of this small protein segment could interact directly with downstream effector proteins to allosterically activate them (i.e. by stabilizing them in an activated conformational state). Given the significant surface exposure of the c-di-GMP ligand in the complex (Figure 3B), the ligand also could participate itself in the relevant intermolecular interactions. The binding of c-di-GMP to the PilZ domain creates a unique molecular surface spanning the ligand, the c-di-GMP switch, and additional protein segments on the adjacent surface of the PilZ domain. Even in the absence of any change in interdomain or intersubunit interactions, the extent of this surface within an isolated PilZ domain is sufficiently large to mediate specific high-affinity regulatory interactions with target proteins. Therefore, the structure of the VCA0042 complex with c-di-GMP provides a clear paradigm for the signal transduction mechanism likely to be used by proteins containing just a single PilZ domain in isolation from any other domain.
The large changes in interdomain and intersubunit interactions that occur upon c-di-GMP binding to VCA0042/PlzD could also be exploited for allosteric regulation of effector proteins. These changes bring the YcgR-N* domain and C-terminal PilZ domain into close apposition with c-di-GMP bound at their mutual interface (Figures 4A, B). An extended region of this new surface covering both domains as well as the ligand itself could interact directly with a target protein to allosterically modulate its activity, making the regulatory process dependent on the specific interdomain interaction geometry stabilized by c-di-GMP binding. The binding of c-di-GMP also induces a dramatic change in the relative orientation of the two PilZ domains in the VCA0042/PlzD dimer. A target protein could bind simultaneously to both PilZ domains specifically in this geometry stabilized by binding c-di-GMP. In this manner, changes in intersubunit interactions induced by ligand binding could also be exploited for regulatory effect, which could be particularly useful for control of target proteins that are themselves oligomeric.
While conservation of surface-exposed residues can provide information regarding the location of likely physiological interaction sites, this approach is hampered in the current system by the low phylogenetic diversity of the sequences within any single group of orthologous proteins sharing a common domain organization with VCA0042/PlzD or YcgR (Figure 1B). Meaningful analyses of conservation require merging sequences from all eight different ortholog groups sharing this domain organization, which is likely to obscure sites that determine the binding specificity within individual groups (i.e. residues that make different interactions in proteins functioning in different physiological systems). Nonetheless, such an analysis of 59 proteins from these eight groups (Supplementary Figure S8 and Table S3) shows enhanced conservation of the protein surfaces immediately flanking the c-di-GMP-binding site as well as in a deep groove at the dimer interface formed largely by the PilZ domains (and therefore absent in apo VCA0042/PlzD). These sites are compatible with the different allosteric activation mechanisms proposed above and provide candidates for evaluation in specific signal transduction systems.
Structural studies of effector complexes will be required to determine which of the potential activation mechanisms and intermolecular interaction interfaces are exploited in different functional systems. Nonetheless, the diversity of available options greatly expands the evolutionary potential of the PilZ domain. All of these mechanisms are ultimately driven by the same conformational change confined to the seven-residue N-terminal c-di-GMP switch, establishing it as a remarkably compact and versatile c-di-GMP-dependent molecular switch.
Materials and methods
X-ray structure determination
Diffraction data were collected on a Quantum-4 CCD detector (ADSC, San Diego, CA) in a single sweep of 360 × 1° oscillations from a crystal maintained at ∼100 K using 0.9793 Å radiation on beamline X4A at the Brookhaven National Synchrotron Light Source. Scaling B-factors of the final frames within 3 Å2 of the initial frames indicated minimal decay. Data were processed using DENZO and SCALEPACK (Otwinowski and Minor, 1997). The crystal structure of the apo protein (PDB ID 1YLN) was used for molecular replacement after division into two models representing either the N-terminal (23–136) or C-terminal (137–248) domains. A standard molecular replacement search was carried out using COMO (Jogl et al, 2001) with an N-terminal domain dimer, and the resulting solution was fixed prior to running a second molecular replacement calculation to find the positions of two individual C-terminal domains. Refinement involved iterations of manual model-building in COOT (Emsley and Cowtan, 2004) followed by computational refinement in CNS (Brünger et al, 1998) using standard stereochemical restraints (Engh and Huber, 1991) in conjunction with a randomly selected Rfree set comprising 10% of the reflections. B-factors were subject to vicinal restraints (1.5–2.0 and 2.0–2.5 Å2 for main-chain and side-chain atoms, respectively). Strong CNS restraints (1050 kJ/Å2, σB=1.5) initially applied throughout the model were ultimately released completely to produce a considerable improvement in Rfree. Water molecules automatically identified using CNS were checked for consistency with 2Fo−Fc maps and H-bonding criteria.
ITC
Measurements were carried out on a VP ITC (Microcal, Northampton, MA) with protein protomer at 6.0 μM in the cell and c-di-GMP at a nominal concentration of 208 μM in the syringe. An initial 1 μl injection was followed by 59 injections of 5 μl each at 240-s intervals. Heat of dilution for c-di-GMP was computed from the last 10 injections and subtracted from raw data before fitting the binding isotherm in ORIGIN 5.0 (Microcal). Details on quantitation of protein and ligand concentrations and the influence of these parameters on ITC data analysis are given in the Supplementary data (on pages 8, 7, and 9, respectively).
Supplementary Material
Supplementary Information
Acknowledgments
We thank S Ashok and V Lee for advice, G Ahlsén and JM Crawford for technical assistance, and R Abramowitz, J Schwanoff, W Hendrickson, and the New York Structural Biology Center for support of NSLS beamline X4A at Brookhaven National Lab. ITC data were collected in the Biophysics Resource of the WM Keck Biotechnology Research Laboratory at Yale. This work was supported by a grant to the Northeast Structural Genomics Consortium from the US National Institutes of Health under the Protein Structure Initiative. SSS was supported in part by a Summer Undergraduate Research Fellowship from Columbia University.
References
- Ades SE (1995) The engrailed homeodomain: determinants of DNA-binding affinity and specificity. PhD Thesis. Department of Biology, Massachusetts Institute of Technology, Cambridge, MA
- Alm RA, Bodero AJ, Free PD, Mattick JS (1996) Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol 178: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikam D, Galperin MY (2006) PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22: 3–6 [DOI] [PubMed] [Google Scholar]
- Ausmees N, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Lindberg M (2001) Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol Lett 204: 163–167 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T (2004) Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA 101: 17084–17089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Allan MG, Folcher M, Jeno P, Grzesiek S, Jenal U (2007) DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci USA 104: 4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Folcher M, Schauerte A, Jenal U (2005) Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280: 30829–30837 [DOI] [PubMed] [Google Scholar]
- Cotter PA, Stibitz S (2007) c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol 10: 17–23 [DOI] [PubMed] [Google Scholar]
- D'Argenio DA, Miller SI (2004) Cyclic di-GMP as a bacterial second messenger. Microbiology 150: 2497–2502 [DOI] [PubMed] [Google Scholar]
- Egli M, Gessner RV, Williams LD, Quigley GJ, van der Marel GA, van Boom JH, Rich A, Frederick CA (1990) Atomic-resolution structure of the cellulose synthase regulator cyclic diguanylic acid. Proc Natl Acad Sci USA 87: 3235–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Engh R, Huber R (1991) Accurate bond and angle parameters for X-ray structure refinement. Acta Crystallogr A 47: 392–400 [Google Scholar]
- Galperin MY (2004) Bacterial signal transduction network in a genomic perspective. Environ Microbiol 6: 552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY (2005) A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol 5: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Nikolskaya AN, Koonin EV (2001) Novel domains of the prokaryotic two-component signal transduction systems. [erratum appears in FEMS Microbiol Lett 2001 204: 213–214]. FEMS Microbiol Lett 203: 11–21 [DOI] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Huang B, Whitchurch CB, Mattick JS (2003) FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J Bacteriol 185: 7068–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS, Drasar BS, Sack RB (1993) The aquatic environment as a reservoir of Vibrio cholerae: a review. J Diarrhoeal Dis Res 11: 197–206 [PubMed] [Google Scholar]
- Jenal U (2004) Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr Opin Microbiol 7: 185–191 [DOI] [PubMed] [Google Scholar]
- Jenal U, Malone J (2006) Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40: 385–407 [DOI] [PubMed] [Google Scholar]
- Jogl G, Tao X, Xu Y, Tong L (2001) COMO: a program for combined molecular replacement. Acta Crystallogr D 57: 1127–1134 [DOI] [PubMed] [Google Scholar]
- Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD (2004) HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol 54: 75–88 [DOI] [PubMed] [Google Scholar]
- Liaw YC, Gao YG, Robinson H, Sheldrick GM, Sliedregt LA, van der Marel GA, van Boom JH, Wang AH (1990) Cyclic diguanylic acid behaves as a host molecule for planar intercalators. FEBS Lett 264: 223–227 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA et al. (2005) CDD: a conserved domain database for protein classification. Nucleic Acids Res 33: D192–D196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS (2002) Type IV pili and twitching motility. Annu Rev Microbiol 56: 289–314 [DOI] [PubMed] [Google Scholar]
- Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S (2007) The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol 65: 876–895 [DOI] [PubMed] [Google Scholar]
- Michalopoulos I, Torrance GM, Gilbert DR, Westhead DR (2004) TOPS: an enhanced database of protein structural topology. Nucleic Acids Res 32: 251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray data diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U (2004) Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18: 715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt JT, Tamayo R, Tischler AD, Camilli A (2007) PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem 282: 12860–12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelot TA, Yee A, Cort JR, Semesi A, Arrowsmith CH, Kennedy MA (2007) NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins 66: 266–271 [DOI] [PubMed] [Google Scholar]
- Romling U, Amikam D (2006) Cyclic di-GMP as a second messenger. Curr Opin Microbiol 9: 218–228 [DOI] [PubMed] [Google Scholar]
- Romling U, Gomelsky M, Galperin MY (2005) C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57: 629–639 [DOI] [PubMed] [Google Scholar]
- Ross P, Aloni Y, Weinhouse C, Michaeli D, Weinberger-Ohana P, Meyer R, Benziman M (1985) An unusual guanyl oligonucleotide regulates cellulose synthesis in Acetobacter xylinum. FEBS Lett 186: 191–196 [DOI] [PubMed] [Google Scholar]
- Ryjenkov DA, Simm R, Romling U, Gomelsky M (2006) The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281: 30310–30314 [DOI] [PubMed] [Google Scholar]
- Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M (2005) Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 187: 1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AJ, Ryjenkov DA, Gomelsky M (2005) The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187: 4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman NC, Rosenberg JM, Rich A (1976) Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci USA 73: 804–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U (2004) GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53: 1123–1134 [DOI] [PubMed] [Google Scholar]
- Tamayo R, Tischler AD, Camilli A (2005) The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J Biol Chem 280: 33324–33330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Andersson CR, Canales SR, Golden SS (2004) PsfR, a factor that stimulates psbAI expression in the cyanobacterium Synechococcus elongatus PCC 7942. Microbiol 150: 1031–1040 [DOI] [PubMed] [Google Scholar]
- Tischler AD, Camilli A (2005) Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect Immun 73: 5873–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information