Abstract
The Tetrahymena thermophila ribosomal DNA (rDNA) replicon contains dispersed cis-acting replication determinants, including reiterated type I elements that associate with sequence-specific, single-stranded binding factors, TIF1 through TIF4. Here, we show that TIF4, previously implicated in cell cycle-controlled DNA replication and rDNA gene amplification, is the T. thermophila origin recognition complex (TtORC). We further demonstrate that TtORC contains an integral RNA subunit that participates in rDNA origin recognition. Remarkably, this RNA, designated 26T, spans the terminal 282 nts of 26S ribosomal RNA. 26T RNA exhibits extensive complementarity to the type I element T-rich strand and binds the rDNA origin in vivo. Mutations that disrupt predicted interactions between 26T RNA and its complementary rDNA target change the in vitro binding specificity of ORC and diminish in vivo rDNA origin utilization. These findings reveal a role for ribosomal RNA in chromosome biology and define a new mechanism for targeting ORC to replication initiation sites.
Keywords: DNA replication, gene amplification, origin recognition complex, ribonucleoprotein complex, ribosomal RNA
Introduction
The initiation of eukaryotic DNA replication is regulated by a conserved heterohexameric origin recognition complex (ORC) (Bell and Stillman, 1992) and additional factors that associate with ORC or bind directly to DNA. Although ORC is conserved in eukaryotes (Gavin et al, 1995), the architecture of replicons and mechanism for recruiting ORC to initiation sites are not. In Saccharomyces cerevisiae, ORC and non-ORC DNA-binding proteins associate in a sequence-specific manner with distinct non-overlapping cis-acting determinants (Marahrens and Stillman, 1992; Lee and Bell, 1997). In stark contrast, Schizosaccharomyces pombe ORC exhibits no apparent specificity in vitro, binding stochastically to long degenerate AT-rich tracts (Kim and Huberman, 1998; Kong and DePamphilis, 2001). Accordingly, virtually any intergenic DNA segment can support autonomous episomal replication in this species (Segurado et al, 2003; Dai et al, 2005), and no functional role has been described for non-ORC DNA-binding proteins.
Although metazoan ORCs also lack sequence specificity in vitro (Vashee et al, 2003; Remus et al, 2004), their replicons are comprised of discrete, dispersed cis-acting determinants (Aladjem and Fanning, 2004; Minami et al, 2006; Gray et al, 2007). In Drosophila melanogaster and the rat, ORC can be tethered to origins by associating with other sequence-specific DNA-binding proteins (Beall et al, 2002; Minami et al, 2006). It remains to be determined whether this is a general targeting mechanism for ORC.
Tetrahymena thermophila is an attractive model system for studying the regulation of eukaryotic DNA replication. The 21-kb ribosomal DNA (rDNA) minichromosome initially forms during development when a copy of the transcriptionally silent germline micronucleus differentiates into a polyploid, transcribed macronucleus in newly formed progeny. At this time, the single integrated rDNA copy is released from its parental chromosome, rearranged into a large inverted repeat and amplified to ∼9000 copies. Macronuclear rDNA is replicated once on average per cell cycle during subsequent vegetative divisions. DNA replication initiates from nucleosome-free regions in the 1.9 kb 5′ non-transcribed spacer (5′ NTS) (Figure 1, Ori) (Zhang et al, 1997). Dispersed type I elements are required for amplification and vegetative replication, and regulate replication fork progression at adjacent pause site elements (PSEs) (reviewed in Tower, 2004). Promoter-proximal type I elements regulate rRNA transcription as well. Separation-of-function alleles disrupt replication or transcription, but not both, suggesting that different trans-acting factors compete for binding to shared cis-acting determinants.
Figure 1.
Schematic of the T. thermophila rDNA minichromosome. ‘Palindromic' rDNA encodes the 17S, 5.8S and 26S rRNAs. Domains 1 and 2 (D1, D2) are an imperfect duplication and contain coordinately regulated origins (Ori). Reiterated type I elements (IA–ID), pause site elements (PSE1–3) and type III elements are shown. RFB, developmentally regulated replication fork barrier; ovals, positioned nucleosomes. DNA segments subjected to PCR amplification in Figures 3 and 6 are shown (D1 Ori, D2 Ori, promoter, 26S rRNA gene middle and 3′ end fragments).
In vitro studies identified four sequence-specific type I element binding factors, TIF1 through TIF4, that are differentially regulated during the cell cycle and development (Mohammad et al, 2000, 2003). An unusual property of these factors is that they bind exclusively to single-stranded DNA. Tif1p binds either the A- or T-rich type I element strand in vitro (Saha and Kapler, 2000). In vivo footprinting studies with wild-type and TIF1-depleted strains demonstrated that Tif1p preferentially binds to the A-rich strand at the rDNA origin and T-rich strand at the rRNA promoter (Saha et al, 2001). In addition to the implications for targeting ORC and transcription machinery to different sites in the rDNA, these studies revealed that the rDNA origin and promoter regions exist in an unwound or single-stranded state in native chromosomes. Remarkably, TIF1p regulates rDNA replication timing by repressing origin firing early in S phase (Morrison et al, 2005).
The ∼550 kD TIF4 complex, which exhibits biochemical similarities to S. cerevisiae ORC, binds in an ATP-dependent, sequence-specific manner to the type I element T-rich strand. Its Orc2p crossreactive subunit, Tt-p69, localizes to the macronucleus during vegetative S phase. Tt-p69 and hence the TIF4 complex, have also been linked to DNA replication in the developing macronucleus, including programmed gene amplification (Mohammad et al, 2003).
We report here that TIF4 is Tetrahymena ORC and document an unprecedented feature of the complex––the presence of an integral RNA subunit. We provide evidence that this RNA base pairs with complementary sequences at the rDNA origin. Mutations that disrupt the base-pairing potential change the in vitro specificity of ORC for DNA and diminish rDNA origin utilization. Most unexpectedly, the ORC RNA subunit corresponds to the terminal fragment of 26S ribosomal RNA. Potential roles for this novel rRNA species in chromosome biology are discussed.
Results
TIF4 is Tetrahymena ORC
The presence of an Orc2p crossreactive subunit in TIF4 suggested that it might be Tetrahymena ORC. BLAST analysis of the T. thermophila genome database (TGD; http://www.tigr.org/tdb) revealed homology to Orc1p through Orc5p. The predicted Orc1 protein (TGD gene prediction: TTHERM_00865050) exhibits a high degree of similarity to human Orc1p (e value: 1.4e−42, 24% sequence identity, 49% similarity). Multiple conserved blocks were detected in the carboxyl-terminal domain, including sequences for ATP binding and hydrolysis (Supplementary Figure S1A; Walker A and B boxes 1 and 3), as well as Orc1p-specific motifs (boxes 2, 4, 5 and 6) (Gavin et al, 1995). More extensive sequence similarity was observed in the distantly related ciliate, Paramecium tetraurelia (Supplementary Figure S1B; e value: 3.2 × 10e−52, 33.2% identity, 51.4% similarity).
To assess whether this Tetrahymena gene is an ORC1 ortholog, we generated strains in which the coding region was either deleted (TD101) or fused to a tandem affinity peptide (TAP) epitope tag (Table I, TD102). The ORC1 gene was depleted by replacing the coding region with a metallothionein promoter-driven neomycin phosphotransferase cassette (Figure 2A). The chimeric ORC1/MTT1-NEO transgene was introduced into the macronucleus of mating cells, where it underwent homologous recombination with the endogenous ‘ORC1' target. Continuous selection for the transgene in the amitotic polyploid macronucleus yielded phenotypic assortants with an ∼80% reduction in wild-type gene dosage (Figure 2A, strain TD101) and ∼60% reduction in steady-state mRNA (Figure 2B). Since complete replacement of the wild-type gene was not achieved, we predict that this gene is essential.
Table 1.
T. thermophila strains used in this study
| Strain | Micronuclear genotype | Macronuclear phenotype |
|---|---|---|
| CU427 | chx1-1/chx1-1 | cycl-s, pm-s |
| CU428 | mpr1-1/mpr1-1 | 6mp-s, pm-s |
| CU522 | btu1-1/btu1-1 | pt-hs |
| CU727 | btu1-1/btu1-1 | pt-hs |
| TD101 (Mic) | CHX1/chx1-1; MPR1/mpr1-1 | pm-r, cycl-r, 6mp-r |
| ORC1/ORC1∷MTT-neo | ORC1 knockdown | |
| TD102 (Mic) | CHX1/chx1-1; MPR1/mpr1-1 | pm-r, cycl-r, 6mp-r |
| ORC1/TAP-tagged ORC1 | Expresses TAP-tagged ORC1 | |
| MTT1/MTT1neo | ||
| TX615 (Mic) | CHX1/chx1-1; MPR1/mpr1-1 | cycl-r, 6mp-r, pm-r |
| MTT1/MTT1neo | ||
| MM201 (Anl) | btu1-1/btu1-1 | pt-r. Expresses 26T-Ext RNA |
| MM202 (Anl) | btu1-1/btu1-1 | pt-r. Expresses wild-type 26T-Apt RNA |
| MM203 (Anl) | btu1-1/btu1-1 | pt-r. Expresses wild-type 26T-Apt RNA |
| TD151 (Anl/Veg) | btu1-1/btu1-1 | pt-r, pm-r. Expresses 26T-Ext RNA and TAP-tagged ORC1 |
| TD152 (Anl/Veg) | btu1-1/btu1-1 | pt-r, pm-r. Expresses 26T-Apt RNA and TAP-tagged ORC1 |
| AS204 (Anl) | btu1-1/btu1-1 | pt-r. Expresses wild-type 26T-Apt RNA |
| AS205 (Anl) | btu1-1/btu1-1 | pt-r. Expresses left mutant 26T-Apt RNA |
| AS206 (Anl) | btu1-1/btu1-1 | pt-r. Expresses right mutant 26T-Apt RNA |
| AS207 (Anl) | btu1-1/btu1-1 | pt-r. Expresses middle mutant 26T-Apt RNA |
| AS208 (Anl) | btu1-1/btu1-1 | pt-r. Expresses full mutant 26T-Apt RNA |
| Anl, non-germline transformation of the newly developing macronuclear anlagen; Mic, micronuclear (germline) transformant; Veg, transformation of the vegetative macronucleus. | ||
| Expression of the chx1-1 and mpr1-1 alleles in the macronucleus confers resistance to cycloheximide (cycl-r) and 6-methylpurine (6mr), respectively. Expression of the btu1-1 allele confers hypersensitivity to paclitaxel (pt-hs), while insertions into this locus confer pt-resistance (pt-r). Expression of the MTT1neo transgene confers resistance to paromomycin (pm-r). MTT1neo was targeted to either the ORC1 locus (ORC1 disruption) or endogenous MTT1 locus (co-transformation vector), as noted in the text. | ||
Figure 2.
Functional analysis of ORC1 knockdown mutants. (A) Disruption of the ORC1 gene in the polyploid macronucleus. Top: schematic of wild-type ORC1 and the ORC1 disruption allele (H3: HinDIII sites). Bottom: Southern blot analysis (wild type: 3.8 kb; disruption: 5.6 kb). WT: strain CU428; KD: ORC1 knockdown strain, TD101. TIF1 served as a control. (B) Northern analysis of WT and ORC1 KD strains. (C) Growth curves of WT (TX615) and ORC1 knockdown (TD101) strains. TX615 encodes an MTT1-NEO transgene targeted to the MTT1 locus, and was grown in the same pm dosage as TD101 (1 mg/ml). (D) Nuclear division in control (WT, TX615) and ORC1 knockdown (KD) cells visualized with acridine orange. Left panel: the stages of cytokinesis were defined by the extent of constriction of the cytokinetic furrow. Right panel: additional examples of aberrant macronuclear divisions. (E) Flow cytometry of control (TX615) and ORC1 knockdown (TD101) strains synchronized by starvation and re-feeding.
Consistent with a role for this gene in cell cycle progression, the doubling time of the TD101 knockdown cultures increased relative to the control (Figure 2C; 4.5 versus 3.0 h). Asynchronous TD101 cultures exhibited an elevated percentage of dividing cells, most of which underwent aberrant macronuclear division in which incompletely divided macronuclei were bisected by the cytokinetic furrow (Figure 2D, left panel). Unequal distribution of DNA to daughters and the formation of extra-macronuclear DNA-staining bodies were also detected, albeit less frequently (Figure 2D, right panel), but were not observed in wild-type controls.
In addition, flow cytometry revealed a delay in S-phase progression in the ORC1 mutant (Figure 2E). Whereas the control strain exhibited an increase in DNA content ∼120 min after re-feeding of G1 synchronized (starved) cultures, a comparable increase was not detected until 60 min later in the mutant. Sequential administration of a double block to cell cycle progression–starvation and release into media containing hydroxyurea was lethal to most ORC1-depleted cells, but had no effect on wild-type controls (data not shown). We conclude that the targeted gene is required for DNA replication and macronuclear S-phase progression.
To address whether Orc1p is a component of the TIF4 rDNA origin binding complex, we introduced a TAP-tagged ORC1 allele into the diploid germline micronucleus of conjugating wild-type cells (CU427 × CU428 cross). The heterozygous strain, TD102, contained comparable levels of wild-type and tagged alleles in the polyploid macronucleus (data not shown). Western blot analysis identified a tagged protein of the predicted size in the transformant (Figure 3A, TAP). As expected for a Tetrahymena ORC subunit, antibodies directed against the TAP-tagged protein specifically enriched for rDNA origin sequences in formaldehyde crosslinked chromatin immunoprecipitates (Figure 3B).
Figure 3.
In vitro and in vivo analysis of TIF4/ORC. (A) Western blot analysis of TAP-tagged Orc1p. Strains: WT, CU428; TAP-tagged ORC1, TD102; TAP/APT (TAP-tagged ORC1 and aptamer-tagged 26T RNA), TD152. Ponceau S staining of nuclear extracts served as a loading control. (B) ChIP analysis of TAP-tagged ORC complexes. PCR of input and immunoprecipitated DNA from crosslinked chromatin preparations without [(−) Ab] or with [(+) Ab] antibody against the Orc1p TAP tag. Data from two representative independent ChIP experiments are shown. A 2.5- to 5-fold enrichment for the Domain 1 rDNA origin fragment was observed. See Figure 1 for PCR primer locations. (C) Standard EMSA analysis with wild-type nuclear extracts (strain CU428) and labeled type I element oligos (C3 rDNA type IB element T-rich strand, T51: 5′ CTCAAAAGTTGCAAAAGTTCGGAAGGTTTACTATT TTTGTTTTTTTTTTT; A-rich strand, A53: 5′ GGCAAAAAAAAAAACAAAAATAGTAAACCTTCCGA ACTTTTGCAACTTTTGAG) or T51:A53 duplex (DS). (D) Western blot EMSA of Orc1p and Orc2p. Nuclear extracts were prepared from wild-type (CU428) and TAP-tagged ORC1 (TD102) strains. DNA substrates were not labeled. Following native gel electrophoresis and transfer, membranes were probed with Orc1p or Orc2p antibodies to monitor their migration. Lanes MN, RN and DN: MNase-, RNase A- and DNase I-digested extracts.
As previously reported (Mohammad et al, 2003), TIF4 binds the type I element T-rich strand, but fails to recognize the type I element A-rich strand or T/A duplex in standard electrophoretic mobility shift analysis (EMSA) (Figure 3C, EMSA with radiolabeled DNA substrates). DNA binding is ATP-dependent, but does not require ATP hydrolysis. In contrast, TIF1p associates with either type I element strand. To examine the DNA-binding properties of Tetrahymena ORC, nuclear extracts were incubated in the presence or absence of unlabeled type I element substrates and electrophoresed under non-denaturing EMSA conditions. Western blotting was then used to visualize the migration of Orc2p (heterologous antisera) and Orc1p (TAP tag probe) under standard EMSA conditions.
In the absence of added DNA, a single band was detected with the respective Orc1p and Orc2p antibodies. Both proteins co-migrated, suggesting that they reside in the same complex (Figure 3D, Orc2p: left panel, lane 1; Orc1p: right panel, lane 2). Their mobilities were retarded to the same degree in the presence of ATP and unlabeled type I element T-rich strand. No change in mobility was observed with the A-rich strand or type I element duplex, analogous to TIF4 (Figure 3D). These data strongly suggest that Orc1p and Orc2p/Tt-p69 are components of the TIF4 complex. Additional biochemical and in vivo functional studies described below reinforce this conclusion. Hereafter, we refer to TIF4 as TtORC or Tetrahymena ORC.
Tetrahymena ORC is a ribonucleoprotein complex
TIF1, TIF2 and TIF3 exhibit a somewhat relaxed DNA specificity, binding to more than one DNA sequence in vitro and in vivo (Mohammad et al, 2000; Saha et al, 2001). The specificity of Tetrahymena ORC for just the type I element T-rich strand suggested that it may utilize a different mechanism for DNA recognition. We reasoned that TtORC might contain an integral RNA subunit that pairs with its single-stranded target, analogous to large sequence-specific nucleic acid-binding complexes, such as telomerase and components of the RNA splicing machinery (Greider and Blackburn, 1989; Staley and Guthrie, 1998).
Consistent with this prediction, formation of the TtORC gel shift complex was ablated when nuclear extracts were treated with micrococcal nuclease (MNase) and the enzyme was subsequently inactivated prior to incubation with substrate DNA (Figure 4A). In contrast, TIF1-binding activity was unaffected. To assess whether the nucleic acid component was RNA, oligonucleotide-affinity purified ORC preparations and S100 extracts were treated with RNase A prior to DNA binding. Whereas an ATP-dependent ORC gel shift complex was observed in mock-treated samples, RNase A digestion completely eliminated DNA binding, while TIF1p was unaffected (Figure 4B).
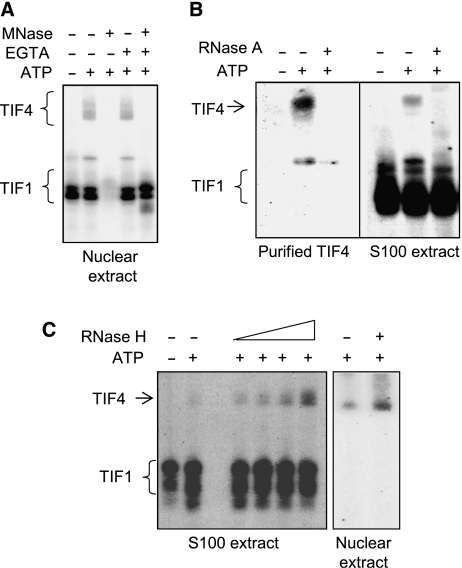
Figure 4.
Tetrahymena ORC is a ribonucleoprotein complex. (A) MNase-sensitive DNA binding. Nuclear extracts were sequentially incubated with MNase and EGTA prior to EMSA with the type I element T strand substrate, C3-ssT51. Unbound DNA was run off the gel to better resolve TIF4/ORC–DNA complexes. In the absence of EGTA, gel shift complexes were not observed due to degradation of substrate DNA. (B) RNase A-sensitive binding to the rDNA type IB element T-rich strand. Oligo affinity-purified ORC or S100 extracts were treated with RNase A prior to EMSA analysis. (C) Enhanced binding to the type I element T-rich strand following RNase H treatment. Gel shift complexes were formed on ice and digested with RNase H (S100 extract: 1–5 U; nuclear extract: 5 U) prior to EMSA.
Western blotting was similarly used to monitor the migration of nuclease-treated ORC complexes under native EMSA gel conditions. The mobility of Orc1p and Orc2p increased following MNase and RNase A treatment, but was unaltered by DNase I (Figure 3D). Orc1p and Orc2p co-migrated under all conditions, suggesting that they remain associated after the RNA is destroyed. The addition of the type I element T-rich strand had no effect on the migration of RNase A-digested ORC complexes (data not shown), consistent with the observed loss of DNA-binding activity in TIF4 gel shift complexes (Figure 4A).
To test whether the ORC-associated RNA forms Watson–Crick base pairs with its cognate DNA target, we treated ORC–DNA complexes with ribonuclease H (RNase H), which requires 4–6 consecutive RNA–DNA base pairs for substrate cleavage. RNase H altered the abundance of the ORC gel shift complex in a dose-dependent manner (Figure 4C, left panel). Unexpectedly, the gel shift signal was reproducibly enhanced rather than diminished by this treatment. While the basis for enhanced DNA binding is not clear, these experiments indicate that Tetrahymena ORC is an ribonucleoprotein (RNP) complex and that RNA–DNA base pairing contributes to sequence-specific rDNA recognition. In vivo experiments with mutant RNAs reinforced this conclusion (see below).
To facilitate cloning of the ORC RNA subunit, we mapped the RNA interaction domain in the type IB element T-rich strand by testing oligonucleotide derivatives for their ability to generate an RNase H-sensitive gel shift. Short type I element substrates (ssT33, ssT37) and longer derivatives that contained sequences upstream of the type IB element (up-ssT51, upNS-ssT51) failed to generate an ATP-dependent ORC gel shift in the absence or presence of RNase H (Supplementary Figure S2; ssT37 and data not shown). In contrast, longer substrates carrying specific or nonspecific sequences downstream of the type IB element formed gel shift complexes (Supplementary Figure S2). The strongest signal was obtained with the C3 rDNA substrate, ssC3-T51, which contains an additional 42 bp downstream of the type IB element that provide C3 rDNA with a replication advantage in heterozygous C3/B rDNA strains (Larson et al, 1986). RNase H digestion enhanced DNA binding to C3 and B rDNA substrates. Since enhancement was also observed with a substrate that contained random sequence downstream of the type IB element (ss-T54), we reasoned that RNA–DNA interactions are restricted to the conserved type I element core.
The ORC RNA subunit is derived from 26S ribosomal RNA
To clone the RNA responsible, ORC complexes were first enriched by sequential purification on conventional and oligonucleotide-affinity resins, yielding a final product that contained ∼10–20 prominent protein bands as visualized by silver staining (Mohammad et al, 2003). RNA was subsequently isolated and reverse transcribed with an oligo-dT primer, which was predicted to pair with an A-rich RNA tract, complementary to 11 consecutive thymidines in the type I element T-rich strand. Ligation-mediated RT–PCR generated a single PCR product that was cloned and sequenced. All of the cDNAs contained a common core sequence, but exhibited minor 5′ heterogeneity (Figure 5A). Omission of the RT step eliminated PCR product formation, ruling out the possibility that the clones were derived from contaminating DNA (data not shown). While we favor the idea that variation in the position of the 5′ end is a cloning artifact, the possibility of natural 5′ heterogeneity cannot be ruled out.
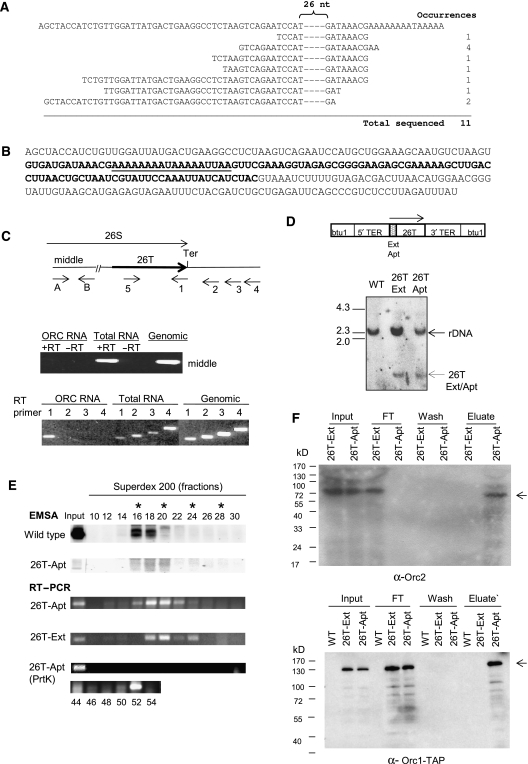
Figure 5.
Characterization of the ORC-associated RNA. (A) Sequence of partial cDNAs derived from oligonucleotide affinity-purified ORC. Top line: DNA sequence of a 26S RNA-coding region segment (sense strand). Remaining sequences: 5′ end of sequenced cDNAs. The dashed region contains 26 nt of complete sequence identity. (B) Complete sequence of 26T RNA starting at the 5′ end of the longest cloned cDNA. The boldfaced segment spans the 26S rRNA D12 expansion region. Underlined residues correspond to the binding site for oligo-dT-directed reverse transcription during cDNA cloning. (C) Characterization of 26T RNA. Upper panel: schematic of the 26S rRNA-coding region and primers used for RT–PCR analysis (Ter: 3′ end of mature 26S rRNA). Middle panel: RT–PCR test for contaminating full-length 26S rRNA. Primers A and B amplify the middle segment of 26S rRNA. Substrates: affinity-purified ORC RNA, total cellular RNA and genomic DNA. The 5′ residue of forward (A) and reverse (B) primers map to nt 5369 and 5624 (EMBL accession number M11155). Lower panel: 3′ end mapping of 26T RNA. RT was performed with primers 1, 2, 3 or 4 (5′ nt positions: 8472, 8534, 8608 and 8756, respectively). cDNAs were amplified following addition of primer 5 (5′ nt position: 8343). Substrates were as noted above. (D) Southern blot analysis of 26T-Ext and 26T-Apt transformants. Upper panel: schematic of transformation constructs. TER: telomerase RNA gene 5′ and 3′ flanking segments. Lower panel: Southern blot analysis of EcoRI-digested DNA from wild-type (CU428), 26T-Ext (MM201) and 26T-Apt (MM202) transformant strains (probe: 26T DNA fragment). (E) Incorporation of transgenic 26T RNAs into an RNP complex. Nuclear extracts from wild-type (CU428), 26T-Ext (MM201) and 26T-Apt (MM202) strains were fractionated on a 24 ml Superdex 200 fast protein liquid chromatography column (GE Healthcare). Fractions (0.3 ml) were assayed for ORC binding to the type I element T-rich strand (ssT51) and 26T RNA. Migration of molecular weight markers are depicted with asterisks (left to right: 665, 430, 230 and 153 kD). The migration of free 26T-Apt RNA was determined by pretreating extracts with proteinase K (RT–PCR, lower panel). (F) Orc1p and Orc2p associate with aptamer-tagged 26T RNA. Nuclear extracts from wild-type (CU428), 26T-Ext (MM201) and 26T-Apt (MM202) strains were incubated with streptavidin (SA) sepharose. Orc2p western blot analysis was performed on bound and unbound fractions. FT: flow through/unbound fraction. For Orc1p analysis, TD151 and TD152 were used. Both strains produce TAP-tagged Orc1p. TD152 (T) encodes 26T-Ext RNA, while TD151 (A) encodes SA-binding 26T-Apt RNA. Arrows: full-length Orc1p and Orc2p. Prior to pull-down analysis, nuclear extracts from the different strains were normalized for protein concentration. Here, 10% of each sample (input, flow through, wash and eluate) was subjected to western blot analysis.
BLAST analysis produced an unexpected finding: the cDNA sequences were identical to the 3′ end of 26S ribosomal RNA. To assure that the starting RNA preparation was not contaminated with intact or partially degraded 26S rRNA, RT–PCR was performed with primers from the middle of 26S rRNA (Figure 5C, primers A and B). These reactions failed to generate a product with affinity-purified ORC RNA, but produced a strong signal with total cellular RNA or genomic DNA (Figure 5C, middle panel).
The extreme 3′ end of novel 26S rRNA subspecies, hereafter designated as 26T (26-terminus), was mapped with RT primers spaced ∼70 nt apart and spanning the 3′ end of mature 26S rRNA (Figure 5C, primers 1–4). Only primer 1, complementary to 3′ end of 26S rRNA, produced an RT–PCR product with RNA derived from oligo affinity-purified ORC (Figure 5C, lower panel). Although this result does not preclude the possibility of 3′ end heterogeneity, it demonstrates that the 3′ end of 26T RNA does not extend beyond the terminus of mature 26S rRNA. The predicted sequence of 26T RNA(s), spanning the 5′ end of sequenced cDNAs and 3′ end of RT–PCR products is shown in Figure 5B. Underlined bases are complementary to type I element T-rich tract, and boldfaced residues delineate the D12 expansion region, which is absent in all prokaryotic 23S rRNAs and most eukaryotic large rRNA subunits (Engberg et al, 1990).
To verify that 26T RNA is a bona fide component of TtORC and not a contaminating 26S rRNA breakdown product, Tetrahymena strains were transformed with 26T RNA derivatives under the control of the telomerase RNA promoter (Figure 5D, schematic) (Engberg et al, 1990). The first construct, 26T-Ext, encoded the wild-type 26T RNA and several extra nucleotides to distinguish it from endogenous 26T and 26S rRNA. The second construct, 26T-Apt(Wt), contained a 44 nt 5′ extension encoding an S1 aptamer RNA sequence that binds with high affinity to streptavidin (SA) (Srisawat and Engelke, 2002). Both constructs were targeted to the paclitaxel (PT)-hypersensitive β-tubulin locus, btu1-1, in the developing macronucleus of progeny derived from a cross between strains CU522 and CU727 (Table I) (Gaertig et al, 1994). PT was used to select for transformants and produce phenotypic assortants with high levels of the btu1-1∷26T gene replacement in the progeny macronucleus. Southern blotting confirmed the presence of 26T-Ext and 26T-Apt(Wt) transgenes in strains MM201 and MM202, respectively (Table I; Figure 5D). RT–PCR verified the production of stable transgenic 26T RNA (Figure 5E, input).
Assembly of transgenic 26T RNA into an RNP complex was assessed by fractionating nuclear extracts on Superdex 200. The peak fractions for 26T-Ext and 26T-Apt(Wt) RNAs had an apparent molecular weight of 500–600 kD (Figure 5E, RT–PCR), coinciding with ORC DNA-binding activity (Figure 5E, EMSA). No free RNA was detected (data not shown). Proteinase K treatment released 26T RNA into low molecular weight fractions, indicating that the tagged RNA resides in an RNP complex.
The association of transgenic 26T RNA with ORC proteins was confirmed using an SA pull-down assay. Heterologous antiserum was used to monitor Orc2p in strain MM202 (Table I). For the analysis of Orc1p, the TAP-tagged ORC1 gene was introduced into the vegetative macronucleus of strains MM201 and MM202, expressing TER1 promoter-driven 26T-Ext and 26T-Apt RNA, respectively (Table I). Vegetative transformants were screened for TAP-tagged Orc1p, and strains with the highest level of tagged Orc1p (TD151 and TD152) were examined further. Nuclear extracts were reacted with SA sepharose and assayed for retention of Orc2p or TAP-tagged Orc1p. Orc1p and Orc2p were exclusively in the flow through (FT) fraction in 26T-Ext strains (MM202 and TD151), which lack the S1 aptamer tag. In contrast, Orc1p and Orc2p bound to SA sepharose in 26T-Apt RNA strains (TD152 and MM202, respectively) (Figure 5F), indicating that transgenic 26T RNA assembles into ORC complexes.
In vivo targeting of Tetrahymena ORC to the rDNA origin
The RNase H sensitivity of TtORC gel shift complexes suggested that 26T RNA forms Watson–Crick base pairs with the type I element T-rich strand. Sequence alignment revealed that 25 out of 30 nts in the type IB element T-rich strand are capable of base pairing with a contiguous internal 26T RNA segment (Figure 6A). The putative interaction domain in the RNA includes the largest predicted single-stranded loop in the D12 expansion region (Supplementary Figure S3) (Engberg et al, 1990).
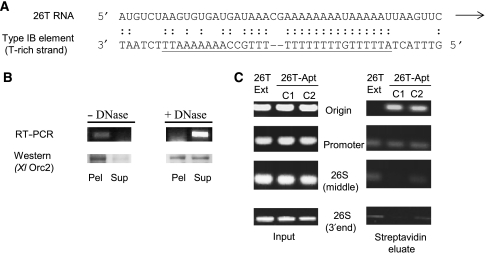
Figure 6.
Sequence alignment and in vitro targeting of 26T RNA to the rDNA origin. (A) Alignment of 26T RNA and the type I element T-rich strand. Underlined DNA residues correspond to sequences present in all four type I elements (IA–ID). (B) Chromatin association of 26T RNA and Orc2p. RT–PCR (26T-Apt RNA) and western blot analysis of mock and DNase I-treated nuclei following centrifugation to separate insoluble (pellet) and soluble (supernatant) fractions (strain MM202). (C) Representative streptavidin chromatin pull-down experiment. Formaldehyde crosslinked chromatin from 26T-Ext (MM201) and 26T-Apt transformants (MM202: C1; MM203: C2) was subjected to PCR amplification following purification on SA sepharose beads. The 5′ end position of forward (F) and reverse complementary (R) PCR primers are designated in their name: origin (F744, R1075), promoter (F1580, R1887), 26S middle (F5369, R5624), 26S 3′ end (F8343, R8472) (see Figure 1 schematic). A minimum three-fold enrichment of the rDNA origin region was observed in four independent experiments with 26T-Apt transformant strains.
Since ORC localizes to replication initiation site in S. cerevisiae ARS1, human lamin B and rat aldolase replicons (Bielinsky and Gerbi, 1998; Abdurashidova et al, 2003; Minami et al, 2006), we predicted that Tetrahymena ORC should be targeted to the rDNA origin and not to other sites complementary to 26T RNA (i.e. the rRNA promoter and 26S RNA 3′ end). To address this issue, we utilized the S1 RNA aptamer tag to study the in vivo association of ORC with rDNA chromatin. We first tested whether transgenic 26T RNA and Orc2p associate with bulk chromatin by partitioning whole-cell nuclear lysates into soluble and pellet (nuclear scaffold/chromatin) fractions. 26T RNA and Orc2p were exclusively in the pellet in untreated samples (Figure 6B, 26T-Apt(Wt); 26T-Ext, data not shown). Consistent with previous analyses of Orc2p/Tt-p69 and histone H3 (Mohammad et al, 2003), ∼50% of Orc2 was rendered soluble by DNase I. 26T RNA was similarly released from nuclei by DNase I digestion (Figure 6B) or high salt extraction (0.75 M NaCl; data not shown). Thus, 26T RNA associates with chromatin in vivo.
We next asked whether 26T RNA was selectively targeted to the rDNA origin. SA sepharose chromatin pull-down assays were performed on formaldehyde crosslinked chromatin. Since formaldehyde crosslinks proteins to DNA or RNA, but does not crosslink nucleic acids to one another, the ability to pull down a given DNA fragment is indicative of the in vivo association of that DNA segment with ORC. PCR amplification with primers spanning the Domain 2 replication origin produced a clear signal in the SA chromatin pull-down fraction of two independent transformants expressing aptamer-tagged RNA, 26T-Apt(Wt) (MM202 and MM203), but not in the untagged 26T-Ext RNA strain (MM201) (Figure 6C, origin; see Figure 1A for primer locations). Repeat experiments on independently crosslinked chromatin preparations produced a 3- to 5-fold or greater enrichment in the 26T-Apt pull-down fraction relative to 26T-Ext controls. No enrichment was observed for a coding region segment that lacked complementarity to 26T RNA (Figure 6C, 26S middle). Although both the origin and promoter regions contain type I elements, the promoter fragment was not enriched in the 26T-Apt(Wt) pellet (Figure 6C). The same result was obtained for the 26S rRNA gene 3′ end, which is fully complementary to 26T RNA. Thus, the ORC RNP complex selectively associates with the rDNA origin in vivo.
The in vitro binding specificity of TtORC is altered in 26T RNA mutants
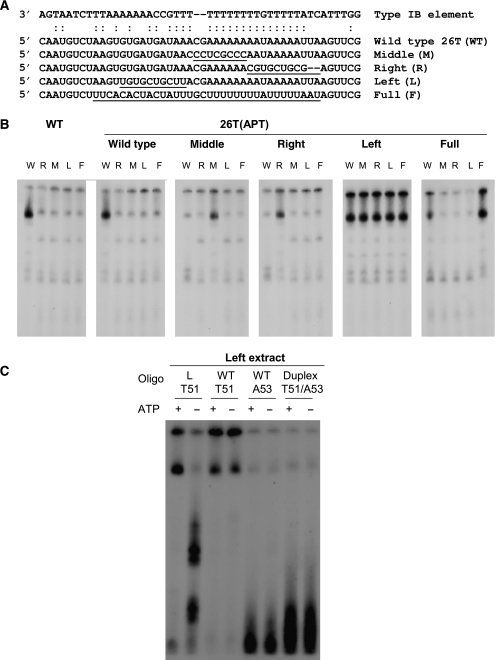
If base pairing between 26T RNA and complementary DNA sequences facilitates rDNA origin recognition, then mutant RNAs that disrupt this interaction should change the in vitro and in vivo properties of TtORC. These predictions were tested by independently expressing four mutant RNAs in Tetrahymena (Figure 7A; 26T right (R), left (L), middle (M) and full (F)). Transformants were tested for ORC binding to wild-type (W) DNA in vitro, and to substrates (R, L, M and F) that restore complementarity to each of the mutant RNAs. Wild-type extracts bound strongly to the wild-type substrate, but weakly recognized all four mutant DNAs (Figure 7B). In contrast, all four mutant ORC complexes bound strongly to the respective mutant DNA substrate that restored RNA–DNA base-pairing potential (Figure 7B). Three mutants ORCs (26T-Apt(R), Apt(M) and Apt(F)) weakly recognized mismatched substrates, analogous to wild-type ORC (Table I, strains AS206, AS207 and AS208). The fourth mutant, 26T-Apt(L) (AS205), bound strongly to all type I element T-strand derivatives. Further analysis of the 26T-Apt(L) mutant revealed a fundamental difference in the recognition of complementary and non-complementary DNA substrates. Binding of 26T-Apt(L) ORC to a substrate that restored complementary to the RNA required ATP, analogous to wild-type ORC–DNA interactions. However, association of the mutant ORC with the wild-type DNA substrate was now ATP-independent (Figure 7C). Although more extensive studies are needed to understand the basis for relaxed binding specificity, the analysis of 26T mutants establishes two important points. First, it confirms that 26T RNA is a component of Tetrahymena ORC. Second, it demonstrates that RNA–DNA base-pairing contribute to sequence-specific DNA recognition.
Figure 7.
Gel shift analysis of 26T RNA base-pairing mutants. (A) Sequence of wild-type (WT) and 26T RNA derivatives (M, R, L and F) carrying mutations in the predicted type I element T-strand base-pairing region. (B) EMSA analysis of nuclear extracts from wild-type and 26T RNA mutants with the wild-type (W) substrate, C3ssT51, and derivatives (M, R, L and F) that re-establish complementary with the respective mutant RNA. (C) Further characterization of the 26T-Apt(L) mutant.
Mutations that alter in vitro DNA recognition perturb rDNA origin function
Similar to depleting ORC1 (Table I, strain TD101), replacement of wild-type 26T RNA with mutant RNA derivatives produced a slow growth phenotype (Table I, strains AS205–208). Continuous PT selection was required to maintain mutant transgenes at high levels, and complete replacement of intact copies of the btu1-1 gene with 26T RNA mutant derivatives was not achieved (data not shown). Accordingly, two populations of ORC complexes could be discerned as few as 15–20 fissions after removal of PT selection in the 26T-R, 26T-M and 26T-F mutants—one that bound to wild-type DNA and another that recognized the complementary mutant DNA substrate (data not shown).
The in vivo effect of 26T RNA mutations on rDNA origin recognition was examined by two-dimensional (2D) gel electrophoresis of replication intermediates (RIs). HinDIII digestion produces a discontinuous RI pattern with an rDNA 5′ NTS probe (Figure 8A, schematic), corresponding to the conversion of bubble-shaped intermediates (ascending RI pattern: initiation within the probed fragment) to Y-shaped intermediates (descending RI pattern), as one of the diverging forks proceeds past the cut site (MacAlpine et al, 1997). No passive replication (complete Y arc) is detected in the 5′ NTS of wild-type C3 and B rDNA strains (Zhang et al, 1997).
Figure 8.
Analysis of rDNA replication intermediates in 26T RNA base-pairing mutants. (A) Schematic of the palindromic rDNA 5′ NTS fragment generated by HinDIII digestion, and possible RI patterns following 2D gel electrophoresis. Simple Y arc: passive replication of 5′ NTS origins; bubble or bubble-to-Y arcs: initiation within the 5′ NTS; composite (simple Y arc and bubbles): active and passive replication of the 5′ NTS. (B) Southern blot analysis of HinDIII-digested DNA resolved on a neutral–neutral 2D gel. Thick right-pointing arrows point to bubble arcs. Thin left-pointing arrows point to simple Y arcs. (C) 2D gel analysis following enrichment for RIs on BND cellulose. (D) Southern blot analysis of total genomic DNA from 26T RNA transformants.
Total genomic DNA from the wild-type aptamer RNA-expressing strain, MM202 (26T-Apt(Wt)), generated a discontinuous RI pattern, indicating that all detectable initiation events occurred within the 5′ NTS (Figure 8B, WT). In contrast, the 26T-right mutant, AS206, exhibited two patterns: a bubble-to-Y arc (right-pointing arrow) and a complete Y arc (left-pointing arrow) (Figure 8B). The complete Y arc indicates that a subset of initiation events occurred distal to the Domains 1 and 2 origins (i.e. proximal to the promoter or within the coding region or 3′ NTS). Passive replication of 5′ NTS origins was more evident when replicating molecules were enriched on benzoylated napthoylated DEAE (BND) cellulose (Figure 8C, complete Y arc: left-pointing arrow).
A more extreme situation was observed in the 26T-left mutant, which exhibited the most relaxed in vitro binding specificity (Figure 7). No bubble arc was detected. Instead, the 5′ NTS origins were passively replicated. In addition, the RI signal in 26T-left and 26T-right mutants was diminished relative to the wild-type control (Figure 8B, compare RI signal intensities relative to the non-replicating (1N) DNA spot). We conclude that these 26T RNA mutations diminish the efficiency of rDNA origin utilization and choice of initiation sites. Despite their effect on origin recognition and utilization, the mutant RNAs did not noticeably alter rDNA abundance (Figure 8D). This observation was not altogether unexpected, as a poorly understood copy-number control mechanism helps maintain rDNA genic balance in the amitotic vegetative macronucleus (Pan and Blackburn, 1995).
Discussion
The specification of eukaryotic replication origins has been the subject of considerable debate. While, ORC is conserved in eukaryotes, the mechanism for recruiting ORC to specific sites in chromosomes is not. Two targeting strategies have been previously documented: sequence-specific DNA binding, and the tethering of ORC to chromosomes by non-ORC DNA-binding proteins (Lee and Bell, 1997; Beall et al, 2002; Minami et al, 2006). Here, we provide a third targeting mechanism: Watson–Crick base pairing between an integral ORC RNA subunit and a single-stranded DNA target.
Like yeast and metazoa, Tetrahymena ORC is a multisubunit complex (Mohammad et al, 2003). Three integral components have been identified thus far: Orc1p, Orc2p and 26T RNA. Bioinformatic analyses provide preliminary evidence for additional conserved proteins (T. thermophila versus human BLAST e values––Orc1p: 10−42; Orc2p: 10−14; Orc3p: 10−2; Orc4p: 10−14; Orc5p: 10−11; GMK, unpublished results). The features of the novel ORC RNA subunit suggest that it serves an important regulatory function (see below).
Previous studies revealed that the Tetrahymena rDNA replicon is dynamically regulated. First, replication origins that mediate gene amplification are subsequently reprogrammed to initiate once per cell cycle in vegetative cells (Zhang et al, 1997). The ability to escape re-replication control during development sets the rDNA replicon apart from the rest of the genome. Second, the ORC-binding sequence––the type I element––is targeted by non-ORC proteins that regulate DNA replication and transcription (Mohammad et al, 2000). For example, TIF1p and ORC bind opposite strands at the rDNA origin (Saha et al, 2001; this work). While ORC is required to initiate DNA replication, TIF1p regulates replication timing by repressing origin activation early in S phase (Morrison et al, 2005).
The most unanticipated feature of Tetrahymena ORC is the presence of an integral RNA subunit. Since all ORC complexes appear to contain RNA and bind the rDNA type I element T-rich strand, it seems likely that they all contain 26T RNA. The ability to ectopically express tagged wild-type and mutant 26T RNAs without perturbing ribosome function allowed us to explore the basis for rDNA origin recognition. Tagged 26T RNA assembles into ORC complexes that are selectively targeted to the rDNA origin (Figures 5E and F and 6C). Since the non-coding strand at the promoter and 26S coding region are also complementary to 26T RNA, selective targeting to the origins cannot be dictated solely by RNA–DNA interactions. Site discrimination may be mediated by protein–DNA contacts or the in vivo association of non-ORC DNA-binding proteins. Local DNA features (single- versus double-stranded character) and chromatin structure may be contributing factors.
While the relative contribution of ORC RNA and protein subunits to origin recognition requires further experimentation, the work described here clearly demonstrates a role for 26T RNA. RNA mutations that disrupt base pairing to wild-type DNA produce ORC complexes that fail to bind the wild-type target sequence in vitro. However, these mutant RNPs strongly associate with substrates that encode compensatory base changes that re-establish Watson–Crick pairing (Figure 7). In vivo analysis of rDNA RIs corroborate the in vitro binding studies. 5′ NTS origins are passively replicated in 26T RNA mutants (Figure 8). Thus, the specificity of TtORC for rDNA origin sequences and site-specific initiation from the rDNA origin are dictated in part by the sequence of 26T RNA.
The DNA-binding site in 26T RNA maps to the 102 nt D12 expansion region (Supplementary Figure S3). Expansion regions are absent in prokaryotic rRNAs and variable in appearance in eukaryotic 26S rRNA counterparts. While expansion sequences typically evolve more rapidly than core, conserved rRNA segments (Michot and Bachellerie, 1987; Dube et al, 1998), the D12 regions of T. thermophila and T. pyriformis do not follow this pattern (D12: 2.9% difference; flanking ∼180 nt: 2.2% difference) (Engberg et al, 1990). Conservation extends to the more distantly related ciliate, Glaucoma chattoni (D12: 7.7% difference; remainder of 26T RNA region: 8.3% difference).
Several observations favor the idea that 26T RNA is processed from an rRNA precursor. The DNA segment upstream of the 26T interval lacks sequence motifs expected for transcription from an internal RNA polymerase I, II or III promoter (Gallagher and Blackburn, 1998; Hargrove et al, 1999). The primary sequence and RNA secondary structure of this region are conserved, suggesting that it is constrained by its role in the ribosome. The 3′ end of 26T RNA appears to be coincident with that of 26S rRNA. These findings argue that 26T RNA is transcribed from the rRNA promoter and subsequently processed from the 35S precursor or mature 26S rRNA.
In unpublished work, we characterized two non-rDNA replicons and determined that they are comprised of modular cis-acting replication determinants (S Datta and GMK, unpublished results). Type I elements are noticeably absent from these autonomously replicating segments. The most parsimonious model predicts that ORC does not utilize RNA–DNA base pairing to associate with non-rDNA origins. If this is correct, then 26T RNA could serve a specialized regulatory role. Two non-mutually exclusive models are considered. In the first model, the different modes of DNA recognition (rDNA origin: RNA+protein; non-rDNA origins: protein only) would establish two classes of replicons, forming a basis for differential regulation. RNA-mediated recruitment could assure that the rDNA origin was efficiently utilized, and facilitate re-replication during vegetative cell cycles to maintain the rDNA at a high copy number. This distinction could be further exploited to selectively amplify rDNA minichromosomes during development.
A second possibility is that the integral ORC RNA couples DNA replication to cell physiology. Accordingly, the production of 26T RNA and hence the formation of ORC holo-complexes would be regulated by the metabolic status of the cell (i.e. low levels of 26T RNA in nutrient-poor media and high levels in nutrient-rich media). While such a means for coupling translation and DNA replication is unprecedented, prior studies in cultured human cells and yeast revealed a connection between ribosome biogenesis and DNA replication. Nucleolar proteins that serve obligate roles in pre-rRNA processing (S. cerevisiae Yph3/Nop7p and human NOC3p) have been linked to replication through their physical association with ORC (Du and Stillman, 2002; Zhang et al, 2002). The presence of rRNA in Tetrahymena ORC may similarly integrate DNA replication and translation. Minimally, our findings demonstrate a new role for ribosomal RNA in chromosome biology.
In a recent study, Krude and colleagues determined that human Y RNAs are required for chromosomal DNA replication in cell-free extracts (Christov et al, 2007). Similar to 26T RNA, Y RNAs assemble in RNP complexes. In contrast to TtORC, disrupting the interaction between hY1 RNA with ro60 does not affect DNA replication. For this and other reasons, we suggest that human Y and Tetrahymena 26T RNAs serve different purposes. These recent discoveries in Tetrahymena and humans expand the functional role of non-coding RNAs to include eukaryotic DNA replication.
Materials and methods
DNA transformation and strain propagation
Standard methods were used for mating, transformation and propagation of Tetrahymena strains (Cassidy-Hanley et al, 1997). RT–PCR was used to deduce the mRNA sequence of the candidate ORC1 gene as previously described (Morrison et al, 2005). The ORC1 knockdown strain, TD101, was generated by replacing the coding region with an MTT1-neo cassette, using flanking ORC1 sequences for targeted homologous recombination. Biolistic micronuclear DNA transformation was used to produce ORC1/ORC1∷MTT1-neo F1 progeny. Phenotypic assortants were selected for increased pm resistance (100–1000 μg/ml), resulting from replacement of endogenous ORC1 with the disrupted transgene in the polyploid macronucleus (reviewed in Turkewitz et al, 2002). The amino terminal TAP-tagged ORC1 strain, TD102 (Puig et al, 2001), was generated by micronuclear co-transformation with an MTT1-neo-MTT1 transgene. pm-resistant transformants were screened for tagged ORC1 by Southern and western blotting (Yakisich et al, 2006). Ectopic expression of 26T RNA was achieved by transcribing the terminal 282 nt of 26S rRNA from the telomerase RNA promoter (Hargrove et al, 1999). This transgene was targeted to the PT-hypersensitive btu1-1 locus in progeny in a CU522 × CU725 cross (Table I) (Gaertig et al, 1994). Transformants were selected for resistance to 30 μM PT (Sigma Chemical, St Louis, MO) and cultured continuously in PT to select for btu1-1∷26T assortants. 26T RNA derivatives were generated by inverse PCR.
Molecular biology and cytology techniques
Standard molecular techniques, including DNA and RNA isolation, PCR, RT–PCR and western blotting were performed, as described (Mohammad et al, 2000; Saha et al, 2001; Yakisich et al, 2006). RNA was prepared using an RNAeasy mini-kit (Qiagen Inc., Chatworth, CA). Western blot analysis of Orc2p was performed with rabbit anti-Xenopus laevis Orc2p serum (Mohammad et al, 2003). For TAP-tagged Orc1p, polyclonal antisera recognizing the IgG epitope was coupled to horseradish peroxidase (Sigma Chemical; product P1291), hence no secondary antibody was needed for chemiluminescent detection (Millipore Corporation, Billerica, MA). Flow cytometry and acridine orange DNA staining of living cells were performed, as described (Morrison et al, 2005).
Extracts, protein purification and in vitro DNA-binding studies
S100 extracts, nuclear extracts and oligo affinity-purified ORC were prepared and subjected to EMSA, as described (Mohammad et al, 2003). TIF4–ORC complexes were sequentially purified on conventional (SP- and Q-Sepharose; Amersham/Pharmacia Biosciences, Piscataway, NJ), and oligo affinity resins (type I element A-strand (minus ATP, FT fraction), type I element T-strand (minus ATP, FT fraction) and type I element T-strand (plus ATP; bound fraction)), as described (Mohammad et al, 2003). Silver staining revealed ∼10–20 prominent proteins in the final preparations. Where noted, nuclear extracts were fractionated on a Sephadex 200 FPLC column (Amersham/Pharmacia Biosciences), calibrated with thyroglobulin, α-ferritin, catalase, lactate dehydrogenase and bovine serum albumin.
For MNase and ribonuclease A (RNase A) (Sigma Chemical) studies, protein preps were treated with nuclease for 15 min at 37°C according to the manufacturer's recommendations, prior to adding labeled or unlabeled DNA (1 U MNase or 12–25 ng RNase A). MNase was inactivated by chelating the cofactor calcium with 1 mM EGTA. DNase I digestions were performed with 5 U of enzyme for 10 min at 37°C (Amersham/Pharmacia Biosciences). For RNase H studies, pre-formed gel shift complexes were incubated with 1–5 U of RNase H (Invitrogen Life Technologies, Carlsbad, CA) for 20 min at 37°C prior to electrophoresis. RNase A was boiled for 20 min to eliminate residual DNase activity. For in vitro DNA binding, wild-type and mutant nuclear extracts were normalized by total protein concentration and incubated with radiolabeled DNAs with comparable specific activities.
Cloning and characterization of 26T RNA
Total RNA was extracted from oligo affinity-purified ORC complexes, using an RNeasy Protect Mini Kit (Qiagen Inc.). 26T cDNA clones were generated with a Gene Race Kit (Invitrogen Life Technologies), using oligo-dT to prime cDNA synthesis. The 3′ terminus of 26T RNA was mapped with RT primers spaced 70 nt apart and spanning the 3′ end of the 26S rRNA.
Chromatin association, chromatin immunoprecipitation and pull-down assays
Whole-cell nuclear extracts were prepared and fractionated as previously described (Mohammad et al, 2003). Chromatin association of Orc2p and 26T RNA was assessed by western blotting with rabbit-anti X. laevis Orc2 antibodies and RT–PCR (using a forward primer in the 26T RNA tag). Chromatin immunoprecipitation (ChIP) was performed on a TAP-tagged ORC1 strain according to the manufacturer's specifications (Upstate Cell Signaling Solutions Inc, Charlotteville, VA). Antisera specific for the IgG epitope tag (Sigma Chemical; product P1291) was used to immunoprecipitate Orc1p. For chromatin pull downs, 10 ml cultures were treated with 1% formaldehyde for 10 min and adjusted to 1% with sodium dodecylsulfate (SDS). Lysates were sonicated to 100–500 bp, diluted 1:10 in 16 mM Tris (pH 8.1), 167 mM NaCl, 1.2 mM EDTA, 1% Triton X-100, 0.01% SDS, pre-cleared with sepharose beads (100 μl) and pre-incubated with 15 U avidin (Sigma Chemical) for 30 min at RT to remove biotinylated proteins. The unbound fraction was incubated with 100 μl of SA sepharose (Amersham/Pharmacia Biosciences), eluted according to the manufacturer's recommendations and subjected to PCR amplification.
2D gel electrophoresis of rDNA RIs
HinDIII-digested total genomic DNA (40 μg) was resolved by neutral–neutral 2D gel electrophoresis and hybridized to an rDNA 5′ NTS probe, as described (Zhang et al, 1997). Where noted, RIs were enriched on BND cellulose (Sigma Chemical).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure Legends
Acknowledgments
We thank Pamela Sandoval for the help in constructing the ORC1 knockout plasmid and Dorothy Shippen for advice and comments on the manuscript. This work was supported by NIH grant GM53572.
References
- Abdurashidova G, Danailov MB, Ochem A, Triolo G, Djeliova V, Radulescu S, Vindigni A, Riva S, Falaschi A (2003) Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J 22: 4294–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aladjem MI, Fanning E (2004) The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep 5: 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR (2002) Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 42: 833–837 [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B (1992) ATP-dependent recognition of eukaryotic origins of DNA-replication by a multiprotein complex. Nature 357: 128–134 [DOI] [PubMed] [Google Scholar]
- Bielinsky AK, Gerbi SA (1998) Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279: 95–98 [DOI] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ (1997) Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146: 135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christov CP, Gardiner TJ, Szuts D, Krude T (2007) Functional requirement for noncoding Y RNAs form human chromosomal DNA replication. Mol Cell Biol 26: 6993–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Chuang RY, Kelly TJ (2005) DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci USA 102: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du YC, Stillman B (2002) Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 109: 835–848 [DOI] [PubMed] [Google Scholar]
- Dube P, Bacher G, Stark H, Mueller F, Zemlin F, van Heel M, Brimacombe R (1998) Correlation of the expansion segments in mammalian rRNA with the fine structure of the 80 S ribosome; a cryoelectron microscopic reconstruction of the rabbit reticulocyte ribosome at 21 angstrom resolution. J Mol Biol 279: 403–421 [DOI] [PubMed] [Google Scholar]
- Engberg J, Nielsen H, Lenaers G, Murayama O, Fujitani H, Higashinakagawa T (1990) Comparison of primary and secondary 26S rRNA structures in two Tetrahymena species: evidence for a strong evolutionary and structural constraint in expansion segments. J Mol Evol 6: 514–521 [DOI] [PubMed] [Google Scholar]
- Gaertig J, Thatcher TH, Gu L, Gorovsky MA (1994) Electroporation-mediated replacement of a positively and negatively selectable beta-tubulin gene in Tetrahymena thermophila. Proc Natl Acad Sci USA 91: 4549–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher RC, Blackburn EH (1998) A promoter mutation affecting replication of the Tetrahymena ribosomal DNA minichromosome. Mol Cell Biol 18: 3021–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin KA, Hidaka M, Stillman B (1995) Conserved initiator proteins in eukaryotes. Science 270: 1667–1671 [DOI] [PubMed] [Google Scholar]
- Gray SJ, Liu G, Altman AL, Small LE, Fanning E (2007) Discrete functional elements required for initiation activity of the Chinese hamster dihydrofolate reductase origin beta at ectopic chromosomal sites. Exp Cell Res 313: 109–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Hargrove BW, Bhattacharyya A, Domitrovich AM, Kapler GM, Kirk K, Shippen DE, Kunkel GR (1999) Identification of an essential proximal sequence element in the promoter of the telomerase RNA gene of Tetrahymena thermophila. Nucleic Acids Res 27: 4269–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Huberman JA (1998) Multiple orientation-dependent, synergistically interacting, similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol Cell Biol 18: 7294–7303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong DC, DePamphilis ML (2001) Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol Cell Biol 21: 8095–8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DD, Blackburn EH, Yaeger PC, Orias E (1986) Control of rDNA replication in Tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell 47: 229–240 [DOI] [PubMed] [Google Scholar]
- Lee D, Bell SP (1997) Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol 17: 7159–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Zhang Z, Kapler GM (1997) Type I elements mediate replication fork pausing at conserved upstream sites in the Tetrahymena thermophila ribosomal DNA minichromosome. Mol Cell Biol 17: 4517–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B (1992) A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255: 817–823 [DOI] [PubMed] [Google Scholar]
- Michot B, Bachellerie JP (1987) Comparisons of large subunit rRNAs reveal some eukaryote-specific elements of secondary structure. Biochimie 69: 11–23 [DOI] [PubMed] [Google Scholar]
- Minami H, Takahashi J, Suto A, Saitoh Y, Tsutsumi K (2006) Binding of AlF-C, an Orc1-binding transcriptional regulator, enhances replicator activity of the rat aldolase B origin. Mol Cell Biol 26: 8770–8780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad M, Saha S, Kapler GM (2000) Three different proteins recognize a multifunctional determinant that controls replication initiation, fork arrest and transcription in Tetrahymena. Nucleic Acids Res 28: 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad M, York RD, Hommel J, Kapler GM (2003) Characterization of a novel origin recognition complex-like complex: implications for DNA recognition, cell cycle control, and locus-specific gene amplification. Mol Cell Biol 23: 5005–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TL, Yakisich JS, Cassidy-Hanley D, Kapler GM (2005) TIF1 represses rDNA replication initiation, but promotes normal S phase progression and chromosome transmission in Tetrahymena. Mol Biol Cell 16: 2624–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan WJ, Blackburn EH (1995) Tandem repeats of the 5′ nontranscribed spacer of Tetrahymena rDNA function as high copy number autonomous replicons in the macronucleus but do not prevent ribosomal-rDNA gene dosage regulation. Nucleic Acids Res 23: 1561–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Remus D, Beall EL, Botchan MR (2004) DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J 23: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Kapler GM (2000) Allele-specific protein–DNA interactions between the single-stranded DNA-binding protein, ssA-TIBF, and DNA replication determinants in Tetrahymena. J Mol Biol 295: 423–439 [DOI] [PubMed] [Google Scholar]
- Saha S, Nicholson A, Kapler GM (2001) Cloning and biochemical analysis of the Tetrahymena origin binding protein TIF1—competitive DNA binding in vitro and in vivo to critical rDNA replication determinants. J Biol Chem 276: 45417–45426 [DOI] [PubMed] [Google Scholar]
- Segurado M, de Luis A, Antequera F (2003) Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep 4: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C, Engelke DR (2002) RNA affinity tags for purification of RNAs and ribonucleoprotein complexes. Methods 26: 156–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Tower J (2004) Developmental gene amplification and origin regulation. Annu Rev Genet 38: 273–304 [DOI] [PubMed] [Google Scholar]
- Turkewitz AP, Orias E, Kapler G (2002) Functional genomics: the coming of age for Tetrahymena thermophila. Trends Genet 18: 35–40 [DOI] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC (2003) Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev 17: 1894–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakisich JS, Sandoval PY, Morrison TL, Kapler GM (2006) TIF1 activates the intra-S-phase checkpoint response in the diploid micronucleus and amitotic polyploid macronucleus of Tetrahymena. Mol Biol Cell 17: 5185–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yu Z, Fu X, Liang C (2002) Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell 109: 849–860 [DOI] [PubMed] [Google Scholar]
- Zhang Z, MacAlpine DM, Kapler GM (1997) Developmental regulation of DNA replication: replication fork barriers and programmed gene amplification in Tetrahymena thermophila. Mol Cell Biol 17: 6147–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure Legends