Abstract
The signaling lipid molecule 15-deoxy-delta 12,14-prostaglandin J2 (15d-PGJ2) has multiple cellular functions, including anti-inflammatory and antineoplastic activities. Here, we report that 15d-PGJ2 blocks translation through inactivation of translational initiation factor eIF4A. Binding of 15d-PGJ2 to eIF4A blocks the interaction between eIF4A and eIF4G that is essential for translation of many mRNAs. Cysteine 264 in eIF4A is the target site of 15d-PGJ2. The antineoplastic activity of 15d-PGJ2 is likely attributed to inhibition of translation. Moreover, inhibition of translation by 15d-PGJ2 results in stress granule (SG) formation, into which TRAF2 is sequestered. The sequestration of TRAF2 contributes to the anti-inflammatory activity of 15d-PGJ2. These findings reveal a novel cross-talk between translation and inflammatory response, and offer new approaches to develop anticancer and anti-inflammatory drugs that target translation factors including eIF4A.
Keywords: eIF4A, 15d-PGJ2, inflammation, proliferation, translation
Introduction
Inflammatory response can be considered a double-edged sword. It protects the body by triggering innate and acquired immunity under stress conditions such as tissue damage and infections, but chronic inflammatory responses can result in diseases such as cardiovascular disease, diabetes, arthritis, Alzheimer's disease, pulmonary disease, and autoimmune disease (Aggarwal et al, 2006). There are sophisticated mechanisms to maintain homeostatic inflammatory responses in animals and avoid adverse effects of inflammatory response (Lawrence et al, 2002).
Amines, complements, cyclic nucleotides, adhesion molecules, cytokines, chemokines, and steroid hormones are involved in regulation of inflammatory responses (Lawrence et al, 2002). Besides these factors, lipid mediators such as prostaglandins (PGs), leukotrienes, lipoxins, and resolvins play important roles in resolution of inflammation. Of various lipid mediators, PGs are potent lipid molecules modulating immunity. The PGs are a family of biologically active molecules with diverse actions depending on the PG type and cellular target. For instance, PGE2 provokes inflammatory responses; however, cyclopentenone PGs (cyPGs) such as 15-deoxy-delta 12,14-prostaglandin J2 (15d-PGJ2) and PGA1 inhibit inflammatory responses. cyPGs contain a cyclopentenone ring structure that forms a covalent bond with a cysteine residue in a target protein through a chemically reactive α,β-unsaturated carbonyl group. Various members of the cyPG family have antineoplastic, anti-inflammatory, and antiviral activities (Straus and Glass, 2001). Recent research has indicated that cyPGs are endogenous anti-inflammatory mediators that promote the resolution of inflammation in vivo (Straus and Glass, 2001; Lawrence et al, 2002). In general, the production of pro-inflammatory PGs such as PGE2 triggers and/or maintains inflammatory responses, and then follows the production of anti-inflammatory PGs to prevent adverse effects of inflammatory responses.
15d-PGJ2 is produced in a variety of cells, including mast cells, T cells, platelets, and alveolar macrophages. Several activities of 15d-PGJ2 have been suggested. 15d-PGJ2 is an agonist of peroxisome proliferator-activated receptor-gamma (PPARγ), which is a transcriptional modulator that represses transcription of pro-inflammatory mRNAs, thereby resulting in resolution of inflammatory responses. Moreover, 15d-PGJ2 blocks pro-inflammatory NF-κB signaling cascades independently of PPARγ through direct interactions with signaling molecules (Straus et al, 2000). Other physiological activities of 15d-PGJ2, such as cytoprotection and inhibition of cell proliferation, have also been reported (Pereira et al, 2006). However, the molecular mechanisms involved in these activities remain obscure.
Translation initiation is a complex process that begins with interaction of the cap-binding protein complex eukaryotic initiation factor 4F (eIF4F) with the mRNA 5′-end cap structure. eIF4F comprises three subunits: eIF4E, a cap-binding protein; eIF4A, an RNA helicase; and eIF4G, a scaffolding protein. eIF4G bridges eIF4F with the 40S ribosomal subunit through an interaction with eIF3 that is associated with the 40S ribosomal subunit. The 40S ribosomal subunit with the associated initiation factors is thought to migrate along the 5′-non-translated region until it encounters the initiation codon AUG. The 40S ribosomal subunit stalls at the initiation codon and the 60S ribosomal subunit joins to form the 80S ribosomal complex. Following assembly of the 80S ribosome at the mRNA initiation codon, elongation of the polypeptide chain commences (Holcik and Sonenberg, 2005).
Translation initiation of most mRNAs is repressed when a cell is under stress conditions, such as heat and oxidation. Blockade of translation by stress signals results in formation of stress granules (SGs) in the cytoplasm. SGs contain most of the components of the 48S translational pre-initiation complex (the small, but not the large, ribosomal subunits, namely eIF4A, eIF3, eIF4E, eIF4G, eIF2, and eIF2B), other RNA-binding proteins such as T-cell-restricted intracellular antigens-1 (TIA-1), T-cell-restricted intracellular antigen-related protein (TIAR), and mRNAs. Unlike other RNA granules, SGs are not observed in cells growing under favorable conditions but are rapidly induced in response to environmental stresses (Anderson and Kedersha, 2006). Transient inhibition of protein synthesis, which induces SG formation, is an important protective mechanism used in cells during various stress conditions such as inflammation (Ma and Hendershot, 2003).
Recently, a new role of SGs has been uncovered. We showed that a signaling molecule TRAF2, which has a key role in tumor necrosis factor α (TNF-α; a pro-inflammatory cytokine) signal transduction, is sequestered into the SGs induced by heat treatment through an interaction with the translational factor eIF4GI (Kim et al, 2005). Owing to SG formation, not only translation but also TNF-α signal transduction processes are blocked under heat-stress conditions. This phenomenon represents a novel relationship between translation and inflammatory signaling (Kim et al, 2005; McDunn and Cobb, 2005).
Here, we present data on another cross-talk between translation and TNF-α signaling. The cyPGs 15d-PGJ2 and PGA1, which have anti-inflammatory activities, induce SG formation. However, PGE2, which has pro-inflammatory activity, does not induce SG formation. The SG formation was triggered by blockade of translational initiation by modification of the translational initiation factor eIF4A. Translational inhibition by 15d-PGJ2 is most likely related to the anti-cell-proliferation activity of 15d-PGJ2. Moreover, TRAF2 was sequestered to the SGs induced by cyPGs in a similar manner as it is to the SGs induced by heat treatment. This indicates that the anti-inflammatory activity of cyPGs is attributed in part to inhibition of translation and SG formation resulting in TRAF2 sequestration.
Results
Cyclopentenone PGs induce SG formation
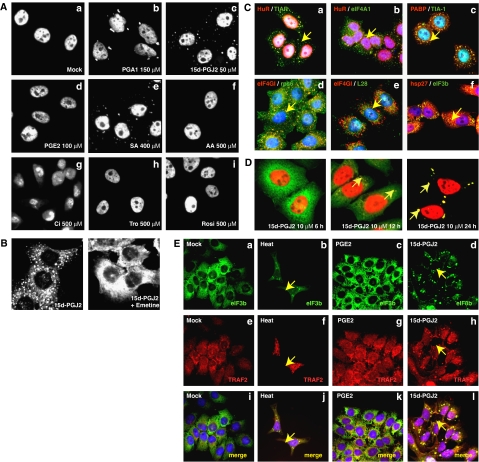
Pro-inflammatory signal transduction can be blocked by sequestration of TRAF2 into SGs (Kim et al, 2005). This indicates that SG formation is a potential regulatory mechanism of inflammatory signaling. Therefore, we tried to identify physiological compounds that induce SG formation. Of the compounds tested (Supplementary Figure S1A), the cyPGs 15d-PGJ2 and PGA1 induced SG formation, which are shown as cytoplasmic speckles in Figure 1A. Our research focused on the effect of 15d-PGJ2 because it was a stronger SG inducer than PGA1 (Figure 1A, compare panel b with c). It should be noted that the anti-inflammatory activity of 15d-PGJ2, in either a PPARγ-dependent or -independent manner, is stronger than that of PGA1 (Straus and Glass, 2001). On the other hand, arachidonic acid (Figure 1A, panel f), which is the precursor of cyPGs, and a pro-inflammatory PG PGE2 (Figure 1A, panel d) did not induce SG formation. Moreover, the PPARγ agonists ciglitazone, troglitazone, and rosiglitazone did not induce SG formation (Figure 1A, panels g–i). In addition, CAY10410, which is a derivative of 15d-PGJ2, did not induce SG formation (Supplementary Figure S1B). These data indicate that induction of SG formation is a specific property of cyPGs and is not common to all PGs, and that this phenomenon is independent of PPARγ activation (see below).
Figure 1.
cyPGs induce SG formation. (A) HeLa cells were mock-treated (Mock) or treated with PGA1, 15d-PGJ2, PGE2, SA, arachidonic acid (AA), ciglitazone (Ci), troglitazone (Tro), and rosiglitazone (Rosi) (at indicated concentrations) for 30 min. Immunocytochemical analyses were performed using TIA-1 antibody. (B) HeLa cells were pretreated for 1 h with 10 μg/ml of emetine and then treated with 15d-PGJ2 (50 μM) for 1 h. (C) HeLa cells were treated with 15d-PGJ2 (50 μM) for 1 h. Immunocytochemical analyses were performed with the indicated antibodies: HuR/TIAR (a), HuR/eIF4A1 (b), PABP/TIA-1 (c), eIF4GI/rps6 (d), eIF4GI/L28 (e), and hsp27/eIF3b (f). Nuclei are shown in blue by Hoechst staining. Arrows indicate SGs. (D) HeLa cells were treated with 15d-PGJ2 (10 μM) for the times indicated. Immunocytochemistry was performed with an eIF3b (green) and HuR (red) antibodies. Arrows indicate SGs. (E) HeLa cells were treated with heat at 44°C (b, f, and j), 50 μM of PGE2 (c, g, and k), or 15d-PGJ2 (d, h, and l) for 1 h. Immunocytochemistry was performed using eIF3b and TRAF2 antibodies. Arrows indicate SGs.
The identity of the cytoplasmic speckles was confirmed by emetine treatment. Emetine freezes ribosomes in the polysomal state and inhibits SG formation (Anderson and Kedersha, 2006). Emetine treatment inhibits 15d-PGJ2-induced SG formation (Figure 1B, right panel) in the same manner as it inhibits those induced by sodium arsenite (SA) or heat (data not shown). This clearly demonstrates that 15d-PGJ2-induced SGs share similar properties to the typical SGs induced by heat or SA.
The components of SGs induced by 15d-PGJ2 were analyzed using an immunocytochemical method (Figure 1C). As expected, known SG marker proteins (TIA-1 and TIAR), an RNA-binding protein (HuR), translational initiation factors (eIF4GI, eIF3b, and poly(A)-binding protein (PABP)), and the 40S ribosomal subunit (as indicated by the rps6 ribosomal protein) were observed in the SGs (Figure 1C). In contrast, the 60S ribosomal subunit, as indicated by the ribosomal protein L28, was not localized to the SGs induced by 15d-PGJ2 (Figure 1C, panel e). Interestingly, heat-shock protein 27 (hsp27), which is localized in the SGs induced by heat but not in the SGs induced by SA (Anderson and Kedersha, 2006), was enriched in the SGs induced by 15d-PGJ2 (Figure 1C, panel f).
We measured the amounts of 15d-PGJ2 required for SG formation. SGs were formed by 10 μM of 15d-PGJ2 after 12–24 h of treatment (Figure 1D). Anti-inflammatory response (Straus et al, 2000; Campo et al, 2002) and other biological activities (Pereira et al, 2006; Aldini et al, 2007; Arnold et al, 2007; Fionda et al, 2007; Hasegawa et al, 2007; Lin et al, 2007) of 15d-PGJ2 were observed under these conditions. Furthermore, we found that 15d-PGJ2 induced SG formation in various cell lines such as a neuronal cell line SH-SY5Y and a macrophage cell line RAW264.7 (Supplementary Figure S2). In subsequent experiments, we treated HeLa cells with 50 μM 15d-PGJ2 for 1 h to induce SGs quickly, unless otherwise indicated.
Subcellular localizations of TRAF2 before (Figure 1E, panels a, e and i) and after induction of SG formation by heat (Figure 1E, panels b, f, and j) or 15d-PGJ2 (Figure 1E, panels d, h, and l) were monitored by an immunocytochemical method. This was because the sequestration of TRAF2 into SGs induced by heat has previously been reported (Kim et al, 2005). Similarly, migration of TRAF2 to SGs induced by 15d-PGJ2 was observed, as indicated by colocalization with eIF3b (yellow dots in Figure 1E, panel l). There was no change in the subcellular localization of TRAF2 with PGE2 treatment (Figure 1E, panels c, g, and k).
We also investigated the localization of RIP, which directly interacts with TRAF2 and conveys TNF-α signal downstream of TRAF2 (Jackson-Bernitsas et al, 2007), and that of IKK α/β which conveys TNF-α signal downstream of RIP and is also known as a target of 15d-PGJ2 (Cheng and Baltimore, 1996). Neither RIP nor IKK α/β was sequestered into SGs (Supplementary Figure S3A). Similar phenomenon was observed in the SGs induced by heat stress (Kim et al, 2005). Moreover, the interaction between RIP and TRAF2 was inhibited by 15d-PGJ2 treatment (Supplementary Figure S3B). These results indicate that the sequestration of TRAF2 by 15d-PGJ2 contributes to the anti-inflammatory activity of this lipid molecule independently of inactivation of IKK and NF-κB by this compound (Straus et al, 2000).
SG formation by 15d-PGJ2 does not need eIF2α phosphorylation, TIA-1 aggregation, and PPARγ activation
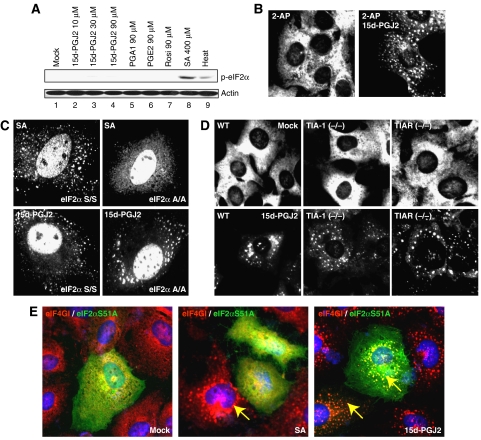
To understand the molecular basis of 15d-PGJ2-induced SG formation, we assessed eIF2α phosphorylation levels using a phospho-eIF2α-specific antibody, because some SG-inducing agents such as SA induce SG formation by phosphorylation of eIF2α (Anderson and Kedersha, 2006). There was no significant increase in eIF2α phosphorylation in the cells treated with either 15d-PGJ2 or PGA1 (Figure 2A, lanes 2–5), although SA-treated and heat-treated cells showed increased levels of phosphorylated eIF2α (Figure 2A, lanes 8 and 9). We also tested the effect of 2-aminopurine (2-AP), a strong PKR (protein kinase, interferon-inducible double-stranded RNA-dependent activator) inhibitor, on blockade of SG formation by 15d-PGJ2. Pretreatment with 2-AP had no effect on 15d-PGJ2-induced SG formation (Figure 2B, right panel). Furthermore, 15d-PGJ2 induced SG formation in a MEF cell with a mutant eIF2α (eIF2α A/A cell) with a S51A knock-in mutation at the PKR target site of the eIF2α gene (McEwen et al, 2005) (Figure 2C, bottom panels). On the other hand, SA-induced SG formation was inhibited in this cell line as reported (McEwen et al, 2005) (Figure 2C, top panels). Furthermore, overproduction of S51A mutant eIF2α inhibited SG formation induced by SA treatment (middle panel in Figure 2E) as reported (Anderson and Kedersha, 2006). On the other hand, overproduction of the mutant eIF2α did not block SG formation by 15d-PGJ2 (right panel in Figure 2E). These results suggest that phosphorylation of eIF2α is not essential for SG formation by 15d-PGJ2. These results are contradictory to a previous report suggesting that 15d-PGJ2 induces phosphorylation of eIF2α through the PKR-mediated pathway (Campo et al, 2002). The discrepancy may be attributed to the difference in conditions and cell lines used in the experiments.
Figure 2.
SG formation by 15d-PGJ2 is independent of eIF2α phosphorylation and TIA-1 aggregation. (A) Phosphorylated eIF2α levels were monitored by western blot analyses using HeLa cell extracts (40 μg) treated with 15d-PGJ2 (lanes 2–4), PGA1 (lane 5), PGE2 (lane 6), Rosi (lane 7), or SA (lane 8) at the indicated concentrations for 30 min or with heat at 44°C for 30 min. (B) HeLa cells grown on cover slips were pretreated with 1 mM of 2-AP or with vehicle for 6 h, and then treated with 50 μM of 15d-PGJ2 for 30 min. Fixed cells were analyzed by immunocytochemistry with an eIF3b antibody. (C) The wild-type and eIF2α A/A mutant MEF cells were treated with 400 μM of SA for 30 min or 50 μM of 15d-PGJ2 for 1 h. Immunocytochemical assays were performed with a TIA-1 antibody. (D) The wild-type, TIA-1 KO, and TIAR KO MEF cells were mock-treated (upper panel) or treated with 15d-PGJ2 (lower panel). Immunocytochemical analyses were performed with an eIF3b antibody. (E) A plasmid encoding FLAG tagged-eIF2α S51A was transfected into HeLa cells. After 48 h of incubation, cells were mock-treated (left), treated with 400 μM of SA (middle) or with 50 μM of 15d-PGJ2 (right) for 30 min. The loci of eIF4GI and eIF2α S51A were visualized by an immunocytochemical method using eIF4GI and FLAG antibodies, respectively.
The prion-like activity of TIA-1 has been reported to function in SG formation (Gilks et al, 2004). The effects of TIA-1 and TIAR on 15d-PGJ2-induced SG formation were investigated using TIA-1 and TIAR KO MEF cell lines. The number of 15d-PGJ2-induced SGs was not reduced in TIA-1 KO cell line (Figure 2D, bottom panels), unlike the level of SGs induced by other agents such as SA (Gilks et al, 2004) (data not shown). This indicates that neither TIA-1 nor TIAR has a key role in 15d-PGJ2-induced SG formation.
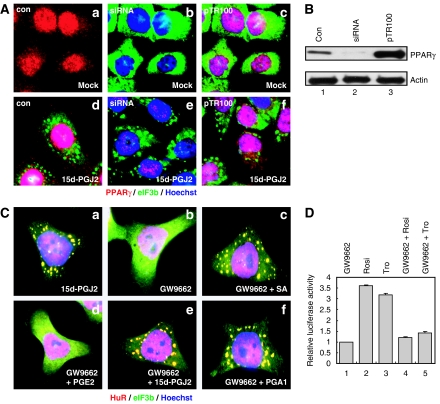
The role of PPARγ in 15d-PGJ2-induced SG formation was investigated because PPARγ is the best-known target molecule of 15d-PGJ2 (Straus and Glass, 2001). Knock-down of PPARγ by a PPARγ-specific siRNA and overexpression of PPARγ (Figure 3B, lanes 2 and 3) had no effect on the SG formation induced by 15d-PGJ2 (Figure 3A, panels e and f). The PPARγ-specific antagonist GW9662 also had no effect on SG formation induced by 15d-PGJ2 and PGA1 (Figure 3C, panels e and f). Under the same conditions, PPARγ-mediated PPRE (PPAR-responsive element) activation was completely blocked by GW9662 (Figure 3D, lanes 4 and 5). These data indicate that PPARγ is not involved in 15d-PGJ2-mediated SG formation.
Figure 3.
SG formation by 15d-PGJ2 is independent of PPARγ. (A) HeLa cells grown on cover slips were transfected with a siRNA against PPARγ (b, e) or a plasmid pTR100-PPARγ expressing high levels of PPARγ (c, f). After transfection, cells were treated with 50 μM of 15d-PGJ2 for 1 h. Immunocytochemical analyses were performed with eIF3b and PPARγ antibodies, shown in green and red, respectively. (B) The amounts of PPARγ in cells transfected with control siRNA (lane 1), siRNA against PPARγ (lane 2) and pTR100-PPARγ (lane 3) were analyzed by western blot assays using a PPARγ antibody. Lysates were normalized by an actin blot. (C) HeLa cells were pretreated with 1 μM of GW9662, an irreversible PPARγ antagonist, for 24 h and then treated with SA (400 μM), PGE2 (50 μM), 15d-PGJ2 (50 μM), or PGA1 (50 μM) for 1 h. Immunocytochemical analyses were performed with eIF3b and HuR antibodies, shown in green and red, respectively. The nuclei are shown in blue by Hoechst staining. (D) 293T cells were transfected with a plasmid (1 μg) expressing a PPARγ reporter gene. After transfection, cells were pretreated or not pretreated with 1 μM of GW9662 for 24 h, before being treated with 10 μM of rosiglitazone (Rosi) or troglitazone (Tro) for 12 h. Columns indicate relative luciferase activities in the cell extracts normalized to a mock-treated control extract. A full color version of this figure is available at the EMBO Journal Online.
15d-PGJ2 inhibits translation
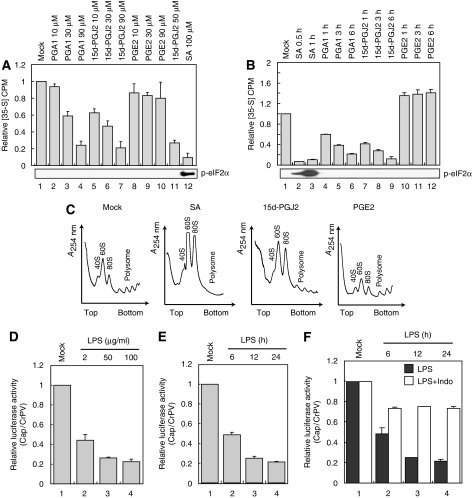
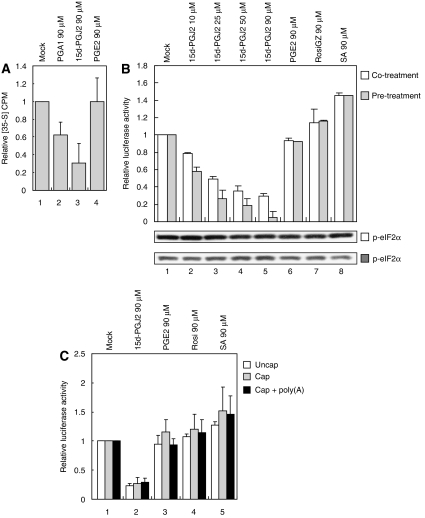
As SG formation is accompanied by translational blockade, the effects of 15d-PGJ2 on protein synthesis were investigated. Metabolic labeling of HeLa cells with [35S]methionine clearly showed that total protein synthesis was inhibited by 15d-PGJ2 in a concentration-dependent manner (Figure 4A, lanes 5–7) and a time-dependent manner (Figure 4B, lanes 7–9). PGA1 had a similar effect as 15d-PGJ2 (Figure 4A, lanes 2–4 and B, lanes 4–6), but PGE2 did not block translation (Figure 4A, lanes 8–10 and B, lanes 10–12). No significant phosphorylation of eIF2α was observed from the cells treated with 15d-PGJ2 (Figure 4A and B, bottom panels).
Figure 4.
15d-PGJ2 and PGA1 inhibit translation in vivo. (A) HeLa cells were grown on 60-mm dishes up to about 70–80% confluence. Cells were mock-treated (lane 1) or treated with PGA1 (lanes 2–4), 15d-PGJ2 (lanes 5–7), or PGE2 (lanes 8–10) at the indicated concentrations for 30 min, then in vivo labeling of newly synthesized proteins was performed as described in Materials and methods. Here, 4200 c.p.m. was obtained from the TCA-precipitated control sample (lane 1). Phosphorylated eIF2α levels were monitored by western blot analyses (bottom panel). (B) Cells were mock-treated (lane 1), treated with SA (400 μM) (lanes 2 and 3), PGA1 (90 μM) (lanes 4–6), 15d-PGJ2 (90 μM) (lanes 7–9), and PGE2 (90 μM) (lanes 10–12) at indicated times. Newly synthesized proteins were measured as (A). Here, 4500 c.p.m. was obtained from the TCA-precipitated control sample (lane 1). Phosphorylated eIF2α levels were monitored by western blot analyses (bottom panel). (C) HeLa cells were mock-treated or treated with SA (400 μM) for 30 min, 15d-PGJ2 (50 μM) for 1 h, or PGE2 (50 μM) for 1 h. Sucrose gradient experiment was performed as described in Materials and methods. The lines show absorbance at 254 nm. (D–F) Effects of LPS on translation in RAW264.7 macrophage cells. (D) RAW264.7 cells were incubated with LPS for 24 h at the indicated concentrations. After the LPS treatment, mRNAs (1 μg) containing Renilla luciferase translated in a cap-dependent manner and mRNAs (1 μg) containing firefly luciferase under the control of cricket paralysis virus (CrPV) IRES were co-transfected into the cells. Luciferase activities were measured 3 h post-transfection. Columns indicate ratios of relative luciferase activities (Renilla luciferase/firefly luciferase) in the cell extracts normalized to that in a mock-treated control extract. Firefly luciferase activities are considered as an indicator of mRNA transfection efficiency since CrPV IRES function is insensitive to 15d-PGJ2 as described in Figure 6B. (E) RAW264.7 cells were incubated with LPS (10 μg/ml) for the times indicated. Transfection of mRNAs and analyses of luciferase activities were performed as described in (D). (F) RAW264.7 cells were pretreated (white columns) or not pretreated (gray columns) with indomethacin (1 μM) for 30 min before being treated with LPS (10 μg/ml). Transfection of mRNAs and analyses of luciferase activities were performed as described in (D).
It is well known that inhibition of translation and SG formation alters the polysome profile. SA treatment induces the disassembly of polysomes, leading to an increase in the extents of ribosomal subunit peaks, indicating the accumulation of ribosomal subunits not participating in translation (Figure 4C, panel SA) (Anderson and Kedersha, 2006). Ribosomal shift to the subunit state was also observed in 15d-PGJ2-treated cells, albeit the magnitude of which was weaker than that seen in SA-treated cells (Figure 4C, panel 15d-PGJ2). As expected, PGE2 did not induce a ribosomal shift (Figure 4C, panel PGE2). SA- and 15d-PGJ2-induced monosome shifts disappeared when cells were pretreated with emetine (data not shown). These data indicate that 15d-PGJ2, similarly to SA, inhibits protein synthesis in vivo.
To confirm that the translational inhibition by 15d-PGJ2 occurs under physiological conditions, we tested the effect of prolonged treatment of lipopolysaccharide (LPS) on RAW264.7 that produces PGD2 and 15d-PGJ2 upon treatment of LPS through a COX-2-dependent pathway (Shibata et al, 2002). Interestingly, cap-dependent translation, but not CrPV IRES-dependent translation, was inhibited in a dose-dependent (Figures 4D and 6B) and a time-dependent manner (Figure 4E). The kinetics of time-dependent translational inhibition (Figure 4E) was similar to that of 15d-PGJ2 production (Supplementary Figure S4) as reported by Shibata et al (2002). Moreover, the translational inhibition by LPS was greatly weakened by a pretreatment of indomethacin, a non-selective COX inhibitor (compare white columns with gray columns in Figure 4F). The effect of indomethacin treatment on translational inhibition induced by LPS is most likely attributed to blockage of 15d-PGJ2 production (Chang et al, 2006). These results suggest that translation inhibition by 15d-PGJ2 occurs at physiological conditions.
Figure 6.
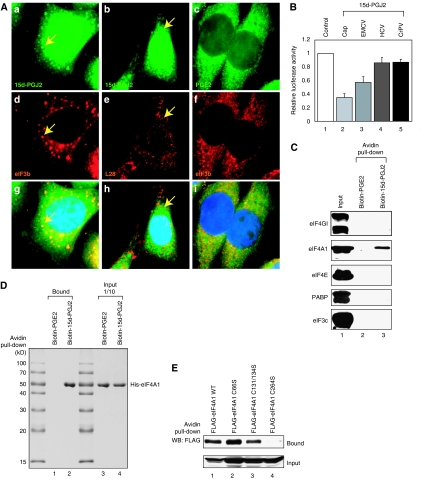
eIF4A is the target of 15d-PGJ2. (A) HeLa cells were grown on cover slips and then treated with biotinylated 15d-PGJ2 (50 μM; a, b, d, e, g, and h) or biotinylated PGE2 (50 μM; c, f, and i) for 1 h. Immunocytochemical analyses were performed with primary antibodies against eIF3b (a, c, d, f, g, and i) and L28 antibodies (b, e, and h). Biotinylated chemicals were visualized with FITC-conjugated streptavidin. Arrows indicate SGs induced by biotinylated 15d-PGJ2. (B) Monocistronic mRNAs with cap structure (lanes 1 and 2), EMCV IRES (lanes 1 and 3), HCV IRES (lanes 1 and 4), and CrPV IRES (lanes 1 and 5) were translated in HeLa lysates for 1 h in the presence (lanes 2–4) or absence (lane 1) of 15d-PGJ2 (50 μM). Various IRES activities (RLUs of 20 000–75 000) were observed from the mock-treated HeLa lysates. Luciferase activities in the translation mixtures containing 15d-PGJ2 were normalized to those in the corresponding translation mixtures without 15d-PGJ2, and are shown as columns (mean values). (C) Cytoplasmic HeLa lysates (1 mg) were treated with 50 μM of biotinylated PGE2 (lane 2) and 50 μM biotinylated 15d-PGJ2 (lane 3) for 1 h at 30°C and then streptavidin pull-down was performed as described in experimental procedures. Resin-bound proteins were analyzed by western blot analyses with antibodies against eIF4GI, eIF4AI, eIF4E, poly(A)-binding protein (PABP), and eIF3c. (D) Purified His–eIF4A1 (6 μg) was incubated with 50 μM of biotinylated PGE2 (lane 1) or biotinylated 15d-PGJ2 (lane 2), and then precipitated by streptavidin–sepharose. The resin-bound proteins were then analyzed by Coomassie blue staining. (E) 293T cells were transfected with the wild-type (WT, lane 1) or mutant (lanes 2–4) FLAG-eIF4A1s. Immunoprecipitation was performed as described in Materials and methods.
The effect of 15d-PGJ2 on translation was also monitored using a HeLa lysate in vitro translation system. PGA1 and 15d-PGJ2 inhibited protein synthesis in vitro (Figure 5A, lanes 2 and 3) but PGE2 did not (Figure 5A, lane 4). Pretreatment of HeLa cell lysates with 15d-PGJ2 increased the inhibitory effect about two-fold with 50 μM and about five-fold with 90 μM (Figure 5B, lanes 4 and 5). Treatment of PGE2 did not block translation (Figure 5B, lane 6). Treatment with rosiglitazone (RosiGZ) and SA slightly increased translation in vitro (Figure 5B, lanes 7 and 8). The molecular basis of this phenomenon remains to be determined.
Figure 5.
PGA1 and 15d-PGJ2 inhibit translation in vitro. (A) β-gal mRNA (40 nM) was translated in HeLa lysates for 1 h in the presence of PGA1 (lane 2), 15d-PGJ2 (lane 3), and PGE2 (lane 4) at indicated concentrations. 35S-labeling experiment was performed as described by Pestova et al (1998). (B) HeLa lysates were pretreated with vehicle (lane 1) or with indicated chemicals (lanes 2–9) for 30 min at indicated concentrations, a capped Renilla luciferase mRNA (40 nM) was added to the translation mixtures, and then incubated at 30°C for 1 h (gray columns). White columns show the effects of the same set of chemicals added together with the reporter mRNA during 1 h in vitro translation. Relative luciferase activities (mean values) are depicted by columns. Phosphorylated eIF2α levels were monitored by western blot analyses (bottom panel). (C) Poly(A)-tailed mRNAs were produced by in vitro transcription of plasmid pRLCMV-poly(A)60. Capped mRNAs were produced by in vitro transcription of plasmids pRLCMV and pRLCMV-poly(A)60 in the presence of 7methyl GTP. In vitro translation reactions were performed with various reporter mRNAs (40 nM) for 1 h in the presence of chemicals (90 μM) indicated on top of the panel. Luciferase activities in the translation mixtures containing various compounds were normalized to those in mock-treated extracts with the corresponding mRNAs, and are shown as columns (mean values).
Basal levels of phosphorylated eIF2α were observed in HeLa cell lysates (Figure 5B, lane 1). No increase of eIF2α phosphorylation was observed from the HeLa cell lysates treated with 15d-PGJ2, PGE2, rosiglitazone, and SA (Figure 5B, lanes 2–8). These results indicate that eIF2α phosphorylation does not occur in HeLa cell extracts even with SA treatment (Figure 5B, lane 8). This would be the reason why translation is not inhibited by SA in the HeLa cell lysates (Figure 5B, lane 8) unlike in the in vivo system where eIF2α is phosphorylated by SA (Figure 4A, lane 12).
Capping and poly(A) addition to the reporter mRNA did not affect the relative inhibitory activity of 15d-PGJ2 on uncapped and poly(A)-tail-less mRNA, even though the translational efficiency of capped and poly(A)-tailed mRNAs was greater than that of the uncapped tail-less mRNA (Figure 5C, lane 2). This indicates that the eIF4E, which is a cap-binding protein, or the PABP may not be involved in the translation inhibition activity of 15d-PGJ2. Pretreatment with 15d-PGJ2 increased the inhibitory effect on translation, so we speculated that a thiol modification of the target protein by the electrophilic carbon of 15d-PGJ2 is involved in the translational inhibition of 15d-PGJ2.
eIF4A is the cytoplasmic target of 15d-PGJ2
Subcellular localization of 15d-PGJ2 was investigated using an immunocytochemical method. Visualization of 15d-PGJ2 was accomplished by treatment with biotinylated 15d-PGJ2 followed by treatment with streptavidin-conjugated fluorescein isothiocyanate (FITC). 15d-PGJ2 molecules localized to both the nucleus and the cytoplasm. Interestingly, 15d-PGJ2 molecules were localized at SGs induced by 15d-PGJ2, as indicated by colocalization with the SG marker eIF3b (Figure 6A, panels a, d, and g with arrows). In contrast, biotinylated PGE2 was rather evenly distributed in the cytoplasm and SGs were not induced by PGE2 (Figure 6A, panels c, f, and i). The large ribosomal subunit, which was visualized by the ribosomal protein L28, was not colocalized with 15d-PGJ2 (Figure 6A, panels b, e, and h). This indicates that 15d-PGJ2-induced SGs contain high levels of 15d-PGJ2, possibly by complexing with a SG component.
The requirements of initiation factors vary in different internal ribosome entry sites (IRESes), so the target of 15d-PGJ2 was investigated by analyzing the effects of 15d-PGJ2 on the activities of various IRESes (Jang, 2006). For example, eIF4A, eIF4B, and eIF4G have important roles in encephalomyocarditis virus (EMCV) IRES-dependent translation (Jang, 2006). The eIF2 ternary complex and eIF3 are needed for hepatitis C virus (HCV) IRES-dependent translation, whereas no translational initiation factor is needed for cricket paralysis virus (CrPV) IRES-dependent translation (Pisarev et al, 2005). Cap-dependent and EMCV IRES-dependent translation was inhibited by 15d-PGJ2 treatment (Figure 6B, lanes 2 and 3), but HCV IRES- and CrPV IRES-dependent translation was not (Figure 6B, lanes 4 and 5). These data indicate that eIF4G and eIF4A are potential targets of 15d-PGJ2.
We therefore performed biotin pull-down experiments using biotinylated 15d-PGJ2. Of the translation factors tested, only eIF4A was precipitated by streptavidin agarose beads from cytoplasmic HeLa cell extracts treated with biotinylated 15d-PGJ2 (Figure 6C, panel eIF4A1). Other eIF4F proteins such as the scaffold protein eIF4G, cap-binding proteins eIF4E, PABP, and eIF3, which bridge the eIF4G and the small ribosomal subunit, did not have a direct interaction with 15d-PGJ2. Direct interaction between eIF4A and 15d-PGJ2 was confirmed using purified eIF4A1 proteins. The recombinant eIF4A1 proteins were precipitated by biotin–15d-PGJ2 but not by biotin–PGE2 (Figure 6D, lanes 1 and 2).
15d-PGJ2 contains an electrophilic carbon center susceptible to undergoing addition reactions (Michael addition) with nucleophiles such as the free sulfhydryl group of cysteine residues in cellular proteins (Straus and Glass, 2001). Human eIF4A1 contains four cysteine residues, 66C, 131C, 134C, and 264C, which are potential target sites of 15d-PGJ2. To determine which cysteine residues are involved in 15d-PGJ2-binding, we monitored effects of cysteine to serine mutations on 15d-PGJ2-binding. A derivative of eIF4A with the C264S mutation could not interact with 15d-PGJ2 (Figure 6E, lane 4); however, other mutant forms of eIF4A are bound to 15d-PGJ2 (lanes 2 and 3 in Figure 6E). These data indicate that 15d-PGJ2 directly binds to the cysteine residue 264 of the eIF4A protein.
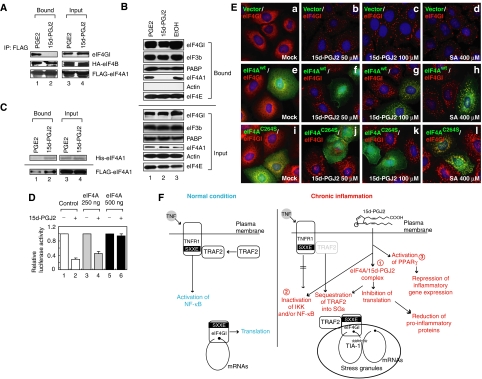
The mechanism by which 15d-PGJ2 inhibits translation was investigated by monitoring the effects of 15d-PGJ2 on translational initiation complex formation. eIF4G is a scaffold protein that recruits eIF4E, eIF4A, eIF3, and PABP into the translational initiation complex. The effect of 15d-PGJ2 on the eIF4GI–eIF4A interaction was monitored by a co-immunoprecipitation assay. eIF4GI was co-precipitated with eIF4A1 from mock-treated (data not shown) or PGE2-treated (Figure 7A, lane 1) cell extracts; however, eIF4GI was not co-precipitated with eIF4A1 from the cell extract treated with 15d-PGJ2 (Figure 7A, lane 2). eIF4B, which directly interacts with eIF4A, was co-precipitated with eIF4A1 regardless of whether the cell extracts had been treated with 15d-PGJ2 (Figure 7A, panel HA-eIF4B in lanes 1 and 2). These data indicate that 15d-PGJ2 blocks the eIF4A–eIF4G interaction but not the eIF4A–eIF4B interaction.
Figure 7.
15d-PGJ2 blocks the interaction between eIF4G and eIF4A. (A) 293T cells were co-transfected with HA-eIF4B and FLAG-eIF4A1. Cells were lysed and then treated with 50 μM of PGE2 or 15d-PGJ2 at 30°C for 1 h. Immunoprecipitation was performed with an anti-FLAG antibody. Western blot analysis was performed with anti-FLAG, anti-HA, and anti-eIF4GI antibodies. (B) Proteins in the eIF4F complex were analyzed using a 7methyl GTP resin and cytoplasmic HeLa lysates treated with vehicle (lane 3), 50 μM of 15d-PGJ2 (lane 2), or 50 μM of PGE2 (lane 1) for 1 h. Resin-bound proteins were washed three times and then analyzed by western blot assays with antibodies against eIF4GI, eIF3b, PABP, eIF4AI, actin, or eIF4E. (C) Purified His-eIF4A protein (2 μg, upper panel) and 293T-cell lysate containing overexpressed FLAG-eIF4A (2 mg, lower panel) were incubated with 50 μM of PGE2 (lanes 1 and 3) or 15d-PGJ2 (lanes 2 and 4) for 1 h. The biotinylated RNA (1 μg) β-globin mRNAs were incubated with the pretreated purified eIF4A1 or cell lysate in the presence of RNasin and nonspecific competitor tRNAs for 1 h. RNA-bound proteins were precipitated by a streptavidin-agarose resin and then visualized by silver staining (upper panel) or western blot analysis with an anti-FLAG antibody (lower panel). (D) In vitro translation was performed in RRL (Promega) with a Renilla luciferase mRNA (40 nM) for 1 h with the additional purified His-eIF4A1 protein (0 ng, lanes 1 and 2; 250 ng, lanes 3 and 4; 500 ng, lanes 5 and 6). 15d-PGJ2 (50 μM) was added to the translation mixtures shown in lanes 2, 4, and 6. Luciferase activities with (lanes 2, 4, and 6) 15d-PGJ2 treatment were compared with those without (lanes 1, 3, and 5) 15d-PGJ2 treatment in the presence of additional eIF4A at particular concentrations and are shown as columns (mean values). (E) HeLa cells were grown on cover slips and transfected with a FLAG vector, plasmid FLAG-eIF4Awt expressing the wild-type eIF4A tagged with FLAG, or plasmid FLAG-eIF4AC264S expressing a C264S mutant eIF4A tagged with FLAG. After 48 h of incubation, cells were treated with the chemicals at the concentrations indicated for 30 min. Immunocytochemical analyses were performed with eIF4GI and FLAG antibodies. (F) Hypothetical model of anti-inflammatory activities of 15d-PGJ2. At the chronic inflammatory stage, 15d-PGJ2 is highly produced by immune cells and inhibits the positive feedback loop of inflammation (Gilroy et al, 2004). There are multiple target molecules of 15d-PGJ2 in the cell. Modifications of some of the target proteins result in anti-inflammatory activity. Pathway 1, 15d-PGJ2 inactivates eIF4A, resulting in inhibition of translation, as described in this paper. This induces SG formation and TRAF2 proteins are sequestered into the SGs. This in turn blocks the key pro-inflammatory TNF-α signaling pathway. Translational inhibition may also reduce the expression of pro-inflammatory proteins. Pathway 2, 15d-PGJ2 directly inactivates pro-inflammatory molecules such as IKK and NF-κB (Straus et al, 2000). Pathway 3, 15d-PGJ2 functions as an agonist of PPARγ that represses transcriptional activation of inflammatory response genes. These anti-inflammatory responses may occur independently or in a concerted manner depending on the concentration of 15d-PGJ2 and on internal and external conditions of target cells.
The effect of 15d-PGJ2 on cap-binding protein complex formation was monitored by analyzing components in the protein complex precipitated with 7methyl GTP resin. eIF4E, eIF4GI and eIF4A1 were found in the precipitates from mock-treated and PGE2-treated HeLa cell extracts (Figure 7B, lanes 3 and 1). eIF4E and eIF4GI were detected in the 7methyl GTP resin-bound protein complex even after 15d-PGJ2 treatment (Figure 7B, lane 2). By contrast, eIF4A1 was not co-precipitated with eIF4GI after 15d-PGJ2 treatment (Figure 7B, lane 2). These data also indicate that 15d-PGJ2 inhibits the eIF4G–eIF4A interaction.
eIF4A has RNA-binding activity (Low et al, 2005). The effect of 15d-PGJ2 on the RNA-binding activity of eIF4A was monitored using β-globin mRNA. Binding of purified eIF4A1 to the β-globin mRNA was increased with 15d-PGJ2 treatment (Figure 7C, panel His-eIF4A in lanes 1 and 2). Similarly, the RNA-binding activity of FLAG-eIF4A protein expressed in mammalian cells was also increased after treatment with 15d-PGJ2, as shown by binding to β-globin mRNA (Figure 7C, panel FLAG-eIF4A in lanes 1 and 2). The implications of this phenomenon are discussed below.
To confirm that eIF4A is the main target of 15d-PGJ2 involved in inhibition of translation, we monitored the effect of eIF4A1 supplementation on the inhibition of translation by 15d-PGJ2. Addition of purified eIF4A1 restored translation activity of the in vitro translation mixture treated with 15d-PGJ2 in a dose-dependent manner (Figure 7D, lanes 2, 4, and 6). Furthermore, the effects of overproduction of eIF4A and its derivative with C264S mutation on the translational inhibition by 15d-PGJ2 were monitored by using HeLa cell transfection. The cells overexpressing wild-type eIF4A were resistant to SG formation by 15d-PGJ2 at the 50 μM (Figure 7E, green cells on panel f) while untransfected cells form SGs at this condition (Figure 7E, red cells on panel f). At higher concentration of 15d-PGJ2 (100 μM), however, SG formation was observed in the cells overexpressing wild-type eIF4A (Figure 7E, yellow dots in green cells on panel g). Importantly, cells overexpressing C264S mutant eIF4A, which does not bind to 15d-PGJ2, were resistant to SG formation by 15d-PGJ2 at both 50 and 100 μM (Figure 7E, green cells on panels j and k). On the contrary, overexpression of eIF4A and its derivative did not inhibit SG formation by SA (Figure 7E, green cells on panels h and l). Taken together, these data strongly indicate that 15d-PGJ2 blocks translation through direct binding to the eIF4A protein.
Discussion
Recently, progress has been made in determining the interplay between translational processes and pro-inflammatory signaling (Kim et al, 2005; McDunn and Cobb, 2005). Here, we report that the anti-inflammatory signaling molecule 15d-PGJ2, which is known to block pro-inflammatory signaling, inhibits translation in vivo (Figure 4) and in vitro (Figure 5). Several lines of evidence suggest that, for translational inhibition, the main target of 15d-PGJ2 is the translational initiation factor eIF4A. First, 15d-PGJ2 directly binds to eIF4A1, as shown by pull-down experiments with biotinylated 15d-PGJ2, HeLa cell extracts (Figure 6C), and purified eIF4A (Figure 6D). Second, eIF4A has previously been identified as a cellular target of 15d-PGJ2 using a proteomic approach; however, the physiological importance of the eIF4A–15d-PGJ2 interaction was not reported (Aldini et al, 2007). Third, the translation inhibitory effect of 15d-PGJ2 was relieved by the addition of purified eIF4A1 (Figure 7D). The amount of purified eIF4A1 required for complete restoration of translation was about 0.5 μM to final that is about 1/100 amount of 15d-PGJ2 by molarity in the reaction mixture. This suggests that restoration of translation is not due to nonspecific inactivation of 15d-PGJ2 by the newly added eIF4A1 in the translation reaction mixture, but is likely to be due to replacement of inactive 15d-PGJ2-conjugated eIF4A1 proteins with functional eIF4A1 proteins. Fourth, SG formation in HeLa cells by 15d-PGJ2, which reflects translational inhibition, was hampered by overproduction of eIF4A and its derivative (C264S mutant) (Figure 7E). The C264S mutant, which lacks the binding site of 15d-PGJ2, showed stronger resistance to SG formation by 15d-PGJ2 than the wild-type eIF4A. This suggests that SG formation by 15d-PGJ2 is induced by the binding of 15d-PGJ2 to eIF4A.
While investigating anti-proliferating agents, Low et al (2005) found that a natural marine compound named pateamine A (PatA) could block translation by inactivating eIF4A. Interestingly, like 15d-PGJ2, this compound also induces formation of SGs independently of eIF2α phosphorylation (Dang et al, 2006) and impairs ribosome binding to mRNA (Supplementary Figure S5) (Bordeleau et al, 2006b; Mazroui et al, 2006). The authors suggested that PatA inhibits translation by blocking the eIF4G–eIF4A interaction (Low et al, 2005). However, Pelletier and colleagues have suggested that RNA-mediated sequestration of eIF4A is the translational inhibitory mechanism of PatA (Bordeleau et al, 2006a). It is possible that sequestration of eIF4A into RNAs may also partly contribute to translational inhibition by 15d-PGJ2 because the RNA-binding activity of purified eIF4A was increased in the presence of 15d-PGJ2 (Figure 7C). However, it should be noted that the eIF4A and eIF4G interaction was also blocked in the presence of RNase (Figure 7A), indicating that 15d-PGJ2 actively blocks this protein–protein interaction.
Investigations into the role of the tumor-suppressor protein named programmed cell death 4 (Pdcd4), which blocks cell proliferation by inhibiting translation, found that modulation of eIF4A can control cellular activities (Yang et al, 2003). The translational inhibition is caused by binding of Pdcd4 to eIF4A, which competitively blocks eIF4A-binding to the C-terminal domain of eIF4G and inhibits helicase activity of eIF4A (Yang et al, 2003). In this respect, the translation inhibitory action of 15d-PGJ2 is likely to contribute to antineoplastic activity of 15d-PGJ2. These indicate that eIF4A is a good target for the regulation of biological activities intracellularly through Pdcd4 and intercellularly through 15d-PGJ2 and that eIF4A is a good therapeutic target for developing anticancer drugs.
The eIF4A amino-acid residue targeted by 15d-PGJ2 was identified by monitoring the 15d-PGJ2-binding capabilities of mutant eIF4As (Figure 6E), and the cysteine at residue 264 was found to be the target site of 15d-PGJ2 (Figure 6E). Interestingly, the cysteine 264 is positioned close to the eIF4A residues R360, R363, and R366 that had previously been shown, by site-directed mutagenesis, to be important for binding to the middle and the C-terminal domains of eIF4G1 (Zakowicz et al, 2005). Moreover, cysteine 264 is next to the residues aspartic acid 265 and glutamic acid 268 that have been shown to be essential for interaction with eIF4G1 (Oberer et al, 2005). Cysteine 264 is located in the α-helix that forms a contact surface with the middle domain of eIF4GI, as shown by nuclear magnetic resonance spectroscopy (Oberer et al, 2005). Therefore, it is possible that PGJ2 covalently bound to eIF4A at the eIF4G contact site sterically hinders the eIF4A–eIF4G interaction.
SGs are formed under various conditions that block translation. SG formation owing to 15d-PGJ2 treatment (Figure 1) is most likely to be due to inhibition of translation by eIF4A–15d-PGJ2 complex formation. Interestingly, TRAF2 proteins, which are scaffold proteins that recruit pro-inflammatory signaling proteins, are sequestered to 15d-PGJ2-induced SGs (Figure 1E) and heat-induced SGs (Figure 1E; also see Kim et al, 2005). Among the TNF-α-signaling molecules tested, only TRAF2 was sequestered into SGs (Supplementary Figure S3A). Furthermore, RIP–TRAF2 interaction, which is required for NF-κB activation mediated by TNF-α (Cheng and Baltimore, 1996), was weakened by 15d-PGJ2 treatment (Supplementary Figure S3B). This may indicate that the sequestration of TRAF2 into SGs contributes, at least in part, to the anti-inflammatory activity of 15d-PGJ2. However, it is difficult to quantify the contribution of TRAF2 sequestration to the anti-inflammatory activity of 15d-PGJ2 because 15d-PGJ2 reduces pro-inflammatory gene expression through activation of PPARγ and inhibits TNF-α signaling by directly inactivating NF-κB and IKK (Straus et al, 2000). Moreover, inhibition of translation by 15d-PGJ2 may also contribute to anti-inflammatory responses by lowering the levels of labile proteins required for maintaining inflammatory responses. The hypothetical process by which chronic inflammatory responses are resolved is shown in Figure 7F.
Here, we report that 15d-PGJ2 inhibits translation by inactivating eIF4A. This activity is most likely related to the anti-proliferation activity of 15d-PGJ2. Further investigations into this activity of 15d-PGJ2 will provide clues for development of anticancer drugs that target eIF4A. Moreover, such research would extend our understanding of the anti-inflammatory activity of 15d-PGJ2.
Materials and methods
Plasmid construction
Plasmid information is in Supplementary data.
Antibodies and chemicals
Antibodies against TIA-1, eIF3b, eIF3c, eIF4A1, eIF4E, HA, HuR, hsp70, L28, TIAR, PABP, PPARγ, RIP, IKKα/β, and rps6 were purchased from Santa Cruz. Antibodies against TRAF2, FLAG, actin, and hsp27 were purchased from BD Pharmingen, Sigma, ICN, StressGen, respectively. Antibodies against eIF2α and phospho-eIF2α were purchased from Cell Signaling Technology. Antibody against eIF4GI was prepared in our laboratory (Kim et al, 2005).
Chemicals PGA1, 15d-PGJ2, biotinylated 15d-PGJ2, PGE2, biotinylated PGE2, arachidonic acid, ciglitazone, troglitazone, rosiglitazone, CAY10410, lipoxin A4, lipoxin B4, epi-lipoxin A4, LPS, and lovastatin were purchased from Cayman Chemical. Sodium arsenite, emetine, indomethacin, and 2-AP were purchased from Sigma. GW9662 was purchased from Calbiochem. TGF-β and IL-10 from R&D Systems.
Immobilized streptavidin agarose was purchased from Pierce. 7m-GTP-sepharose 4B, and Protein G agarose were purchased from GE Healthcare.
Ribosomal pull-down
Ribosomal pull-downs were performed as described by Colon-Ramos et al (2006). Biotinylated β-globin mRNAs synthesized from plasmid pcDNA3-7B-ARE-MS2bs were incubated in a rabbit reticulocyte lysate (RRL) (Promega) in the presence or absence of 15d-PGJ2. After the translation reactions, 10 μg/ml of cycloheximide was added to stop the translation, and then reaction mixtures were incubated with 50 μl of streptavidin–sepharose beads at 4°C for 1 h. Precipitated resins were washed three times, resolved by SDS–PAGE and then transferred to a nitrocellulose membrane.
Pull-down with streptavidin
DNA-transfected HeLa or 293T cells were lysed by soaking in NP-40 lysis buffer (0.5% NP-40, 50 mM HEPES (pH 7.4), 250 mM NaCl, 2 mM EDTA, 2 mM sodium orthovanadate, 2.5 mM β-glycerophosphate, 1 μg/ml aprotinin, 1 μg/ml antipain, 1 μg/ml bestatin, 1 μg/ml pepstatinA and 1 mM PMSF). Lysates were clarified by centrifugation at 14 000 g at 4°C for 15 min and then incubated with 50 μM of biotinylated PGE2 or biotinylated 15d-PGJ2 at 30°C for 1 h. After incubation, lysates were clarified by centrifugation at 14 000 g at 4°C for 5 min and then incubated with 50 μl slurry of immobilized streptavidin agarose at 4°C for 1 h. Precipitated resins were washed three times with the lysis buffer, resolved by SDS–PAGE and then transferred to a nitrocellulose membrane.
In the 15d-PGJ2-binding experiment with purified eIF4A1 and biotinylated 15d-PGJ2, 6 μg of eIF4A were used in 400 μl of the NP-40 lysis buffer.
Analysis of components of eIF4F complex
Cells were lysed with the NP-40 lysis buffer. Cell lysates were incubated with ethanol (EtOH), PGE2, or 15d-PGJ2 at 30°C for 1 h. After incubation, lysates were clarified by centrifugation at 14 000 g at 4°C for 5 min and then incubated with 50 μl slurry of 7m-GTP-sepharose 4B at 4°C for 1 h. Precipitated proteins bound to resin were washed three times with the lysis buffer, resolved by SDS–PAGE and then transferred to a nitrocellulose membrane.
Immunoprecipitation
293T cells transfected with DNAs were lysed using the NP-40 lysis buffer. The lysates were clarified by centrifugation at 14 000 g for 15 min. Anti-FLAG monoclonal antibody (4 μg) was incubated with 20 μl of Protein G agarose for 1 h in 1 ml NP-40 lysis buffer at 4°C. Lysates were pre-cleared with 10 μl of protein G agarose at 4°C for 30 min. After pre-clearing, cell lysates were treated with 50 μM of EtOH, PGE2, or 15d-PGJ2 at 30°C for 1 h, followed by centrifugation. Then protein G agarose-conjugated antibodies were incubated with the pre-cleared lysates at 4°C for 1 h. Precipitates were washed three times with lysis buffer and analyzed by SDS–PAGE.
In vitro transcription and pull-down with biotinylated RNAs
For in vitro transcription, monocistronic reporter plasmids were digested by HpaI and then analyzed by the T7 polymerase reaction. pcDNA3-7B-ARE-MS2bs (kindly provided by Dr Satoshi Yamasaki, Brigham and Women's Hospital) digested by XbaI before use in the T7 polymerase reaction in the presence of biotinylated UTP. Before incubation with the biotinylated RNAs, cell lysates were incubates with 50 μM PGE2 or 15d-PGJ2 at 30°C for 1 h, followed by clarification with centrifugation. RNA-affinity chromatography was performed with purified His-eIF4A or 293T cell lysates transfected with FLAG-eIF4A, as described elsewhere (Kim et al, 2004).
Preparation of HeLa cell lysates and in vitro translation
In vitro translation reactions using HeLa cell lysates and RRL are described elsewhere (Kim et al, 2004).
Ribosome profiling with sucrose gradient
Cells were treated with various agents and for various times, as indicated in the figure legends. Experiments were performed as described elsewhere (Kedersha et al, 2000) using a 0.5–1.5 M sucrose gradient.
Fluorescence microscopy
The immunocytochemical analyses of proteins were performed as described elsewhere (Kim et al, 2005).
Monitoring newly synthesized proteins with [35S] labeling
HeLa cells were treated with various agents and for various times, as indicated in the figure legends. HeLa Cells on 60-mm culture dishes were then washed twice with phosphate-buffered saline (PBS) and incubated in methionine-free Dulbecco's Modified eagle's medium (DMEM) (BMS) medium for 1 h. Cells were incubated for 30 min after supplementation with [35S]methionine ([35S]Met) (500 mCi/ml; NEN Life Science Products), washed twice with ice-cold PBS, harvested, and then lysed with the NP-40 lysis buffer. The cell lysates were centrifuged and the protein concentrations in the cell lysates were measured using the Bradford assay method. To quantify newly synthesized proteins, cell lysates labeled with [35S]-Met were precipitated with 10% trichloroacetic acid (TCA) (w/v), and the precipitated proteins were then dissolved in water and analyzed by a liquid scintillation assay (Packard).
Luciferase assay
Luciferase assays were performed as described elsewhere (Kim et al, 2005).
Cell cultures and transient transfection
MEF TIA (−/−), MEF TIAR (−/−) cells, MEF eIF2α S51A cells were grown as described elsewhere (Gilks et al, 2004). RAW 264.7 cells, SH-SY5Y cells, HeLa cells and 293T cells were grown as described elsewhere (Kim et al, 2005).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure Legends
Acknowledgments
We thank Dr Nancy Kedersha and Paul Anderson (Brigham and Women's Hospital) for providing the TIA(−/−) and TIAR(−/−) MEF cell lines, Dr Randal Kaufman and Dr Sung Hoon Back (University of Michigan Medical Center) for the MEF eIF2α S51A cell lines, Dr Nadejda Korneeva (Louisiana State University) for E. coli purified eIF4A1 protein, Dr Yongjun Dang and Dr Jian Liu (Johns Hopkins) for the FLAG-eIF4A1 clone, Dr Peter Sarnow (Stanford University) for the CrPV IRES reporter, and Dr Todd Leff (Wayne State University) for the PPRE reporter and PPARγ clone. This work was supported in part by grants from MOST and NCRC (R15-2004-033-05001-0), a grant FPR05B 1-310 of the 21C Frontier Functional Proteomics Project from KMST, a grant R17-2007-089-01001-0 of NRL from KOSEF, and a grant from POSCO.
References
- Aggarwal BB, Shishodia S, Sandur SK, Pandey MK, Sethi G (2006) Inflammation and cancer: how hot is the link? Biochem Pharmacol 72: 1605–1621 [DOI] [PubMed] [Google Scholar]
- Aldini G, Carini M, Vistoli G, Shibata T, Kusano Y, Gamberoni L, Dalle-Donne I, Milzani A, Uchida K (2007) Identification of actin as a 15-deoxy-delta(12,14)-prostaglandin J(2) target in neuroblastoma cells: mass spectrometric, computational, and functional approaches to investigate the effect on cytoskeletal derangement. Biochemistry 46: 2707–2718 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N (2006) RNA granules. J Cell Biol 172: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold R, Neumann M, Konig W (2007) Peroxisome proliferator-activated receptor-gamma agonists inhibit respiratory syncytial virus-induced expression of intercellular adhesion molecule-1 in human lung epithelial cells. Immunology 121: 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Cencic R, Lindqvist L, Oberer M, Northcote P, Wagner G, Pelletier J (2006a) RNA-mediated sequestration of the RNA helicase eIF4A by pateamine A inhibits translation initiation. Chem Biol 13: 1287–1295 [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J (2006b) Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2: 213–220 [DOI] [PubMed] [Google Scholar]
- Campo PA, Das S, Hsiang CH, Bui T, Samuel CE, Straus DS (2002) Translational regulation of cyclin D1 by 15-deoxy-delta(12,14)-prostaglandin J(2). Cell Growth Differ 13: 409–420 [PubMed] [Google Scholar]
- Chang YC, Li PC, Chen BC, Chang MS, Wang JL, Chiu WT, Lin CH (2006) Lipoteichoic acid-induced nitric oxide synthase expression in RAW 264.7 macrophages is mediated by cyclooxygenase-2, prostaglandin E2, protein kinase A, p38 MAPK, and nuclear factor-kappaB pathways. Cell Signal 18: 1235–1243 [DOI] [PubMed] [Google Scholar]
- Cheng G, Baltimore D (1996) TANK, a co-inducer with TRAF2 of TNF- and CD 40L-mediated NF-kappaB activation. Genes Dev 10: 963–973 [DOI] [PubMed] [Google Scholar]
- Colon-Ramos DA, Shenvi CL, Weitzel DH, Gan EC, Matts R, Cate J, Kornbluth S (2006) Direct ribosomal binding by a cellular inhibitor of translation. Nat Struct Mol Biol 13: 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO (2006) Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem 281: 32870–32878 [DOI] [PubMed] [Google Scholar]
- Fionda C, Nappi F, Piccoli M, Frati L, Santoni A, Cippitelli M (2007) Inhibition of trail gene expression by cyclopentenonic prostaglandin 15-deoxy-{Delta}12,14-PGJ2 in T lymphocytes. Mol Pharmacol 2: 2. [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P (2004) Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy DW, Lawrence T, Perretti M, Rossi AG (2004) Inflammatory resolution: new opportunities for drug discovery. Nat Rev Drug Discov 3: 401–416 [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Yamada Y, Komiyama K, Hayashi M, Ishibashi M, Sunazuka T, Izuhara T, Sugahara K, Tsuruda K, Masuda M, Takasu N, Tsukasaki K, Tomonaga M, Kamihira S (2007) A novel natural compound, a cycloanthranilylproline derivative (Fuligocandin B), sensitizes leukemia cells to apoptosis induced by tumor necrosis factor related apoptosis-inducing ligand (TRAIL) through 15-deoxy-{Delta}12, 14 prostaglandin J2 production. Blood 110: 1664–1674 [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N (2005) Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327 [DOI] [PubMed] [Google Scholar]
- Jackson-Bernitsas DG, Ichikawa H, Takada Y, Myers JN, Lin XL, Darnay BG, Chaturvedi MM, Aggarwal BB (2007) Evidence that TNF-TNFR1-TRADD-TRAF2-RIP-TAK1-IKK pathway mediates constitutive NF-kappaB activation and proliferation in human head and neck squamous cell carcinoma. Oncogene 26: 1385–1397 [DOI] [PubMed] [Google Scholar]
- Jang SK (2006) Internal initiation: IRES elements of picornaviruses and hepatitis C virus. Virus Res 119: 2–15 [DOI] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P (2000) Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Paek KY, Ha SH, Cho S, Choi K, Kim CS, Ryu SH, Jang SK (2004) A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol Cell Biol 24: 7878–7890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WJ, Back SH, Kim V, Ryu I, Jang SK (2005) Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol Cell Biol 25: 2450–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, Willoughby DA, Gilroy DW (2002) Anti-inflammatory lipid mediators and insights into the resolution of inflammation. Nat Rev Immunol 2: 787–795 [DOI] [PubMed] [Google Scholar]
- Lin TH, Yang RS, Tang CH, Lin CP, Fu WM (2007) PPARgamma inhibits osteogenesis via the down-regulation of the expression of COX-2 and iNOS in rats. Bone 4: 4. [DOI] [PubMed] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO (2005) Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell 20: 709–722 [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM (2003) Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem 278: 34864–34873 [DOI] [PubMed] [Google Scholar]
- Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J (2006) Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol Biol Cell 17: 4212–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDunn JE, Cobb JP (2005) That which does not kill you makes you stronger: a molecular mechanism for preconditioning. Sci STKE 2005: pe34. [DOI] [PubMed] [Google Scholar]
- McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ (2005) Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 280: 16925–16933 [DOI] [PubMed] [Google Scholar]
- Oberer M, Marintchev A, Wagner G (2005) Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev 19: 2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MP, Hurtado O, Cardenas A, Bosca L, Castillo J, Davalos A, Vivancos J, Serena J, Lorenzo P, Lizasoain I, Moro MA (2006) Rosiglitazone and 15-deoxy-delta12,14-prostaglandin J2 cause potent neuroprotection after experimental stroke through noncompletely overlapping mechanisms. J Cereb Blood Flow Metab 26: 218–229 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT (1998) A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Shirokikh NE, Hellen CUT (2005) Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. Comptes Rendus Biologies 328: 589–605 [DOI] [PubMed] [Google Scholar]
- Shibata T, Kondo M, Osawa T, Shibata N, Kobayashi M, Uchida K (2002) 15-Deoxy-delta 12,14-prostaglandin J2. A prostaglandin D2 metabolite generated during inflammatory processes. J Biol Chem 277: 10459–10466 [DOI] [PubMed] [Google Scholar]
- Straus DS, Glass CK (2001) Cyclopentenone prostaglandins: new insights on biological activities and cellular targets. Med Res Rev 21: 185–210 [DOI] [PubMed] [Google Scholar]
- Straus DS, Pascual G, Li M, Welch JS, Ricote M, Hsiang CH, Sengchanthalangsy LL, Ghosh G, Glass CK (2000) 15-Deoxy-delta 12,14-prostaglandin J2 inhibits multiple steps in the NF-kappa B signaling pathway. Proc Natl Acad Sci USA 97: 4844–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH (2003) The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 23: 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakowicz H, Yang H-S, Stark C, Wlodawer A, Laronde-Leblanc N, Colburn NH (2005) Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI. RNA 11: 261–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure Legends