Abstract
The small Ras-like GTPase Rap1 has been identified as a regulator of integrin activation and cadherin-mediated cell–cell contacts. Surprisingly, null mutants of RAP-1 in Caenorhabditis elegans are viable and fertile. In a synthetic lethal RNAi screen with C. elegans rap-1 mutants, the Ras-like GTPase ral-1 emerged as one of seven genes specifically required for viability. Depletion of exoc-8 and sec-5, encoding two putative RAL-1 effectors and members of the exocyst complex, also caused lethality of rap-1 mutants, but did not affect wild-type worms. The RAP-1 and the RAL-1/exocyst pathway appear to coordinate hypodermal cell movement and elongation during embryonic development. They mediate their effect in part through targeting the α-catenin homologue HMP-1 to the lateral membrane. Genetic interactions show that the RAP-1 and RAL-1/exocyst pathway also act in parallel during larval stages. Together these data provide in vivo evidence for the exocyst complex as a downstream RAL-1 effector in cell migration.
Keywords: adherens junction, embryogenesis, Ral, Rap1, synthetic lethality
Introduction
Cell migration is an important aspect of various biological processes like development and immune responses. Several members of the Ras-like family of GTPases are involved in the control of cell migration. For example, Rap1 affects migration by enhancing integrin-mediated cell–matrix attachment in many different cell types in vitro (Caron et al, 2000; Katagiri et al, 2000). In addition, Rap1 may inhibit migration of cells by stimulating the formation of cell–cell contacts via E-cadherin in adherens junctions (Hogan et al, 2004; Price et al, 2004). Also VE-cadherin, present in endothelial cells, is under the control of Rap1 (Fukuhara et al, 2006). The current view is that Rap1 acts locally via different effectors to mediate its effect on integrins and cadherins. For example, the Rap1-binding proteins RIAM, RAPL, TIAM and VAV have all been invoked as Rap1 effectors functioning in enhancement of cell–matrix contacts, whereas AF-6 and Cdc42 are prime candidates for regulators of cadherin in adherens junctions (for a review, see Bos, 2005; Kooistra et al, 2007)). However, Rap1 can induce cell polarity in suspension cells (Shimonaka et al, 2003) and this polarizing activity of Rap1 may also contribute to the effects seen on integrins and cadherins. A second Ras-like GTPase involved in cell migration is Ral (Suzuki et al, 2000; Rosse et al, 2006). In its active, GTP-bound form, Ral binds to various effectors that play specific roles in migration. For example, Ral can induce filopodia formation by interacting with the actin-bundling protein filamin (Ohta et al, 1999). Filopodia are often found at the leading edge of migrating cells and outgrowing neurites. Furthermore, active Ral can bind to Sec5 (Brymora et al, 2001) and Exo84 (Moskalenko et al, 2003; Zhang et al, 2005), two proteins present in the exocyst complex that mediates targeting of E-cadherin to the basolateral plasma membrane. Binding of Ral to Sec5 and Exo84, which are most likely present in distinct subcomplexes of the exocyst, is mutually exclusive. Thus, Ral may enhance the formation of a functional exocyst complex at a specific subcellular site by bringing together different subcomplexes of the exocyst, containing either Sec5 or Exo84 (Jin et al, 2005). The exocyst complex was originally discovered in yeast as a protein complex required for polarized secretion at the tip of newly formed buds (TerBush et al, 1996). In vertebrate cells, the exocyst complex mediates polarized delivery of membranes from the Golgi apparatus and recycling endosomes to the basolateral membrane, but not to the apical membrane (Lipschutz and Mostov, 2002; Prigent et al, 2003; Yeaman et al, 2004). Among proteins targeted to the basolateral membranes by the exocyst complex are the LDL receptor and E-cadherin (Grindstaff et al, 1998; Langevin et al, 2005; Shipitsin and Feig, 2004). Targeting of E-cadherin to the basolateral membrane is of special importance in epithelial cells that are about to establish cell–cell contacts with other epithelial cells. Once E-cadherin is stabilized at the membrane by homotypic interactions with E-cadherin on neighboring cells, it may help in targeting more exocyst to the lateral membrane, thereby enhancing the formation of cell–cell contacts and elaborating cell polarity.
In vivo, the requirement for Ras-like GTPases in cell migration is clearly illustrated by the mutant phenotype of Rap1 mutant flies. Embryos, devoid of Rap1 protein, display defects in ventral furrow closure, head involution and migration of mesodermal and presumptive primordial germ cells (Asha et al, 1999). Thus, both morphogenetic events of complete cell layers as well as individual cell migration are disturbed. Also the migration of a group of specialized follicle cells, called border cells, is under the control of Ras-like GTPases in Drosophila. Migration of border cells represents a cadherin-dependent mode of cell movement and requires signaling via Ras and via Ral (Lee et al, 1996). It is presently unclear which Ral effector is required in this process.
A major migration event in Caenorhabditis elegans is that of hypodermal cells during embryonic development. Hypodermal cells are the epidermal cells in C. elegans. They arise as a series of six rows at the dorsal side of the embryo. The two dorsal-most arrays of cells undergo a process called dorsal intercalation, which organizes them into a single dorsal row. After this process has begun, the ventral-most cells on each side start to migrate ventrally to envelope the embryo. In this process of ventral enclosure, the two most anteriorly located cells, named leading cells, are crucial and the first to establish cell–cell contacts with their contralateral neighbors (Williams-Masson et al, 1998; for a review Simske and Hardin, 2001). In between the dorsal and ventral cells, a row of lateral epidermal cells, called seam cells, is present. After ventral enclosure is completed, the ovoid embryo elongates about fourfold. This process relies on circumferentially oriented actin bundles that are linked at the apical site of adherens junctions in hypodermal cells. In embryos mutant for the α-catenin homologue HMP-1 or the β-catenin homologue HMP-2, the circumferential actin bundles in dorsal hypodermal cells detach from adherens junctions, which probably explains the elongation phenotype seen in these animals (Costa et al, 1998).
So far, relatively few gene products have been identified, which play a crucial role in ventral migration of hypodermal cells. Among them are HMP-1, HMP-2 and the cadherin HMR-1, which are physically interacting in the cadherin–catenin complex (CCC). In HMR-1 homozygous embryos, or embryos devoid of maternal HMP-1 or -2 protein, the leading cells do not migrate completely to the ventral side (Costa et al, 1998). Other proteins involved are the APC-related protein APR-1 that may also be present in the CCC, the inositol 1,4,5-triphosphate receptor ITR-1 and ephrin receptors and ligands (George et al, 1998; Hoier et al, 2000; Thomas-Virnig et al, 2004). These latter proteins are required in the neuroblast cells, over which the hypodermal cells migrate. Also the cytoskeletal regulatory WAVE/WASP proteins are required for normal hypodermal cell migration (Withee et al, 2004). The interconnectivity of these different genes has not been established. Remarkably, no role for any of the C. elegans integrins has been established in hypodermal cell migration (Cox and Hardin, 2004).
In the present study, we have performed a synthetic lethal RNAi screen with a C. elegans rap-1 mutant, which revealed that rap-1 mutants are highly sensitive to diminished signaling via the RAL-1/exocyst pathway. During embryogenesis, the Ras-like GTPases RAP-1 and RAL-1 act in concert to orchestrate hypodermal cell migration and sorting. Interfering in the Ral-1/exocyst pathway in a rap-1 mutant background leads to loss of the CCC at adherens junctions. Interestingly, the observed phenotype is more severe than that of CCC mutants alone, indicating additional roles of the Ral-1/exocyst pathway. Furthermore, our screen has identified various other genes that may be involved in the RAL-1/exocyst pathway or otherwise are required for viability of rap-1 mutants.
Results
RAP-1 and RAP-2 are not required during C. elegans embryonic development
In addition to our previously described rap-1(pk2082) allele, a second rap-1 allele (tm861) has been isolated, in which a 549 bp deletion removes exon 3 and part of exon 4, leaving only the first 42 amino acids of RAP-1 intact (Figure 1). Both homozygous mutant strains are viable and fertile, although progression through larval stages is delayed (Pellis-van Berkel et al, 2005; data not shown). Like most rap-1(pk2082);rap-2(gk11) animals, the majority of rap-1(tm861);rap-2(gk11) animals die during late larval stages. The small fraction of double homozygous adults, derived from either rap-1(pk2082/+);rap-2(gk11) or rap-1(tm861/+);rap-2(gk11) animals, is egg-laying defective. Surprisingly, inside such animals viable offspring is present. We, therefore, isolated embryos from N2 or rap-1(tm861);rap-2(gk11) animals and determined the percentage of non-hatched embryos. Although the percentage of non-hatched rap-1(tm861);rap-2(gk11) embryos was higher (19%, n=340) as compared with wild-type embryos (3%, n=365), the majority of double mutants hatched normally. Of the embryos that did not hatch, the majority (73%) had elongated more than twofold. This showed that whereas RAP-1 or RAP-2 is required for normal viability in late larval stages, there is no strict requirement for RAP-1 and RAP-2 during embryogenesis.
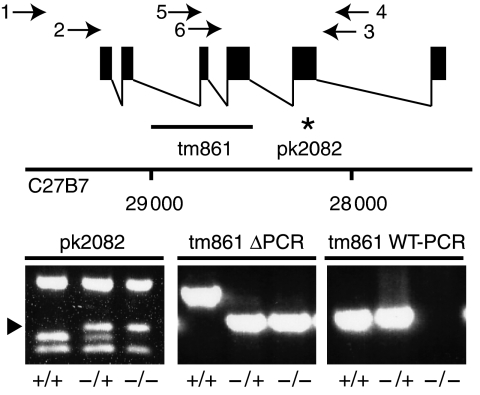
Figure 1.
Schematic representation of the genomic organization of the rap-1 locus and detection of mutant alleles. Filled boxes indicate exons and the line underneath the graphic representation indicates the position of the deletion in rap-1(tm861) animals and the star indicates the amber codon (amino acid 130) in the pk2082 allele. Numbers indicate the position of the rap-1 locus on the C27B7 cosmid. Below are PCR reactions shown, performed on single-worm lysates that demonstrate the presence of the wild-type allele in +/+ and +/− worms and the absence of the wild-type allele in −/− worms. An arrowhead indicates a 170 bp Sau3A product that is characteristic for the pk2082 allele. Arrows indicate the position of the PCR primers used for detection of the deletion and wild-type alleles.
Synthetic lethal screen with rap-1 mutants
The normal development of rap-1 mutants in C. elegans contrasts the situation in Drosophila and this suggests that other signaling pathways functionally compensate for the rap-1 pathway. To investigate this option, a genome-wide synthetic lethal RNAi screen was performed using the Ahringer library (Kamath et al, 2003; van Haaften et al, 2004). Apart from wild type and rap-1(pk2082) mutants, rap-2(gk11) and epac-1(pk1313) mutants were included in this screen. epac-1(pk1313) mutants carry a deletion in the homologue of the cAMP-dependent Rap-specific guanine nucleotide exchange factor (GEF) Epac (T20G5.5; unpublished results) (de Rooij et al, 1998). Foods that caused specific lethality with one of the mutant strains, were rescreened using the same mutants in the case of rap-2 and epac-1 or with both rap-1(pk2082) and rap-1(tm861) (see Supplementary data). In this rescreen, no genes emerged that caused clear and reproducible synthetic lethality with rap-2 or epac-1. In contrast, seven genes caused specific synthetic lethality of rap-1 mutants (Table I). These include the MKP7 homologue vhp-1 (F08B1.1)(Mizuno et al, 2004) and a gene with unknown function C01B7.1. Interestingly, ral-1 and exoc-8 were found that encode homologues of vertebrate exocyst complex members. Finally, the genes encoding the phosphatase sur-6 (Sieburth et al, 1999), the vesicle-sorting protein phi-24 (Howard et al, 2001) and him-3, known to be involved in meiosis, gave synthetic lethality, but with variable results for rap-1(tm861). To exclude the possibility that an additional mutation present in both rap-1 strains was responsible for the observed synthetic lethality, two independent rap-1(tm861) strains, with rap-1 under control of the general let-858 promoter, were generated. Although the level of RAP-1 was too low for detection on blot, we observed a partial rescue of the phenotype when these worms were subjected to exoc-8(RNAi) food as L1-stage animals (Table II).
Table 1.
Overview of genes found in the synthetic lethal screen with rap-1(pk2082)
| Localization | Gene | Mammalian homologue | N2 | rap-1 (pk2082) | rap-2 (gk11) | epac-1 (pk1313) |
|---|---|---|---|---|---|---|
| 90G7 | ral-1 | RalA, RalB | 21 (549) | 0 (0) | 36 (702) | 19 (344) |
| Y53G8AR.3 | 45 (1068) | 0 (0) | 23 (408) | 36 (640) | ||
| 25G8 | exoc-8 | EXOC8 | 38 (984) | 2 (26) | 55 (1087) | 21 (377) |
| Y105E8B.2 | 55 (1314) | 14 (189) | 69 (1204) | 97 (1721) | ||
| 2C2 | phi-24 | CHMP1B | 146 (2198) | 22 (433) | 93 (2300) | 139 (2699) |
| F23C8.6 | 88 (1038) | 30 (256) | 83 (1141) | 50 (813) | ||
| 26 (826) | 9 (158) | 68 (1200) | — | |||
| 16E8 | sur-6 | PR55/B | 59 (892) | 4 (75) | 44 (1101) | 47 (912) |
| F26E4.1 | 47 (550) | 0 (0) | 51 (696) | 53 (614) | ||
| 47 (1533) | 1 (13) | 55 (973) | 46 (740) | |||
| 44D3 | vhp-1 | MKP7 | 91 (2165) | 0 (0) | 38 (935) | 163 (2350) |
| F08B1.1 | 166 (3150) | 8 (72) | 24 (327) | 95 (2282) | ||
| 101G7 | him-3 | HORMAD1 | 54 (819) | 14 (276) | 58 (1424) | 51 (984) |
| ZK381.1 | 108 (1259) | 35 (296) | 68 (1203) | 126 (1472) | ||
| 107 (3479) | 7 (121) | — | 55 (884) | |||
| 104B8 | ARP10 | 137 (2068) | 27 (529) | 79 (1934) | 109 (2129) | |
| C49H3.8 | 74 (2393) | 80 (681) | 100 (1378) | 123 (1437) | ||
| — | 35 (583) | 71 (1257) | 111 (1785) | |||
| 146B2 | Zn-finger | 117 (1760) | 9 (168) | 62 (1545) | 99 (1928) | |
| C01B7.1 | Protein | 102 (1208) | 7 (64) | 96 (1319) | 90 (1054) | |
| 43 (1373) | — | 17 (299) | 20 (330) | |||
| Numbers represent the percentage of progeny relative to those found on control RNAi for each strain as determined in rescreening experiments. Numbers in brackets are total numbers of worms per well at day 7. Successive lines represent independent experiments. | ||||||
Table 2.
rap-1(tm861); dpy-20 animals carrying transgenic rap-1 (rescue 1 and 2) are less sensitive to exoc-8(RNAi) than rap-1(tm861); dpy-20 animals
| Control RNAi | exoc-8 (RNAi)-1 | Control RNAi (%) | |
|---|---|---|---|
| Wild type | 62 | 70 | 113 |
| rap-1(tm861); dpy-20 | 57 | 10 | 17 |
| Rescue 1 | 61 | 36 | 59 |
| Rescue 2 | 65 | 86 | 132 |
| Numbers in the first and second column are absolute numbers of viable progeny after a 16-h egg lay. Numbers are the mean value of progeny of hermaphrodites (n=15 for rescue strain 1 and 2, n=10 for wild-type and rap-1(tm861); dpy-20). The third column shows the percentage of the amount of progeny on exoc-8(RNAi) in comparison with animals subjected to control RNAi. | |||
We focused on RAL-1 and the exocyst member EXOC-8, as their vertebrate homologues RalA and Exo84 have previously been reported to directly interact: in its active GTP-bound form, RalA binds to Exo84. Therefore, we retested the effects of RNAi of other exocyst complex members on the viability of both rap-1 mutants (Supplementary Table I). No or only very limited effects were scored for sec-6, sec-8, sec-15, sec-3 and exoc-7. Although not found to be synthetic lethal in the screen, quantification showed less progeny on sec-10(RNAi). Finally, also sec-5(RNAi) resulted in specific synthetic lethality with both rap-1 alleles.
In vertebrates, Sec5 is identified as a member of the exocyst complex and also as a direct downstream effector of Ral. Other Ral effectors include RalBP, involved in endo- and exocytosis, and the actin-bundling protein filamin. For each of these effectors, a single homologue was found in the C. elegans genome (rlbp-1 (T23G11.5), flna-1 (C23F12.1)). In addition, a single homologue for the family of RALGEFs is found, RGL-1 (F28B4.2). We tested the effect of RNAi feeding of L1 larvae on plate for each of these genes. ral-1 and sec-5(RNAi) prevented rap-1(pk2082) from producing viable offspring, whereas rlbp-1(RNAi) had no effect. flna-1(RNAi) resulted in a high number of sick animals, but this was also seen with wild-type animals (Supplementary Figure 1). Together, these results suggest that full functionality of the ral-1/sec-5/exoc-8 pathway is required in rap-1 mutants, but not in wild-type worms. Further evidence comes from the observation that also rgl-1(RNAi) reduces the number of rap-1 offspring, whereas no effect was seen on wild-type worms (Supplementary Figure 1A; data not shown). A drawback of RNAi studies is that the efficacy of RNAi can be variable. However, when the same set of RNAi constructs was tested in a more RNAi-sensitive background using the rrf-3 strain, similar results were observed (Supplementary Figure 1B, C). Strikingly, a further reduction was seen in the number of rap-1;rrf-3 mutants on rgl-1(RNAi).
Comparative analysis of ral-1/sec-5/exoc-8(RNAi) in rap-1 mutants
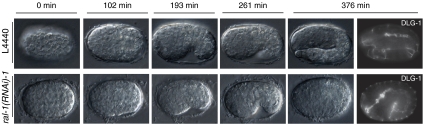
To obtain more definitive proof that ral-1, sec-5 and exoc-8 operate in the same pathway in C. elegans, we compared the phenotypes of rap-1 mutants after RNAi feeding on plate. When rap-1 L1 larvae are subjected to RNAi for any of these genes, most animals make it to adulthood but are sluggish and largely sterile. In contrast, when L4 larvae were used at the start of RNAi, the resulting adults appeared healthy. Initially, they produce viable offspring but then start to shed many embryos that do not hatch. Inspection of these embryos showed a phenotypic series that can be roughly classified into two types (Figure 2). In class I embryos, the hypodermis still covers the entire embryo, but little morphogenesis is seen following the comma stage. Class II embryos appeared more disorganized and hypodermal cells are clustered and do not envelope the embryo. Hypodermal cell junctions have been characterized in much detail and consist of a single electron-dense junction, in which an apical and basal domain can be discerned by immunofluorescence. The apical domain of the junction harbors the CCC, whereas the basal domain contains the membrane-associated protein DLG-1 and its binding partner AJM-1. We used DLG-1 and AJM-1 localization to visualize adherens junctions in the hypodermis and developing gut, which allows the simultaneous evaluation of the development of both tissues relative to each other (Segbert et al, 2004). The pattern and organization of hypodermal cells in rap-1 mutant worms on control RNAi is identical to that of wild-type worms and the dorsal, seam and ventral cells form regular arrays of cells (Figure 2A; reviewed by Simske and Hardin, 2001). In contrast, many of the class I embryos obtained with ral-1, sec-5 or exoc-8(RNAi) are characterized by a failure of cells to align in the stereotyped pattern of dorsal, seam and ventral cells (Figure 2B–D; Supplementary Figure 2). 3D-animation indicates that this results from defects in both dorsal intercalation and ventral migration (data not shown). In other class I embryos, migration appears to have stopped before reaching the ventral midline. Analysis of pharyngeal and intestinal cells of these embryos showed that these tissues are not affected (Figure 2F, G), although in rare cases abnormalities were seen (Figure 2H). Overall, the phenotypes of ral-1, sec-5 or exoc-8(RNAi) are very similar. In class II embryos, DLG-1 and AJM-1 staining showed the presence of disorganized patches of hypodermal cells at various sites, but both proteins still colocalized (Figure 2I–L, sec-5 and exoc-8(RNAi); data not shown). The intestine of these embryos was frequently recognizable on the exterior, indicating that despite the severe morphogenetic defect, cell specification of this tissue had occurred normally (arrowhead Figure 2K). To study cell specification of hypodermal cells in class II embryos, we used the seam cell marker SCM∷GFP, which is specifically expressed in 10 hypodermal seam cells on each lateral side from the 1.5-fold stage onwards (Koh and Rothman, 2001). GFP-positive cells were detected in the aberrantly localized hypodermal cell clusters following ral-1, sec-5 or exoc-8(RNAi) feeding of rap-1(tm861) animals, showing that specification of at least a subset of hypodermal cell fates had occurred in these embryos (Figure 3, showing a single focal plane). However, the number of SCM∷GFP-positive cells was reduced in some cases. Whether this is the consequence of altered cell division, defects in specification or loss to cell death is presently not clear. In conclusion, the phenotype of rap-1 mutants on ral-1, sec-5 and exoc-8(RNAi) is consistent with the fact that these latter three genes operate in a single pathway involved in hypodermal cell organization.
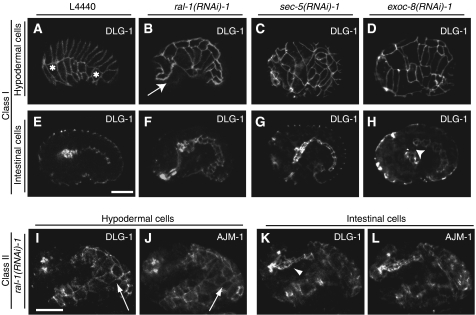
Figure 2.
Phenotype of rap-1 mutant embryos carrying the dlg-1∷GFP gene subjected to control, ral-1, sec-5 or exoc-8(RNAi). Phenotypes of rap-1 mutant embryos carrying the dlg-1∷GFP marker (FZ271) that were derived from animals subjected to RNAi for ral-1, sec-5 or exoc-8. DLG-1 indicates dlg-1∷GFP expression, AJM-1 indicates immunofluorescence staining using the MH27 antibody. Genes used for RNAi are indicated above each panel. L4440 is the empty vector control RNAi. (A–H) show class I embryos, (I–L) class II embryos. (A–D) and (I–J) show focal planes to visualize hypodermal cells, (E–H) and (K–L) are focal planes at the level of the gut. The most anterior and posterior visible seam cells are marked with *. In all cases, AJM staining was performed (only shown for class II, subjected to ral-1 (RNAi)), which demonstrated clear colocalization with DLG-1∷GFP). Class I embryos are characterized by halted migration of hypodermal cells as examplified by a ral-1(RNAi) embryo (arrow in B) or misalignment of hypodermal cells as shown here for a sec-5 and exoc-8(RNAi) embryo (C, D). Halted migration can be discerned from early stages of migration by comparison of the developmental stage of the gut. Note that during normal hypodermal cell migration (A, E), the gut has not yet extended along the entire length of the embryos, in contrast to the gut of the ral-1(RNAi) embryo, which has established clear adherens junctions and shows a more mature pharynx (B, F). In some cases, a gap between intestinal and pharyngeal cells is observed (arrowhead H). In class II embryos, hypodermal cells do not envelope the entire embryo (arrow I, J) and a clearly recognizable gut structure is found on the outside of the embryo (arrowhead K). All embryos are oriented such that their pharynx is on the left side. Scale bar: 10 μm.
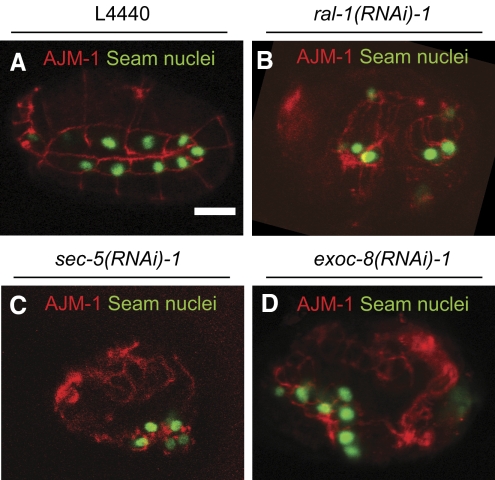
Figure 3.
Phenotype of rap-1 mutant embryos carrying the SCM∷GFP gene subjected to control, ral-1, sec-5 or exoc-8(RNAi) (A–D respectively). Expression of the seam cell marker SCM∷GFP in nuclei of the disorganized hypodermis of class II embryos. Hypodermal cell borders are visualized using the AJM-1 antibody (red). Only one focal plane is shown per condition, seam nuclei are also present in other slices (data not shown). Scale bar: 10 μm.
Localization of HMP-1
Certain aspects of the RNAi phenotypes like the arrested migration of hypodermal cells seemed consistent with a defect in CCC function. We, therefore, investigated whether HMP-1 was normally located at adherens junctions. HMP-1 colocalized with DLG-1 in wild type on control, ral-1, sec-5 or exoc-8 (RNAi) and rap-1 mutants on control RNAi. In contrast, in almost all rap-1 mutant embryos on ral-1, sec-5 or exoc-8(RNAi), a diffuse distribution of HMP-1 was observed (Figure 4). So these gene products are required for targeting HMP-1 to adherens junctions or for stabilizing it there. This diffuse staining was clearly distinct from that of EEA-1 (early endosome) and RAB-11 (recycling endosome) staining (Andrews and Ahringer, 2007; Poteryaev et al, 2007; Supplementary Figure 3). Genetic disruption of CCC members has been shown to result in elongation defects. We, therefore, performed a limited time lapse analysis of rap-1 mutants on ral-1(RNAi). Elongation in such embryos is almost completely blocked, whereas the development of the intestine and pharynx is not disturbed (Figure 5). Loss of HMR-1 or maternal and embryonic HMP-1 or -2 causes migration defects of hypodermal cells, but has not been reported to cause dorsal intercalation defects or to result in an abnormal alignment of dorsal, lateral or ventral cells (Costa et al, 1998). In this respect, the defects seen in rap-1 mutants after ral-1, sec-5 or exoc-8(RNAi) appear more severe. Possibly, deletion of rap-1 enhances the phenotype of hmr-1 mutants. Alternatively, the RAL-1/exocyst pathway targets more proteins than those of the CCC to the membrane, which are also involved in cell migration. To investigate this, we constructed hmr-1;rap-1 double mutants. Double homozygous embryos, derived from rap-1(tm861);hmr-1(zu389/+) hermaphrodites did not differ from those derived from hmr-1(zu389/+) hermaphrodites (Supplementary Figure 4). Due to the fragility of hmr-1 embryos, we were not able to stain sufficient numbers of embryos for adherens junction markers. As loss of maternal HMP-1 protein from hmp-1 homozygous embryos strongly enhances their phenotype to resemble that of hmr-1 embryos, we also investigated the effect of loss of RAP-1 in hmp-1(zu278) embryos. Again, double homozygous embryos could not be discerned from hmp-1 mutants (Supplementary Figure 4). Together, these results indicate that the aberrant organization of hypodermal cells in rap-1 embryos seen after ral-1, exoc-8 or sec-5(RNAi) is not a simple compound phenotype of rap-1 and members of the CCC.
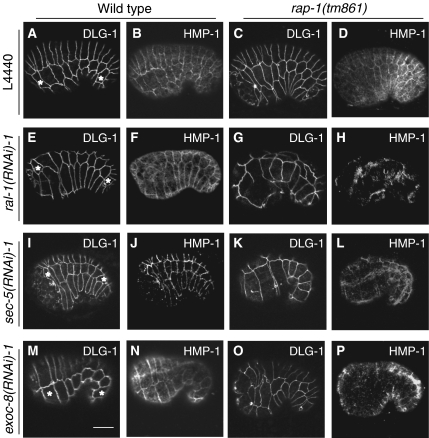
Figure 4.
Localization of HMP-1 in wild-type or rap-1 mutant embryos subjected to control, ral-1, sec-5 or exoc-8(RNAi). dlg-1∷GFP and HMP-1 staining in a lateral view of wild type carrying dlg-1∷GFP (FZ224) (A, B, E, F, I, J, M, N) and rap-1 mutant embryos (FZ271) (C, D, G, H, K, L, O, P), derived from animals subjected to either control L4440 (A–D), ral-1(RNAi) (E–H), sec-5(RNAi) (I–L) and exoc-8(RNAi) (M–P). Pictures show hypodermal cells in which DLG-1 indicates dlg-1∷GFP expression and HMP-1 indicates immunofluorescence staining using the P1E11 antibody. The most anterior and posterior visible seam cells are marked with *. Scale bar: 10 μm.
Figure 5.
Time-lapse analysis of rap-1 mutant embryos subjected to control and ral-1(RNAi). Time lapse recordings of embryos derived from dlg-1∷GFP;rap-1(tm861) mutant (FZ271) embryos on control L4440 (upper panels) or ral-1(RNAi) (lower panels). Last timepoint also shows DLG-1∷GFP expression in the gut of the same embryo.
Genetic interactions between the RAP-1 and RAL-1/exocyst pathway
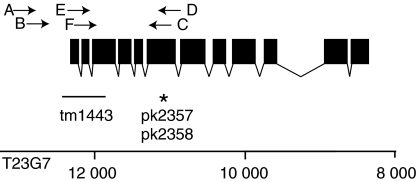
To obtain additional proof for genetic interactions between the RAP-1 and RAL-1 pathways in C. elegans, we studied three independent mutants. sec-5(pk2357) and sec-5(pk2358) carry the same amber mutation at amino acid position 389 and were obtained with target selected mutagenesis (Cuppen et al, 2007; Figure 6). Animals homozygous for sec-5(pk2357) or sec-5(pk2358) become adult but produce only few embryos that elongate normally but never progress to the L2 stage. These sec-5 mutations are hypomorphic based on the fact that when placed over a deficiency (mnDf67) most adults do not produce elongated embryos. The sec-5(tm1443) allele is most likely a null, as it has a 385 bp deletion removing the first two exons and part of the third (Figure 6). Indeed, sec-5(tm1443) mutants have a more severe phenotype: homozygotes never produce progeny and die as late L4-stage larvae.
Figure 6.
Schematic representation of the genomic organization of the sec-5 locus. Filled boxes indicate exons and the line underneath the graphic representation indicates the position of the deletion in sec-5(tm1443) animals and the star indicates the stop codon (amino acid 389) in the pk2357 and pk2358 allele. Numbers indicate the position of the rap-1 locus on the T23G7 cosmid. Arrows indicate the position of the PCR primers used for detection of the deletion and wild-type alleles.
When introduced into a rap-1(tm861) mutant background, the phenotype of all sec-5 alleles was clearly enhanced, showing RAP-1 and SEC-5 act in different pathways. sec-5(pk2357);rap-1(tm861) double homozygotes arrested as late L3 early L4 larvae (n=23, confirmed by PCR). Importantly, sec-5(tm1443);rap-1(tm861) die at the L2 (25%) or L3 (72%) stage. This phenotype is less severe than that of rap-1(tm861) animals on sec-5(RNAi). This is most likely due to maternal input of SEC-5, as sec-5(tm1443) homozygous embryos shed by heterozygous animals on sec-5(RNAi) die as embryo (data not shown).
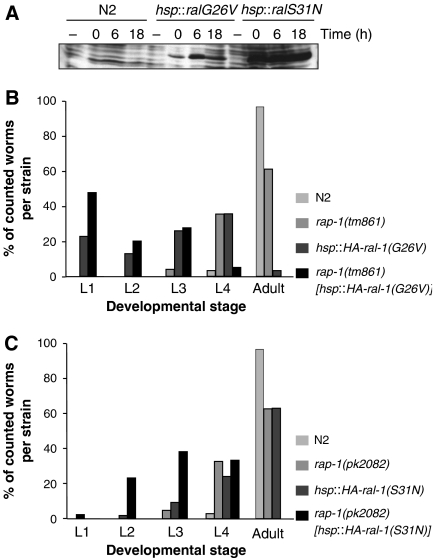
For ral-1, we did not identify mutants with a stop codon. As an alternative, we generated transgenic animals, in which the dominant negative RAL-1(S31N) or the constitutively GTP-bound RAL-1(G26V) are under the control of a heat-shock promoter. Despite being constitutively GTP-bound, mutations corresponding to RAL-1(G26V) (like Ral G23V in mammals), have been described as interfering mutants (Moskalenko et al, 2002; Feig, 2003). Expression of either mutant protein (Figure 7A) did not severely affect these transgenic animals, although it took slightly longer to reach adulthood as compared to heat shocked, wild-type worms. In contrast, expression of RAL-1 mutant proteins in a rap-1 mutant background induced a clear larval arrest, which was strongest for RAL-1(G26V) (Figure 7B, C). Notably, whereas expression of HA-RAL-1(G26V) only delays development in wild-type worms, rap-1 worms expressing the same protein never reach adulthood. The effects of expression of Ral-1(G26V) in a rap-2(gk11) background were not significantly different from those observed in wild-type worms, showing the specificity of the effect of RAL-1(G26V) on rap-1 animals (data not shown). Together these data show that the RAP-1 and RAL-1/exocyst pathway also interact during other stages of the life cycle.
Figure 7.
Effect of heat-shock-driven expression of RAL-1 proteins in wild-type and rap-1 mutant backgrounds. (A) Western blot of total lysates from L1 larvae. Larvae were lysed without heat shock (−) or at 0 h (0), 6 h (6) or 18 h (18) after a 2-h heat shock. Probing was done with the 12CA5 antibody, which recognizes the HA-tag at the N-terminus of overexpressed proteins. (B, C) Embryos subjected to heat shock were allowed to develop 5 days before determination of developmental stage. Percentage of animals at the different developmental stages of the total number of hatched animals is shown for overexpression RAL-1(G26V) and RAL-1(S31N) in (B) and (C), respectively.
Discussion
Synthetic lethal screens are an efficient approach towards identification of signaling pathways that function in a redundant fashion to mediate critical functions during development or any other stage of an organism's life cycle (Tong et al, 2001; van Haaften et al, 2004; Withee et al, 2004). Here, we have used an RNAi feeding-based method to screen for signaling routes that function in conjunction with the RAP-1 pathway in C. elegans. Seven genes were identified, whose full function is required for two distinct rap-1 mutants to produce viable offspring. These genes are synthetic lethal with two independent rap-1 mutations, strongly suggesting that mutations in rap-1 rather than a closely linked mutation is responsible for the observed effect. Indeed, we obtained a partial rescue of the phenotype on exoc-8(RNAi) with lines expressing rap-1 from a heterologous promoter. Synthetic lethality on ral-1(RNAi) that is generally more robust, was not rescued, which we contributed to the low levels of RAP-1 expression from this transgene (data not shown). High levels of RAP-1 expression appear not to be tolerated in C. elegans as is also seen for Rap1 in tissue culture cells (data not shown).
Not all genes may function in the same pathway (see below), but based on literature, it seems likely that ral-1, sec-5 and exoc-8 do so; in vertebrate cells, active Ral directly binds to the exocyst members Sec5 or Exo84 (Brymora et al, 2001; Moskalenko et al, 2003), which results in targeting of proteins, including E-cadherin, to the basolateral membrane (Moskalenko et al, 2002; Shipitsin and Feig, 2004). The fact that Ral binds to Sec5 or Exo84 in a mutually exclusive manner may hint to two distinct pathways. In addition, Ral collaborates with Sec5 in the formation of filopodia (Sugihara et al, 2002). Analysis of embryos derived from rap-1 mutant worms, grown on ral-1, sec-5 or exoc-8(RNAi), showed that they displayed virtually identical phenotypes. This strongly suggests that also in C. elegans these genes function together in hypodermal cell migration.
Rap1 has been claimed to activate the Ral pathway in Drosophila by direct interaction with the RalGEF RGL (Mirey et al, 2003). Also C. elegans RAP-1 and RGL-1 interact in a yeast two-hybrid assay (J Riedl and F Zwartkruis, unpublished results). Although our studies did not exclude the option that RAP-1 contributes to activation of RAL-1 in C. elegans, our current hypothesis is that RAP-1 has a distinct function in parallel to that of RAL-1. This is based on the fact that RAP-1 null mutants have only a very mild phenotype under normal conditions, indicating that RAP-1 is not a major activator of the RAL-1/exocyst pathway. Furthermore, interfering at the level of RAL-1 by expression of mutant RAL-1 proteins has a strong effect in rap-1 mutants but not in wild-type animals. It is possible that RAP-1 converges with RAL-1 pathway at the level of the exocyst complex or below. As no interactions of RAP-1 with the exocyst have been documented, we favor the latter option. In agreement with this, we find that RAP-1 clearly enhances the phenotype of sec-5 mutations.
The defects observed in rap-1 embryos following ral-1, sec-5 or exoc-8(RNAi) are most prominent in hypodermal cell migration during ventral enclosure. It may be that in the most severely affected class II embryos also gastrulation movements are abnormal, but this has not been investigated in any detail. Various causes may underlie the improper alignment and defective migration of hypodermal cells. For example, hypodermal cells need to be specified correctly to accommodate to their correct position. Abnormal cell specification, leading to defects in dorsal intercalation, ventral enclosure and elongation, has been proposed to underlie the phenotype of apr-1 mutants (Hoier et al, 2000). However, cell specification is unlikely to cause the migration defects found in ral-1(RNAi), rap-1 embryos. First, even in class II embryos, expression of a seam cell marker could be detected, which demonstrates that at least some specification within the hypodermis has occurred. Second, we did not observe any abnormal patterning of hypodermal cells before the onset of migration, in contrast to what was reported for apr-1 mutants (data not shown). Finally, the Ral-1/exocyst pathway was found to regulate migration in tissue culture cells, which is unlikely to depend on cell fate specification (Rosse et al, 2006).
A more likely explanation for the observed phenotype is the inability of hypodermal cells to change their adhesive properties that can support the extensive reorganization of cell–cell contacts required for dorsal intercalation and ventral migration. Cell–cell adhesion is of fundamental importance in cell sorting, migration and cell shape changes. This is exemplified in Drosophila, where loss of DE-cadherin in border cells blocks migration and in follicle cells has dramatic effects on cell sorting (Lee et al, 1996; Niewiadomska et al, 1999; Pacquelet and Rorth, 2005). Even loss of DE-cadherin from part of the cell circumference, as seen in clones of Rap1 mutant cells following cell division in the wing, changes their sorting behavior (Knox and Brown, 2002). Indeed, our analysis of proteins present in adherens junctions revealed the absence of HMP-1 and thus most likely the complete CCC (Costa et al, 1998). This indicates that cell adhesive properties have changed. Loss of HMP-1 from adherens junctions upon interfering in the RAL-1/exocyst route is in line with recent studies in vertebrate tissue culture cells and Drosophila (Shipitsin and Feig, 2004; Classen et al, 2005; Langevin et al, 2005). It should be noted, however, that in C. elegans, loss of HMP-1 from adherens junctions upon ral-1(RNAi) was only seen in rap-1 worms and not in wild-type worms. Thus, upon ral-1 depletion, RAP-1 is crucial for targeting the CCC to the lateral membrane or stabilizing it. In mammalian cells, Rap1 has been shown to promote E-cadherin interaction at newly forming adherens junctions in tissue culture cells, whereas it seems not required for the maintenance of mature junctions (Hogan et al, 2004; Price et al, 2004). Possibly, Rap1 acts by inhibiting endocytosis of E-cadherin proteins, which are not yet ligated to E-cadherin on other cells. To do so, Rap1 is suggested to interact via the actin binding protein AF-6 with p120 catenin, an established stabilizer of adherens junctions (Hoshino et al, 2005). Although it is attractive to explain the lack of HMP-1 in adherens junctions in terms of the combined effect of diminished targeting via the RAL-1/exocyst complex and enhanced internalization, due to lack of RAP-1, the situation may be more complex. It should be kept in mind that during reorganization of cell–cell contacts, cadherin is recycled via endosomes (Bryant and Stow, 2004). Indeed, disturbing the function of these endosomes in the Drosophila wing during the period when cells organize themselves into hexagonal arrays, results in a loss of DE-cadherin at selective cell contact sites (Classen et al, 2005). Consequently, loss of HMP-1 may not result from effects of RAP-1 on endocytosis, but on sorting events in recycling endosomes or exit from such endosomes. The situation is further complicated by the observation that the exocyst member Sec5 functions not only in exocytosis, but also in endocytosis (Sommer et al, 2005; Sonnichsen et al, 2005). Indeed, it is currently unclear if and how the exocyst complex interacts with the recycling endosomes. In this respect, phi-24 may be an interesting hit from our screen, as this protein is homologous to the endosomal protein CHMP1 (Howard et al, 2001), which is involved in vesicular sorting.
In contrast to HMP-1, DLG-1 and AJM-1 localized normally to adherens junctions and this is consistent with the finding that the localization of these latter proteins occurs independently of the CCC (Costa et al, 1998; Koppen et al, 2001). In addition, it shows that the RAL-1 and RAP-1 pathways affect only the transport of a subclass of proteins present at the basolateral membrane. Differential effects of inhibition of the exocyst complex on transport to the basolateral membrane have previously been shown in Drosophila, where DE-cadherin transport is blocked but delivery of the septate junction protein Coracle is normal (Langevin et al, 2005). Importantly, class I embryos differ from hmr-1 embryos in that the shape and alignment of hypodermal cells is clearly abnormal. Therefore, the synthetic lethal phenotype observed cannot be simply attributed to loss of the CCC at the adherens junction. Double mutant analysis demonstrates that the phenotype of hmr-1 or hmp-1 embryos does not become more severe if rap-1 is simultaneously absent. Consequently, the RAL-1/exocyst pathway is not only required for targeting the CCC, but has also additional functions.
It will be interesting to learn if and how the other genes uncovered in the synthetic lethal screen fit in the RAL-1/exocyst pathway. At least one gene, vhp-1, has been indirectly linked to Ral signaling. VHP-1 is a phosphatase for the stress-induced kinases KGB-1 and PMK-1, which are homologous to Jnk and p38 MAPK, respectively (Mizuno et al, 2004). Previously, Ral has been found to activate the Jnk-pathway in tissue culture cells (de Ruiter et al, 2000), whereas in Drosophila it was found to act as a negative regulator of Jnk (Sawamoto et al, 1999). Therefore, vhp-1 may act together with RAL-1 in other processes, possibly with different RAL-1 effectors.
In conclusion, a synthetic lethal screen has been used as a starting point to learn more about genes, that function in a redundant fashion with RAP-1. Our data demonstrate a functional overlap for the Ras-like GTPases RAP-1 and RAL-1 during various phases of the life cycle. Using RNAi to time the interference in the RAL-1/exocyst pathway, we identify both GTPases as novel elements in C elegans hypodermal cell migration.
Materials and methods
Worms
General methods for culturing and manipulating worms used were as described previously (Lewis and Fleming, 1995). Worms were cultured on NGM plates at 20°C. Strains, constructs and detection of mutants are described in Supplementary data.
Transgenic animals were obtained by injection of plasmid DNA into the gonads of dpy-20(e1362) animals, or rap-1(tm861);dpy-20(e1362) (Mello et al, 1991). FZ311 and FZ312 were made with pPD103.5 containing untagged cDNA sequence of rap-1 (C27B7.8). Transgenic arrays were integrated by irradiating animals with 40 Gy of gamma radiation from a 137 Cs source (Way et al, 1991). Target-selected mutagenesis for obtaining sec-5 or ral-1 mutants was performed as described previously (Cuppen et al, 2007). Mutant animals were outcrossed at least four times to N2 animals before phenotypic analysis. Determination of viability of N2 and rap-1(tm861);rap-2(gk11) was performed by cutting gravid adults in water and collecting the embryos with a drawn-out pipette. Embryos were counted and non-hatched embryos were scored after 24 h.
The synthetic lethal screen was performed essentially as described previously (van Haaften et al, 2004). However, we used 20–25 L1 larvae per well in 100 μl of M9+ buffer, to which 50 μl of induced bacterial suspension was added. Scoring was done after 7 days either by eye for genome scale screen or by counting the total amount of progeny per well and comparing it to the amount of progeny on control RNAi (L4440).
Antibody staining and time-lapse recording
Antibody staining of embryos was performed as described previously (Bossinger et al, 2001). Antibodies used are: α-HMP-1 (clone P1E11, Chemicon), α-AJM-1 (provided by O Bossinger), mabMH27, mouse, hybridoma supernatant), EEA-1 (provided by B Grant), RAB-11 (provided by A Spang) and the Alexa anti-mouse 568 secondary antibody (Jackson ImmRes Lab). Embryos were visualized under a Zeiss Axiokop 2. For time-lapse recordings, L4 animals were put on an RNAi feeding plate on day 1. On day 3, adults were moved to a fresh plate and embryos were collected after a 2-h egg lay. Embryos were mounted on a 3% agarose pad in M9+ buffer, covered with a coverslip and sealed with Vaseline.
Heat shock protocol and Western blotting
For heat shock experiments, hermaphrodites were allowed to lay eggs for 2 h (day 0). At 1 h after removal of the adults, the plates were incubated at 33°C for 30 min. On days 1, 2 and 3 a 2-h heat shock was given. Detection of HA-tagged RAL-1 in lysates of heat-shocked animals was done by immunoblotting using the 12CA5mAb against HA (Pellis-van Berkel et al, 2005).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Data
Supplementary Table I
Supplementary Information
Acknowledgments
We thank R Vulders for technical assistance, Dr S Mitani for the rap-1(tm861) and sec-5(tm1443) mutant, Dr M Labouesse for the DLG-1∷GFP strain, Dr J Ahringer, Dr A Spang and Dr B Grant for antibodies. Some nematode strains used were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR) and we thank T Stiernagle for sending these. We thank Dr O Bossinger for valuable discussions and teaching us antibody labeling of embryos, Dr A Fire for making available heat shock constructs and J Mudde for technical assistance in target-selected mutagenesis. This work was supported by the CBG (EF, WP-vB, FZ).
References
- Andrews R, Ahringer J (2007) Asymmetry of early endosome distribution in C. elegans embryos. PLoS ONE 2: e493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha H, de Ruiter ND, Wang MG, Hariharan IK (1999) The Rap1 GTPase functions as a regulator of morphogenesis in vivo. EMBO J 18: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL (2005) Linking Rap to cell adhesion. Curr Opin Cell Biol 17: 123–128 [DOI] [PubMed] [Google Scholar]
- Bossinger O, Klebes A, Segbert C, Theres C, Knust E (2001) Zonula adherens formation in Caenorhabditis elegans requires dlg-1, the homologue of the Drosophila gene discs large. Dev Biol 230: 29–42 [DOI] [PubMed] [Google Scholar]
- Bryant DM, Stow JL (2004) The ins and outs of E-cadherin trafficking. Trends Cell Biol 14: 427–434 [DOI] [PubMed] [Google Scholar]
- Brymora A, Valova VA, Larsen MR, Roufogalis BD, Robinson PJ (2001) The brain exocyst complex interacts with RalA in a GTP-dependent manner: identification of a novel mammalian Sec3 gene and a second Sec15 gene. J Biol Chem 276: 29792–29797 [DOI] [PubMed] [Google Scholar]
- Caron E, Self AJ, Hall A (2000) The GTPase Rap1 controls functional activation of macrophage integrin alphaMbeta2 by LPS and other inflammatory mediators. Curr Biol 10: 974–978 [DOI] [PubMed] [Google Scholar]
- Classen AK, Anderson KI, Marois E, Eaton S (2005) Hexagonal packing of Drosophila wing epithelial cells by the planar cell polarity pathway. Dev Cell 9: 805–817 [DOI] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR (1998) A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol 141: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox EA, Hardin J (2004) Sticky worms: adhesion complexes in C. elegans. J Cell Sci 117: 1885–1897 [DOI] [PubMed] [Google Scholar]
- Cuppen E, Gort E, Hazendonk E, Mudde J, van de Belt J, Nijman IJ, Guryev V, Plasterk RH (2007) Efficient target-selected mutagenesis in Caenorhabditis elegans: toward a knockout for every gene. Genome Res 17: 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J, Zwartkruis FJ, Verheijen MH, Cool RH, Nijman SM, Wittinghofer A, Bos JL (1998) Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396: 474–477 [DOI] [PubMed] [Google Scholar]
- de Ruiter ND, Wolthuis RM, van Dam H, Burgering BM, Bos JL (2000) Ras-dependent regulation of c-Jun phosphorylation is mediated by the Ral guanine nucleotide exchange factor-Ral pathway. Mol Cell Biol 20: 8480–8488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA (2003) Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol 13: 419–425 [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Sakurai A, Yamagishi A, Sako K, Mochizuki N (2006) Vascular endothelial cadherin-mediated cell–cell adhesion regulated by a small GTPase, Rap1. J Biochem Mol Biol 39: 132–139 [DOI] [PubMed] [Google Scholar]
- George SE, Simokat K, Hardin J, Chisholm AD (1998) The VAB-1 Eph receptor tyrosine kinase functions in neural and epithelial morphogenesis in C. elegans. Cell 92: 633–643 [DOI] [PubMed] [Google Scholar]
- Grindstaff KK, Yeaman C, Anandasabapathy N, Hsu SC, Rodriguez-Boulan E, Scheller RH, Nelson WJ (1998) Sec6/8 complex is recruited to cell–cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell 93: 731–740 [DOI] [PubMed] [Google Scholar]
- Hogan C, Serpente N, Cogram P, Hosking CR, Bialucha CU, Feller SM, Braga VM, Birchmeier W, Fujita Y (2004) Rap1 regulates the formation of E-cadherin-based cell–cell contacts. Mol Cell Biol 24: 6690–6700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoier EF, Mohler WA, Kim SK, Hajnal A (2000) The Caenorhabditis elegans APC-related gene apr-1 is required for epithelial cell migration and Hox gene expression. Genes Dev 14: 874–886 [PMC free article] [PubMed] [Google Scholar]
- Hoshino T, Sakisaka T, Baba T, Yamada T, Kimura T, Takai Y (2005) Regulation of E-cadherin endocytosis by nectin through afadin, Rap1, and p120ctn. J Biol Chem 280: 24095–24103 [DOI] [PubMed] [Google Scholar]
- Howard TL, Stauffer DR, Degnin CR, Hollenberg SM (2001) CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J Cell Sci 114: 2395–2404 [DOI] [PubMed] [Google Scholar]
- Jin R, Junutula JR, Matern HT, Ervin KE, Scheller RH, Brunger AT (2005) Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J 24: 2064–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Katagiri K, Hattori M, Minato N, Irie S, Takatsu K, Kinashi T (2000) Rap1 is a potent activation signal for leukocyte function-associated antigen 1 distinct from protein kinase C and phosphatidylinositol-3-OH kinase. Mol Cell Biol 20: 1956–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox AL, Brown NH (2002) Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science 295: 1285–1288 [DOI] [PubMed] [Google Scholar]
- Koh K, Rothman JH (2001) ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development 128: 2867–2880 [DOI] [PubMed] [Google Scholar]
- Kooistra MR, Dube N, Bos JL (2007) Rap1: a key regulator in cell–cell junction formation. J Cell Sci 120: 17–22 [DOI] [PubMed] [Google Scholar]
- Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, Hardin JD (2001) Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol 3: 983–991 [DOI] [PubMed] [Google Scholar]
- Langevin J, Morgan MJ, Sibarita JB, Aresta S, Murthy M, Schwarz T, Camonis J, Bellaiche Y (2005) Drosophila exocyst components Sec5, Sec6, and Sec15 regulate DE-Cadherin trafficking from recycling endosomes to the plasma membrane. Dev Cell 9: 355–376 [DOI] [PubMed] [Google Scholar]
- Lee T, Feig L, Montell DJ (1996) Two distinct roles for Ras in a developmentally regulated cell migration. Development 122: 409–418 [DOI] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT (1995) Basic culture methods. Methods Cell Biol 48: 3–29 [PubMed] [Google Scholar]
- Lipschutz JH, Mostov KE (2002) Exocytosis: the many masters of the exocyst. Curr Biol 12: R212–R214 [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J 10: 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirey G, Balakireva M, L'Hoste S, Rosse C, Voegeling S, Camonis J (2003) A Ral guanine exchange factor-Ral pathway is conserved in Drosophila melanogaster and sheds new light on the connectivity of the Ral, Ras, and Rap pathways. Mol Cell Biol 23: 1112–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Hisamoto N, Terada T, Kondo T, Adachi M, Nishida E, Kim DH, Ausubel FM, Matsumoto K (2004) The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J 23: 2226–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA (2002) The exocyst is a Ral effector complex. Nat Cell Biol 4: 66–72 [DOI] [PubMed] [Google Scholar]
- Moskalenko S, Tong C, Rosse C, Mirey G, Formstecher E, Daviet L, Camonis J, White MA (2003) Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem 278: 51743–51748 [DOI] [PubMed] [Google Scholar]
- Niewiadomska P, Godt D, Tepass U (1999) DE-Cadherin is required for intercellular motility during Drosophila oogenesis. J Cell Biol 144: 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP (1999) The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci USA 96: 2122–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacquelet A, Rorth P (2005) Regulatory mechanisms required for DE-cadherin function in cell migration and other types of adhesion. J Cell Biol 170: 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellis-van Berkel W, Verheijen MH, Cuppen E, Asahina M, de Rooij J, Jansen G, Plasterk RH, Bos JL, Zwartkruis FJ (2005) Requirement of the Caenorhabditis elegans RapGEF pxf-1 and rap-1 for epithelial integrity. Mol Biol Cell 16: 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D, Fares H, Bowerman B, Spang A (2007) Caenorhabditis elegans SAND-1 is essential for RAB-7 function in endosomal traffic. EMBO J 26: 301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price LS, Hajdo-Milasinovic A, Zhao J, Zwartkruis FJ, Collard JG, Bos JL (2004) Rap1 regulates E-cadherin-mediated cell–cell adhesion. J Biol Chem 279: 35127–35132 [DOI] [PubMed] [Google Scholar]
- Prigent M, Dubois T, Raposo G, Derrien V, Tenza D, Rosse C, Camonis J, Chavrier P (2003) ARF6 controls post-endocytic recycling through its downstream exocyst complex effector. J Cell Biol 163: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosse C, Hatzoglou A, Parrini MC, White MA, Chavrier P, Camonis J (2006) RalB mobilizes the exocyst to drive cell migration. Mol Cell Biol 26: 727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto K, Winge P, Koyama S, Hirota Y, Yamada C, Miyao S, Yoshikawa S, Jin MH, Kikuchi A, Okano H (1999) The Drosophila Ral GTPase regulates developmental cell shape changes through the Jun NH(2)-terminal kinase pathway. J Cell Biol 146: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segbert C, Johnson K, Theres C, van Furden D, Bossinger O (2004) Molecular and functional analysis of apical junction formation in the gut epithelium of Caenorhabditis elegans. Dev Biol 266: 17–26 [DOI] [PubMed] [Google Scholar]
- Shimonaka M, Katagiri K, Nakayama T, Fujita N, Tsuruo T, Yoshie O, Kinashi T (2003) Rap1 translates chemokine signals to integrin activation, cell polarization, and motility across vascular endothelium under flow. J Cell Biol 161: 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M, Feig LA (2004) RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol Cell Biol 24: 5746–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth DS, Sundaram M, Howard RM, Han M (1999) A PP2A regulatory subunit positively regulates Ras-mediated signaling during Caenorhabditis elegans vulval induction. Genes Dev 13: 2562–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simske JS, Hardin J (2001) Getting into shape: epidermal morphogenesis in Caenorhabditis elegans embryos. BioEssays 23: 12–23 [DOI] [PubMed] [Google Scholar]
- Sommer B, Oprins A, Rabouille C, Munro S (2005) The exocyst component Sec5 is present on endocytic vesicles in the oocyte of Drosophila melanogaster. J Cell Biol 169: 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R et al. (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469 [DOI] [PubMed] [Google Scholar]
- Sugihara K, Asano S, Tanaka K, Iwamatsu A, Okawa K, Ohta Y (2002) The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat Cell Biol 4: 73–78 [DOI] [PubMed] [Google Scholar]
- Suzuki J, Yamazaki Y, Li G, Kaziro Y, Koide H (2000) Involvement of Ras and Ral in chemotactic migration of skeletal myoblasts. Mol Cell Biol 20: 4658–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P (1996) The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J 15: 6483–6494 [PMC free article] [PubMed] [Google Scholar]
- Thomas-Virnig CL, Sims PA, Simske JS, Hardin J (2004) The inositol 1,4,5-trisphosphate receptor regulates epidermal cell migration in Caenorhabditis elegans. Curr Biol 14: 1882–1887 [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- van Haaften G, Vastenhouw NL, Nollen EA, Plasterk RH, Tijsterman M (2004) Gene interactions in the DNA damage-response pathway identified by genome-wide RNA-interference analysis of synthetic lethality. Proc Natl Acad Sci USA 101: 12992–12996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way JC, Wang L, Run JQ, Wang A (1991) The mec-3 gene contains cis-acting elements mediating positive and negative regulation in cells produced by asymmetric cell division in Caenorhabditis elegans. Genes Dev 5: 2199–2211 [DOI] [PubMed] [Google Scholar]
- Williams-Masson EM, Heid PJ, Lavin CA, Hardin J (1998) The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev Biol 204: 263–276 [DOI] [PubMed] [Google Scholar]
- Withee J, Galligan B, Hawkins N, Garriga G (2004) Caenorhabditis elegans WASP and Ena/VASP proteins play compensatory roles in morphogenesis and neuronal cell migration. Genetics 167: 1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman C, Grindstaff KK, Nelson WJ (2004) Mechanism of recruiting Sec6/8 (exocyst) complex to the apical junctional complex during polarization of epithelial cells. J Cell Sci 117: 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zajac A, Zhang J, Wang P, Li M, Murray J, TerBush D, Guo W (2005) The critical role of Exo84p in the organization and polarized localization of the exocyst complex. J Biol Chem 280: 20356–20364 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Data
Supplementary Table I
Supplementary Information