Abstract
Proneural basic helix–loop–helix proteins are key regulators of neurogenesis but their ‘proneural' function is not well understood, partly because primary targets have not been systematically defined. Here, we identified direct transcriptional targets of the bHLH proteins Neurogenin and NeuroD and found that primary roles of these transcription factors are to induce regulators of transcription, signal transduction, and cytoskeletal rearrangement for neuronal differentiation and migration. We determined targets induced in both Xenopus and mouse, which represent evolutionarily conserved core mediators of Neurogenin and NeuroD activities. We defined consensus sequences for Neurogenin and NeuroD binding and identified responsive enhancers in seven shared target genes. These enhancers commonly contained clustered, conserved consensus-binding sites and drove neural-restricted transgene expression in Xenopus embryos. We then used this enhancer signature in a genome-wide computational approach to predict additional Neurogenin/NeuroD target genes involved in neurogenesis. Taken together, these data demonstrate that Neurogenin and NeuroD preferentially recognize neurogenesis-related targets through an enhancer signature of clustered consensus-binding sites and regulate neurogenesis by activating a core set of transcription factors, which build a robust network controlling neurogenesis.
Keywords: E-box, enhancer, NeuroD, neurogenesis, Neurogenin
Introduction
Transcription factors regulate many biological processes, including cell-fate determination and differentiation during embryonic development. With the completion of genome sequencing in many organisms, a major remaining challenge is to globally define transcriptional regulatory networks underlying complex biological processes. This requires identifying primary targets directly controlled by each transcription factor and then defining how expression of these targets is regulated with the proper specificity in a particular biological context.
Transcription factors of the neural basic helix–loop–helix (bHLH) class, including the Neurogenins (Ngn1/2/3) and NeuroD, are key regulators of vertebrate neurogenesis. The Ngns are required for neuronal commitment, as evidenced by loss of cranial and spinal sensory ganglia and ventral spinal cord neurons in Ngn1 or Ngn2 single- or Ngn1/2 double-mutant mice (Fode et al, 1998; Ma et al, 1998, 1999; Scardigli et al, 2001). Among the downstream mediators of Ngn activity is NeuroD, which is expressed in essentially all areas of the brain, spinal cord, peripheral ganglia, and sense organs that express Ngn1, 2, or 3 (Sommer et al, 1996). NeuroD expression diminishes as neurons mature except in the cerebellum and hippocampus, where its expression is maintained throughout adulthood (Miyata et al, 1999). In contrast to NeuroD's extensive expression within the nervous system, NeuroD-null mice display defects in restricted areas. The NeuroD-null phenotype includes loss of cerebellar and hippocampal granule cells, inner ear sensory neurons, and retinal photoreceptor cells but other regions do not exhibit gross cellular deficits (Miyata et al, 1999; Kim et al, 2001; Pennesi et al, 2003). Taken together, these features suggest that NeuroD is a major mediator of Ngn activities that may act redundantly with other molecules to regulate neuronal differentiation.
bHLH transcription factors act as heterodimers with ubiquitously expressed E proteins and bind sequences with the general consensus CANNTG, termed E-boxes, in their target genes (Bertrand et al, 2002 and references therein). bHLH proteins have different preferences for the two central nucleotides (CANNTG) and so can recognize distinct consensus sequences and additional nucleotides flanking the E-box on either side also contribute to specificity of bHLH–E-box interactions. However, consensus sequences for most neural bHLH proteins have not been systematically determined.
While the expression and activities of the neural bHLH transcription factors have been extensively characterized in many organisms, molecular mechanisms underlying their ability to regulate neurogenesis are not well understood. This is in large part because primary target genes and transcriptional programs that are directly regulated by neural bHLH proteins have not been systematically defined. It is also not clear which regulatory sequence features enable neural bHLH proteins to distinguish among many potential targets in the genome to specifically activate targets relevant to neurogenesis. This is significant as E-box sequences are present in regulatory regions of many non-neural target genes and also occur frequently in the genome by chance.
Here, we addressed these questions by identifying direct transcriptional targets of the Atonal-related neural bHLH proteins Ngnr1 and NeuroD. We used Xenopus animal cap explants, which are multi-potent naive ectoderm that can differentiate into ectodermal, endodermal, or mesodermal cell derivatives. To control transcription factor activity, we used Ngnr1 and NeuroD proteins fused to the human glucocorticoid receptor (GR) hormone-binding domain. In the absence of the ligand dexamethasone (DEX), these fusion proteins are sequestered in the cytoplasm and remain transcriptionally inactive (Kolm and Sive, 1995). Addition of DEX allows GR-fused transcription factors to translocate into the nucleus and regulate target genes. We induced Ngnr1-GR and NeuroD-GR activities in the presence of the protein synthesis inhibitor cycloheximide, which blocks translation and prevents induction of secondary targets, whose expression is regulated by primary target proteins. This strategy effectively defines primary target genes activated directly by transcription factors (for example, Pozzoli et al, 2001; Logan et al, 2005).
Using this approach, we identified direct transcriptional targets of Ngnr1 and NeuroD in neurogenesis. We also determined evolutionarily conserved target genes from Xenopus to mammals, which represent a core machinery to generate Ngn and NeuroD-regulated neurons. We then analyzed these targets, enabling us to (i) understand primary roles of these key transcription factors in neurogenesis, (ii) define a transcriptional network controlling Ngn- and NeuroD-dependent neurogenesis, and (iii) define and characterize enhancers regulated by Ngn and NeuroD in their target genes. Finally, we used the sequence features of these enhancers for genome-wide identification of additional Ngn/NeuroD target genes relevant to neurogenesis.
Results
Identification of direct targets of Ngnr1 and NeuroD in Xenopus ectoderm
To identify Ngnr1 and NeuroD direct targets, we expressed Ngnr1-GR and NeuroD-GR fusion proteins in Xenopus laevis ectodermal explants. In the presence of a protein synthesis inhibitor, Ngnr1 and NeuroD activity was induced for 2.5 h from the early gastrula stage and gene expression was analyzed on Affymetrix Xenopus laevis Genome Arrays. Ngnr1 or NeuroD target genes were determined by comparing DEX-treated versus -untreated Ngnr1-GR- or NeuroD-GR-injected samples. β-Galactosidase-injected, DEX-treated samples (control) were also compared to exclude genes induced by DEX (see Materials and methods and Supplementary data). We validated the microarray results using quantitative real-time RT–PCR (qRT–PCR) to define 57 and 62 genes as direct targets of Ngnr1 and NeuroD, respectively (Table I).
Table 1.
Direct transcriptional targets of Ngnr1 and NeuroD identified in Xenopus
| UniGene number | Gene title | Induction by X-Ngnr1 | Induction by X-NeuroD | Function | ||
|---|---|---|---|---|---|---|
| Microarray | qRT–PCR | Microarray | qRT–PCR | |||
| Genes induced by both Ngnr1 and NeuroD | ||||||
| Xl.13309 | Snail | 23.81 | 88.87 | Transcription factor | ||
| Xl.879 | Twist homolog 1 (Twist1-A) | 8.59 | 8.7 | 15.57 | 667.4 | Transcription factor |
| Xl.1263 | Xenopus atonal homolog (NeuroD4, Xath3) | 6.38 | 3.1 | 14.89 | 296.0 | Transcription factor |
| Xl.450 | Achaete-scute complex-like 1 (Ascl-1A) | 4.99 | 9.8 | 9.54 | 103.5 | Transcription factor |
| Xl.23642 | Zinc finger protein 238 (RP58, Znf238, Zfp238) | 6.82 | 8.7 | 8.73 | 189.8 | Transcription factor |
| Xl.34267 | Transcribed locus | 7.10 | 7.66 | Unknown | ||
| Xl.53293/ Xl.76708 | Transcribed locus, moderately similar to NP_062738.1 angiomotin-like 2 (Amotl2) (Mus musculus) | 4.15 | 5.45 | Unknown | ||
| Xl.68 | Similar to Drosophila delta-like 1 (Delta-2) | 3.51 | 5.4 | 5.00 | 6.5 | Signaling |
| Xl.15502 | Transcribed locus, moderately similar to NP_001087914.1 protein | 3.95 | 4.41 | Unknown | ||
| Xl.16456 | Hypothetical protein MGC68858 (Cbfa2t2-B, MTGR1b) | 2.17 | 3.8 | 3.84 | 6.9 | Transcription factor |
| Xl.958 | Smad-interacting protein 1 (SIP1, Zfhx1b) | 3.38 | 3.79 | Transcription factor | ||
| Xl.3150 | Follistatin-related protein (FRP, Fstl1) | 1.82 | 3.1 | 3.58 | 7.4 | Signaling |
| Xl.34006 | Transcribed locus | 3.87 | 3.48 | Unknown | ||
| Xl.547 | Early B-cell factor 2 (Ebf2-A) | 3.62 | 66.4 | 3.48 | 58.6 | Transcription factor |
| Xl.57163/ Xl.31433 | C2-HC-type zinc finger protein X-MyT1 | 2.04 | 3.29 | 6.8 | Transcription factor | |
| Xl.5041 | Hypothetical protein LOC443700. Similar to solute carrier family 43, member 2/L-type amino-acid transporter 4 (Slc43a2/LAT4) | 1.83 | 3.29 | Amino-acid transporter | ||
| Xl.4522 | Homeobox transcription factor Iroquois 3 (Xiro3, Irx3) | 2.39 | 3.28 | 5.4 | Transcription factor | |
| Xl.15305 | MGC81642 protein. Similar to CIDE-c (FSP27) | 4.05 | 3.4 | 3.10 | 4.9 | Apoptosis |
| Xl.237 | Early B-cell factor 2 (Ebf2-B) | 3.52 | 3.08 | Transcription factor | ||
| Xl.16835 | Clone 31 polo-like kinase 3 (Plx3, Fnk, Plk3) | 2.87 | 3.8 | 2.67 | 4.7 | Signaling |
| Xl.71723 | Transcribed locus, weakly similar to XP_513615.1 similar to maba1 | 1.88 | 2.37 | Unknown | ||
| Xl.9030 | cDNA clone 10C6 (similar to Hes-6) | 1.74 | 2.22 | 4.8 | Transcription factor | |
| Xl.52150 | Transcribed locus | 2.25 | 2.02 | Unknown | ||
| Xl.4960 | Similar to core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (cbfa2t2-A-prov, MTGR1) | 1.84 | 1.90 | 5.8 | Transcription factor | |
| Xl.6849 | Transcribed locus, weakly similar to NP_060261.1 chromosome 14 open reading frame 58 | 2.41 | 3.2 | 1.85 | 3.7 | Unknown |
| Xl.11203 | Transcribed locus | 2.04 | 1.84 | Unknown | ||
| Genes induced by Ngnr1 | ||||||
| Xl.9429 | Transcribed locus | 8.24 | Unknown | |||
| Xl.16264 | MGC80588 protein. Similar to Selenoprotein M | 4.03 | Unknown | |||
| Xl.330 | Neurogenic differentiation 1 (NeuroD-1A) | 3.53 | 14.2 | Transcription factor | ||
| Xl.21807 | Guanine nucleotide binding protein 13, gamma (Gng-13) | 3.41 | 3.5 | Signaling | ||
| Xl.633 | Frizzled homolog 7 (Fzd7) | 3.02 | 1.2 | Signaling/cell polarity | ||
| Xl.63639 | Transcribed locus, moderately similar to NP_080083.2 solute carrier family 16 (monocarboxylic acid transporters), member 9 | 2.95 | Transporter | |||
| Xl.34898 | Transcribed locus, weakly similar to XP_417553.1 similar to hairy and enhancer of split 5 | 2.85 | Transcription factor | |||
| Xl.7751 | Mesendoderm nuclear factor (menF) | 2.64 | Unknown | |||
| Xl.68131 | Hypothetical LOC494650, similar to dihydropyrimidinase-related protein-3 (Dpysl3) | 2.50 | Nucleic acid metabolism | |||
| Xl.12606 | Transcribed locus | 2.44 | Unknown | |||
| Xl.6174 | Riddle 2 (Rdd2) | 2.42 | 2.3 | Unknown | ||
| Xl.12769 | Transcribed locus | 2.39 | Unknown | |||
| Xl.58208 | Hypothetical LOC495465, similar to phosphomannomutase1 (Pmm1) | 2.37 | Mannose biosynthesis | |||
| Xl.53396 | Transcribed locus | 2.34 | Unknown | |||
| Xl.554 | Calponin 2 (Cnn2) | 2.31 | 3.4 | Cytoskeleton/cell migration | ||
| Xl.21493 | Heat shock protein 70 (Hsp70) | 2.23 | Protein folding | |||
| Xl.18615 | Transcribed locus, weakly similar to XP_425354.1 similar to testis-specific protein kinase 1 (Gallus gallus) | 2.19 | Unknown | |||
| Xl.467 | Similar to RNA-binding region (RNP1, RRM) containing 1 | 2.15 | Unknown | |||
| Xl.25073 | Hypothetical protein MGC131362 | 2.06 | Unknown | |||
| Xl.23649 | Angiotensin receptor-related protein (Agtrl1) | 2.06 | 2.5 | Signaling | ||
| Xl.6457 | Transcribed locus, moderately similar to XP_001088371.1 similar to paraspeckle protein 1 isoform 1 (Macaca mulatta) | 2.00 | Unknown | |||
| Xl.74698 | Transcribed locus | 1.98 | Unknown | |||
| Xl.15669 | Hypothetical protein Loc 733196 | 1.98 | Unknown | |||
| Xl.15844 | Solute carrier family 7 (cationic amino-acid transporter, y+ system), member 8 (Slc7a8) | 1.97 | Amino-acid transporter | |||
| Xl.5942 | CDNA clone IMAGE:7012207, partial cds | 1.93 | Unknown | |||
| Xl.9596 | Transcribed locus, weakly similar to XP_001105794.1 similar to poly(ADP-ribose) polymerase family, member 15 (Macaca mulatta) | 1.93 | Unknown | |||
| Xl.8912 | Similar to GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein (Grasp) | 1.86 | 1.4 | Signaling | ||
| Xl.4618 | Clone S10-37-A10 mRNA sequence splicing factor, arginine/serine-rich 2 (Sfrs2-prov) | 1.82 | RNA binding/ splicing | |||
| Xl.20945 | Eukaryotic initiation factor 5 (eIF-5) | 1.79 | Protein synthesis | |||
| Xl.6143 | Hypothetical protein MGC64314, small GTPase Ran-binding protein 1 (Ranbp1) | 1.76 | Intracellular transport | |||
| Xl.72130 | Transcribed locus | 1.73 | 3.0 | Unknown | ||
| Genes induced by NeuroD | ||||||
| Xl.437 | Transcription factor xHEN1 | 120.71 | Transcription factor | |||
| Xl.1274/ Xl.21449 | Similar to POU domain gene (POU50)/Nrl-34 Pou-homeobox protein (nrl-34A) | 14.54 | 236.5 | Transcription factor | ||
| Xl.49528 | Hypothetical LOC495039 | 4.33 | Unknown | |||
| Xl.15208 | Transcribed locus, weakly similar to XP_001072266.1 similar to protein C8orf4 (thyroid cancer protein 1) (TC-1) (Rattus norvegicus) | 3.91 | Unknown | |||
| Xl.34957 | BTB domain protein (P7E4) | 3.80 | Unknown | |||
| Xl.14733 | Transcribed locus | 3.68 | Unknown | |||
| Xl.24147 | Transcribed locus, moderately similar to NP_036466.1 myosin X | 3.67 | Motor/cytoskeleton | |||
| Xl.5764 | Hypothetical protein MGC78892. Similar to solute carrier family 7/L-amino-acid transporter (Slc7a7) | 3.22 | Amino-acid transporter | |||
| Xl.71386 | Hypothetical protein MGC81163. (similar to UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase 3, B4galt3) | 3.07 | 28.8 | Carbohydrate metabolism | ||
| Xl.8440 | Similar to enhancer of split related (ESR1) | 2.80 | Transcription factor | |||
| Xl.54916 | CDNA clone IMAGE: 6636225, partial cds (similar to delta-like 1) | 2.12 | 7.7 | Signaling | ||
| Xl.35785 | Transcribed locus | 2.09 | Unknown | |||
| Xl.25379 | Transcribed locus, weakly similar to XP_415598.1 PREDICTED: hypothetical protein XP_415598 | 1.89 | Unknown | |||
| Xl.1035 | Ribonucleoprotein (Elavl3, elrC) | 1.88 | 3.6 | RNA binding | ||
| Xl.34 | Angiotensin receptor-like 1b | 1.86 | 3.4 | Unknown | ||
| Xl.47730 | MGC80820 protein | 1.83 | 1.3 | Unknown | ||
| Xl.45029 | MGC84173 protein. Similar to secretory carrier-associated membrane protein 2 (Scamp2) | 1.81 | 2.0 | Vesicular transport | ||
| Xl.23523 | Hypothetical protein MGC53446, similar to zinc finger protein 238 isoform 2 | 1.77 | Transcription factor | |||
| Xl.24275 | MGC82057 protein (similar to spermidine/ spermine N(1)-acetyltransferase 1) | 1.76 | Acetyltransferase | |||
| Xl.72351 | Transcribed locus | 1.71 | Unknown | |||
| Xl.586 | Enhancer of split-related protein-7 (ESR-7) | 1.70 | 2.9 | Transcription factor | ||
| Xl.5263 | MGC82028 protein | 1.70 | 1.2 | Unknown | ||
| Xl.7556 | LIM protein prickle (Prickle1) | 1.69 | 2.9 | Cell polarity/ migration | ||
| Xl.12125 | XGadd45-gamma protein (Gadd45g) | 1.68 | 3.8 | Cell cycle | ||
| Xl.16395 | Hypothetical protein MGC53682 (similar to Gadd45-gamma) | 1.67 | Cell cycle | |||
| Xl.77004 | Transcribed locus | 1.66 | Unknown | |||
| Xl.5886 | Transcribed locus, moderately similar to XP_001091614.1 similar to response gene to complement 32 (Macaca mulatta) | 1.64 | 2.4 | Unknown | ||
| Xl.51207 | MGC81672 protein | 1.59 | 2.4 | Unknown | ||
| Xl.77402/ Xl.50498 | Transcribed locus/Mxi1 (LOC734207) | 1.58 | Unknown | |||
| Xl.49084 | Hypothetical LOC494829, moderately similar to drebrin 1 (Dbn1) | 1.58 | Cytoskeleton | |||
| Xl.7910 | Hypothetical protein MGC115585 | 1.58 | 2.3 | Unknown | ||
| Xl.1717 | Pyruvate dehydrogenase kinase, isoenzyme 2 (PDK2) | 1.57 | 2.6 | Carbohydrate metabolism | ||
| Xl.1419 | Hypothetical protein MGC68691 | 1.56 | 4.7 | Unknown | ||
| Xl.4807 | Hypothetical protein MGC68463 (similar to growth arrest-specific 6, Gas-6) | 1.55 | 2.3 | Cell cycle/migration | ||
| Xl.34014 | Transcribed locus | 1.55 | Unknown | |||
| Xl.8188 | Hypothetical protein MGC81174, similar to LIM and senescent cell antigen-like domains 1 (Lims1, PINCH1) | 1.54 | 2.3 | Cell adhesion/ signaling | ||
Of these, 26 genes were shared direct targets of both Ngnr1 and NeuroD. In total, 52 of the 93 target genes were described previously, while 41 were novel or uncharacterized. Most known genes (41 of 52; 79%) were previously reported to either have a role in or be expressed in neural tissues (including this study; Figure 1). Therefore, as predicted, neural genes are highly enriched in our list. We also obtained genes previously shown to respond to Ngn or NeuroD in various species. For example, Math3, NeuroD, HEN1 (Nhlh1), Dll1 (Delta-1), and Hes-6 expression was reduced or lost in Ngn1 and Ngn2 single-mutant mice (Fode et al, 1998; Ma et al, 1998; Koyano-Nakagawa et al, 2000) and Ebf2, MTGR1, Elavl3, Gadd45g, MyT1, Hes-6, and ESR1 were previously defined as either direct or indirect targets of Ngnr1 and/or NeuroD in Xenopus (Bellefroid et al, 1996; Koyano-Nakagawa et al, 2000; Lamar and Kintner, 2005; Logan et al, 2005). Thus our approach effectively determined direct transcriptional targets of Ngn and NeuroD and demonstrated that several genes previously shown to act downstream of Ngn and/or NeuroD are under their direct transcriptional control.
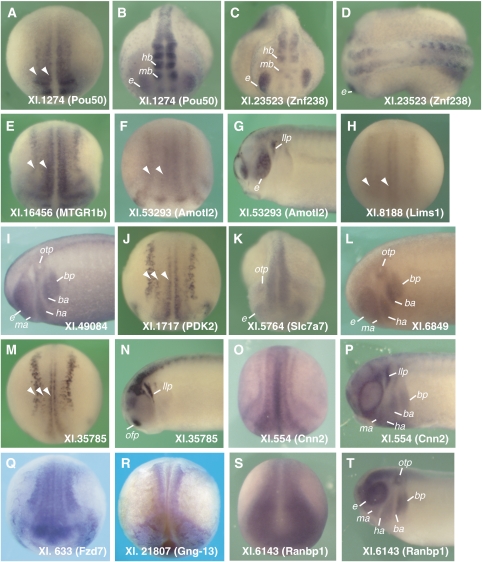
Figure 1.
Expression patterns of novel Ngn and NeuroD target genes. Expression was examined by in situ hybridization of Xenopus embryos at stages 16 (A, E, F, H, J, M, O, Q, R, S) and 22–23 (B–D, G, I, K, L, N, P, T). Embryos are oriented in dorsal view, anterior down (A, E, F, H, J, M, O, Q, R), anterior view (B, C, K, S), and dorsal (D) or lateral (I, L, P, T) views with anterior to the left. Expression was detected in primary neurons (arrowheads), mid-brain (mb), hind-brain (hb), eye (e), branchial arches (posterior (bp), anterior (ba), hyoid (ha), or mandibular (ma) arches), or cranial placodes (otic (otp), olfactory (ofp), or lateral line (llp) placodes). Of the 20 genes analyzed, 16 were expressed predominantly in neural structures (Xl.14733 and Xl.34267 not shown), while four others (not shown) were ubiquitous (Xl.71386), were expressed in other tissues (Xl.15305), or gave a weak signal (Xl.34, Xl.45029).
Analysis of direct targets of Ngnr1 and NeuroD
In total, 33 of 57 Ngnr1 targets were either characterized previously or had strong sequence homology to known genes. Fifteen of these (45%) were transcription factors, including NeuroD, NeuroD4 (Ath3), Ebf2, MTGR1, and Hes6, which were previously shown to regulate neurogenesis (Lee et al, 1995; Bellefroid et al, 1996; Dubois et al, 1998; Perron et al, 1999; Koyano-Nakagawa et al, 2000; Pozzoli et al, 2001; Garcia-Dominguez et al, 2003; Koyano-Nakagawa and Kintner, 2005). Ngnr1 also induced seven direct targets (21%) involved in signal transduction. These data indicate that the major role of Ngnr1 in neurogenesis is to induce regulators of gene expression and signaling, consistent with Ngnr1's role as a proneural gene that acts at the commitment/determination step.
For NeuroD, 37 of 62 target genes had known functions and, similar to results for Ngnr1, 18 genes (49%) were transcription factors, while five genes (14%) were involved in signal transduction. In contrast, we did not find genes directly involved in neuronal function such as neuropeptides, neurotransmitter receptors, or channels. These data suggest that NeuroD may regulate neuronal differentiation by inducing transcription factor targets, such as Xath3, Ebf3, HEN1, and MTGR1, which then subsequently induce genes controlling neuronal function. Alternatively, the cell context used for our screen may have enriched for targets regulated by NeuroD during its early expression in committed and differentiating primary neuronal progenitors, while being unfavorable to detect NeuroD's later targets in neurons (see also Discussion).
We additionally identified 43 Ngnr1 and 43 NeuroD direct targets that were either novel or had not been previously characterized as having a role in neurogenesis. These included transcriptional regulators, signaling molecules, regulators of cell migration, and/or the actin cytoskeleton, and genes with no attributed homology or function. Interestingly, we defined Calponin2, Frizzled homolog 7, Prickle, Gas-6, Lims1, Myosin 10, Dbn1, Plk3, and Amotl2 as direct targets of Ngnr1 and/or NeuroD. All of these molecules or closely related proteins have defined roles in regulating the actin cytoskeleton and/or mediating morphological and migratory events in cells including neurons (Supplementary Figure S3). During neurogenesis, neuronal cell migration and differentiation temporally coincide but it was unclear whether these processes were regulated independently or coordinately. Our finding of direct targets that regulate both neuronal cell fate and morphology/migration suggests that Ngn and NeuroD directly coordinate these processes at the transcriptional level. This is consistent with results of Ge et al (2006), suggesting that Ngn can control the expression of several cell migration regulators.
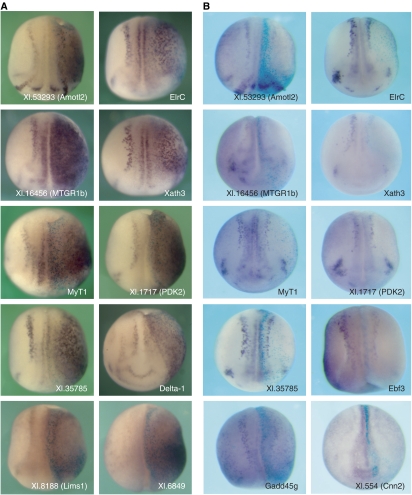
We next examined expression patterns of novel or uncharacterized Ngnr1 and NeuroD targets. As expected, expression of the majority of genes tested (16 of 20) was predominantly in the embryonic nervous system (Figure 1). Ngnr1 and NeuroD target genes were commonly expressed in the primary neurons at neurula stages (Figure 1A, E, F, H, J, M, O, and Q) and in the central nervous system (e.g. brain, spinal cord, and eye) at tailbud stages (Figure 1B–D, G, I, K, L, N, and T). Expression was also frequently found in the neural plate at neurula stages (Figure 1A, Q–S) and, at tailbud stages, and in the cranial placodes and branchial arches, which contain placodal ectoderm or neural crest derivatives (Figure 1G, I, K, L, N, P, and T). In an embryonic context, NeuroD strongly induced all 10 target genes that we tested (Figure 2A). Conversely, lowering Ngnr1 activity in embryos reduced or eliminated the neural-specific expression of 9 of the 11 target genes we tested (Figure 2B). These data demonstrate that Ngnr1 and NeuroD induce or are required to activate the expression of these target genes in vivo.
Figure 2.
NeuroD and Ngnr1 regulate their target genes in Xenopus embryos. One bilateral half of each embryo was injected with either (A) NeuroD mRNA or (B) antisense morpholino oligonucleotides (MO) directed against Ngnr1 and with β-galactosidase RNA as a lineage tracer (blue stain) by injecting one cell at the two-cell stage as described (Supplementary data). Embryos were raised to neurula stages (stages 15–16) and in situ hybridization with the indicated probes was performed. (A) NeuroD induced ectopic expression of all 10 genes tested on the injected side (oriented to the right). (B) The Ngnr1 MO reduced the expression of 9 of the 11 target genes we tested (shown), while expression of two target genes was only marginally reduced or unaffected (Cnn2 (shown) and Pou50 (not shown)). Dorsal views with anterior down are shown, except for Cnn2, which is an anterior view.
Taken together, these results demonstrate that the approach of using hormone-inducible forms of Ngnr1 and NeuroD and naive ectoderm effectively identified direct targets of these transcription factors and that primary roles of Ngn and NeuroD include inducing regulators of gene transcription, signal transduction, and cytoskeletal rearrangement for neuronal differentiation and migration. Additionally, we found many novel target genes, which are expressed in the nervous system and may play roles in neuronal differentiation, migration, and/or function.
Induction of Ngn and NeuroD targets in mammalian cells
We examined whether Ngnr1 and NeuroD direct target genes identified in Xenopus ectoderm were also regulated by their mouse orthologs in mammalian cells. For this purpose, we used P19 embryonic carcinoma cells, which are induced to differentiate into neurons by overexpressing proneural bHLH proteins (Farah et al, 2000). We transfected P19 cells with mouse Ngn2 or NeuroD and examined the expression of mouse orthologs or closest homologs of 16 Ngn and 30 NeuroD Xenopus target genes by qRT–PCR (Table II). Mouse Ngn2 induced 7 of 16 (43.8%) and NeuroD 14 of 30 (47%) genes tested by >2-fold in P19 cells. Four genes (25%) for Ngn2 and six genes (20%) for NeuroD were also induced 1.5- to 2.0-fold. Most Xenopus target genes that were not or were only weakly induced were already highly expressed in P19 cells before transfection (see Table II, Ct values). Thus, Ngn2 and NeuroD overexpression may not further increase their expression in P19 cells but those genes may be induced in other cellular contexts. With respect to this, we compared NCBI expression profiles for 12 targets that were strongly induced by NeuroD and eight targets that were not induced (Supplementary Figure S4). Most strongly induced genes were highly enriched in brain, dorsal root ganglion, eye, and/or spinal cord in vivo and were expressed in few other tissue types. In comparison, many non-induced target genes were more broadly expressed. This suggests that the P19 cell context may be more conducive to Ngn2 and NeuroD induction of targets with neural restricted expression in vivo relative to those with broader expression profiles.
Table 2.
Induction of Ngn and NeuroD targets in mammalian P19 cells
| UniGene for Xenopus genes | UniGene for mouse orthologs | Gene title | Induction by Ngn2a | Induction by NeuroD1a | Range of Ct values in qRT–PCRb |
|---|---|---|---|---|---|
| Xl.437 | Mm.2474 | Nescient helix loop helix 1 (Nhlh1, HEN1)c | 229.1 | 198.0 | 29–33 |
| Xl.879 | Mm.3280 | Twist gene homolog 1 (Drosophila) | 0.8 | 0.8 | 24–26 |
| Xl.1263 | Mm.10695 | Neurogenic differentiation 4 (NeuroD4, Math3, NeuroM)c | 22.6 | 123.7 | 31–34 |
| Xl.1274/Xl.21449d | Mm.297371 | POU domain, class 3, transcription factor 1 (Pou3f1, Oct6)c | ND | 3.5 | 26–28 |
| Mm.129387 | POU domain, class 3, transcription factor 2 (Pou3f2, Brn2)c | ND | 5.6 | 33–34 | |
| Xl.450 | Mm.136217 | Achaete–scute complex homolog-like 1 (Ascl1) | 1.8 | 1.8 | 34–37 |
| Xl.23642 | Mm.330700 | Zinc finger protein 238 (RP58)c | 1.7 | 1.7 | 24–25 |
| Xl.53293/Xl.76708 | Mm.21145 | Angiomotin-like 2 (Amotl2) | 1.7 | 1.7 | 21–24 |
| Xl.16456/Xl.4960 | Mm.29914 | Core-binding factor, runt domain, alpha subunit 2, translocated to, 2 homolog (Cbfa2t2h, MTGR1)c | 1.3 | 1.5 | 22–25 |
| Xl.34957 | Mm.440140 | BTB (POZ) domain containing 6 (Btbd6) | ND | 1.0 | 28–31 |
| Xl.958 | Mm.440702 | Zinc finger homeobox 1b (Zfhx1b, SIP1)c | ND | 1.0 | 20–21 |
| Xl.24147 | Mm.60590 | Myosin 10 | ND | 1.9 | 24–25 |
| Xl.547/Xl.237 | Mm.319947 | Early B-cell factor 2 (Ebf2) | 84.5 | 84.4 | 28–31 |
| Xl.57163/Xl.31433 | Mm.130005 | Myelin transcription factor 1 (MYT1, NZF-2b)c | 1.7 | 1.8 | 26–28 |
| Xl.5041 | Mm.11186 | Solute carrier family 43, member 2/L-type amino-acid transporter 4 (Slc43a2/LAT4) | ND | 0.7 | 24–25 |
| Xl.4522 | Mm.238044 | Iroquois-related homeobox 3 (Irx3)c | 1.1 | 1.1 | 24–26 |
| Xl.5764 | Mm.142455 | Solute carrier family 7 (cationic amino-acid transporter, y+ system), member 7 (Slc7a7) | ND | 0.9 | 20–22 |
| Xl.15305 | Mm.10026 | Cell death-inducing DFFA-like effector c (CIDE-c, FSP27) | 0.9 | 1.4 | 29–31 |
| Xl.71386 | Mm.274011 | UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 3 (B4galt3) | ND | 1.1 | 22–24 |
| Xl.8440 | Mm.137268 | Hairy and enhancer of split 5 (Hes5) | 2.7 | 8.1 | 30–37 |
| Xl.16835 | Mm.259022 | Polo-like kinase 3 (Plx3) mRNA (Plk3)c | ND | 0.8 | 32–34 |
| Xl.54916 | Mm.4875 | Delta-like 1 (Dll1)c | 6.2 | 3.1 | 33–35 |
| Xl.1035 | Mm.347707 | ELAV-like 3 (HuC, Elavl3)c | 5.1 | 9.4 | 29–31 |
| Xl.24275 | Mm.2734 | Spermidine/spermine N1-acetyltransferase 1 (Sat1) | ND | 2.9 | 33–36 |
| Xl.7556 | Mm.150314 | Prickle1c | 1.0 | 1.2 | 22–23 |
| Xl.12125 | Mm.281298 | Growth arrest and DNA-damage-inducible 45 gamma (Gadd45g) | ND | 7.3 | 28–30 |
| Xl.5886 | Mm.29811 | Response gene to complement 32 (Rgc32) | ND | 2.0 | 33–35 |
| Xl.51207 | Mm.130063 | RIKEN cDNA 9330182L06 gene | ND | 3.7 | 25–27 |
| Xl.1717 | Mm.29768 | Pyruvate dehydrogenase kinase, isoenzyme 2 (Pdk2) | ND | 2.0 | 24–25 |
| Xl.632e | Mm.258708 | Early B-cell factor 3 (Ebf3) | 10.2 | 26.1 | 32–34 |
| Average fold change of three independent experiments determined by qRT–PCR, ND indicates not determined | |||||
| qRT–PCR threshold cycle numbers (Ct) represent when target cDNA begins to be exponentially amplified during PCR and inversely correlate with target cDNA abundance. Target genes from the Xenopus microarrays that were not or weakly induced were often already highly expressed in P19 cells and Ngn/NeuroD transfection did not further increase expression | |||||
| These genes were also predicted as Ngn/NeuroD targets in a computational, genome-wide analysis with the PAP program. See text for further details | |||||
| Two mouse genes (Pou3f1/Oct6 and Pou3f2/Brn2) showed highest homology to POU50 (Xl.1274/Xl.21449) depending on the alignment method (Clustal W or Clustal V). Thus, it was unclear which gene was a true ortholog of Xenopus POU50 and we tested both genes | |||||
| Xl.632 Ebf3 was not identified in our microarray experiments (see Supplementary methods) but is a known NeuroD target and we confirmed its induction by NeuroD by qRT–PCR in Xenopus. Thus, we included Ebf3 in this list | |||||
Therefore, approximately half (and probably more) of the target genes identified in Xenopus showed conserved induction in mammalian cells, suggesting that Ngn and NeuroD regulate similar target genes in multiple vertebrates and that these targets are evolutionarily conserved core mediators of Ngn and NeuroD activities in neurogenesis.
Determining consensus sequences for Ngn and NeuroD binding
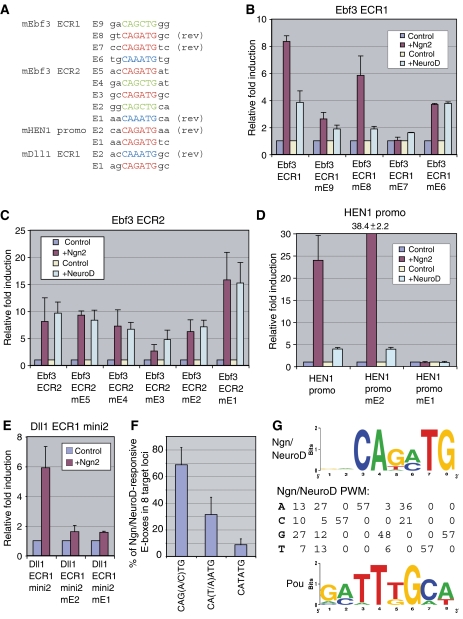
To identify Ngn/NeuroD regulatory enhancers in the target loci defined above, we focused on genes induced in both Xenopus and mouse and initially used the criteria that potential enhancers should show some cross-species conservation and contain >1 E-box, since transcription factors including bHLH proteins often utilize multiple, clustered binding sites (Weintraub et al, 1990). We used the evolutionarily conserved region (ECR) browser to compare genomic sequences for Dll1, Ebf3, Elavl3, Gadd45g, NeuroD4 (Ath3), and HEN1 (Nhlh1) between multiple vertebrates and detect E-box containing ECRs, which represent potential enhancers (Ovcharenko et al, 2004). We then tested 2–3 candidate enhancers per locus (16 total ECRs) for Ngn2 and/or NeuroD responsiveness by luciferase assay in P19 cells (Supplementary Figure S5, A and B). We set a three-fold cutoff for responsiveness because enhancers showing >3-fold induction were consistently enriched for Ngn2 and NeuroD binding in chromatin immunoprecipitation (ChIP) assays, while those with lower induction fold-change values were not (Supplementary Figure S8 and data not shown). By these criteria, Ngn and NeuroD induced expression of the same reporter constructs (Ebf3 ECR1, Ebf3 ECR2, HEN1 promo, and Dll1 ECR1mini2), except for Dll1 ECR1mini2, which was only induced by Ngn. While Ngn regulation of NeuroD could indirectly contribute to Ngn's ability to induce these enhancers, we found that Ngn bound these enhancers by ChIP (Supplementary Figure S8), indicating that Ngn can directly regulate these enhancers. These data suggested that Ngn and NeuroD may use similar binding sites and enhancers to regulate their target genes.
We next analyzed which E-boxes in these ECRs were required for Ngn and NeuroD-mediated expression. There are 10 possible E-box sequences, considering four nucleotides for each N in the E-box (CANNTG) and disregarding orientation (Supplementary Figure S2C). The 16 ECRs tested above contained in total 42 E-boxes, 29 in non-responsive ECRs and 13 in the four Ngn2/NeuroD-responsive ECRs. However, only 3 of 10 possible E-box sequence types (CAGCTG, CAGATG, or CAAATG) were present in the responsive ECRs, suggesting Ngn and NeuroD binding preferences (Figure 3A). Other types of E-boxes were found only in non-functional ECRs (Supplementary Figure S2C). We next disrupted individual E-boxes: disrupting CAGATG, CAGCTG, and CAAATG E-boxes abolished or reduced induction by Ngn2, while induction by NeuroD was affected by mutating CAGCTG and CAGATG but not CAAATG E-boxes (Figure 3B–E).
Figure 3.
Defining Ngn/NeuroD consensus sequences. E-boxes in four Ngn/NeuroD-responsive enhancers (A) were disrupted (by E-box mutation to the restriction site CTCGAG) and Ngn/NeuroD responsiveness of wild-type versus mutated enhancers was measured by luciferase assay (B–E). Fold change values were normalized to luciferase activity without Ngn/NeuroD transfection. Rev, reverse complementary sequence. (F) Relative frequencies of conserved Ngn2/NeuroD-responsive E-boxes were defined for eight target loci (summary of data from Supplementary Figure S7). (G) A POU site and E-box sequences were identified as enriched motifs in Ngn/NeuroD targets using CompareProspector. DNA sequence logos (for the POU site and E-box) and PWM (for the E-box) are shown.
As a parallel, independent approach to assess Ngn- and NeuroD-binding sequences, we built 10 artificial enhancer-luciferase reporter constructs. Each contained a different E-box sequence, present as three copies with the same intervening sequences for all constructs. In luciferase assays, Ngn2 and NeuroD robustly induced the three E-box types defined experimentally above (CAG[A/C]TG and CAAATG). In addition to these three E-box types, CATATG was also responsive to Ngn and NeuroD while the six other E-box types were unresponsive (Supplementary Figure S6). Taken together with the results from 16 ECRs, these data suggest that Ngn and NeuroD recognize similar E-box sequences.
Finally, we used CompareProspector (Liu et al, 2004) to computationally define conserved, enriched sequence motifs within 25 ECRs from 14 Ngn and NeuroD target genes in human and mouse (Ascl1, Dll1, Ebf2, Ebf3, Elavl3, Gadd45g, HEN1, MTGR1, Myo10, MyT1, NeuroD4, PDK2, Pou3f1, and Zfp238; for details, see Supplementary data). In this search, the E-box sequence CAG[A/C]TG (degenerate consensus considering bases with more than 25% abundance) was identified as the second rank. The Ngn/NeuroD E-box consensus sequence logo and position weight matrix (PWM) are shown in Figure 3G. These sites were also frequently found in a clustered pattern (Supplementary Figure S7B). The top-ranking motif was GATTTGCA (Figure 3G) (G[A/C]TT[G/T]GC[A/T]: degenerate consensus), which resembles a consensus-binding sequence for class 2 POU transcription factors (e.g. Oct-1 and Oct-2) (TESS: Transcription Element Search System, http://www.cbil.upenn.edu/tess, weight matrix: M00210). CAAATG and CATATG E-boxes, which were able to respond to Ngn2 and NeuroD in the context of an artificial enhancer (Supplementary Figure S6), were also found in this analysis but they were under-represented relative to CAG[A/C]TG E-boxes. When we manually searched eight Ngn/NeuroD target loci (Dll1, Ebf2, Ebf3, MTGR1, MyT1, NeuroD4, HEN1 (Nhlh1), and Zfp238) for conserved E-boxes of these four types, we found that CAG[C/A]TG accounted for 69%, while CAAATG and CATATG comprised 22 and 9% of the E-boxes present in these loci, respectively (Figure 3F and Supplementary Figure S7). These data suggest that while CATATG E-boxes can mediate Ngn- and NeuroD-transcriptional responses, this sequence is not frequently used in the genome for Ngn- and NeuroD-mediated target activation. In summary, E-boxes with the consensus CAG[C/A]TG were over-represented in our target loci and were required for Ngn and NeuroD activation of these loci, with minor contributions from CA[A/T]ATG E-boxes.
Identifying Ngn/NeuroD regulatory enhancers in target genes
Previously, defining ECRs with clusters of generic E-boxes (CANNTG) had successfully predicted some enhancers within target loci but was not efficient (Supplementary Figure S5). Therefore, we tested whether using the Ngn/NeuroD consensus sequences improved enhancer prediction. We used the Ngn/NeuroD PWM, which was obtained from CompareProspector and reflects frequencies of the four E-boxes able to respond to Ngn2 and NeuroD (Figure 3G), in combination with the Enhancer Element Locator (EEL) (Hallikas et al, 2006) and ECR Browser. This approach greatly reduced the ‘noise' of generic E-boxes (CANNTG), allowing us to analyze wider regions of each locus. Using a generic E-box sequence, as above, we had frequently obtained more than three candidate enhancers within 10 kb of the transcription start site. In contrast, using the Ngn/NeuroD PWM usually predicted only a few high-scoring enhancers per locus, including introns, untranslated regions and 50 kb each of 5′ and 3′ flanking sequences in the analysis. We applied this approach to the Dll1, Ebf2, Ebf3, HEN1, MTGR1, MyT1, NeuroD4, and Zfp238 loci. The approach successfully identified two previously defined enhancers within the Ebf3 locus (Ebf3 ECR1 and ECR2) and also predicted 12 new candidate enhancers within the Dll1, Ebf2, MTGR1, MyT1, NeuroD4 and Zfp238 loci (Supplementary Figure S2).
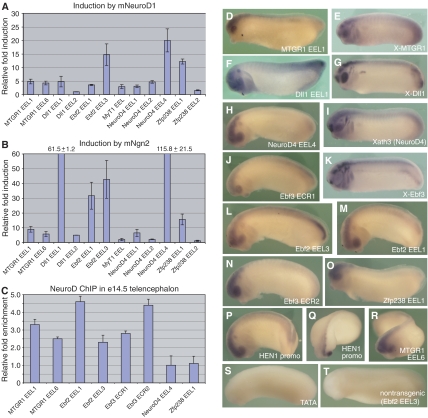
We used luciferase assays to test whether these 12 putative enhancers could mediate transcriptional responses to Ngn and NeuroD. Nine of the 12 enhancer constructs (75%) were induced >3-fold by both Ngn2 and NeuroD (Figure 4A and B). This is a major improvement in enhancer prediction compared with our prior results, where only four (25%) or three (19%) candidate enhancers of 16 were induced >3-fold by Ngn2 and NeuroD, respectively (Supplementary Figure S5, A and B). Ngn2 induced enhancer activation to a greater degree than NeuroD, suggesting that although Ngn2 and NeuroD can regulate shared target genes through the same enhancers, Ngn activates transcription more efficiently than NeuroD at these enhancers.
Figure 4.
Ngn- and NeuroD-bound regulatory elements drive neural-restricted transgene expression. (A, B) Enhancers predicted by Enhancer Element Locator (EEL) were tested in luciferase assays after cotransfection of Ngn/NeuroD and reporter vectors into P19 cells as before. (C) NeuroD binding to target gene enhancers in embryonic brain tissue was assayed by qChIP. Enrichment was calculated as the ratio of specific antibody to IgG after normalization with GAPDH (See Supplementary data). (D, F, H, J, L–R) Transgenic Xenopus embryos carrying enhancer-luciferase reporter constructs were analyzed by in situ hybridization for luciferase at stages 22–23 and compared to endogenous expression of some Ngn/NeuroD target genes (E, G, I, K) and to control embryos carrying the empty pTATA reporter vector (S) or non-transgenic siblings (T).
We next used quantitative ChIP (qChIP) to test whether Ngn2 and/or NeuroD bound these enhancers. In P19 cells transfected with myc-tagged Ngn2 or NeuroD, all eight functional enhancers we tested within the Ebf2, Ebf3, MTGR1, NeuroD4, and Zfp238 loci showed enrichment for Ngn2 and NeuroD, while the non-functional HEN1 ECR1 was not enriched above background levels (1.1-fold) (Supplementary Figure S8). Furthermore, six of the eight functional enhancers (75%) were occupied by endogenous NeuroD in telencephalon tissue from e14.5 mouse embryos (Figure 4C). Thus, Ngn2 and NeuroD directly bind these enhancers in vivo and can induce their expression.
We also tested whether these conserved Ngn- and NeuroD-responsive enhancers drove endogenous expression patterns of the target genes in embryos. We generated transgenic Xenopus embryos carrying 10 enhancer constructs, which efficiently mediated Ngn2 and NeuroD induction in luciferase assays, and analyzed transgene expression by in situ hybridization for the luciferase reporter. All 10 enhancers tested partly recapitulated their endogenous expression patterns (Figure 4D–R). Expression patterns of Xenopus Dll1, Ebf3, MTGR1, and NeuroD4 are shown for comparison (Figure 4E, G, I, and K). Interestingly, all 10 enhancers drove transgene expression within the brain and eye in a very similar pattern (Figure 4D, F, H, J, L–R), while embryos carrying Ebf2 EEL3 or Dll1 EEL1 additionally expressed the transgene in the tailbud (Figure 4F and L). This suggests that our Ngn/NeuroD regulatory elements, which were selected for enrichment for Ngn/NeuroD consensus sites, might be sufficient to drive gene expression in the brain and eye, while expression in other territories requires additional information. As negative controls, we generated transgenic embryos carrying the same TATA-luciferase vector without an introduced enhancer (Figure 4S) or the pBS vector (data not shown) and we also compared transgenic versus non-transgenic embryos (for example, Figure 4, compare L and T); these did not show localized transgene expression. Figure 4Q shows a ‘half-transgenic' embryo, where the transgene integrated after the first cleavage division and thus is expressed only in one side of the embryo (Kroll and Amaya, 1996), further confirming that this in situ pattern is specific to transgene expression. Taken together, these data demonstrate that these conserved Ngn- and NeuroD-responsive regulatory enhancers contain sequence information sufficient to direct restricted expression to the embryonic brain and eye in vivo.
Genome-wide prediction of Ngn and NeuroD direct target genes
Above, we found that evolutionary conserved non-coding sequences with greater than one Ngn/NeuroD consensus site were robustly activated by Ngn and NeuroD. To determine whether this regulatory signature could predict additional Ngn and NeuroD target genes, we used the Promoter Analysis Pipeline (PAP) program (Chang et al, 2006) and Ngn/NeuroD PWM (Figure 3G). PAP uses evolutionary conservation and enrichment for a PWM for genome-wide prediction of coregulated genes. PAP predicted 347 potential Ngn/NeuroD target genes (cutoff, P<0.01; Supplementary Figure S9A), including many experimentally defined targets (13 of the 30 targets in Table II). To test the specificity of PAP predictions, we also analyzed two heterologous E-boxes in TRANSFAC that are diverged from the Ngn/NeuroD PWM (E-box, M01034 and c-Myc/Max, M00118, PWMs in Supplementary data) and predicted 365 and 127 targets, respectively (Supplementary Figure S9, B and C). These CACGTG-type E-boxes did not respond to Ngn or NeuroD (Supplementary Figures S2 and S6).
We used functional clustering to compare PAP-predicted target genes for the Ngn/NeuroD PWM versus the heterologous E-box PWMs (Supplementary Figure S10 and Supplementary data). For PAP-predicted Ngn/NeuroD target genes, nervous system development was the top gene ontology (GO) term, followed by cell differentiation, tyrosine kinase signaling pathway, and development. In contrast, neither of the heterologous E-boxes predicted nervous system development as a top GO term (Supplementary Figure S10). Ngn/NeuroD target genes related to neural development or development were also largely non-overlapping with targets predicted for these heterologous E-boxes (M01034 or M00118; five and zero overlapping targets, respectively). For each PWM, functional clustering and genes in top-scoring clusters are listed in Supplementary Figure S10. Therefore, PAP predicted a distinct set of putative Ngn/NeuroD targets genome-wide and loci involved in neural development are most frequently enriched for conserved Ngn/NeuroD consensus sites. To test whether PAP-predicted target genes were indeed Ngn/NeuroD targets, we tested whether NeuroD induced their expression. We tested 51 PAP-predicted targets not previously analyzed in our experimental work. Of these, 20 (39%) responded to NeuroD >2-fold and another 10 (20%) responded >1.5-fold (Table III). As for the experimental targets, most PAP-predicted targets that were uninduced in P19 cells were already highly expressed and NeuroD transfection did not increase expression. Also as before, most genes that were robustly induced by NeuroD in P19 cells had highly neural-restricted expression in vivo (Supplementary Figure S4). These results verify that the computational whole genome prediction effectively identified additional NeuroD target genes.
Table 3.
NeuroD induction of target genes predicted by Promoter Analysis Pipeline (PAP)
| Entrez Gene ID | Gene title | P-value (h/m) in PAP | qRT–PCR (average fold change) | Average Ct value |
|---|---|---|---|---|
| 90 | Anaplastic lymphoma kinase (ALK) | 0 | 1.5 | 28.73 |
| 463 | AT-binding transcription factor 1 (ATBF1) | 0 | 1.1 | 21.04 |
| 84913 | Atonal homolog 8 (ATOH8) | 0 | 3.0 | 28.77 |
| 55799 | Calcium channel, voltage-dependent, alpha 2/delta 3 subunit (CACNA2D3) | 0 | 2.0 | 28.40 |
| 1295 | Collagen, type VIII, alpha 1 (COL8A1) | 0.0022 | 2.4 | 28.46 |
| 55118 | Cartilage acidic protein 1(CRTAC1) | 0 | 2.1 | 27.67 |
| 1600 | Disabled homolog 1 (DAB1) | 0 | 0.8 | 21.24 |
| 1630 | Deleted in colorectal carcinoma (DCC) | 0.0001 | 18.5 | 28.16 |
| 1641 | Doublecortex (DCX) | 0 | 3.6 | 22.53 |
| 1785 | Dynamin 2 (Dnm2) | 0.0013 | 1.2 | 21.17 |
| 1896 | Ecdysplasin A (EDA) | 0.0005 | 0.8 | 24.97 |
| 2042 | EPH receptor A3 (EPHA3) | 0.0003 | 12.8 | 27.82 |
| 2047 | EPH receptor B1 (EPHB1) | 0.0004 | 1.4 | 22.15 |
| 2048 | EPH receptor B2 (EPHB2) | 0 | 1.8 | 22.26 |
| 2104 | Estrogen-related receptor gamma (ESRRG) | 0 | 2.1 | 24.10 |
| 2138 | Eyes absent homolog1 (EYA1) | 0.0004 | 1.9 | 21.12 |
| 8522 | Growth arrest specific7 (GAS7) | 0.0064 | 1.0 | 22.31 |
| 3382 | Islet cell autoantigen1 (ICA1) | 0.0043 | 0.9 | 22.74 |
| 26280 | Interleukin 1 receptor accessory protein-like 2 (IL1RAPL2) | 0 | 1.2 | 31.59 |
| 3752 | Potassium voltage-gated channel, Shal-related subfamily, member 3 (KCND3) | 0 | 33.3 | 29.28 |
| 4045 | Limbic system-associated membrane protein (LSAMP) | 0.0001 | 1.9 | 28.91 |
| 4684 | Neural cell adhesion molecule1 (NCAM1) | 0 | 1.5 | 21.18 |
| 4851 | Notch homolog 1, translocation-associated (NOTCH1) | 0.002 | 5.1 | 24.45 |
| 8828 | Neuropilin2 (NRP2) | 0 | 2.3 | 24.84 |
| 9378 | Neurexin 1 (Nrxn1) | 0 | 0.7 | 22.27 |
| 9423 | Netrin 1 (NTN1) | 0.0001 | 1.9 | 27.55 |
| 4916 | Neurotropic tyrosine kinase type 3 (NTRK3) | 0.0001 | 1.1 | 23.75 |
| 57451 | odz, odd Oz/ten-m homolog 2 (ODZ2) | 0 | 0.7 | 24.76 |
| 5076 | Paired box gene 2 (PAX2) | 0.0001 | 2.1 | 28.68 |
| 79960 | PHD finger protein 17 (PHF17) | 0.0006 | 1.0 | 20.53 |
| 5362 | Plexin A2 (PLXNA2) | 0 | 7.7 | 25.41 |
| 5579 | Protein kinase C, beta 1 (PRKCB1) | 2.3 | 34.13 | |
| 5629 | Prospero-related homeobox 1 (PROX1) | 0 | 1.3 | 21.89 |
| 601309 | Patched homolog 1 (PTCH1) | 1.3 | 21.14 | |
| 6091 | Roundabout, axon guidance receptor, homolog 1 (ROBO1) | 0 | 4.0 | 21.39 |
| 146760 | Reticulon 4 receptor-like 1 (RTN4RL1) | 0 | 1.3 | 22.32 |
| 862 | Runt-related transcription factor 1; translocated to, 1 (RUNX1T1) | 0 | 1.1 | 21.19 |
| 10371 | Sema domain, immunogloblin (Ig) domain, short basic domain, secreted, (semaphorin) 3A (SEMA3A) | 0 | 1.3 | 24.23 |
| 56920 | Sema domain, immunogloblin domain (Ig), short basic domain, secreted, (semaphorin) 3G (SEMA3G) | 0.0005 | 1.8 | 28.70 |
| 80031 | Sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D (SEMA6D) | 0.0003 | 2.3 | 25.65 |
| 9353 | Slit homolog 2 (SLIT2) | 0.0002 | 0.9 | 22.17 |
| 6586 | Slit homolog 3 (SLIT3) | 0 | 0.9 | 23.15 |
| 9705 | Suppression of tumorigenicity 18 (ST18) | 0 | 27.7 | 28.81 |
| 8224 | Synapsin III (SYN3) | 0 | 1.2 | 28.60 |
| 132204 | Synaptoporin (SYNPR) | 0.0002 | 6.7 | 27.65 |
| 6857 | Synaptotagmin I (SYT1) | 0.0012 | 2.8 | 24.13 |
| 6925 | Transcription factor4 (TCF4) | 0 | 1.2 | 21.24 |
| 57616 | Teashirt family zinc finger 3 (TSHZ3) | 1.6 | 24.68 | |
| 8633 | Unc 5c homolog (UNC5C) | 0.0032 | 6.0 | 27.97 |
| 80326 | Wingless-type MMTV integration site family, member 10A (Wnt10a) | 0.0014 | 1.9 | 32.73 |
| 7482 | Wingless-type MMTV integration site family, member 2B (Wnt2b) | 0.002 | 1.6 | 26.11 |
Discussion
Identification of direct transcriptional targets of Neurogenin and NeuroD
Transcription factors are key regulators of many cellular processes, but defining how they perform their functions has been limited by a lack of knowledge of their direct target genes. Microarray technologies enable genome-wide transcription factor target gene identification by comparing mRNA abundance between two sample types (e.g. wild-type mice versus those with a targeted gene disruption) but this technology does not distinguish between direct and indirect targets. Recently, ChIP coupled with microarray (ChIP-on-chip) has also been used to define direct transcription factor targets. However, this approach does not incorporate information regarding gene expression changes, and only a fraction of genes identified in ChIP-on-chip show changed expression upon transcription factor introduction or removal. This suggests either that some sites represent false positives or could reflect target locus misidentification, since current genome annotation is imperfect and vertebrate enhancers are also frequently located many kilobases away from transcription start sites or even within other loci.
As an alternative to these approaches, we employed a modified expression profiling approach to identify direct transcription factor targets. By using hormone-inducible forms of transcription factors in Xenopus naive ectoderm, we could acutely induce target gene expression and detect only rapid transcriptional responses occurring within a 2.5-h window. We also induced transcription factor activity in the presence of a translational inhibitor, which avoided detection of indirect targets expressed in response to primary target activities or following cell-fate changes. Using this approach, we successfully identified direct targets, which were validated as Ngn and NeuroD responsive at high frequency. Ngn and NeuroD shared many target genes, and most were responsive in both Xenopus embryonic tissue and mammalian cells. We then analyzed the genomic loci of these targets to define Ngn and NeuroD consensus-binding sequences and regulatory enhancers that drove expression both in mouse P19 cells and in the Xenopus nervous system. We further used these enhancer features for genome-wide computational screening and successfully predicted additional Ngn/NeuroD direct target genes. Since Xenopus embryonic ectoderm is multipotent and can differentiate into most ectodermal, endodermal, and mesodermal cell derivatives, we propose that this approach could be used to identify direct targets for a wide range of other transcription factors.
A Ngn- and NeuroD-regulated molecular network for neurogenesis
Ngn and NeuroD induced 26 shared targets, including most of the transcription factors, suggesting that they act through a common set of transcription factors to induce neuronal differentiation. In developing embryos, Ngn1 and 2 are transiently expressed in the ventricular zone at the commitment stage, and this is followed by NeuroD expression during differentiation. Our data indicate that Ngn and NeuroD recognize very similar E-box sequences. Thus, Ngn activity could initiate target gene expression during neuronal commitment, while NeuroD activity maintains the later expression of these genes using the same enhancers, to sustain commitment to neuronal fates. It has been a long-standing enigma that NeuroD is only expressed after the time of neuronal commitment in vivo, yet can activate the entire neurogenesis program upon overexpression. Our finding that NeuroD directly binds enhancers in genes encoding key transcriptional regulators of neurogenesis may account for this perhaps surprising ability of NeuroD to broadly regulate neurogenesis when overexpressed.
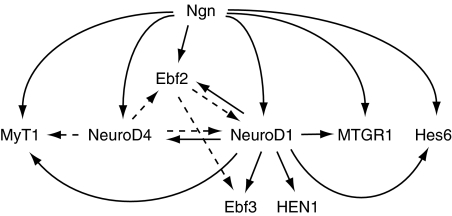
We propose that primary transcription factor targets of Ngn and NeuroD (Ebf2, Ebf3, HEN1 (Nhlh1), Hes6, MTGR1, MyT1, NeuroD, NeuroD4 (Ath3), and potentially Znf238) represent a core transcriptional network mediating Ngn- and NeuroD-regulated neurogenesis (Figure 5). Ngn initiates neuronal differentiation, and NeuroD is a key Ngn-regulated transcriptional node. There are many reciprocal and redundant regulatory relationships between these transcription factors. For example, we found that NeuroD induces Ebf2, Ebf3, MyT1, and NeuroD4, but Ebf2 also activates Ebf3 and NeuroD (Dubois et al, 1998; Pozzoli et al, 2001) and NeuroD4 (Ath3) induces Ebf2, MyT1, and NeuroD (Perron et al, 1999). This may account for the limited defects observed in NeuroD-null mice. In contrast, to our knowledge, reciprocal regulation of Ngn expression by its primary targets has not been observed. These data suggest that Ngn acts at the top of a regulatory cascade to initiate neurogenesis, while at least two or three primary Ngn transcription factor targets (Ebf2, NeuroD, and NeuroD4) then act reciprocally and potentially redundantly to generate a robust network controlling neuronal differentiation and the expression of transcription factors, such as Ebf3, Hes6, MTGR1, and MyT1 also supports this differentiation program.
Figure 5.
Model for Ngn and NeuroD regulation of their target genes during neurogenesis. Solid arrows indicate direct transcriptional targets and dotted arrows were used when direct–indirect regulatory relationship was not determined.
Although we focused on shared targets of Ngn and NeuroD in this study, our microarray experiment identified many distinct, non-shared targets. We found that Ngn and NeuroD recognize very similar E-box sequences, but their binding preferences were not identical as shown in the mutagenesis analyses in Figure 3. Slight differences in the DNA-binding properties of Ngn and NeuroD may explain their differential induction of these targets. Alternatively, Ngn and NeuroD may interact with distinct cofactors in different cellular contexts and this may account for the induction of these non-shared targets. Understanding the nature of genes induced only by Ngn or only by NeuroD and how these targets are differentially activated by these two transcription factors will help to elucidate how Ngn and NeuroD fulfill their distinct biological functions in vivo.
A minimal enhancer signature for activation of Ngn and NeuroD targets in neural tissue
Our approach defined direct transcriptional targets, which should contain Ngn and/or NeuroD regulatory elements. We also found many shared targets, suggesting that Ngn and NeuroD may recognize common enhancers or consensus sites in those targets. We initially focused on targets induced in both Xenopus and mouse, which may employ conserved Ngn/NeuroD regulatory elements, and we tested clustered E-boxes around target loci for enhancer activity, since transcription factors often utilize clustered binding sites. This defined some enhancers but was inefficient since E-boxes occur frequently by chance. We then experimentally determined that Ngn and NeuroD bind a similar consensus sequence and we used these PWMs to predict a few high-scoring enhancers within large genomic regions encompassing and surrounding each target gene locus. These predicted enhancers frequently responded to Ngn and NeuroD in mouse cells and most were also occupied by endogenous NeuroD in embryonic brain tissue. In addition, all 10 enhancers drove reporter gene expression in the brain and eye of transgenic Xenopus embryos. Thus, enhancers containing conserved, clustered consensus sites are sufficient both to respond to Ngn and NeuroD in mammalian cells and to drive neural-restricted expression in embryos.
We further used these enhancer features for computational genome-wide prediction of other Ngn/NeuroD target loci and defined 347 putative direct targets, many of which were induced by NeuroD in P19 cells. Our experimental approach defined a core Ngn and NeuroD-regulated neurogenesis program in Xenopus and mammals but was unable to define some classes of Ngn/NeuroD target genes, including those induced only in specialized neuronal contexts (e.g. the mammalian neocortex) or whose induction required a cofactor absent in Xenopus neural ectoderm. As described previously, we potentially also missed detecting some NeuroD targets induced only during later neuronal differentiation or Ngn/NeuroD targets whose induction occurred in a temporally delayed manner, for example through a feed-forward mechanism as is used during myogenic bHLH activation of target genes (Tapscott, 2005). Interestingly, our computational approach identified 6 of 26 genes previously identified as misregulated in Ngn2 or Ngn1/2 mutant mouse neocortices, including AKT3, Dcc, Elavl4, Negr1, NeuroD2, Robo1, as well as the previously defined Ngn2 direct target Dcx (Mattar et al, 2004; Schuurmans et al, 2004; Ge et al, 2006). Our data suggest that these genes are direct Ngn/NeuroD targets and that our computational approach defined additional target genes missed in our microarray experiments.
In our search for the over-represented sequence motifs within 14 Ngn and NeuroD target genes using CompareProspector, the top-ranking motif was GATTTGCA (Figure 3G), which resembles a consensus-binding sequence for class 2 POU transcription factors (e.g. Oct-1 and Oct-2). Indeed, many of our enhancers also contain this motif in close proximity to Ngn/NeuroD E-boxes (Supplementary Figure S2A). This is reminiscent of the recent finding that Mash1 synergizes with POU class 3 proteins by direct binding of Mash/Brn proteins to adjacent cis-sequences in target genes (Castro et al, 2006). We found that disrupting POU sites did not affect Ngn and NeuroD responsiveness of our enhancers in P19 cells (data not shown). This may indicate differences in Ngn and NeuroD versus Mash1 target gene regulation. Alternatively, POU sites may contribute to Ngn/NeuroD-mediated target activation in specific tissues in vivo, although they are not required for enhancer activation in our cell-based assay. Further study is required to clarify this.
We propose that clustered, conserved consensus sites constitute a ‘minimal neural enhancer signature' through which Ngn and NeuroD selectively activate target genes in neural tissue. Enhancers with these features robustly responded to Ngn and NeuroD in mammalian cells and drove expression in the brain and eye of transgenic Xenopus embryos. Furthermore, neural development-related genes throughout the genome were preferentially enriched for this enhancer signature and many were indeed regulated by NeuroD. In vivo, this enhancer signature may allow Ngn and NeuroD to activate their target genes specifically in neural tissue, so that these targets can mediate Ngn and NeuroD's general neuronal commitment and differentiation promoting activities.
While this enhancer signature is sufficient for Ngn- and NeuroD-mediated gene expression in neural tissues, it is insufficient to drive expression in some other endogenous expression domains. For example, in transgenic Xenopus embryos, our 10 Ngn/NeuroD-responsive enhancers all drove gene expression in the brain and eye, where corresponding genes are commonly expressed, but could not drive expression in territories like the cranial placodes where the endogenous genes show differential expression. Thus, these regulatory elements, which were selected for enrichment for Ngn/NeuroD consensus sites, may lack other transcription factor-binding sites needed to drive expression in these tissues.
Our data also suggest that non-neural bHLH target genes do not contain the same regulatory signature. For example, neither our experimental or computational approaches predicted NeuroD's targets in the pancreas (insulin (INS), PDX-1/IPF1, secretin, glucokinase (GCK), IGRP/G6PC2, Pax6, and SUR1; Chae et al, 2004 and references therein; Naya et al, 1995; Mutoh et al, 1997; Sharma et al, 1997). Therefore, NeuroD appears to use different enhancer signatures to induce targets in non-neural tissues. Likewise, there was little overlap between our computationally predicted NeuroD targets and previously defined targets of the myogenic bHLH transcription factors, MyoD and Myogenin, although these have a somewhat similar E-box core nucleotide preference (Bergstrom et al, 2002; Cao et al, 2006).
In summary, our data indicate that clustered, conserved consensus sites represent a minimal neural enhancer signature that is used for Ngn and NeuroD recognition of neural development-related target genes. Ngn and NeuroD bind these enhancers to activate targets that execute core programs regulating neurogenesis.
Materials and methods
Capped RNA preparation
Xenopus Ngnr1, NeuroD and GR-fusion variants were previously described (Lee et al, 1995; Ma et al, 1996; Perron et al, 1999; Pozzoli et al, 2001). Capped RNAs were made by in vitro transcription with the mMessage mMachine SP6 kit (Ambion) and these templates: CS2MT-XNeuroD, CS2MT-XNeuroD-GR, CS2MT-XNgnr1-GR, and CS2NLS-βGal.
Microarray analysis
Xenopus embryos were obtained by in vitro fertilization and raised as described (Seo et al, 2005). To minimize nonspecific target gene induction, we predetermined minimal doses of Ngnr1 and NeuroD RNAs that moderately induced neurogenesis but did not overtly affect gastrulation or morphology of embryos. Both blastomeres of two-cell-stage pigmented embryos were injected with X-Ngnr-GR (10 pg), NeuroD-GR (30 pg), or β-galactosidase (50 pg) RNAs and raised until stages 8–9. Xenopus animal caps (50–60) per sample were isolated and raised in 0.7 × MMR at 25°C. When sibling embryos were at stages 10–10.5, explants were pretreated with cycloheximide (final concentration 10 μg/ml) for 30 min and DEX (final 10 μM) was added. Explants were incubated at 25°C for 2.5 h and frozen in liquid nitrogen (when sibling embryos were at stage 12.0–12.5). Total RNA (20 μg) per sample, prepared with Trizol (Invitrogen), was used for probe synthesis and hybridization to Affymetrix Xenopus laevis Genome Arrays (Washington University Genechip facility). Result was analyzed with dChip software (http://biosun1.harvard.edu/complab/dchip/). See Supplementary data for details.
qRT–PCR
Xenopus explants and total RNAs were obtained as above. P19 cells were cultured as described (Seo et al, 2005) and transfected with 400 ng of US2MT-mNgn2 or US2MT-mNeuroD and 2.1 μg of US2MT in six-well plates with FuGene6 (Roche). In negative controls, cells were transfected with 2.5 μg of US2MT. At 44–48 h after transfection, total RNA was extracted with Trizol (Invitrogen) and 1 μg/20 μl reaction used for cDNA synthesis with oligo(dT) primers (Invitrogen) and SuperScript II reverse transcriptase. qRT–PCR was performed with the MyiQ real-time PCR Detection System (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad) or Platinum SYBR Green Supermix (Invitrogen). Primer sequences are in Supplementary Figure S1. Relative gene expression was calculated following normalization with EF1α for Xenopus or RPL19 for P19 cells. PCR was carried out in triplicate and entire experiments were repeated three times with independently prepared RNA samples. Tables I, II and III show an average of three experiments. For each primer pair, the melt curve was analyzed and the PCR product was examined on a 2% agarose gel to ensure that a single fragment of the predicted molecular weight was amplified.
Microinjection and whole-mount in situ hybridization
To examine target gene induction by NeuroD, one blastomere of two-cell-stage albino embryos was injected with 30 pg each of NeuroD and β-galactosidase RNAs. Embryos were grown to stages 16–17 (Nieuwkoop and Faber, 1967), fixed, X-Gal stained, and in situ hybridized as described (Seo et al, 2005). cDNA clones indicated in Figure 1 were purchased from OpenBiosystems or ATCC.
Cloning of target promoter/enhancer regions, luciferase constructs, and mutagenesis
Enhancers were amplified with KlenTaqLA (DNA Pol. Tech. Inc.) and mouse genomic DNA as a template and inserted into the E1X3-TATA luciferase reporter plasmid (Huang et al, 2000) after removing its E-boxes. Site-directed mutagenesis used the QuikChange Site-Directed Mutagenesis protocol (Stratagene) and Pfu Turbo polymerase (Stratagene). E-box sequences (CANNTG) were changed to CTCGAG (XhoI site) and mutants were screened by XhoI digestion and sequence confirmed. For primer sequences and ECR/EEL sequences, see Supplementary Figures S1 and S2. See text and Supplementary data for further information.
Luciferase assay
P19 cells were transfected in 12-well plates with 0.5 μg of indicated luciferase reporters and 50 ng of CS2-βGal with or without 250 ng of US2MT-mNgn2 or US2MT-mNeuroD. pUS2MT plasmid was added as needed to adjust total DNA to 1.2 μg. After 44–48 h of incubation, lysates were analyzed with the luciferase and β-galactosidase enzyme assay systems (Promega) according to the manufacturer's instructions. Samples were assayed in duplicate and experiments were repeated three times.
ChIP
Telencephalon tissue was dissected from e14.5 mouse embryos and 50–60 mg of material was immunoprecipitated with 5 μg of NeuroD (Santa Cruz (sc)-1084) or isotype-matched IgG antibodies, after standard protocols (Upstate; details in Supplementary data).
Transgenic Xenopus embryos
Transgenic Xenopus embryos were generated using the REMI (Kroll and Amaya, 1996) and SceI meganuclease methods (Ogino et al, 2006; Pan et al, 2006). Details in Supplementary data.
Supplementary Material
Supplementary data
Acknowledgments
We thank Drs Tomas Pieler, Timothy Grammer, and David Turner for kindly providing plasmids (IS-EF1α-eGFP, IS-pBS, and US2) and Jim Skeath and David Gottlieb for their thoughtful comments on the manuscript. We are also grateful to Dominique Thomson for his help in qRTP–CR and cell culture. LWC was supported by NIH grants HG00249 and GM63340 to GD Stormo. This work was supported by grants from the NIH (GM66815-01), the American Cancer Society (RSG-06-148-01-DDC), and the March of Dimes (#1-FY06-374) to KLK.
References
- Bellefroid EJ, Bourguignon C, Hollemann T, Ma Q, Anderson DJ, Kintner C, Pieler T (1996) X-MyT1, a Xenopus C2HC-type zinc finger protein with a regulatory function in neuronal differentiation. Cell 87: 1191–1202 [DOI] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ (2002) Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell 9: 587–600 [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F (2002) Proneural genes and the specification of neural cell types. Nat Rev Neurosci 3: 517–530 [DOI] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ (2006) Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J 25: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, Matter JM, Guillemot F (2006) Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell 11: 831–844 [DOI] [PubMed] [Google Scholar]
- Chae JH, Stein GH, Lee JE (2004) NeuroD: the predicted and the surprising. Mol Cell 18: 271–288 [PubMed] [Google Scholar]
- Chang LW, Nagarajan R, Magee JA, Milbrandt J, Stormo GD (2006) A systematic model to predict transcriptional regulatory mechanisms based on overrepresentation of transcription factor binding profiles. Genome Res 16: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois L, Bally-Cuif L, Crozatier M, Moreau J, Paquereau L, Vincent A (1998) XCoe2, a transcription factor of the Col/Olf-1/EBF family involved in the specification of primary neurons in Xenopus. Curr Biol 8: 199–209 [DOI] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL (2000) Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 127: 693–702 [DOI] [PubMed] [Google Scholar]
- Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F (1998) The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron 20: 483–494 [DOI] [PubMed] [Google Scholar]
- Garcia-Dominguez M, Poquet C, Garel S, Charnay P (2003) Ebf gene function is required for coupling neuronal differentiation and cell cycle exit. Development 130: 6013–6025 [DOI] [PubMed] [Google Scholar]
- Ge W, He F, Kim KJ, Blanchi B, Coskun V, Nguyen L, Wu X, Zhao J, Heng JI-T, Martinowich K, Tao J, Wu H, Castro D, Sobeih MM, Corfas G, Gleeson JG, Greenberg ME, Guillemot F, Sun YE (2006) Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc Natl Acad Sci USA 103: 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J (2006) Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124: 47–59 [DOI] [PubMed] [Google Scholar]
- Huang HP, Liu M, El-Hodiri HM, Chu K, Jamrich M, Tsai MJ (2000) Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Mol Cell Biol 20: 3292–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fritzsch B, Serls A, Bakel LA, Huang EJ, Reichardt LF, Barth DS, Lee JE (2001) NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development 128: 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolm PJ, Sive HL (1995) Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol 171: 267–272 [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N, Kintner C (2005) The expression and function of MTG/ETO family proteins during neurogenesis. Dev Biol 278: 22–34 [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N, Kim J, Anderson D, Kintner C (2000) Hes6 acts in a positive feedback loop with the neurogenins to promote neuronal differentiation. Development 127: 4203–4216 [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E (1996) Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 122: 3173–3183 [DOI] [PubMed] [Google Scholar]
- Lamar E, Kintner C (2005) The Notch targets Esr1 and Esr10 are differentially regulated in Xenopus neural precursors. Development 132: 3619–3630 [DOI] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H (1995) Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix–loop–helix protein. Science 268: 836–844 [DOI] [PubMed] [Google Scholar]
- Liu Y, Wei L, Batzoglou S, Brutlag DL, Liu JS, Liu XS (2004) A suite of web-based programs to search for transcriptional regulatory motifs. Nucleic Acids Res 32: W204–W207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan MA, Steele MR, Van Raay TJ, Vetter ML (2005) Identification of shared transcriptional targets for the proneural bHLH factors Xath5 and XNeuroD. Dev Biol 285: 570–583 [DOI] [PubMed] [Google Scholar]
- Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ (1998) Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 20: 469–482 [DOI] [PubMed] [Google Scholar]
- Ma Q, Fode C, Guillemot F, Anderson DJ (1999) Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev 13: 1717–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ (1996) Identification of neurogenin, a vertebrate neuronal determination gene. Cell 87: 43–52 [DOI] [PubMed] [Google Scholar]
- Mattar P, Britz O, Johannes C, Nieto M, Ma L, Rebeyka A, Klenin N, Polleux F, Guillemot F, Schuurmans C (2004) A screen for downstream effectors of Neurogenin2 in the embryonic neocortex. Dev Biol 273: 373–389 [DOI] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE (1999) NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev 13: 1647–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh H, Fung BP, Naya FJ, Tsai MJ, Nishitani J, Leiter AB (1997) The basic helix–loop–helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc Natl Acad Sci USA 94: 3560–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Stellrecht CM, Tsai MJ (1995) Tissue-specific regulation of the insulin gene by a novel basic helix–loop–helix transcription factor. Genes Dev 9: 1009–1019 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J (1967) Normal Table of Xenopus laevis (daudin). Amsterdam: North-Holland Publishing Company [Google Scholar]
- Ogino H, McConnell WB, Grainger RM (2006) Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech Dev 123: 103–113 [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L (2004) ECR browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res 32: W280–W286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, Chen Y, Loeber J, Henningfeld K, Pieler T (2006) I-SceI meganuclease-mediated transgenesis in Xenopus. Dev Dyn 235: 247–252 [DOI] [PubMed] [Google Scholar]
- Pennesi ME, Cho JH, Yang Z, Wu SH, Zhang J, Wu SM, Tsai MJ (2003) BETA2/NeuroD1 null mice: a new model for transcription factor-dependent photoreceptor degeneration. J Neurosci 23: 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron M, Opdecamp K, Butler K, Harris WA, Bellefroid EJ (1999) X-ngnr-1 and Xath3 promote ectopic expression of sensory neuron markers in the neurula ectoderm and have distinct inducing properties in the retina. Proc Natl Acad Sci USA 96: 14996–15001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzoli O, Bosetti A, Croci L, Consalez GG, Vetter ML (2001) Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Dev Biol 233: 495–512 [DOI] [PubMed] [Google Scholar]
- Scardigli R, Schuurmans C, Gradwohl G, Guillemot F (2001) Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron 31: 203–217 [DOI] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, Cunningham JM, Dyck R, Walsh C, Campbell K, Polleux F, Guillemot F (2004) Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J 23: 2892–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Richardson GA, Kroll KL (2005) The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development 132: 105–115 [DOI] [PubMed] [Google Scholar]
- Sharma S, Jhala US, Johnson T, Ferreri K, Leonard J, Montminy M (1997) Hormonal regulation of an islet-specific enhancer in the pancreatic homeobox gene STF-1. Mol Cell Biol 17: 2598–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer L, Ma Q, Anderson DJ (1996) Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci 8: 221–241 [DOI] [PubMed] [Google Scholar]
- Tapscott SJ (2005) The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132: 2685–2695 [DOI] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Lockshon D, Lassar A (1990) MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci USA 87: 5623–5627 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data