Abstract
Post-translational modifications of histones are involved in transcript initiation and elongation. Methylation of lysine 36 of histone H3 (H3K36me) resides promoter distal at transcribed regions in Saccharomyces cerevisiae and is thought to prevent spurious initiation through recruitment of histone-deacetylase activity. Here, we report surprising complexity in distribution, regulation and readout of H3K36me in Drosophila involving two histone methyltransferases (HMTases). Dimethylation of H3K36 peaks adjacent to promoters and requires dMes-4, whereas trimethylation accumulates toward the 3′ end of genes and relies on dHypb. Reduction of H3K36me3 is lethal in Drosophila larvae and leads to elevated levels of acetylation, specifically at lysine 16 of histone H4 (H4K16ac). In contrast, reduction of both di- and trimethylation decreases lysine 16 acetylation. Thus di- and trimethylation of H3K36 have opposite effects on H4K16 acetylation, which we propose enable dynamic changes in chromatin compaction during transcript elongation.
Keywords: chromatin, histone methyltransferases, histone modifications, transcription, transcription elongation
Introduction
Nucleosomal packaging provides a barrier for protein binding to DNA and for the processivity of DNA and RNA polymerases. Changes in chromatin structure involve ATP-dependent remodeling of nucleosomes and numerous post-translational modifications of histones (Felsenfeld and Groudine, 2003). Depending on the modification and the targeted residue, these alterations can affect sequence accessibility for DNA-binding proteins or create recognition modules for effector proteins with defined functions (Jenuwein and Allis, 2001; Peters and Schubeler, 2005). Indeed, chromatin modifications appear involved in all steps of transcription, such as initiation, elongation and termination (Sims et al, 2004). A large body of work links histone H3 modifications, such as lysine 4 methylation and lysine 9 and 14 acetylation to RNA polymerase recruitment at the promoter. Recent genome-wide studies revealed that these active modifications are present at transcribed genes, yet occur preferentially at the promoter and adjacent downstream regions. This suggests a shared chromatin profile consisting of several histone tail modifications involved in early events of transcription (Robert et al, 2004; Schubeler et al, 2004; Pokholok et al, 2005; Barski et al, 2007).
Although extensive knowledge exists on promoter-proximal events, less is known about the cross talk between the elongating polymerase and chromatin. Several chromatin-associated proteins are implicated in aiding transcriptional elongation. Some affect histone acetylation (Wittschieben et al, 1999; Winkler et al, 2002), whereas others show nucleosomal remodeling or histone chaperone activity (Belotserkovskaya et al, 2003; Kaplan et al, 2003; Mason and Struhl, 2003; Morillon et al, 2003). These findings suggest that nucleosomes are acetylated and mobilized during transcriptional elongation. Moreover, high levels of transcription cause nucleosomal displacement, leading to reduced nucleosomal density, which in metazoa appears to be compensated in part by depositing nucleosomes that contain the variant histone H3.3 (Lee et al, 2004; Mito et al, 2005; Schwartz and Ahmad, 2005; Wirbelauer et al, 2005). Thus, although chromatin opening is required for gene activation at promoters, transcription itself causes chromatin disruption, which appears to be compensated by reestablishing a compact chromatin state.
Methylation of lysine 36 of histone H3 (H3K36me) is the only covalent histone modification reported to be enriched in the 3′ end of active genes (Bannister et al, 2005; Kizer et al, 2005; Pokholok et al, 2005; Rao et al, 2005; Barski et al, 2007; Mikkelsen et al, 2007). Therefore, understanding the regulation of this modification is likely to shed light on the interplay between chromatin and transcriptional elongation. In S. cerevisiae, all lysine 36 methylation is mediated by Set2 histone methyltransferase (HMTase), which is directed to active genes through interaction with elongation-competent RNA polymerase II (Krogan et al, 2003; Li et al, 2003; Xiao et al, 2003; Kizer et al, 2005). In turn, Set2-methylated nucleosomes signal for cooperative binding of two subunits of the Rpd3S complex (Li et al, 2007a), resulting in recruitment of histone-deacetylase activity to the body of active genes (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005). Thus, H3K36 methylation has been proposed to be involved in the maintenance of repressive chromatin structure. Indeed, loss of Rpd3 recruitment by deletion of SET2 results in an increase of spurious intragenic initiation events (Carrozza et al, 2005; Li et al, 2007b). This supports a function for H3K36me in compensating for transcription-coupled disruption and hyperacetylation of chromatin, which otherwise could unmask cryptic promoters. However, because deacetylation interferes with transcriptional initiation, mechanisms need to be in place to ensure that H3K36me-mediated HDAC recruitment does not occur in vicinity of active promoters.
Little is known about potentially different chromosomal distributions or functions of the mono-, di- or trimethylated states of H3K36. Interestingly, two recently identified histone demethylases in the human genome have been shown to demethylate either preferentially H3K36me2 (Tsukada et al, 2005) or H3K36me3 (Klose et al, 2006; Whetstine et al, 2006) opening the possibility of enzymatically regulated differential turnover of H3K36 methylation states.
To investigate function and regulation of this residue in a higher eukaryote, we have characterized H3K36 methylation in Drosophila melanogaster. We show that dimethylation and trimethylation of H3K36 have distinct chromosomal localization, and we suggest that these methylation states rely on separate HMTases. Importantly, we find that H3K36 methylation states show antagonistic cross talk to H4 acetylation at lysine 16 (H4K16ac), which has been shown to directly influence packaging of higher-order chromatin (Dorigo et al, 2003; Shogren-Knaak et al, 2006).
These findings suggest opposing functions for H3K36 methylation states in Drosophila to regulate chromatin acetylation and presumably compaction during transcriptional elongation in higher eukaryotes.
Results
Global distribution of H3K36 di- and trimethylation in Drosophila cells
To address if H3K36me associates with repressive or permissive chromatin in metazoa, we determined the nuclear localization of H3K36me2 and H3K36me3 in Drosophila Kc cells. Similar to other euchromatic marks such as H3K4 methylation (Wirbelauer et al, 2005), both H3K36 methylation states are largely excluded from the transcriptionally inert heterochromatin but are highly enriched in the transcriptionally active euchromatic regions of the nucleus (Figure 1A).
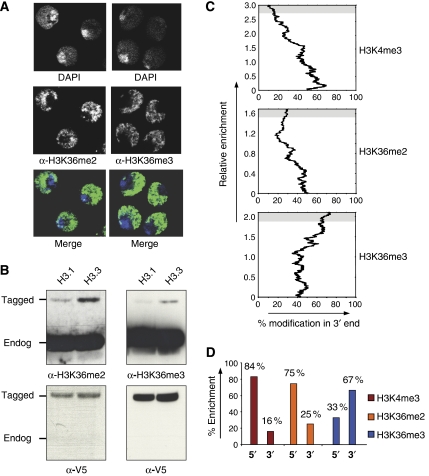
Figure 1.
Nuclear and chromosomal localization of H3K36me2 and H3K36me3. (A) Drosophila Kc cells were stained with DAPI in combination with antibodies specific to H3K36me2 and H3K36me3. Merged pictures are pseudo-colored with antibody staining in green and DAPI staining in blue. The heterochromatic chromocenter is visible in these cells as a region with high signal for DAPI, whereas the euchromatin shows weaker DAPI staining. Both methylation states of H3K36 strongly stain euchromatin and are excluded from the transcriptionally inert chromocenter. (B) H3K36me2 and H3K36me3 are enriched at the variant histone H3.3. Histones were isolated from Kc cells expressing either epitope-tagged H3.1 or H3.3 (Wirbelauer et al, 2005) and analyzed by western blot. This analysis finds H3K36me2 and H3K36me3 enriched at H3.3 compared to canonical H3.1. Detection of the V5-epitope tag serves as a loading control. (C) Chromosome-wide analysis of the distribution of H3K4me3 and H3K36me2/me3 relative to the 5′ or 3′ end of coding regions. Following ChIP, DNA from input and bound fraction were co-hybridized to a microarray representing the complete Drosophila chromosome 2L (see Materials and methods). To determine the relative enrichment for either the 5′or 3′ end of genes, we focused on tiles that were within genes and either contained the 5′ or 3′ end. We ignored those that were intergenic or mid-genic. The 5′ and 3′ end tiles (2105 for H3K4me3, 2041 for H3K36me2, 2123 for H3K36me3) were ranked according to their ChIP enrichment, and then we asked if tiles that are highly enriched are biased toward the 5′ or 3′ end. Note that enrichment of 3′ end tiles implies absence from 5′ tiles. Shown is a moving average (%, n=100) of tiles in the 3′ end relative to enrichment. This illustrates a preferential 5′ position of tiles enriched for H3K4me3 (85% of enriched regions reside at a 5′ end of a gene) and a 3′ bias for tiles enriched for H3K36me3 (67% of enriched tiles are at the 3′ end). H3K36me2 shows a 5′ bias in this analysis similar to H3K4me3, yet less pronounced. (D) Summary of chromosomal array with 2 kb resolution. The bar chart shows the abundance in 5′ or 3′ positions for those sequences with the highest enrichment in each modification (upper 10% of tiles, illustrated with a gray background in C).
This nuclear localization reflects the presence of both H3K36 methylation states at active genes, as we find both enriched at the ectopically expressed histone variant H3.3 (Figure 1B). H3.3 is deposited at active genes, which are subject to transcription-coupled nucleosomal displacement events (Mito et al, 2005; Schwartz and Ahmad, 2005; Wirbelauer et al, 2005). This confirms previous observations by mass spectrometry of endogenous H3.3 in Drosophila Kc cells (McKittrick et al, 2004) and suggests that both H3K36 methylation states are enriched at sites of active transcription.
To investigate the chromosomal distribution of lysine 36 methylation, we performed chromatin-immunoprecipitation (ChIP) using antisera specific for H3K36me2 or H3K36me3. DNA from enriched chromatin was compared to input chromatin by comparative hybridization to a DNA microarray representing chromosome 2L of the Drosophila genome in a 2 kb tiling resolution (MacAlpine et al, 2004; Schubeler et al, 2004). In addition, we determined the chromosomal distribution of H3K4me3, which we and others have previously shown to occur promoter proximal at active genes (Bernstein et al, 2005; Pokholok et al, 2005; Wirbelauer et al, 2005). The resulting distribution on chromosome 2L confirms these previous studies, as we find preferential enrichment of H3K4me3 at the 5′ end of active genes (Figure 1C and D). H3K36me3, in contrast, shows a different localization, as it is highly enriched toward the 3′ end of active genes as previously reported for S. cerevisiae (Pokholok et al, 2005). The distribution of H3K36me2 with preferential distribution toward the 5′ end is remarkably different, although this distribution is less pronounced than that of H3K4me3 (Figure 1C and D). Together, this chromosome-wide analysis revealed different distributions of di- and trimethylated lysine 36, with trimethylation localizing toward the 3′ end and dimethylation being adjacent to the promoter.
Before proceeding with further analysis, we confirmed the specificity of the antibodies. We did not detect cross-reactivity against different methylation states of H3K36 when tested against peptides, suggesting that both antibodies are selective for either the di- or trimethylated state of this residue (Supplementary Figure 1A). We note that not all tested commercial antibodies showed equally high levels of discrimination in this assay (Supplementary Figure 1A), which might explain why a differential distribution of di- and trimethylation of H3K36 has not been reported previously. To test for potential cross-reactivity to regions of histone H3 outside of the peptides used, we tested both antibodies against ectopically expressed H3.3 in which H3K36 had been mutated to alanine. This point mutation leads to a loss of detection for each antibody (Supplementary Figure 1B), indicating that both are specific for defined H3K36 methylation states in the context of full-length histone H3.
Next, we determined the distribution of H3K36 methylation states at a subset of genes using real-time PCR (RT–PCR) and previously published amplicons with a spatial resolution of approximately 750 bp (as compared to over 2000 bp of the microarray) (Wirbelauer et al, 2005). This enabled us to relate the observed enrichments to our existing datasets of other histone tail modifications, RNA polymerase II (RNA-Pol II) and the replacement histone H3.3 (Wirbelauer et al, 2005). This analysis confirmed that H3K36me3 is biased toward the 3′ end of active genes (Figure 2A and B). It also verified a different distribution for H3K36me2, as this mark was most abundant at a region between the 5′ peak of H3K4me3 and the 3′ peak of H3K36me3 (Figure 2B). The observed distribution is not cell line-specific, as similar results were obtained at the same set of genes in a second Drosophila cell line, SL2 (data not shown).
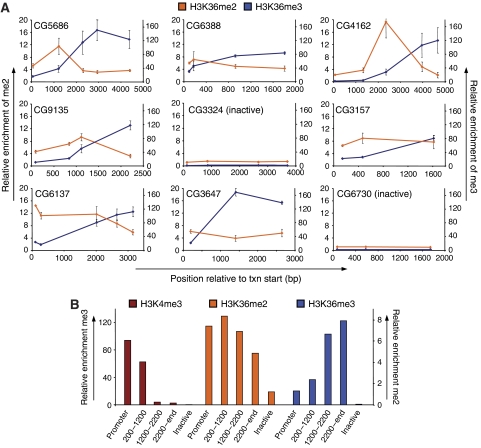
Figure 2.
High-resolution analysis of di- and trimethylation of H3K36 at individual genes. (A) ChIP analysis in Drosophila Kc cells along the body of several genes using antibodies specific for H3K36me2 or H3K36me3 and quantification by RT–PCR. Enrichments were normalized to nucleosomal abundance determined with an antibody against the C-terminus of H3. Shown is average and standard deviation from at least three independent repeats starting with cells at different passages. X-axis reflects the base-pair position relative to the transcriptional start site. Y-axis reflects enrichment (bound/input normalized to an intergenic control). H3K36me2 (orange line, scale on the left), H3K36me3 (blue line, scale on the right). Numbers in graphs are gene IDs according to Flybase. (B) Summary of individual genes. Probes at active genes shown in A were grouped according to the distance from the transcriptional start site (promoter n=5, 200–1200 bp n=8, 1200–2200 bp n=4, 2200-end bp n=7). All probes at inactive genes were grouped separately (inactive n=7). For each group, the average enrichment for each modification was calculated. Values for H3K4me3 and H3K36me3 are according to the left scale, values for H3K36me2 are according to the right scale. This representation illustrates a strong 5′ bias for H3K4me3, an intermediate 5′ bias for H3K36me2 and a strong 3′ bias for H3K36me3.
We conclude that three chromatin signatures can be distinguished along active genes based on H3 tail modifications. A promoter-proximal region of high H3K4 methylation, an intermediate region characterized by a peak in H3K36me2 and a further 3′ region characterized by high H3K36me3. Thus, different K36 methylation states mark discrete regions of transcribed genes.
Two proteins mediate H3K36 methylation in Drosophila
To gain further insights into the underlying enzymatic regulation and function, we sought to identify the proteins responsible for H3K36 methylation based on homology to the SET domain sequence of the single H3K36 HMTase in S. cerevisiae (Set2) (Supplementary Figure 2A). We performed an RNAi screen against putative HMTases and used bulk analysis of H3K36me2 and H3K36me3 levels by western blot as a readout for loss of function. This identified two SET domain-containing proteins (CG4976 and CG1716) (Figure 3A) that upon knockdown showed reduced levels of H3K36 methylation (Figure 3B and D). Thus, we find that at least two putative HMTases are involved in H3K36 methylation in flies. To ensure specificity of the RNAi, we raised specific antibodies against both proteins (see Material and methods), which confirmed efficient protein reduction upon addition of dsRNA (Figure 3C). We named CG1716 as ‘Drosophila Hypb' (dHypb) based on homology to the human HMTase HYPB (Sun et al, 2005). CG4976, in contrast, shows homology to the nuclear-receptor-binding SET-domain-containing protein (NSD) family of SET domain proteins (Supplementary Figure 2B) and has previously been annotated as Drosophila Mes-4 (dMes-4) based on its similarity to a SET domain-containing protein in the C. elegans genome. In worms, Mes-4 is required for H3K36 methylation at autosomes in early embryo and is necessary for germline viability (Bender et al, 2006).
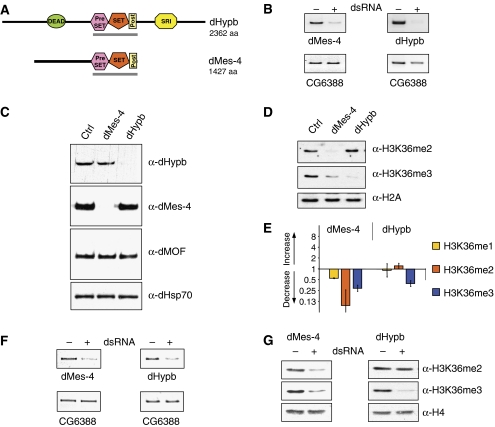
Figure 3.
Identification of Drosophila SET domain proteins involved in H3K36 methylation. (A) Domain structure of full-length HMTase proteins as predicted by the SMART software (EMBL). DEAD=ATP dependant helicase domain, SRI=Set2 Rpb1 interacting domain. The gray bar indicates protein fragments tested for HMTase activity in vitro. (B) Validation of mRNA knockdown in cultured Kc cells. RT–PCR of target message in the absence (−) or presence (+) of dsRNA reveals efficient mRNA knockdown. The gene expression of CG6388 is unaffected in both knockdowns and serves as a loading control. (C) Validation of protein reduction. Western blot using antibodies specific for dMes-4 and dHypb in the absence (−) or presence (+) of dsRNA reveals efficient reduction of the targeted proteins in Kc cells. dHsp70 and dMOF are unaffected by RNAi knockdown and serve as loading control. (D) Reductions of dMes-4 and dHypb have specific effects on global levels of H3K36 methylation. Loss of dHypb results in the reduction of H3K36me3 and coinciding increase of H3K36me2. Knockdown of dMes-4 leads to a reduction in both H3K36me2 and H3K36me3. H2A serves as loading control. Global levels of H3 and H4 were unaffected (Supplementary Figure 5A). (E) Mass spectrometry analysis of H3K36-methylated peptides. MS-MS analysis of mono-, di- or trimethylated H3K36 moieties following knockdown of putative HMTases in Drosophila Kc cells. The bar chart displays fold changes in the abundance of H3K36 methylation states relative to untreated control cells. (F) Knockdown of putative Drosophila H3K36 HMTases in vivo. RT–PCR from larvae uninduced (−) or induced (+) for targeted knockdown of either dMes-4 or dHypb mRNA in vivo. Reduced transcript abundance is detected in the presence of GAL4 driver under the control of a ubiquitously expressed (tubulin) promoter. CG6388 mRNA levels serve as loading control. (G) Western blot analysis of H3K36 methylation states in fly larvae mirror the observations in cultured cells. Reduction of dMes-4 message results in the reduction of H3K36 di- and trimethylation, whereas dHypb RNAi specifically downregulates H3K36me3. Levels of total H4 serve as loading control.
When we compared the levels of di- versus trimethylation of H3K36 upon reduction of dHypb or dMes-4, we noted a striking difference. RNAi against dMes-4 reduces di- and trimethylation, suggesting that the activity of this enzyme is required for both methylation states (Figure 3D). Knockdown of dHypb, in contrast, results in the downregulation of trimethylation alone, whereas levels of dimethylation slightly increase. This result is in disagreement with the recent study that reported reduction of dimethylation following RNAi knockdown of CG1716 in flies (Stabell et al, 2007). However, we point out that the authors relied for detection on an antibody that we found cross-reactive with trimethylated lysine 36 peptide (Supplementary Figure 1A), possibly accounting for this discrepancy.
To validate the differential effects on lysine 36 methylation by an antibody-independent approach, we analyzed histones isolated from either control or RNAi-treated cells by mass spectrometry. We compared the levels of H3K27 and H3K36 methylation within the peptides comprising amino acids 27–40 of histone H3. To determine the changes in methylation of H3K36, we analyzed the mono-, di- and trimethylated isoforms and measured the levels of H3K36 methylation relative to the methylation at H3K27 by nanospray MS/MS (see Materials and methods). This confirmed the downregulation of both di- and trimethylation of H3K36 upon loss of dMes-4, whereas loss of dHypb reduced trimethylation alone (Figure 3E). In addition, we observed a modest increase of K36me2 in the dHypb knockdown in line with the results obtained by western blot; however, the low abundance of K36me2 relative to K27me2 precludes a robust quantification.
dHypb is essential for fly development
To examine whether changes in H3K36 methylation states would influence organismal development, we generated transgenic fly lines harboring an RNAi construct complementary to either dMes-4 or dHypb message under the control of a GAL4-inducible promoter (see Materials and methods). Transcription of the respective RNAi construct was triggered by crossing in a fly strain that expresses the GAL4 activator ubiquitously under the control of the tubulin promoter. Induction of the dMes-4 RNAi construct led to a detectable reduction of target mRNA (Figure 3F), yet only in a subset of fly lines. Those with reduced expression of dMes-4 mRNA showed a decrease of both K36me2 and K36me3 at larval stages, confirming the results from cell culture (Figure 3G). In the case of dHypb, induction of the RNAi transgene efficiently reduced dHypb mRNA (Figure 3F) and led to decreased levels of H3K36me3, confirming the results in cultured cells (Figure 3G and D). Moreover, postzygotic depletion of dHypb levels was lethal at the larvae–pupae transition with 100% penetrance observed in multiple independent integration sites of the RNAi construct (see Materials and methods) in agreement with a recent report (Stabell et al, 2007). We conclude that reducing HMTase levels in flies mirror the chromatin effects seen in cultured cells and that a strong reduction of dHypb is lethal, suggesting that dHypb-mediated H3K36me3 is essential for development.
dHypb shows HMTase activity at histone H3 in vitro
To test the enzymatic activity of both enzymes in vitro, we expressed fragments containing pre-SET, SET and post-SET domains as GST-tagged fusion proteins using Baculovirus infection of insect cells (Supplementary Figure 3B). Recombinant proteins were purified and incubated with radioactively labeled SAM as methyldonor and histones purified from calf thymus as a substrate. HMTase activity was reproducibly detected for dHypb by measuring radioactive incorporation into histones. Subsequent gel separation of labeled products revealed that dHypb methylates preferentially histone H3 (Figure 4A). Unlike dHypb, dMes-4 displayed only weak activity under various conditions tested (data not shown). Although this might reflect the lack of necessary cofactors, it precluded us from further defining dMes-4 activity in vitro.
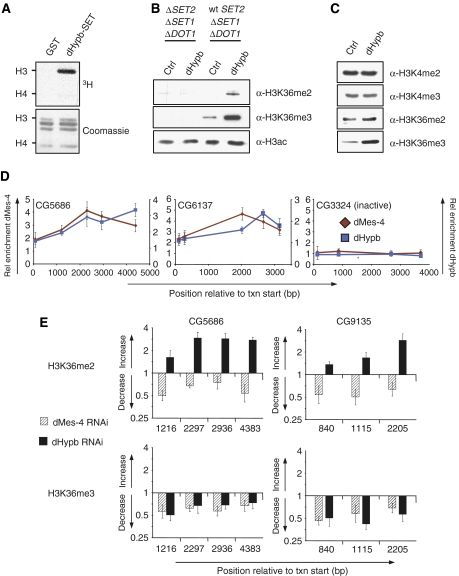
Figure 4.
dHypb methylates lysine 36 in vitro and colocalizes with dMes-4 at active genes. (A) dHypb shows histone-methyltransferase activity in vitro. Recombinant protein fragments containing pre- and post-SET domain (dHypb: aa 1351–1553, shown as gray bar in Figure 3A) were incubated with radioactive SAM and histones from calf thymus as substrates. Shown in the upper panel is the reaction product that was separated by SDS–PAGE and incorporated radioactivity measured by exposure to film. The lower panel displays Coomassie-stained SDS–PAGE gel and serves as loading control. Recombinant dHypb-SET shows HMTase activity to histone H3 in this assay. (B) dHypb methylates H3K36 in vitro. Western blot analysis displays H3K36 di- and trimethylation levels of HMTase assay with recombinant full-length dHypb using mutant yeast nuclear extracts deficient for H3K4me, K36me and K79me (ΔSET2, ΔSET1, ΔDOT1) or K4me and K79me (wt SET2, Δset1, Δdot1) as a substrate. A dHypb-dependent increase of di- and trimethylation is obtained only with chromatin substrate from wt SET2 strain, suggesting that dHypb requires premethylated lysine 36 substrate for its activity. (C) Western blot analysis of Drosophila Kc-overexpressing dHypb shows a specific increase in trimethylation. A similar experiment with full-length dMes-4 in Kc cells did not reveal robust changes in H3K36 methylation (data not shown). (D) ChIP analysis using antibodies generated against endogenous dMes-4 and dHypb along the body of two active genes (CG6137 and CG5686) and one inactive gene (CG3324). Shown is average and standard deviation from at least three independent repeats. X-axis reflects the base-pair position relative to the transcriptional start site. Y-axis reflects enrichment (bound/input normalized to an intergenic control). (E) dMes-4 and dHypb define distinct methylation states in Kc cells. Levels of H3K36 methylation states in RNAi and control cells were compared with ChIP followed by RT–PCR analysis. Shown is the ratio of H3K36me enrichments (fold change, Y-axis) of RNAi over control cells relative to the position from the transcription start site (X-axis) of two actively transcribed genes. RNAi against dHypb leads to a reduction of H3K36me3 in the coding region with a coinciding increase of H3K36me2 predominantly toward the 3′ end. Loss of dMes-4 leads to a reduction of both H3K36me2 and H3K36me3.
To gain insights into dHypb specificity, we incubated recombinant full-length protein with nuclear extracts from yeast strains that lack K4me, K36me and K79me of histone H3 or K4me and K79me only (Figure 4B and Material and methods). Subsequent western blotting revealed a dHypb-dependent increase in di- and trimethylation of lysine 36, yet only when supplied with yeast chromatin positive for mono-, di- and trimethylation of H3K36 (Figure 4B). In contrast, no methylation was detected in mutant extracts deficient for any lysine 36 methylation. Whereas these results demonstrate that dHypb mediates H3K36 methylation in vitro, they also indicate that its activity relies on the presence of premethylated lysine 36 substrate.
To address if a similar specificity can be observed in vivo, we ectopically expressed dHypb in Drosophila Kc cells. Upon overexpression, we found only H3K36me3 levels to be increased (Figure 4C and Supplementary Figure 3C), which is in agreement with the reduction of trimethylation observed upon knockdown of dHypb.
In conclusion, we found that dHypb methylates H3K36 in vitro, yet requires premethylated lysine 36 substrate for its activity. Combined with the specific changes of trimethylation observed in vivo, our results suggest that dHypb is responsible for the trimethylated state of lysine 36 in Drosophila.
dMes-4 and dHypb localize at sites of H3K36 methylation at active genes
As both dMes-4 and dHypb show distinct effects on H3K36me2 and H3K36me3 upon knockdown in vivo, we asked if both act at the same genes. To test for chromosomal binding, we carried out ChIP using antibodies specific for endogenous dMes-4 or dHypb and quantified enrichments by RT–PCR. This revealed that both enzymes colocalize to actively transcribed genes, while being absent from an inactive gene (Figure 4D). The distributions of both HMTases are largely overlapping, with highest relative abundances downstream of the promoter at sites of di- and trimethylation of H3K36 (compare Figure 2A).
H3K36 methylation states occur enzyme-specific at individual genes
To test if recruitment of either enzyme mediates defined methylation states, we reduced the level of either one of the two enzymes by RNAi and determined the local effect on H3K36me by ChIP analysis at individual genes. Reduction of dHypb at selected genes results in reduced presence of trimethylation and coincident increase of dimethylation (Figure 4E). This finding suggests that the slight upregulation of bulk H3K36me2 levels detected by western blot and mass spectrometry (Figure 3D and E) reflects an increase at intragenic regions, which are trimethylated in untreated cells. In contrast, reduction of the endogenous levels of dMes-4 coincides with a decrease of both H3K36me2 and H3K36me3 not only in bulk (Figure 3D), but also at individual genes (Figure 4E). One possible interpretation of these results is that both enzymes act consecutively at the same set of genes to mediate distinct patterns of di- and trimethylation of H3K36.
Specific cross talk between H4 lysine 16 acetylation and H3K36 methylation states
In S. cerevisiae, H3K36 methylation has been reported to recruit an HDAC-containing complex, resulting in deacetylation of the 3′ end of actively transcribed genes (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005). No direct comparison between localization of di- and trimethylation of H3K36 has been reported in budding yeast; however, loss of the only H3K36 methylase Set2 results in hyperacetylation of both histones H3 and H4 within reading frames (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005).
We therefore examined whether H3K36 methylation in flies shows a similar cross talk to histone acetylation and whether the di- and trimethylated states of H3K36 in Drosophila have separate functions in this pathway. Reduction of H3K36 trimethylation by dHypb knockdown in female Kc cells resulted in significant upregulation of bulk levels of H4 acetylation at lysine 16 (Figure 5A). Although this hyperacetylation is reminiscent of global hyperacetylation reported in yeast SET2 mutants (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005; Li et al, 2007b), it appeared to be specific for H4K16, as no change was detected at other lysine residues tested (Figure 5A and Supplementary Figure 3A). In contrast, reduction of H3K36 di- and trimethylation upon knockdown of dMes-4 had the inverse effect, as acetylation at lysine 16 of histone H4 was globally decreased. Together, these data reveal a cross talk between methylation of H3K36 and acetylation of H4 in D. melanogaster, however, in a more intricate way than what has been reported for S. cerevisiae. Notably, the knockdown of dMes-4 resulted in the reduction of both di- and trimethylation similar to a SET2 mutant, yet it did not result in increased H4 acetylation, which appears to require decline of trimethylation in the presence of dimethylation.
Figure 5.
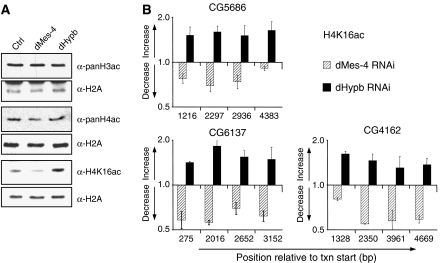
Differential cross talk between H3K36 methylation states and H4K16 acetylation. (A) Knockdown dMes-4 or dHypb has specific effects on the acetylation of H4K16. Levels of H3 and H4 acetylation in bulk histones were measured upon RNAi by western blot analysis. In Drosophila Kc cells, knockdown of dMes-4 results in a reduction, whereas knockdown of dHypb leads to an increase of H4K16 acetylation. No changes were detected at other residues on H4 (Supplementary Figure 3A). (B) ChIP analysis of H4K16 acetylation changes upon knockdown of dMes-4 and dHypb. Shown is the ratio of H4K16ac enrichments (fold change, Y-axis) of RNAi over control cells relative to the position from the transcription start site (X-axis) of three actively transcribed genes. These results suggest that the changes in bulk acetylation of histones occur at the same chromosomal positions that change levels of H3K36 methylation states.
Next, we measured H4K16ac by ChIP to test if changes in global levels reflect acetylation differences at genes that lose H3K36me after HMTase knockdown. In untreated Kc cells, H4K16 acetylation peaked at the promoter and was present to a lesser extent throughout coding regions (Supplementary Figure 5). After knockdown of dHypb, H4K16ac increased at sequences that showed reduced levels of H3K36 trimethylation. In contrast, acetylation decreased at chromosomal positions, which also had reduced di- and trimethylation after dMes-4 RNAi (Figure 5B). These results are in agreement with bulk changes observed by western blot analysis. Importantly, the acetylation changes occur preferentially at H4K16, as other residues tested in ChIP appear unaffected (H3K9/K14ac, H4K8ac, H4K12ac; Supplementary Figure 4 and data not shown).
We conclude that H3K36me2 and H3K36me3 inversely influence H4K16 acetylation at transcribed regions in the Drosophila genome.
Discussion
In this study, we provide a comprehensive analysis of the distribution and regulation of the only described chromatin modification associated with transcriptional elongation in D. melanogaster. Our results suggest a novel regulatory pathway involving two enzymes, different localization of di- and trimethylation of H3K36 and cross talk of these methylation states to acetylation of lysine 16 of histone H4 (Figure 6A and B), a modification previously shown to prevent formation of compact chromatin.
Figure 6.
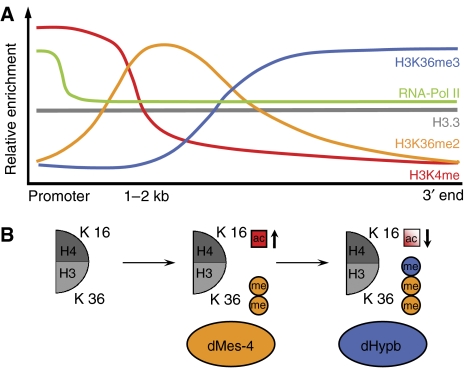
Summary model of H3K36 distribution and function in Drosophila. (A) Summary of the distribution of several H3 tail modifications along an exemplified active gene. A peak in promoter-proximal marks such as H3K4me followed by a region high in H3K36me2 and a 3′-specific region marked by high levels of H3K36me3, both depending on separate HMTases (this study). (B) Model of a differential cross talk to H4K16 acetylation at autosomes. dMes-4-dependent H3K36me2 leads to increased H4K16 acetylation, which is compensated by dHypb-catalyzed H3K36me3.
Spatially defined H3K36 methylation states
Analogous to previously described euchromatic histone modifications, di- and trimethylation of H3K36 are enriched at transcribed genes; yet both show unique distributions downstream of 5′-biased modifications such as H3K4me (Bernstein et al, 2005; Pokholok et al, 2005; Wirbelauer et al, 2005). H3K36me3 peaks in the 3′ end, whereas H3K36me2 shows an intermediate distribution (Figure 6A). A previous analysis of H3K36me2 and H3K36me3 in chicken erythrocyte genes did not deduce a differential distribution; however, only two genes were compared (Bannister et al, 2005). Thus, whereas the actual conservation of H3K36 methylation patterns in other metazoa needs to be tested, the spatial and functional differences that we report for Drosophila might be shared by eukaryotes that encode multiple H3K36 methylases.
We, furthermore, note that despite the topographic differences between H3K36me2, H3K36me3 and H3K4me3, all tested modifications are equally enriched at the replacement histone H3.3 (Figures 1B and 6A and Wirbelauer et al, 2005). This is in agreement with our previous observation that the distribution of this variant histone throughout coding regions does not mirror the complex pattern of underlying tail modifications (Wirbelauer et al, 2005).
Two enzymes mediate H3K36 methylation states
The consequences of protein knockdown suggest that dMes-4 is required for dimethylation and that dHypb mediates trimethylation (Figure 3D and E). This model is supported by the in vitro activity of dHypb (Figure 4A and B) and localization of both enzymes to the same genes (Figure 4D). Thus, a stepwise mechanism for lysine 36 methylation seems plausible in which dMes-4 mediates dimethylation, which is substrate for dHypb-mediated trimethylation. Methylation-state-specific HMTases have been described for H3K76 involved in cell-cycle regulation of Trypanosoma brucei (Janzen et al, 2006). Our findings suggest similar enzyme specificity for H3K36 methylation at active genes. As such, it is different from the regulation of H3K4 methylation, where trimethylation depends on the presence of cofactors (Steward et al, 2006). We also note that both H3K4 di- and trimethylation peak at the 5′ end, whereas H3K36 di- and trimethylation mark distinct intragenic regions, which supports a differential recruitment of the HMTases along the transcribed gene (Figure 6). The absence of lysine 36 methylation at the promoter might reflect acetylation at this residue (H3K36ac), which has recently been reported in S. cerevisiae. In yeast, Gcn5-dependent H3K36 acetylation predominantly localizes to active promoters and inversely correlates with methylation, suggesting that its presence precludes methylation from the 5′ end of the transcription unit (Morris et al, 2006).
Work in budding yeast suggests that targeting of Set2 involves physical interaction with the phosphorylated CTD of RNA polymerase II via a Set2 Rpb1 interacting (SRI) domain (Kizer et al, 2005). Indeed, domain prediction algorithms identify a similar SRI domain in the C-terminal region of dHypb (Figure 3A), and a recent study reports physical interaction with hyperphosphorylated RNA polymerase (Stabell et al, 2007). Moreover, loss of Set2 leads to elevated levels of histone acetylation similar to a loss of dHypb. Together, these observations suggest that targeting of H3K36me3 involves interaction with elongating RNA polymerase and that dHypb is the functional Drosophila equivalent to yeast Set2.
dMes-4, in contrast, shows homology to human and mouse NSD1, WHSC1 (also referred to as NSD2 or MMSET) and WHSC1-L (NSD3) (Supplementary Figure 2B). This family of NSDs has been implicated in several human malignancies, including protein fusions in cancer (Jaju et al, 2001; Wang et al, 2007). Moreover, haploinsufficiency of NSD1 causes Sotos syndrome, a neurological disorder (Kurotaki et al, 2002). NSD1 has been reported to methylate H3K36 in vitro (Rayasam et al, 2003) and to interact with transcription factors (Huang et al, 1998). It is conceivable that the mammalian NSD class of proteins shares activity and function that we report for dMes-4.
Cross talk of H3K36 methylation states to histone acetylation suggests opposing roles in regulating chromatin compaction on autosomes
Our results indicate that, in metazoa, di- and trimethylation of H3K36 exert different effects on the level of histone acetylation throughout coding regions (Figure 6B). Reduction of trimethylation coincides with specific upregulation of H4 acetylation. This trans-histone effect on acetylation is reminiscent of the phenotype of a SET2 deletion in budding yeast (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005). Here, we show that, in Drosophila, upregulation occurs preferentially at lysine 16, a residue that plays a particular role in chromatin compaction, as in vitro and in vivo evidence established that its acetylation is sufficient to prevent formation of higher-order structure (Corona et al, 2002; Dorigo et al, 2003; Shogren-Knaak et al, 2006). This points toward a function of H3K36me3 in mediating a more compact chromatin structure at the 3′ end of Drosophila genes either actively by recruiting an HDAC in analogy to yeast or passively by interfering with the acetylation of H4K16.
In light of these results, it is unexpected that reduction of di- and trimethylation of H3K36 through knockdown of dMes-4 does not increase H4 acetylation. Hence, the increase of dimethylation in the dHypb knockdown is required for hyperacetylation of H4, indicating that dimethylation has a function distinct from trimethylation and possibly recruits an H4K16 HAT activity. The HAT MOF has been shown in human cells to be responsible for H4K16ac (Smith et al, 2005; Taipale et al, 2005) and is assumed to be present in various complexes. Interestingly, human MOF has recently been shown to interact with the H3K4 methylases MLL (Dou et al, 2005), suggesting that parallel chromatin pathways can recruit H4K16 acetylation to different regions along the gene.
Together, our results suggest that H3K36 methylation states regulate acetylation levels of H4K16 at transcribed genes. Such opposing behavior provides a rationale for independent regulation by dMes-4 and dHypb and, moreover, could justify regulated demethylation of this residue. Indeed, histone demethylases specific for H3K36me2 (Tsukada et al, 2005) and H3K36me3 (Whetstine et al, 2006) have recently been described in the human genome, making a scenario of precise targeting and removal of individual H3K36 methylation states possible. Such network of setting and reverting-defined K36 methylation states with opposing effects on chromatin structure could be utilized to mediate cycles of chromatin opening and compaction that coincide with passage of the polymerase. If this is indeed the case, this could facilitate dynamic chromatin changes to temporarily allow polymerase passage or access for RNA-processing activities involved in splicing, termination or transport.
Materials and methods
ChIP and RT–PCR
ChIPs of histone modifications and PCR quantifications were carried out as described (Wirbelauer et al, 2005). Minor modifications and further details are provided in Supplementary data.
Microarray hybridization and analysis
Input and antibody-bound DNA were separately amplified and labeled as described (Schübeler et al, 2004), except using indirect labeling via aminoallyl-modified nucleotides. Labeled DNA was co-hybridized to a chromosomal tiling array representing chromosome 2L of the Drosophila genome in a 2 kb tiling resolution as described (MacAlpine et al, 2004). Two repeats (including dye swap) starting from independent ChIP experiments were carried out. After hybridization and washing, slides were scanned (Axon) and fluorescent reads were analyzed according to the standard normalization and filtering criteria in the GenePix software package (Axon). Repeats showed high reproducibility (H3K4me3 R=0.98, H3K36me2 R=0.91, H3K36me3 R=0.92), so that average value from two experiments could be used in further analysis. Complete datasets are available at GEO database (accession number GSE9414).
Antibodies
A detailed description of purchased and generated antibodies is provided in Supplementary data.
Mass spectrometry
A total of 5–10 pmol of acid-extracted histones were separated by SDS–PAGE. Coomassie blue-stained bands corresponding to histone H3 were excised and subjected to chemical modification to derive free amino groups of lysine residues (Greiner et al, 2005). Digestions were carried out overnight with sequencing-grade trypsin (Promega, Madison, WI, USA), according to the manufacturer's protocol. MALDI spectra were acquired (Bonaldi et al, 2004) for identification and determination of global changes. To compare methylation levels of H3K27 and H3K36, doubly charged peptides of H3 containing amino acids 27–40 to collision-induced decay were subjected to fragmentation and fragment spectra were analyzed in a Q-STAR XL mass spectrometer. For quantification, the relative intensities of the y8 and the y9 fragment ions that either carried methyl groups (K36me) or not (K27me) were compared.
Preparation of Kc cell nuclei and SDS–PAGE western blot analysis
Detailed protocols are provided in Supplementary data.
Fly strains and transgenes
Details are provided in Supplementary data.
HMTase assays
Detailed protocols are provided in Supplementary data.
Note added in proof
During the review of this manuscript, Larschan et al reported that H3K36 trimethylation mediated by dHypb has an additional function for dosage compensation of the male X-chromosome in Drosophila.
Supplementary Material
Supplementary Data
Acknowledgments
We thank Elena Aritonovska for assistance, Tianke Wang for sharing unpublished results, FMI core facilities for generation of antisera (Susanne Schenk) and peptide synthesis (Franz Fischer), Michael Rebhahn for support in bioinformatics, Floor Frederiks for help with yeast strains, members of the Schübeler lab for advice during the course of the projects and comments on the manuscript and Susan Gasser and Matthew Lorincz for critical reading of the manuscript. We acknowledge support by the Novartis Research Foundation (to AP and DS) and the EU 6th framework program (NOE ‘The Epigenome' LSHG-CT-2004-503433 to AP, FvL and DS and LSHG-CT-2006-037415 to AI and DS). SPB is supported by the Howard Hughes Medical Institute and the National Institutes of Health (GM52339). MS and AS are predoctoral fellows of the Boehringer Ingelheim foundation.
References
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T (2005) Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280: 17732–17736 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D (2003) FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093 [DOI] [PubMed] [Google Scholar]
- Bender LB, Suh J, Carroll CR, Fong Y, Fingerman IM, Briggs SD, Cao R, Zhang Y, Reinke V, Strome S (2006) MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 133: 3907–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR, Schreiber SL, Lander ES (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Bonaldi T, Imhof A, Regula JT (2004) A combination of different mass spectroscopic techniques for the analysis of dynamic changes of histone modifications. Proteomics 4: 1382–1396 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Corona DF, Clapier CR, Becker PB, Tamkun JW (2002) Modulation of ISWI function by site-specific histone acetylation. EMBO Rep 3: 242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Bystricky K, Richmond TJ (2003) Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol 327: 85–96 [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG (2005) Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121: 873–885 [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M (2003) Controlling the double helix. Nature 421: 448–453 [DOI] [PubMed] [Google Scholar]
- Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A (2005) Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol 1: 143–145 [DOI] [PubMed] [Google Scholar]
- Huang N, vom Baur E, Garnier JM, Lerouge T, Vonesch JL, Lutz Y, Chambon P, Losson R (1998) Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J 17: 3398–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaju RJ, Fidler C, Haas OA, Strickson AJ, Watkins F, Clark K, Cross NC, Cheng JF, Aplan PD, Kearney L, Boultwood J, Wainscoat JS (2001) A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 98: 1264–1267 [DOI] [PubMed] [Google Scholar]
- Janzen CJ, Hake SB, Lowell JE, Cross GA (2006) Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol Cell 23: 497–507 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F (2003) Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD (2005) A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 25: 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y (2006) The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442: 312–316 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J (2003) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T, Ohashi H, Naritomi K, Tsukahara M, Makita Y, Sugimoto T, Sonoda T, Hasegawa T, Chinen Y, Tomita Ha HA, Kinoshita A, Mizuguchi T, Yoshiura Ki K, Ohta T, Kishino T et al. (2002) Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet 30: 365–366 [DOI] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI (2007) MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28: 121–133 [DOI] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD (2004) Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL (2007a) Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL (2007b) Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21: 1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR III, Workman JL (2003) The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278: 8897–8903 [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP (2004) Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18: 3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K (2003) The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol 23: 8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick E, Gafken PR, Ahmad K, Henikoff S (2004) Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci USA 101: 1525–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S (2005) Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet 37: 1090–1097 [DOI] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, O'Sullivan J, Kent N, Proudfoot N, Mellor J (2003) Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115: 425–435 [DOI] [PubMed] [Google Scholar]
- Morris SA, Rao B, Garcia BA, Hake SB, Diaz RL, Shabanowitz J, Hunt DF, Allis CD, Lieb JD, Strahl BD (2006) Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J Biol Chem 282: 7632–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Schubeler D (2005) Methylation of histones: playing memory with DNA. Curr Opin Cell Biol 17: 230–238 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Rao B, Shibata Y, Strahl BD, Lieb JD (2005) Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol 25: 9447–9459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayasam GV, Wendling O, Angrand PO, Mark M, Niederreither K, Song L, Lerouge T, Hager GL, Chambon P, Losson R (2003) NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J 22: 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA (2004) Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell 16: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M (2004) The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BE, Ahmad K (2005) Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev 19: 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Belotserkovskaya R, Reinberg D (2004) Elongation by RNA polymerase II: the short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC (2005) A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol 25: 9175–9188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabell M, Larsson J, Aalen RB, Lambertsson A (2007) Drosophila dSet2 functions in H3-K36 methylation and is required for development. Biochem Biophys Res Commun 359: 784–789 [DOI] [PubMed] [Google Scholar]
- Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A (2006) Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 13: 852–854 [DOI] [PubMed] [Google Scholar]
- Sun XJ, Wei J, Wu XY, Hu M, Wang L, Wang HH, Zhang QH, Chen SJ, Huang QH, Chen Z (2005) Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem 280: 35261–35271 [DOI] [PubMed] [Google Scholar]
- Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A (2005) hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol 25: 6798–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada YI, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2005) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Wang GG, Cai L, Pasillas MP, Kamps MP (2007) NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 9: 804–812 [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125: 467–481 [DOI] [PubMed] [Google Scholar]
- Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2002) Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci USA 99: 3517–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbelauer C, Bell O, Schubeler D (2005) Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev 19: 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ (1999) A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell 4: 123–128 [DOI] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD (2003) Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev 17: 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data