Abstract
Kinetochores are complex protein machines that link chromosomes to spindle microtubules and contain a structural core composed of two conserved protein–protein interaction networks: the well-characterized KMN (KNL1/MIND/NDC80) and the recently identified CENP-A NAC/CAD. Here we show that the CENP-A NAC/CAD subunits can be assigned to one of two different functional classes; depletion of Class I proteins (Mcm21RCENP−O and Fta1RCENP−L) causes a failure in bipolar spindle assembly. In contrast, depletion of Class II proteins (CENP-H, Chl4RCENP−N, CENP-I and Sim4RCENP−K) prevents binding of Class I proteins and causes chromosome congression defects, but does not perturb spindle formation. Co-depletion of Class I and Class II proteins restores spindle bipolarity, suggesting that Class I proteins regulate or counteract the function of Class II proteins. We also demonstrate that CENP-A NAC/CAD and KMN regulate kinetochore–microtubule attachments independently, even though CENP-A NAC/CAD can modulate NDC80 levels at kinetochores. Based on our results, we propose that the cooperative action of CENP-A NAC/CAD subunits and the KMN network drives efficient chromosome segregation and bipolar spindle assembly during mitosis.
Keywords: CENP-H, FtalR, Mcm21R, mitosis, spindle
Introduction
Human kinetochores are complex molecular machines that assemble on a centromere that spans mega bases of DNA and ensure bi-orientation, congression and disjunction of sister chromatids during mitosis (Cleveland et al, 2003). Kinetochores execute three main functions: (1) they form a structure that is compatible with tight, but dynamic binding to the plus-end of spindle microtubules (MTs); (2) they modulate MT dynamics and kinesin-like proteins to generate the forces necessary to drive chromosome movement and (3) they generate spindle checkpoint signals that block cells before initiation of anaphase in the presence of unattached or tension-free kinetochores (Musacchio and Hardwick, 2002; Musacchio and Salmon, 2007). Bioinformatic and biochemical studies have demonstrated that kinetochores contain a structural core composed of two conserved protein–protein interaction networks called KMN and CENP-A NAC/CAD (De Wulf et al, 2003; Nekrasov et al, 2003; Cheeseman et al, 2004, 2006; Obuse et al, 2004; Liu et al, 2005; Foltz et al, 2006; Meraldi et al, 2006; Okada et al, 2006). The KMN network is composed of three complexes that are required for distinct functions: the NDC80 complex that is essential for MT–kinetochore attachment and spindle checkpoint signaling (Kline-Smith et al, 2005), the MIND/Mis12 complex that is essential for spindle checkpoint signaling and contributes to kinetochore-based force generation (Kline et al, 2006; McAinsh et al, 2006), and KNL-1/Spc105 that links MIND and NDC80 (Cheeseman et al, 2006). Importantly, the KMN network binds to MTs directly in vitro and is conserved in all eukaryotes (Cheeseman et al, 2006; Meraldi et al, 2006; Wei et al, 2007).
The CENP-A NAC/CAD network is only partially conserved with corresponding protein-protein interaction networks identified in Saccharomyces cerevisiae (CTF19 complex) and Schizosaccharomyces pombe (Sim4 complex; Cheeseman et al, 2002; Liu et al, 2005). These three networks require the centromeric histone-H3 variant CENP-A for kinetochore binding. Moreover, bioinformatic analysis reveals that they are built up from a set of common orthologues (Figure 1A; Supplementary Figures S1 and S2; Meraldi et al, 2006). CENP-A NAC/CAD components have been subdivided into either NAC proteins (nucleosome-associated complex; CENP-C, CENP-H, CENP-50CENP−U, CENP-M, CENP-T and Chl4RCENP−N) or CAD proteins (CENP-A Distal; CENP-I, Mcm21RCENP−O, Fta1RCENP−L, Sim4RCENP−K, CENP-P, CENP-Q, CENP-R and CENP-S). This designation is based on their biochemical isolation: NAC proteins were purified in association with CENP-A nucleosomes, whereas CAD proteins could only be purified with NAC proteins (Foltz et al, 2006). Work with S. cerevisiae has demonstrated that the counterpart of CENP-A NAC/CAD, the CTF19 complex, consists of several stable biochemical subcomplexes that denote distinct functional units of the kinetochore (De Wulf et al, 2003; McAinsh et al, 2003). However, it is unknown whether designation of proteins as ‘NAC' or ‘CAD' reflects a similar functional organization, since the exact function(s) of CENP-A NAC/CAD proteins is uncertain; depletion of NAC and CAD proteins in human or chicken cells causes chromosome congression defects (Liu et al, 2003; Minoshima et al, 2005; Foltz et al, 2006; Okada et al, 2006). At the same time, depletion of the CAD protein Mcm21RCENP−O leads to kinetochore–MT attachment defects that cause monopolar spindles (McAinsh et al, 2006). The human CENP-A NAC/CAD network has also been proposed to play a role in loading CENP-A at centromeres; however, the depletion of CENP-A NAC/CAD subunits does not affect the recruitment of endogenous CENP-A (Fukagawa et al, 2001; Okada et al, 2006), in contrast to other bona fide CENP-A loading factors such as hMis18alpha, Mis18beta and M18BP1 (Fujita et al, 2007).
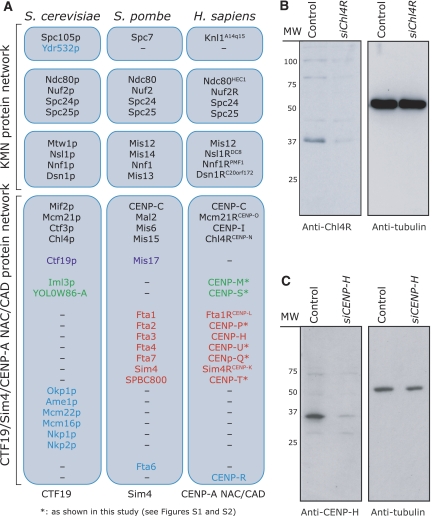
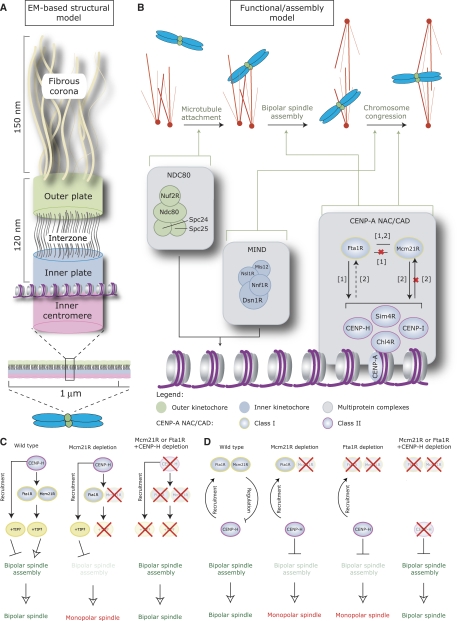
Figure 1ac.
Localization of CENP-H and Chl4RCENP−N. (A) Schematic diagram illustrating the conservation of KMN and CENP-A NAC/CAD protein interaction network subunits in S. cerevisiae, S. pombe and Homo sapiens. Proteins conserved in all organisms (black text), absent in S. cerevisiae (red text), absent in S. pombe (green text) or restricted to a single organism (blue text) are indicated. (B, C) Immunoblots of whole-cell lysates from cells transfected with control (siLaminA), siChl4R or siCENP-H RNA and probed with antibodies as indicated.
It is also unclear to what degree the CENP-A NAC/CAD network interacts with the KMN network. Some studies reported that CENP-A NAC/CAD is required for loading of the MIND and NDC80 subunits onto kinetochores (Hori et al, 2003; Liu et al, 2006; Okada et al, 2006; Kwon et al, 2007), while other studies found no such effect (Foltz et al, 2006; McAinsh et al, 2006). Another group even observed the opposite effect in which CENP-A NAC/CAD components displayed reduced kinetochore binding in the absence of the MIND complex (Kline et al, 2006). Thus, an important challenge in the kinetochore field is to define the precise function of CENP-A NAC/CAD and to understand how it interacts with the core KMN network at the functional level. We have therefore addressed the following three key questions in this paper by combining functional cell biological assays, small interfering RNA (siRNA)-mediated protein depletion, high-resolution microscopy and live-cell imaging: (1) are components of the human CENP-A NAC/CAD network responsible for different kinetochore functions? (2) How would these different CENP-A NAC/CAD components cooperate with one another to modulate kinetochore function? (3) How does CENP-A NAC/CAD functionally interact with the KMN network?
Results
Loading of CENP-A NAC/CAD kinetochore components during the cell cycle
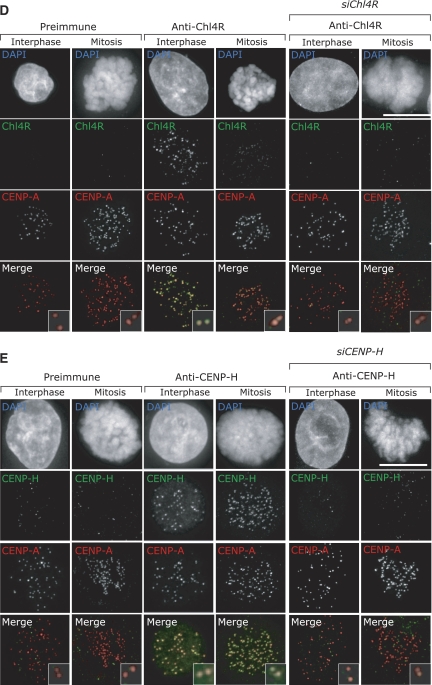
To investigate the function and organization of CENP-A NAC/CAD, we first focused on two NAC proteins (CENP-H and Chl4RCENP−N) and one CAD protein (Mcm21RCENP−O). We chose these proteins because different phenotypes have been reported for the depletion of CENP-H and Chl4R (congression defects; Fukagawa et al, 2001; Foltz et al, 2006), or Mcm21R (monopolar spindles; McAinsh et al, 2006). We first raised polyclonal antibodies against human CENP-H and Chl4R (antibodies against Mcm21R were available; McAinsh et al, 2006). By immunoblotting, CENP-H antisera recognized a 34-kDa band in whole-cell extracts and Chl4R antisera recognized a 38-kDa band. Depletion of CENP-H or Chl4R with siRNA abolished the 34- or 38-kDa bands, confirming the specificity of our antibodies (Figure 1B and C). These antibodies were next tested by immunofluorescence on HeLa cells in conjunction with CENP-A antisera (kinetochore marker) and DAPI (DNA marker). Previous studies demonstrated that CENP-H localizes to kinetochores (Fukagawa et al, 2001), but Chl4R has been only examined as a GFP-tagged fusion protein (Foltz et al, 2006). Both CENP-H and Chl4R antisera (but not the corresponding preimmune sera) recognized kinetochores in interphase and mitotic cells (Figure 1D and E). This staining was specific, as in both cases depletion of CENP-H or Chl4R by siRNA reduced the kinetochore signal more than 20-fold in over 95% of the cells, whereas a control siRNA had no effect (Figure 1D and E, and data not shown). These results validated the specificity of our CENP-H and Chl4R antibodies and siRNAs.
Figure 1de.
(D, E) Immunofluorescence images of mitotic or interphase HeLa cells transfected with control or siChl4R RNA and stained with DAPI (DNA), CENP-A antisera (kinetochores; red) and Chl4R preimmune or antisera (green) (D); or transfected with control or siCENP-H RNA and stained with DAPI (DNA), CENP-A antisera (kinetochores; red) CENP-H pre-immune or antisera (green) (E). Scale bar=10 μm.
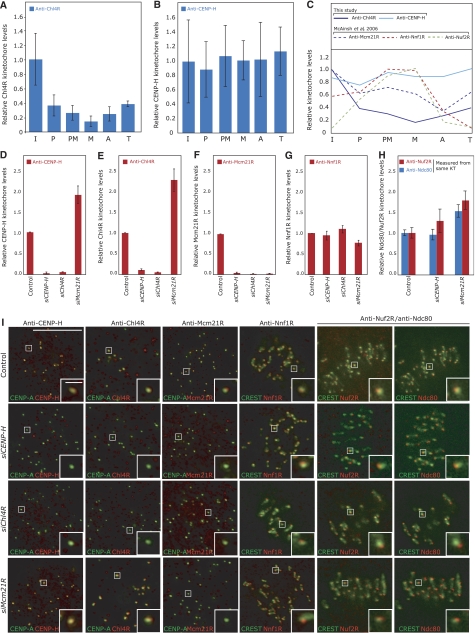
Previous studies have found that CENP-H levels at kinetochores are constant during the cell cycle (Fukagawa et al, 2001), whereas Mcm21R levels are highest during interphase and diminished by 50% during mitosis (McAinsh et al, 2006; Figure 2C). This suggested that different members of CENP-A NAC/CAD might accumulate with distinct dynamics on kinetochores during the cell cycle. We therefore quantified the Chl4R levels on kinetochores for each cell-cycle stage by 3D-deconvolution microscopy with respect to CREST levels, which are constant throughout the cell cycle and act as an internal control, and compared them to CENP-H levels (Hoffman et al, 2001; see Materials and methods for details). We found that the levels of kinetochore-bound Chl4R decreased markedly as cells entered mitosis (>80% decrease at metaphase), and increased again as cells exit mitosis, whereas CENP-H levels remained constant (Figures 1D, 2A–C). To exclude that this decrease was caused by epitope masking, we confirmed the results with a second polyclonal Chl4R rabbit antibody (Supplementary Figure S3A and B). Thus, the dynamic accumulation of Chl4R onto kinetochores during the cell cycle resembles Mcm21R but is distinct from CENP-H.
Figure 2.
Cell-cycle loading and recruitment dependencies of Chl4RCENP−N, CENP-H and Mcm21RCENP−O. Levels of (A) Chl4R or (B) CENP-H on kinetochores were determined from deconvolved 3D reconstructions of cells stained with DAPI (DNA), CREST antisera or CENP-A antibodies (kinetochores) and Chl4R or CENP-H antisera. The intensity of the kinetochore signal was determined in interphase (I), prophase (P), prometaphase (PM), metaphase (M), anaphase (A) or telophase (T) relative to CREST/CENP-A after background correction (see Materials and methods for details). (C) Comparison of cell-cycle loading profiles for CENP-H and Chl4R with Mcm21R, Nuf2R (NDC80 complex) and Nnf1R (MIND complex; McAinsh et al, 2006). (D–H) Levels of CENP-H, Chl4R, Mcm21R, Nnf1R, Nuf2R and Ndc80 on kinetochores were determined in control, siCENP-H-, siChl4R- or siMcm21R-transfected cells from deconvolved 3D reconstructions of cells stained with DAPI (DNA), CREST antisera or CENP-A antibodies (kinetochores) and the corresponding antisera. The intensity of kinetochore signal was determined relative to CREST/CENP-A after background correction. (I) Representative images of experiments described in (D–H) with CREST/CENP-A (green) and anti-CENP-H, Chl4R, Mcm21R, Nnf1R, Nuf2R or Ndc80 (red). Note that Ndc80 (mouse) and Nuf2R (rabbit) antibody staining was carried out on the same cells. Scale bar=10 μm, scale bar in zoom=1 μm.
Binding dependencies between CENP-A NAC/CAD, MIND and NDC80 subunits
We next tested by quantitative immunofluorescence to what extent Mcm21R, CENP-H or Chl4R are required for the kinetochore binding of one another and of MIND (Nnf1R) and NDC80 (Ndc80 and Nuf2R) components. Depletion of CENP-H or Chl4R had little effect on the levels of MIND and NDC80, but abolished the kinetochore association of both CENP-A NAC/CAD proteins (Figure 2D–I). Surprisingly, depletion of Mcm21R caused up to a two-fold increase in the levels of kinetochore-bound Chl4R, CENP-H, Ndc80 and Nuf2R while leaving Nnf1R levels unaffected (Figure 2D–H). These increases in kinetochore-bound levels are unlikely to be caused by epitope masking, as in each case we obtained similar results with several antibodies (antibodies against two proteins in the NDC80 complex, two polyclonal antibodies for CENP-H and two polyclonal antibodies for Chl4R; Figure 2; Supplementary Figure S3D). Based on these results we conclude that CENP-A NAC/CAD contains at least two classes of proteins: Mcm21R belongs to a first class of proteins (Class I), which appears to modulate the kinetochore-bound levels of the NDC80 complex, and a second class (Class II) of protein that includes CENP-H and Chl4R. Class II proteins are not required for binding of the MIND and NDC80 complexes, but are required for kinetochore binding of at least two other CENP-A NAC/CAD subunits.
Functional independence of CENP-A NAC/CAD, MIND and NDC80
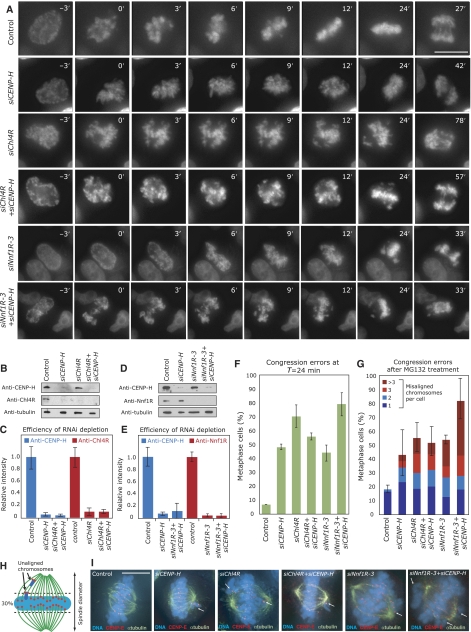
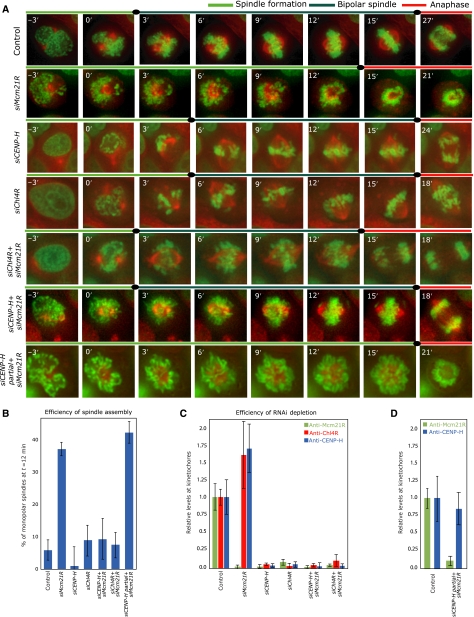
We next investigated how the dependency relationships between the CENP-A NAC/CAD, MIND and NDC80 complexes reflect their kinetochore functions. Our dependency experiments predict that MIND functions independently of Chl4R/CENP-H, while Chl4R and CENP-H should be interdependent at the functional level. Previous studies have reported that depletion of CENP-A NAC/CAD proteins causes chromosome congression defects (Fukagawa et al, 2001; Foltz et al, 2006; Kline et al, 2006; McAinsh et al, 2006). To test their functional relationship, we quantitatively compared the effect of Chl4R, CENP-H and Nnf1R depletion on chromosome congression, using single and double siRNA transfections. Immunofluorescence and immunoblotting demonstrated that Chl4R, CENP-H and Nnf1R were depleted to the same level in double transfections as in single siRNA transfections (Figure 3B–E; Supplementary Figure S4). As a first functional assay, we performed live-cell imaging of HeLa cells stably expressing Histone2B-GFP to monitor chromosome congression. Specifically, depleted cells were imaged every 3 min for 8 h (Figure 3A), with nuclear breakdown (NBD) set as T=0 min in each cell and the time of chromosome congression (defined as the time at which the last unaligned chromosome had congressed to the metaphase plate) recorded relative to NBD. When control cells (n=45) were monitored, 93% of cells completed chromosome congression by T=24 min (Figure 3A and D). In contrast, 50% of siCENP-H (n=95) and 70% of siChl4R (n=80)-transfected cells had not yet completed congression by T=24 min (Figure 3A and D). Importantly, when cells were transfected with both siChl4R and siCENP-H oligonucleotides (n=96), we observed no additive effect when compared to either single depletion (55% cells with uncongressed chromosomes at T=24 min; Figure 3A and D). Nnf1R depletion by itself caused congression errors in 46% of the cells at T=24 min (n=104; Figure 3A and D). Strikingly, co-depletion of CENP-H and Nnf1R produced an additive effect, as 80% of the cells had not yet completed chromosome congression by T=24 min (n=186; Figure 3A and D).
Figure 3.
CENP-H/Chl4RCENP−N and Nnf1R have separate roles in chromosome congression. (A) Successive frames every 3 min from live-cell movies of H2B-GFP-expressing HeLa cells transfected with control, siCENP-H, siChl4R or siChl4R+siCENP-H, siNnf1R-3 or siCENP-H+siNnf1R-3 RNA. NBD was set as T=0 min. Scale bar=10 μm. (B, D) Immunoblots showing protein levels of CENP-H, Chl4R or Nnf1R after treatment with siRNA as indicated. (C, E) Protein levels of CENP-H, Chl4R and Nnf1R as measured by quantitative immunofluorescence as in Figure 2D–H (see Supplementary Figure S4 for representative images). (F) Percentage of cells, treated as described in panel A, with uncongressed chromosomes at T=24 min. (G) Percentage of cells with one (dark blue), two (blue), three (red) or more than three (dark red) uncongressed chromosomes following 30-min treatment with MG132 as determined from images such as shown in (I) Error bars indicate the s.d. for the total number percentage of cells with uncongressed chromosomes. (H) Chromosomes in metaphase cells were counted as unaligned if they were located outside of the central 30% of the mitotic spindle, or if their kinetochores were aligned perpendicular to the spindle axis. (I) Representative images of cells treated for 30 min with MG132 and stained with DAPI (DNA; blue), anti-α-tubulin (green) and CENP-E antisera (red). Scale bar=10 μm, scale bar in zoom=1μm.
To confirm these results and quantify the number of unaligned chromosomes at higher resolution, we used immunofluorescence on HeLa cells treated with MG132 for 30 min before fixation. The proteasome inhibitor MG132 blocks cells at multiple points in the cell cycle, including an arrest at the metaphase–anaphase transition independent of the spindle checkpoint (Rock et al, 1994). The fixed cells were stained with DAPI and sera against α-tubulin (MT marker) and CENP-E (kinetochore marker). In cells containing a metaphase plate indicating arrest at the metaphase–anaphase transition, we quantified the number of uncongressed chromosomes. Chromosomes were considered uncongressed if they were outside of a rectangular area encompassing the central 30% of the spindle (Figure 3H). We found that CENP-H or Chl4R depletion caused chromosome congression defects in 43 and 56% of metaphase cells, respectively (versus 18.5% following control depletion; Figure 3G and I). Simultaneous depletion of CENP-H and Chl4R did not result in any additive effect (53% of cells with unaligned chromosomes; Figure 3G and I). In contrast, co-depletion of CENP-H and Nnf1R resulted in an additive effect when compared with single depletions (80% of cells with unaligned chromosomes following siNnf1R+siCENP-H treatment versus 54% in Nnf1R-depleted cells; Figure 3G and I). We therefore conclude that CENP-H and Chl4R are required to efficiently align the chromosomes on a metaphase plate as part of a single process that is distinct from MIND function.
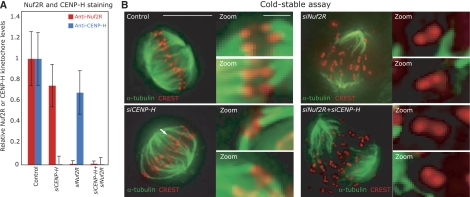
Our dependency experiments also indicated that the NDC80 complex remains kinetochore bound following depletion of CENP-H. To confirm that NDC80 is still functional in the absence of CENP-A NAC/CAD, we tested whether kinetochores in control, siCENP-H, siNuf2R or siCENP-H+siNuf2R treated cells bind to MTs in a cold-stable assay. This assay is based on the observation that abrupt cooling of cells to 4°C causes the depolymerization of MTs that are not bound to kinetochores, while kinetochore-bound MTs (k-fibers) remain stable (Salmon and Begg, 1980). We first confirmed by quantitative immunofluorescence that Nuf2R and CENP-H were depleted to the same level in double transfections as in single siRNA transfections (Figure 4A). Cells were then treated with ice-cold medium and processed for immunofluorescence using α-tubulin and CREST antibodies. Mitotic control cells subjected to such a treatment maintained a high number of stable k-fibers, demonstrating a robust MT–kinetochore attachment (Figure 4B). In contrast, most MTs of Nuf2R-depleted mitotic cells were depolymerized, as previously found (Figure 4B; Kline-Smith et al, 2005). Importantly, the MTs were stable in CENP-H-depleted cells but depolymerized in cells co-depleted for CENP-H and Nuf2R (Figure 4B). The fact that CENP-H-depleted cells only lose stable MT–kinetochore attachment following Nuf2R depletion confirms that CENP-H depletion alone does not abrogate the MT-binding function of the NDC80 complex at kinetochores, and that active NDC80 is loaded on kinetochores independently of CENP-H.
Figure 4.
The Ndc80 complex is functional in the absence of CENP-H/Chl4RCENP−N. (A) Quantification of Nuf2R and CENP-H levels on kinetochores in cells treated with control, siCENP-H, siNuf2R or siCENP-H+siNuf2R RNAs (as described in Figure 2). (B) Cells treated with siRNAs as described in panel A were subjected to cold treatment for 10 min before fixation and staining with anti-α-tubulin (green) and CREST antisera (red). Scale bar=10 μm.
CENP-H-Chl4R depletion suppresses the monopolar spindle phenotype caused by Mcm21R depletion
How does the phenotype of CENP-H-Chl4R depletion (Class II) relate to that of Mcm21R (Class I)-depleted cells? The striking feature of Mcm21R-depleted cells is that their kinetochores inhibit bipolar spindle formation, causing an increase in monopolar spindles (McAinsh et al, 2006). Consistently, monitoring of spindle assembly in HeLa cells stably expressing H2B-GFP (chromosome marker) and α-tubulin-mRFP (spindle marker) revealed that 38% of Mcm21R-depleted cells fail to establish a bipolar spindle within the first 12 min after NBD (versus 5% in control depleted cells; Figure 5A and B). However, live-cell imaging also showed that CENP-H or Chl4R depletion does not perturb bipolar spindle assembly (Figure 5A and B), even though CENP-H-, Chl4R- or Mcm21R-depleted cells have similar low levels of Mcm21R at kinetochores (Figure 5C; Supplementary Figure S5). This discrepancy could be due to three possible causes: (1) a cytoplasmic pool of Mcm21R could rescue bipolar spindle assembly in CENP-H-Chl4R-depleted cells, (2) excessive levels of kinetochore-bound CENP-H-Chl4R in Mcm21R-depleted cells could interfere with bipolar spindle formation (Figures 2D and E, 5C) and (3) the mere presence of CENP-H/Chl4R on kinetochores lacking Mcm21R could be sufficient to disrupt bipolar spindle assembly.
Figure 5.
Chl4RCENP−N or CENP-H depletion rescues the bipolar spindle defect in Mcm21RCENP−O-depleted cells. (A) Successive frames every 3 min from live-cell movies of H2B-GFP/α-tubulin-mRFP-expressing HeLa cells transfected with control, siMcm21R, siCENP-H, siChl4R, siMcm21R+Chl4R, siMcm21R+siCENP-H or partial siCENP-H (50% siRNA)+siMcm21R RNA. The composite images for H2B-GFP (green) and α-tubulin-mRFP (red) are shown. Scale bar=10 μm. (B) Quantification of bipolar spindle formation errors at T=12 min after NBD in cells treated with siRNA as indicated. (C) Protein levels of Mcm21R, CENP-H and Chl4R in cells treated with siRNA as indicated as measured by quantitative immunofluorescence. (D) Protein levels of Mcm21R and CENP-H in cells treated with control or partial siCENP-H (50% siRNA)+siMcm21R RNAs.
To distinguish between these possibilities, we first depleted both CENP-H or Chl4R and Mcm21R by siRNA, and followed spindle assembly by time-lapse microscopy. We found that the vast majority (91% compared with 95% in control cells) of double siChl4R+siMcm21R- or siCENP-H+siMcm21R-transfected cells assembled a bipolar spindle within the first 12 min after NBD (Figure 5A and B). Quantification of Mcm21R levels by immunofluorescence and immunoblotting in single and double depleted cells confirmed that this rescue was not due to incomplete depletion of Mcm21R (Figure 5C; Supplementary Figure S5). We therefore can exclude that cytoplasmic Mcm21R rescues bipolar spindle assembly, and conclude that the monopolar spindles observed in Mcm21R depleted cells require CENP-H/Chl4R at kinetochores. We next specifically removed the excessive amount of kinetochore-bound CENP-H in cells depleted of Mcm21R, using a partial CENP-H depletion combined with a full Mcm21R depletion (Figure 5D; for experimental details see Materials and methods). We found that 42% of the cells failed to form a bipolar spindle within 12 min after NBD (Figure 5A and B). We conclude that in the absence of Mcm21R even normal levels of CENP-H are sufficient to disrupt bipolar spindle formation.
CENP-A NAC/CAD modulates NDC80 levels at kinetochores
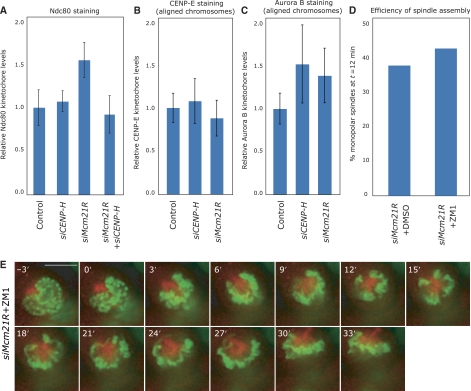
While the MT-binding NDC80 complex is loaded independently of CENP-A NAC/CAD, our dependency experiments also showed that the levels of the NDC80 were elevated in Mcm21R-depleted cells and normal in CENP-H-depleted cells (Figures 2H, I and 6A). Co-depletion of Mcm21R and CENP-H restored normal Ndc80 levels at kinetochores, indicating that kinetochore binding by Ndc80 can be modulated by CENP-A NAC/CAD (Figure 6A). This effect was specific for NDC80, as we found no evidence that the levels of another MT-binding kinetochore protein, CENP-E kinesin, were regulated by CENP-H or Mcm21R (Figure 6B). A key regulator of the NDC80 complex is the Aurora B kinase (DeLuca et al, 2006). It is therefore possible that an increase in the kinetochore binding of NDC80 reflects a misregulation of Aurora B, which could potentially cause monopolar spindles. However, we again found no evidence that CENP-A NAC/CAD regulates the levels of Aurora B at kinetochores (Figure 6C). In addition, treatment of Mcm21R-depleted cells with the Aurora B kinase inhibitor ZM1 at concentrations that inhibited cytokinesis (data not shown; see also Ditchfield et al, 2003) did not rescue the monopolar spindle phenotype (Figure 6D and E). Importantly ZM1 treatment by itself did not cause monopolar spindles (data not shown; Ditchfield et al, 2003). This indicates that failure in bipolar spindle formation in Mcm21R-depleted cells is not mediated by Aurora B activity. Overall, we conclude that CENP-A NAC/CAD can modulate NDC80 levels at kinetochores, even though at this point we have no evidence that these changes are linked to the monopolar spindles in Mcm21R-depleted cells.
Figure 6.
The CENP-A NAC/CAD controls the levels of kinetochore-bound NDC80 complex. (A–C) Protein levels of Ndc80, CENP-E and Aurora B in cells treated with siRNAs as indicated by quantitative immunofluorescence as described in Figure 2. (D) Quantification of bipolar spindle formation errors at T=12 min after NBD in cells treated with siMcm21R RNA and DMSO or 2 μM ZM1. (E) Successive frames every 3 min from live-cell movies of H2B-GFP/α-tubulin-mRFP-expressing HeLa cells transfected with siMcm21R RNA and 2 μM ZM1. The composite images for H2B-GFP (green) and α-tubulin-mRFP (red) are shown. Scale bar=10 μm.
Classification of further CENP-A NAC/CAD subunits
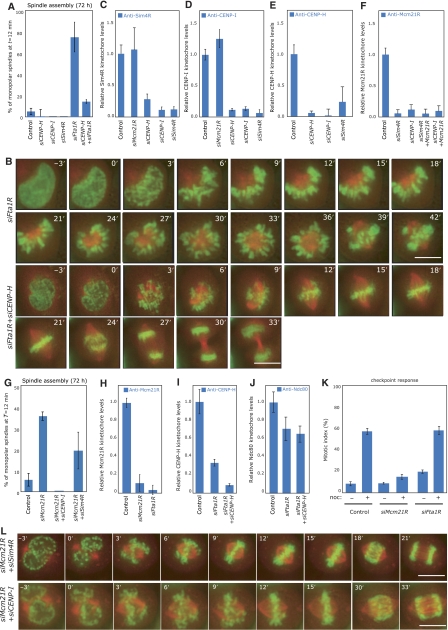
Finally, we investigated whether any further CAD subunits of the CENP-A NAC/CAD complex were required for bipolar spindle formation. We first treated cells with siRNAs against the CAD components Fta1RCENP−L, Sim4RCENP−K or CENP-I, and validated the specificity of our RNAi depletions by immunofluorescence or western blot analysis (see Supplementary Figure S6). Next, by live-cell imaging we tested whether these depletions affected chromosome congression and/or bipolar spindle formation. Depletion of all three proteins resulted in chromosome congression errors as previously shown (data not shown; Foltz et al, 2006; Okada et al, 2006), whereas only Fta1R depletion impaired bipolar spindle formation resulting in monopolar spindles in 74% of mitotic cells (Figure 7A and B). To exclude that this phenotype was caused by off-target effects, we confirmed using immunofluorescence in fixed cells that a second siRNA targeted against Fta1R caused the accumulation of monopolar spindles (Supplementary Figure S6H). These data suggest that Fta1R, like Mcm21R, may be a Class I protein that is required for bipolar spindle assembly, while Sim4R and CENP-I, along with CENP-H and Chl4R, are Class II proteins.
Figure 7.
Classification of further CENP-A NAC/CAD components. (A) Quantification of bipolar spindle formation errors at T=12 min after NBD in cells treated with control, siCENP-H, siCENP-I, siSim4R, siFta1R or siFta1R+siCENP-H RNAs. For comparison, values for control and siCENP-H-treated cells were transferred from Figure 5B. (B) Successive frames every 3 min from live-cell movies of H2B-GFP/α-tubulin-mRFP-expressing HeLa cells transfected with siFta1R or siFta1R+siCENP-H RNAs. The composite images for H2B-GFP (green) and α-tubulin-mRFP (red) are shown. Scale bar=10 μm. (C–E) Protein levels of Sim4R (C), CENP-I (D) and CENP-H (E) on kinetochores following treatment with control, siMcm21R, siCENP-H, siCENP-I or siSim4R RNA by quantitative immunofluorescence. (F) Protein levels of Mcm21R on kinetochores following treatment with control, siSim4R, siCENP-I, siSim4R+siMcm21R or siCENP-I+siMcm21R RNA by quantitative immunofluorescence. (G) Quantification of bipolar spindle formation errors at T=12 min after NBD in cells treated with control, siMcm21R, siMcm21R+siCENP-I or siMcm21R+siSim4R RNA. For comparison, values for control and siMcm21R-treated cells were transferred from Figure 5B (H) Protein levels of Mcm21R on kinetochores following treatment with control, siMcm21R or siFta1R RNA by quantitative immunofluorescence. (I, J) Protein levels of CENP-H (I) or Ndc80 (J) on kinetochores following treatment with control, siFta1R or siFta1R+siCENP-H RNA. (K) Percentage mitotic arrest in control, siMcm21R- or siFta1R-transfected cells with (+) or without (−) 16 h nocodazole treatment. (L) Successive frames every 3 min from live-cell movies of H2B-GFP/α-tubulin-mRFP-expressing HeLa cells transfected with siMcm21R+siSim4R or siMcm21R+siCENP-I RNA. The composite images for H2B-GFP (green) and α-tubulin-mRFP (red) are shown. Scale bar=10 μm.
A first prediction of such a classification is that Sim4R and CENP-I should be required for the binding of Class I and Class II proteins to kinetochores. Indeed, Sim4R, CENP-I, CENP-H and Mcm21R failed to bind kinetochores in the absence of either Sim4R or CENP-I (Figure 7C–F). Moreover, we found that Sim4R and CENP-I, like CENP-H and Chl4R, did not require Mcm21R to bind to kinetochores (Figure 7C–F). A second, more functional prediction is that depletion of Sim4R or CENP-I should rescue the spindle assembly defect in Mcm21R-depleted cells. We found that the vast majority (83 and 100% compared with 95% in control cells) of double siSim4R+siMcm21R- or siCENP-I+Mcm21R-transfected cells assembled a bipolar spindle within the first 12 min after NBD (Figure 7G and L). We thus conclude that CENP-I and Sim4R together with CENP-H and Chl4R function as Class II proteins at kinetochores.
We next tested whether failure in bipolar spindle formation in Fta1R-depleted cells occurs through a mechanism similar to that found in Mcm21R-depleted cells. This appears to be the case as simultaneous depletion of Mcm21R and Fta1R did not have an additive effect on the percentage of monopolar spindles in a fixed-cell assay (Supplementary Figure S6H). Moreover, co-depletion of CENP-H in Fta1R-depleted cells strongly reduced the occurrence of monopolar spindles at T=12 min (14 versus 75% in siFta1R-treated cells; Figure 7A and B). We therefore conclude that Fta1R is a Class I protein in that its depletion causes a CENP-H-dependent failure in bipolar spindle assembly. We did however notice a number of key differences between Fta1R- and Mcm21R-depleted cells; although Mcm21R requires Fta1R to bind kinetochores (Figure 7C), we found that the levels of kinetochore-bound CENP-H and Ndc80 were not increased as in Mcm21R-depleted cells, but rather reduced to 33 and 70%, respectively (Figure 7I and J). This indicated that Fta1R depletion does not affect kinetochores in the same way as Mcm21R depletion. We furthermore observed that Fta1R-depleted cells with monopolar spindles failed to enter anaphase, suggesting an active spindle checkpoint, in contrast to Mcm21R-depleted cells with monopolar spindles, which are known to lack a functional spindle checkpoint (McAinsh et al, 2006). To confirm this difference we treated control, Fta1R- and Mcm21R-depleted cells for 16 h with nocodazole, a drug that depolymerizes MTs leading to spindle checkpoint activation and a high mitotic index. While Mcm21R-depleted cells failed to arrest in mitosis (mitotic index of 12%), Fta1R and control depleted cells had an intact spindle checkpoint response (mitotic index of 60 and 58%; Figure 7K). Overall we conclude that even though Fta1R and Mcm21R depletions perturb kinetochore function in different ways, depletion of either of these Class I proteins impairs bipolar spindle formation in a CENP-H-dependent manner.
Discussion
In this paper we have used a combination of siRNA-mediated protein depletion, quantitative immunofluorescence and live-cell imaging to determine the functional organization of the CENP-A NAC/CAD protein–protein interaction network. Recent studies have subdivided this network into separate subcomplexes using data from experiments based on dependency (Okada et al, 2006) or affinity purification (Foltz et al, 2006). However, the reported number and subunit composition of such subcomplexes was not in agreement. By taking a function-based approach we show that CENP-A NAC/CAD subunits can be assigned to one of at least two distinct functional classes (Figure 8B); depletion of Class I proteins (Mcm21RCENP−O or Fta1RCENP−L) causes a failure in bipolar spindle assembly in a CENP-H-dependent manner. Interestingly, there are also differences between Fta1R and Mcm21R suggesting that Class I proteins can be further subdivided: (1) Mcm21R depends on Fta1R for kinetochore binding, whereas the reverse is not true (Figure 7H; Okada et al, 2006); (2) a functional spindle checkpoint is only abrogated after Mcm21R depletion; (3) depletion of Fta1R or Mcm21R differentially affects the subunit composition of the kinetochore. Depletion of Class II proteins (CENP-H, Chl4RCENP−N, CENP-I and Sim4RCENP−K) causes chromosome congression defects, but does not perturb bipolar spindle formation. Therefore, depletion of the CAD proteins CENP-I and Sim4R phenocopies the depletion of the NAC proteins Chl4RCENP−N or CENP-H. These findings demonstrate that the simple separation of proteins into ‘NAC' or ‘CAD' does not reflect the functional organization of the network. In contrast, the dependency experiments by Okada and co-workers fits much better with the functional organization of the CENP-A NAC/CAD described in this study. Interestingly, biochemical fractionation of soluble mitotic cell extracts reveals that CENP-A-NAC/CAD is assembled from a set of smaller subcomplexes (Supplementary Figure S7). These data together with our cell-cycle fluctuation analysis suggests a more complex and dynamic organization that may involve cell-cycle-dependent assembly of soluble subcomplexes into larger ‘complexes' that form the CENP-A NAC/CAD network on centromeres. Clearly, detailed biochemical and in vitro reconstitution-based experiments will be required to dissect these complicated assembly pathways. The combination of such biochemical experiments with a functional classification, such as that described in this study, will then allow the construction of an accurate structure–function model of the CENP-A NAC/CAD network.
Figure 8.
Model for the structure and function of the CENP-A NAC/CAD interaction network. (A) Model of vertebrate kinetochore organization based on electron micrographs and adapted from Emanuele et al (2007) and McAinsh et al (2006). (B) Functional-assembly model for core kinetochore complexes. Solid black arrows represent dependencies; a red ‘X' indicates a lack of dependency and dotted black arrows a ‘partial' dependency (for references see [1] Okada et al, 2006 and [2] McAinsh et al, 2006). Green arrows indicate whether a complex contributes to MT attachment, bipolar spindle assembly or chromosome congression. (C, D) Possible models for the effect of Mcm21R, CENP-H and Fta1R on bipolar spindle assembly (red crosses indicate the depletion of a protein); (C) the first model predicts that Mcm21R/Fta1R and CENP-H recruit a different set of plus-end MT-binding proteins to kinetochores. Loss of Mcm21R/Fta1R causes an imbalance in plus-end-MT-binding activity, which impairs bipolar spindle formation. Concomitant loss of CENP-H corrects this balance and restores bipolar spindle assembly; (D) the second model proposes that in wild-type cells Class II proteins (such as CENP-H) are antagonized by Mcm21R and Fta1R. Depletion of Mcm21R or Fta1R causes an alteration in CENP-H protein activity, leading to monopolar spindle formation. Depletion of CENP-H and Mcm21R or Fta1R re-establishes bipolar spindle assembly.
Internal regulation of CENP-A NAC/CAD function
How do these classes of proteins functionally interact within the CENP-A NAC/CAD network? Our data indicate that the Class II proteins, such as CENP-H, CENP-I, Chl4R or Sim4R, are required for binding of Mcm21R at kinetochores, whereas Mcm21R seems to control or counteract the function of Class II proteins on kinetochores. Importantly, the absence of such a control impairs bipolar spindle assembly during mitosis. This is the first example of an antagonistic relationship among two kinetochore proteins/complexes, and has important implications for the understanding of kinetochore function; depletion of a kinetochore protein does not necessarily reveal the function of that particular protein, but can create misfunctioning kinetochores. This is the case for Mcm21R- or Fta1R-depleted kinetochores, which interfere with bipolar spindle assembly in a dominant-negative manner (McAinsh et al, 2006). Crucial challenges will be to understand how Mcm21R antagonizes Class II proteins, and how even low amounts of CENP-H, as seen in Fta1R-depleted cells, can impair spindle formation. One possibility is that Class I and Class II proteins are required for the loading of different plus-end MT-binding proteins, and that the Class I and II depletion phenotypes reflect different combinations of plus-end MT-binding proteins (for details see model in Figure 8C). A second possibility is that Mcm21R binds and directly impairs the function of a Class II protein, building in effect a negative regulatory loop (see model in Figure 8D). Such a negative regulatory mechanism could modulate flexible and dynamic kinetochore–MT attachments to ensure correct chromosome segregation. Our data indicate that Mcm21R efficiently binds to Sim4R in a yeast two-hybrid assay (Supplementary Figure S8). However, additional biochemical data will be required to dissect the molecular and functional interactions between Class I and Class II proteins. One crucial aspect will also be to determine whether these proteins are themselves regulated during mitosis. In this regard, it is interesting that the NAC protein CENP-U/CENP-50/PBIP1 is phosphorylated by the protein kinase Plk1 during mitosis (Kang et al, 2006). Future investigations will be required to determine whether other CENP-A NAC/CAD components are subject to post-translational modifications, and how such modifications might contribute to the functional regulation of this protein network.
Functional interaction between CENP-A NAC/CAD and KMN networks
We found that the CENP-A NAC/CAD network plays an important role in regulating MT–kinetochore attachment and promotes efficient chromosome alignment on the metaphase plate, but is dispensable for MT–attachment per se. Our data indicate that CENP-A NAC/CAD is not required for loading of the KMN network and that it regulates chromosome congression independently of the MIND complex (see Figure 8B). It is important to note that these findings contradict previous models in which CENP-A NAC/CAD is essential for the loading of the KMN subcomplexes MIND and NDC80 onto kinetochores (Liu et al, 2006; Okada et al, 2006; Kwon et al, 2007). At the functional level, such a ‘linear' model of kinetochore assembly is inconsistent with both our own and other CENP-A NAC/CAD knockdown phenotypes; while NDC80 depletion results in MT attachment failure, loss of CENP-H, CENP-I, Chl4R, CENP-U or CENP-T allows the formation of a partial metaphase plate, which requires stable attachment and bi-orientation of multiple sister kinetochore pairs (Fukagawa et al, 2001; Nishihashi et al, 2002; Liu et al, 2003; Minoshima et al, 2005; Foltz et al, 2006; Okada et al, 2006). Using a combination of CENP-H and Nuf2R siRNA depletions, we demonstrate that these stable kinetochore—MT attachments require the presence of functional NDC80. Our dependency and epistatic analysis also indicate separate kinetochore functions for CENP-H and Nnf1R, even though their single depletion phenotypes look qualitatively similar. We therefore conclude that CENP-A NAC/CAD is not essential for the loading or the function of the MIND or NDC80 complexes, but rather that it might fine-tune NDC80 function (see below). We note that the closest equivalent of CENP-A NAC/CAD, the Schizosaccharomyces pombe Sim4 complex, behaves in a similar manner; it is dispensable for loading of the KMN complex, as both MIND and NDC80 are recruited to kinetochores in cells with inactivated Sim4 complex subunits (Hayashi et al, 2004; Saitoh et al, 2005), and genetic evidence suggests cross talk between the KMN and Sim4 complexes (Kerres et al, 2004, 2006).
What could be the function of CENP-A NAC/CAD if it is not a KMN loading factor? Our data indicate that CENP-A NAC/CAD can modulate the levels of the MT-binding NDC80 complex at kinetochores. We propose that the CENP-A NAC/CAD network functions as a fine-tuning regulator, which controls MT attachment through other protein complexes such as NDC80. Such a system might reflect the requirement for alterations in kinetochore–MT attachments during the series of stochastic chromosome movements that take place within the complex mechanical system of the mitotic spindle.
We believe that the major goal of future research will be to understand at the molecular level how the CENP-A NAC/CAD network regulates its own activities and fine-tunes the activity of the KMN network and other kinetochore components. One key challenge will be to derive better assays to differentiate the function of complexes with qualitatively similar RNAi depletion phenotypes, for example, MIND and CENP-H. Overall this will allow us to understand how the two core kinetochore protein networks, CENP-A NAC/CAD and KMN, act together as dynamic molecular machines that ensure faithful MT–kinetochore attachment, bipolar spindle assembly and chromosome segregation.
Materials and methods
Cell culture and siRNA, MG132, ZM1 and nocodazole treatment
HEK293, HeLa, HeLa H2B-EGFP and HeLa H2B-EGFP/αTubulin RFP were grown as described (McAinsh et al, 2006). siRNA oligonucleotides (Qiagen or Invitrogen) are listed in Supplementary data and were transfected as described (Elbashir et al, 2001). For all double siRNA experiments, 20 nM of each oligonucleotide was used (e.g., 20 nM siMcm21R+20 nM siLaminA), with 40 nM siLaminA being used for the control. For the partial CENP-H siRNA treatment, we used 10 nM siCENP-H+20 nM siLaminA or siMcm21R. For MG132 experiments, HeLa cells were treated with 1 μM MG132 for 30 min before fixing for immunofluorescence. For treatment with the Aurora B kinase inhibitor ZM1 (Tocris), cells were treated with 2 μM for 1 h before live-cell imaging. To measure spindle checkpoint activity, cells were treated for 16 h with 100 ng/ml nocodazole and the percentage of cells arrested in mitosis determined by phase-contrast microscopy.
Antibody production
Chl4R cDNA was obtained from ATCC; Sim4R and CENP-I full-length cDNAs were obtained from Open Biosystems and a CENP-H ORF was synthesized as an Escherichia coli codon-optimized version (GENEART). Polyclonal rabbit antibodies (NEOMPS) were raised against His6-tagged Chl4R, a His6-tagged fragment of CENP-I (AA 1–202), an untagged Sim4R or GST-tagged CENP-H expressed in E. coli, and purified from inclusion bodies. Peptide antibodies against Aurora B were raised in rabbits against a KENSYPWPYGRQGC peptide linked to Keyhole Limpet Hemocyanin (Covance). Rabbit anti-Chl4R antibodies were affinity purified against GST-tagged Chl4R, which was expressed in E. coli, purified under native conditions from glutathione–Sepharose beads (GE Healthcare) and covalently bound to an AminoLink Plus Immobilization column (Pierce). The specific antibodies were eluted with 100 mM glycine at pH 2.5.
Immunofluorescence microscopy and live-cell imaging
Cells were fixed at room temperature either for 2.5 min in methanol/acetone (1:1) or for 10 min in PTEMF (20 mM Pipes (pH 6.8), 10 mM EGTA, 1 mM MgCl2, 0.2% Triton X-100, 4% formaldehyde). For cold-stable assays, cells were incubated in ice-cold medium 10 min before fixation. Primary antibodies are listed in Supplementary data. Cross-adsorbed secondary antibodies were used (Molecular Probes). 3D image stacks were acquired in 0.2-μm steps using a × 100 or × 60 oil NA 1.4 objective on an Olympus Deltavision microscope (API) equipped with a DAPI-FITC-Rhod/TR-CY5 filter set (Chroma) and a Coolsnap HQ camera. The 3D image stacks were deconvolved with SoftWorx (API). For quantitative measurements, kinetochore signals were determined and quantified with SoftWorx using the formula
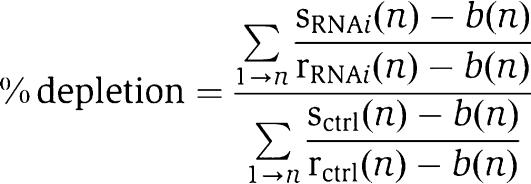 |
with s (signal), b (background) and r (reference signal). For each measurement, levels in at least five cells (50 kinetochores) were determined. For Ndc80, Nuf2R, Nnf1R, CENP-E and Aurora B, signal intensities were measured in mitotic cells. For CENP-H, Mcm21R and Chl4R, CENP-I and Sim4R, intensities were measured in interphase cells as they display the strongest signal. We do however emphasize that we obtained qualitatively the same results when measuring in mitotic cells. For live-cell imaging cells were monitored at 37°C in LabTech II (LabTech) chambers in Leibovitz's L-15 medium containing 10% fetal calf serum (GibcoBRL). Images were acquired every 3 min for 8 h using either a × 20 NA 0.75 or a × 40 oil NA 1.3 objective on a Olympus Deltavision microscope (API) equipped with a GFP/dsRed (Semrock) or DAPI-FITC-Rhod/TR-CY5 (Chroma) filter set.
Immunoblotting
Whole-cell lysate preparation and immunoblotting were carried out as described (Meraldi et al, 2004), using primary antibodies as listed in Supplementary data and anti-mouse and anti-rabbit HRP-conjugated secondary antibodies (Amersham).
Supplementary Material
Supplementary Information
Acknowledgments
We thank the Light Microscopy Centre of ETH Zurich for technical support, René Holtackers for generating CENP-H, CENP-I and Sim4R antisera and Tatsuo Fukagawa for CENP-H plasmids. We are grateful to Yves Barral, Monica Gotta and Anne Straube for critical reading of the manuscript, and members of the McAinsh and Meraldi labs for helpful discussions. Work in the McAinsh laboratory was supported by Marie Curie Cancer Care (ADM, SEM and JRW), and in the Meraldi laboratory by Swiss National Science Foundation grant 3100A0-107912/1 (PM and SB), the Swiss National Science Foundation Förderungsprofessur (PM) and the EURYI award from the European Science Foundation (PM, ACA and MB).
References
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR III, Chan CS, Drubin DG, Barnes G (2002) Phosphoregulation of kinetochore–microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163–172 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR III, Oegema K, Desai A (2004) A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev 18: 2255–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK (2003) Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev 17: 2902–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED (2006) Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127: 969–982 [DOI] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS (2003) Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol 161: 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, McCleland ML, Satinover DL, Stukenberg PT (2007) Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol Biol Cell 16: 4882–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, Cleveland DW (2006) The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8: 458–469 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M (2007) Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 12: 17–30 [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Mikami Y, Nishihashi A, Regnier V, Haraguchi T, Hiraoka Y, Sugata N, Todokoro K, Brown W, Ikemura T (2001) CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J 20: 4603–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M (2004) Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729 [DOI] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED (2001) Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell 12: 1995–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Haraguchi T, Hiraoka Y, Kimura H, Fukagawa T (2003) Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J Cell Sci 116: 3347–3362 [DOI] [PubMed] [Google Scholar]
- Kang YH, Park JE, Yu LR, Soung NK, Yun SM, Bang JK, Seong YS, Yu H, Garfield S, Veenstra TD, Lee KS (2006) Self-regulated Plk1 recruitment to kinetochores by the Plk1–PBIP1 interaction is critical for proper chromosome segregation. Mol Cell 24: 409–422 [DOI] [PubMed] [Google Scholar]
- Kerres A, Jakopec V, Beuter C, Karig I, Pohlmann J, Pidoux A, Allshire R, Fleig U (2006) Fta2, an essential fission yeast kinetochore component, interacts closely with the conserved Mal2 protein. Mol Biol Cell 17: 4167–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerres A, Vietmeier-Decker C, Ortiz J, Karig I, Beuter C, Hegemann J, Lechner J, Fleig U (2004) The fission yeast kinetochore component Spc7 associates with the EB1 family member Mal3 and is required for kinetochore–spindle association. Mol Biol Cell 15: 5255–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A (2006) The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol 173: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith SL, Sandall S, Desai A (2005) Kinetochore–spindle microtubule interactions during mitosis. Curr Opin Cell Biol 17: 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon MS, Hori T, Okada M, Fukagawa T (2007) CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol Biol Cell 18: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ST, Hittle JC, Jablonski SA, Campbell MS, Yoda K, Yen TJ (2003) Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat Cell Biol 5: 341–345 [DOI] [PubMed] [Google Scholar]
- Liu ST, Rattner JB, Jablonski SA, Yen TJ (2006) Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol 175: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, McLeod I, Anderson S, Yates JR III, He X (2005) Molecular analysis of kinetochore architecture in fission yeast. EMBO J 24: 2919–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh AD, Meraldi P, Draviam VM, Toso A, Sorger PK (2006) The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J 25: 4033–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh AD, Tytell JD, Sorger PK (2003) Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol 19: 519–539 [DOI] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK (2004) Timing and checkpoints in the regulation of mitotic progression. Dev Cell 7: 45–60 [DOI] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 7: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima Y, Hori T, Okada M, Kimura H, Haraguchi T, Hiraoka Y, Bao YC, Kawashima T, Kitamura T, Fukagawa T (2005) The constitutive centromere component CENP-50 is required for recovery from spindle damage. Mol Cell Biol 25: 10315–10328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Hardwick KG (2002) The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol 3: 731–741 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV (2003) Interactions between centromere complexes in Saccharomyces cerevisiae. Mol Biol Cell 14: 4931–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihashi A, Haraguchi T, Hiraoka Y, Ikemura T, Regnier V, Dodson H, Earnshaw WC, Fukagawa T (2002) CENP-I is essential for centromere function in vertebrate cells. Dev Cell 2: 463–476 [DOI] [PubMed] [Google Scholar]
- Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M (2004) A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol 6: 1135–1141 [DOI] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR III, Desai A, Fukagawa T (2006) The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 8: 446–457 [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78: 761–771 [DOI] [PubMed] [Google Scholar]
- Saitoh S, Ishii K, Kobayashi Y, Takahashi K (2005) Spindle checkpoint signaling requires the mis6 kinetochore subcomplex, which interacts with mad2 and mitotic spindles. Mol Biol Cell 16: 3666–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ED, Begg DA (1980) Functional implications of cold-stable microtubules in kinetochore fibers of insect spermatocytes during anaphase. J Cell Biol 85: 853–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC (2007) The Ndc80/HEC1 complex is a contact point for kinetochore–microtubule attachment. Nat Struct Mol Biol 14: 54–59 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information