Abstract
The establishment of HIV proviral latency requires the creation of repressive chromatin structures that impair the initiation of transcription and restrict RNAP II elongation. We have found that C-promoter binding factor-1 (CBF-1), a CSL (CBF-1, Su(H) and Lag-1)-type transcription factor and key effector of the Notch signaling pathway, is a remarkably potent and specific inhibitor of the HIV-1 LTR promoter. Knockdown of endogenous CBF-1 using specific small hairpin RNAs expressed on lentiviral vectors results in the partial reactivation of latent HIV proviruses, recruitment of RNAP II, loss of histone deacetylases and the concomitant acetylation of histones. An important property of any repressor utilized to establish HIV latency is that it must become displaced or deactivated upon T-cell activation. Consistent with this hypothesis, CBF-1 mRNA and protein levels are highest in quiescent or unstimulated T cells but decline rapidly in response to proliferative stimulation such as activation of the T-cell receptor or treatment with TNF-α. We conclude that CBF-1 is a previously overlooked factor that induces transcriptional silencing during the establishment of HIV latency.
Keywords: chromatin restriction, histone deacetylases (HDAC), histone methylation, HIV latency, HIV transcription
Introduction
The introduction of highly active antiretroviral therapy in the mid-1990s led to a dramatic increase in patient longevity due to the ability of antiretroviral drugs to suppress HIV replication to below threshold detection levels (<50 copies HIV RNA/ml) (Ho et al, 1995; Wei et al, 1995). Unfortunately, despite the intensive therapy, active viral replication immediately resumes after the interruption of antiviral treatment due to reactivation of latent viral reservoirs (Chun et al, 2000). The ability of HIV to enter and exit from latency is therefore one of the major features of the virus life cycle that prevents the clearance of the virus from infected individuals and limits the effectiveness of current antiviral therapy.
The prevailing consensus is that no single molecular mechanism is responsible for latency. Instead, an absence of the cellular initiation factors NF-κB and NFAT (Nabel and Baltimore, 1987; Kinoshita et al, 1997), a restriction in the levels of the HIV transactivator protein Tat (Lin et al, 2003) and a reduction in the level of the Tat-activated elongation factor P-TEFb (Ghose et al, 2001) are all believed to contribute to the shutdown of viral transcription.
The establishment of specific restrictive chromatin structures at the HIV LTR is believed to be the primary event leading to the restriction in Tat levels during the establishment of latency. Important work from the Verdin laboratory has shown that the chromatin structure of the HIV LTR contains two well-ordered nucleosomes called Nuc-0 and Nuc-1 (Verdin et al, 1993; Emiliani et al, 1996). Nuc-0 is positioned immediately upstream of the enhancer (−415 to −255), while Nuc-1 is very close to the viral RNA start site (+10 to +155). Reactivation of HIV transcription requires histone acetylation and remodeling of the critical Nuc-1 by SWI/SNF (Lusic et al, 2003; Henderson et al, 2004; Mahmoudi et al, 2006; Treand et al, 2006).
Recent studies have shown that the presence of histone deacetylases (HDACs) at the HIV LTR is strongly correlated with transcriptional repression leading to latency. Initial interest in this mechanism was stimulated by the work of Margolis and his colleagues who demonstrated that the transcription factor YY1 can act as a repressor of HIV transcription by recruiting HDAC-1 to the provirus (Coull et al, 2000; He and Margolis, 2002). Subsequently, it was found that drugs that inhibit HDAC activity such as trichostatin A and valproic acid (Ylisastigui et al, 2004; Lehrman et al, 2005) are effective inducers of HIV transcription in latently infected cells. Recently, Williams et al (2006) have reported that removal of p50 by shRNA results in a loss of HDACs from the HIV LTR and reactivation of transcription from latent proviruses.
Entry of HIV into latency also requires the establishment of heterochromatic structures on the HIV provirus. Two recent reports demonstrate that the histone methyltransferase Suv39H1, histone H3 methylated on K9 and K27, and the repressive HP1 proteins all accumulate on transcriptionally inactive proviruses (du Chéné et al, 2007; Marban et al, 2007). Furthermore, HIV-1 reactivation could be achieved after removal of HP1γ by RNA interference (du Chéné et al, 2007).
Although multiple factors are required to establish viral latency, the reactivation of latent proviruses can be achieved by the simple induction of NF-κB (West et al, 2001; Scripture-Adams et al, 2002; Brooks et al, 2003). NF-κB induces multiple changes at the promoter of latent proviruses, including the recruitment of TFIIH and RNAP II (Kim et al, 2006). NF-κB also induces changes in the local chromatin structure at the HIV LTR by recruiting histone acetyltransferases, such as CBP and p300 to the HIV-1 LTR (Gerritsen et al, 1997; Krogan et al, 2002) and chromatin remodeling factors (Lusic et al, 2003; Henderson et al, 2004; Mahmoudi et al, 2006; Treand et al, 2006).
The initial signals used to create specific repressive chromatin structures at the HIV promoter are largely unknown. We have been investigating whether gene-specific repressors can target the HIV LTR. Our initial studies have focused on C-promoter binding factor-1 (CBF-1), the mammalian representative of the CSL family. CSL family proteins (CBF-1 and Su(H) in Drosophila melanogaster and Lag-1 in Caenorhabditis elegans) are essential primary effectors of the Notch signaling pathway and are known to be able to recruit HDAC corepressor complexes to many different cellular and viral promoters carrying appropriate DNA-binding sites (Lai, 2002).
Here, we demonstrate that CBF-1 is a specific and potent inhibitor of the HIV-1 promoter, both in the presence and absence of Tat. Using chromatin immunoprecipitation (ChIP) assays, we demonstrated that CBF-1 strongly reduces the amount of RNAP II at the promoter, recruits HDACs to the LTR and nearly totally abolishes acetylation of core histones at Nuc-1. Knockdown of endogenous CBF-1 by shRNA results in the partial reactivation of latent HIV proviruses, recruitment of RNAP II to the proviral promoter, loss of histone deacetylases and the concomitant acetylation of histones. Thus, CBF-1 is a previously overlooked T-cell-specific regulatory factor that can silence HIV transcription and promote HIV entry into latency.
Results
CBF-1 is a potent repressor of HIV-1 transcription
Although HDACs can act as potent repressors of the HIV LTR, it is unknown how these factors are recruited to the provirus. Surprisingly, our preliminary experiments showed that overexpression of HDACs in trans did not result in HIV transcriptional silencing. This prompted us to look for mediator proteins that could be used to recruit HDACs. We focused our attention on CBF-1 because this protein is known to be expressed in T cells and our sequence analysis of the HIV identified several putative CBF-1-binding sites.
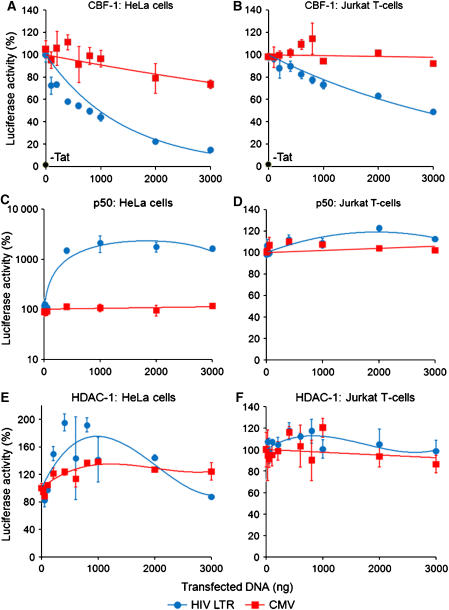
As shown in Figure 1, HeLa and Jurkat T cells infected with lentiviral vectors carrying luciferase reporter genes were used to measure the impact of CBF-1 expression in trans on the promoter activity of the HIV-1 LTR. The cells were infected with either pHR′P-Luc, a lentiviral vector carrying the luciferase reporter gene under the control of the HIV LTR, or as a control, with pHR′P-SIN18-Luc, a self-inactivating (SIN) lentiviral vector carrying a luciferase reporter transcribed by an internal human cytomegalovirus (CMV) immediate-early promoter. The cells were then transfected with up to 3 μg of pcDNA3-CBF-1, a plasmid expressing CBF-1 under the control of the CMV promoter together with 200 ng of pcDNA3-Tat, a plasmid expressing Tat.
Figure 1.
Inhibition of HIV transcription by CBF-1. HeLa (A) and Jurkat T cells (B) carrying latent proviruses expressing the luciferase reporter gene (pHR′-P-Luc; HIV) were co-transfected with between 0 and 3000 ng of a plasmid expressing CBF-1 protein (pcDNA3-CBF-1) along with 200 ng of a Tat-expressing plasmid (pcDNA3-Tat). Control cells were infected with pHR′P-SIN-18-Luc (CMV). In parallel experiments, latently infected HeLa (C) and Jurkat cells (D) were co-transfected with between 0 and 3000 ng of a plasmid expressing NF-κB p50 (pRSV-p50) (West et al, 2001) and 200 ng of pcDNA3-Tat. Latently infected HeLa (E) and Jurkat cells (F) were also co-transfected with between 0 and 3000 ng of a plasmid expressing HDAC-1 and 200ng of pcDNA3-Tat. After 48 h, cell extracts were checked for luciferase activity. Each data set was normalized, with 100% equal to the activity of cells transfected by pcDNA3-Tat alone. As an additional control, basal luciferase levels in the absence of Tat are shown in (A, B).
The repression of HIV transcription by CBF-1 is remarkably potent. In both cell lines, there was a clear dose-dependent inhibition of LTR promoter activation by CBF-1 that could be fitted to an exponential decay function. In the HeLa cells, half-maximal inhibition was reached at approximately 800 ng (Figure 1A), whereas in Jurkat T cells (Figure 1B) half-maximal inhibition was observed at 3 μg of CBF-1 plasmid DNA because of the lower transfection efficiency. This was confirmed by direct measurements of CBF-1 protein expression (Supplementary Figure S1). In contrast, luciferase expression under CMV promoter was only weakly inhibited by CBF-1. At 3 μg of CBF-1 DNA, CMV transcription was inhibited by less than 20% in HeLa cells and less than 10% in the Jurkat T cells. CBF-1 is also able to repress basal HIV-1 transcription in the absence of Tat, indicating that it functions at the level of transcriptional initiation (Supplementary Figure S2).
In contrast to CBF-1, NF-κB p50, which was previously reported to repress HIV transcription in latently infected cells (Williams et al, 2006), was a powerful inducer of HIV transcription in HeLa cells, leading to a 12-fold increase in luciferase activity. As expected, the control CMV promoter, which lacks NF-κB-binding sites, was unaffected by p50. In Jurkat T cells, p50 did not significantly induce HIV transcription. The failure of p50 to induce HIV transcription in Jurkat T cells could either be due to the occupation of the HIV NF-κB-binding sites by other factors in this cell line or the absence of appropriate cofactors.
As shown in Figure 1E and F, overexpression of HDACs did not lead to the repression of HIV LTR, indicating that specific mediator proteins, such as CBF-1, are required to allow HDAC interactions with the provirus.
HIV-1 LTR contains two distinct high-affinity CBF-1-binding sites
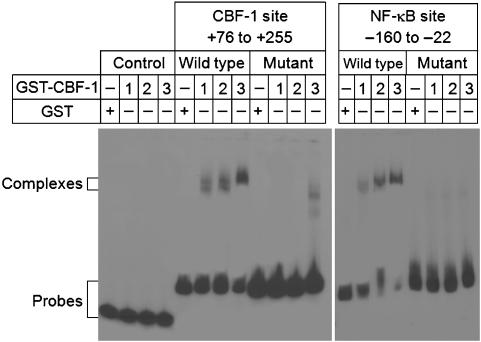
Trans-repression by CBF-1 requires its binding to specific high-affinity DNA-binding sequences which are related to the TGGGAA consensus sequence. To confirm that the LTR carries binding sites for CBF-1, we performed gel-retardation assays using purified GST-CBF-1 protein kindly provided by Dr Katherine Jones (Salk). As shown in Figure 2, CBF-1 is able to bind with high affinity to sequences overlapping the NF-κB elements of LTR-enhancer region (AGGGAC and GGGGAC). Mutation of the GGG sequence to CTC (West et al, 2001), which blocks NF-κB binding, also abolished CBF-1 binding. CBF-1 is also able to bind to a putative CBF-1-binding site (TGGGAA) at position +148 to +153. Introduction of point mutations in the putative CBF-1-binding site (from TGGGAA to ACTATC) abolished CBF-1 binding. As negative control, we chose a fragment of pcDNA3 vector, which does not contain any putative CBF-1-binding site. As expected, this fragment was unable to bind GST-CBF-1 protein. Thus, our results clearly demonstrate that there are two specific binding sites for CBF-1 located in the HIV-1 LTR.
Figure 2.
Complex formation between HIV LTR and CBF-1 protein. Gel-retardation assays were performed using 0–300 ng purified GST-CBF-1 protein and fragments of the HIV LTR. Left: CBF-1 fragment (+76 to +255); right: NF-κB fragment (−160 to −22). LTR fragments with mutated CBF-1-binding sites were used as specificity controls. A fragment of the pcDNA3 vector was used as negative control for CBF-1 binding (far left: control). GST alone was used as control for protein.
Establishment of latently infected Jurkat T-cell lines
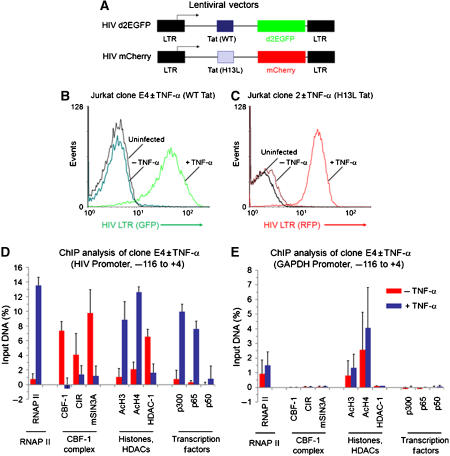
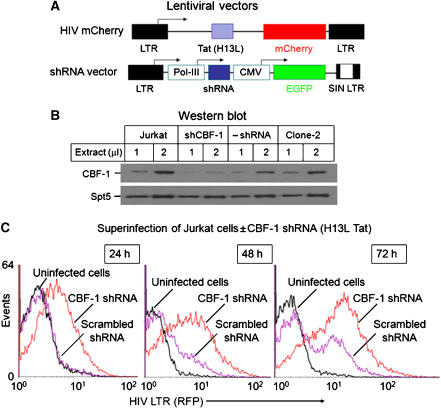
To develop a biochemically tractable experimental system for the study of the impact of CBF-1 on HIV transcription, we have used lentiviral vectors to generate T-cell lines carrying integrated proviruses expressing fluorescent protein reporter genes. An important feature of the viruses that we have constructed (Figure 3A) is that, like HIV itself, they are activated in cis by the regulatory proteins Tat and Rev (Kim et al, 2006). In these vectors, either the d2EGFP or mCherry fluorescent proteins were inserted in place of the Nef gene and were used as reporters to monitor HIV transcription.
Figure 3.
CBF-1 blocks HIV transcription in latently infected cells by recruiting HDACs. (A) Lentiviral vectors. (B) Induction of HIV transcription by TNF-α clone E4 cells. The cells were latently infected with the HIV d2EGFP virus (wild-type Tat) and induced for 18 h with 10 ng/ml TNF-α. (C) Induction of HIV transcription by TNF-α in Jurkat clone 2 cells. The cells were latently infected with the HIV mCherry virus (H13L Tat). (D) The distribution of transcription factors on the HIV LTR in clone E4 cells was analyzed by quantitative chromatin immunoprecipitation (ChIP) assays using the indicated antibodies before and after TNF-α stimulation. (E) Control ChIP experiment using the GAPDH gene. Red bars: unstimulated cells; purple bars: +20 ng/ml TNF-α for 24 h.
As described previously by Kim et al (2006), immediately after infection of the cells with the lentiviral vectors there is a high level of gene expression, which falls progressively over the next few weeks until the majority of the initially infected cells become latently infected and no longer express the fluorescent reporter protein. The gradual entry of proviruses into latency was initially observed by Li et al (1996) and later confirmed by Weinberger and colleagues (Weinberger et al, 2005; Weinberger and Shenk, 2006). The experiments in this paper were performed using viruses carrying either the wild-type Tat gene or viruses carrying the H13L mutation, a partially attenuated Tat variant originally identified in latently infected cells (Emiliani et al, 1998; Reza et al, 2003) that produces a high frequency of latently infected cells. Similar latently infected cell lines such as the J-Lat 6.3 cells (Jordan et al, 2003; Williams et al, 2006, 2007) and J89GFP and THP89GFP cells (Kutsch et al, 2002, 2003) have been developed using lentiviruses carrying fluorescent protein markers.
Two examples of latently infected clones are shown in Figure 3B and C. The Jurkat E4 clone carries a wild-type Tat and the d2EGFP reporter, whereas the Jurkat-mCherry-clone 2 was isolated from a shutdown population of cells infected with the H13L virus. Both the clonal cell lines show an extremely high degree of shutdown and reactivation. More than 95% of the latently infected E4 or clone 2 cells show no detectable reporter gene expression. An advantage of these clones compared to the J-Lat 6.3 model for HIV latency (Jordan et al, 2003), which has been extensively used by Warner Greene and colleagues (Williams et al, 2006, 2007), is that HIV transcription can be induced in over 90% of the cells following treatment of the cells with TNF-α to activate NF-κB, whereas only 30% of the J-Lat 6.3 cells can be reactivated under these conditions. This demonstrates that none of the cells in the E4 and clone 2 lines have lost the latent proviruses.
CBF-1 recruits HDACs and corepressor complexes to the LTR of latent proviruses
CBF-1 typically exerts its transcriptional repressive effect by recruiting HDAC-containing corepressor complexes. To confirm that both CBF-1 and corepressor complex(es) were present at the HIV-1 LTR, ChIP assays were performed using the Jurkat E4 clone (Figure 3D). The immunoprecipitated DNA was measured using primers directed to the promoter region of LTR (−116 to +4 with respect to transcription start site). Since the chromatin fragments used in this experiment were randomly sheared to approximately 400 nt length, the majority of the signal detected in this experiment can be attributed to proteins bound within the −200 to +200 region of the HIV LTR. As shown in Figure 3D, the latently infected clone E4 cells have low, but detectable, levels of RNAP II at the promoter, whereas CBF-1 and its corepressor subunits CIR and mSIN3A are all present at high levels. As expected for a repressed provirus, HDAC-1 is present at high levels, whereas acetylated histone H3 and H4 are present at only very low levels. Similarly, the transcriptional activator p300, and the NF-κB subunits p65 and p50 are present at only baseline levels. As demonstrated below (Figure 6), the recruitment of HDACs to the HIV LTR requires CBF-1.
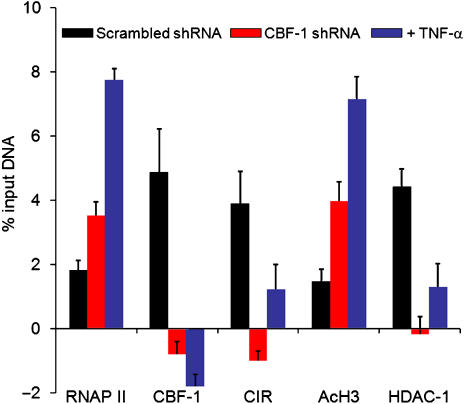
Figure 6.
Knockdown of CBF-1 by shRNA leads to enhanced histone acetylation. The distribution of transcription factors on the HIV LTR in HIV mCherry clone 2 cells superinfected with lentiviral vectors carrying shRNA was analyzed by quantitative chromatin immunoprecipitation (ChIP) assays using the indicated antibodies before and after TNF-α stimulation. Black bars: empty vector (minus shRNA); red bars: CBF-1 shRNA; purple bars: cells treated with 20 ng/ml TNF-α for 24 h.
To provide a control for the specificity of the ChIP assay, we also examined the GAPDH gene (Figure 3E) using the same immunoprecipitated samples that were used to study the HIV promoter (Figure 3D). RNAP II and acetylated histones were present on the GAPDH gene at relatively constant levels before and after TNF-α treatment. As expected, CBF-1 and its cofactors as well as p300 and the NF-κB subunits p65 and p50 were all absent.
We have also observed that the latent provirus in clone E4 cells carries heterochromatic markers, including K9- and K27-methylated histone H3, and the HP1α and HIP1γ proteins, in agreement with the results of du Chéné et al (2007) and Marban et al (2007) (data not shown).
If the HIV LTR is transcriptionally repressed due to CBF-1, then it is likely that it is displaced from the promoter following NF-κB induction. To test this hypothesis, ChIP assays were performed on the Jurkat E4 cells treated with TNF-α (Figure 3D). Induction of the latently infected cells by treatment with TNF-α increased RNAP II at the promoter over 14-fold. In contrast, CBF-1, CIR, mSIN3A and HDAC-1 levels fell to baseline levels as RNAP II levels rose. As expected, TNF-α induction resulted in p65 and p300 recruitment to the promoter. Similar results were obtained when Jurkat clone 2 (Figure 6) or Jurkat cells latently infected by pHR′-Luc (Supplementary Figure S3) were induced by TNF-α.
Thus, in three different clonal cell lines carrying latent proviruses, CBF-1 and its cofactors were present at the HIV LTR prior to induction of transcription by NF-κB. It is important to note that since CBF-1 exerts its effects by direct interaction with the HIV LTR, uniform results were obtained using proviruses that carried either the wild-type Tat (clone E4), the attenuated H13L Tat (clone 2) or no Tat (pHR′-Luc).
CBF-1 inhibits HIV transcription in newly infected cells
To measure the impact of CBF-1 during the early stages of viral infection, we prepared Jurkat cell lines in which CBF-1 was knocked down by shRNA expressed by lentiviral vectors (Figure 4A). As shown in Figure 4B, endogenous CBF-1 was reduced by over 75% in the Jurkat-CBF-1-shRNA cells compared to the control cells not expressing the shRNA.
Figure 4.
CBF-1 limits HIV gene expression in newly infected Jurkat T cells. (A) Structure of lentiviral vectors. (B) Western blot showing nuclear levels of CBF-1 in control cells and cells expressing shRNA to CBF-1. Top panel: CBF-1 antibody; bottom panel: Spt-5 antibody loading control. (C) Flow cytometric analysis of Jurkat T-cell lines with lentiviral vectors. Red line: CBF-1 shRNA; purple line: scrambled shRNA control; black line: uninfected cells. After 24 h, the cells were superinfected with HIV mCherry at an m.o.i. of 1.5 and monitored for mCherry expression over the next 72 h.
To evaluate the effects of CBF-1 knockdown on HIV transcription, we compared a matched pair of cell lines carrying lentiviral vectors that expressed either shRNA to CBF-1 or scrambled shRNA control (Figure 4C). The cells were superinfected with the HIV mCherry virus and analyzed by flow cytometry between 24 and 72 h post-infection. At each time point, the CBF-1-ablated cells showed a 3- to 4-fold higher proportion of mCherry expressing cells than the control cells. Comparable results are shown in Supplementary Figures S4–S6 where HIV viruses carrying either the wild-type Tat or H13L Tat were used and the data are shown as a two-dimensional plot.
We conclude that endogenous levels of CBF-1 are sufficiently high to be able to restrict HIV transcription in newly infected T cells.
Knockdown of CBF-1 in resting cells partially activates latent proviruses and induces an ‘open' chromatin structure at the HIV LTR
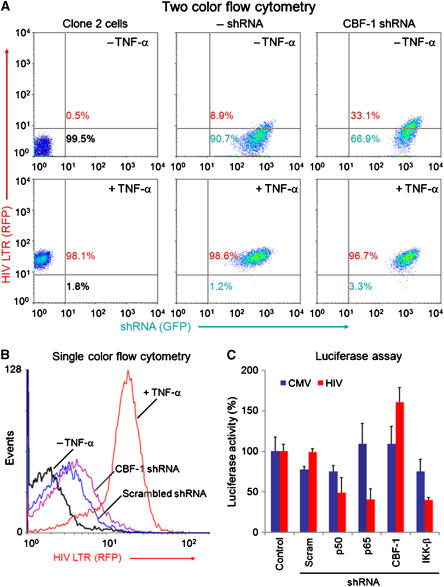
If CBF-1 plays a significant role in HIV silencing, then it should be possible to at least partially reactivate the latent proviruses by knockdown of the endogenous CBF-1 using shRNA. To test this hypothesis, the latently infected Jurkat-clone 2 cell line (mCherry) was superinfected with lentiviral vectors carrying either shRNA against CBF-1 or an empty vector control (Figure 5). The shRNA vectors carried the EGFP reporter under the control of the CMV promoter to provide a marker for the cells expressing shRNA (Figure 4A).
Figure 5.
Knockdown of CBF-1 by shRNA leads to partial induction of latent HIV. (A) Two-color flow cytometric analysis. Jurkat T cells latently infected by HIV mCherry (clone 2 cells) were superinfected by shRNA vectors. Left: clone 2 cells; middle: empty vector control; right: CBF-1 shRNA; top panels: unstimulated cells; bottom panels: cells treated with 20 ng/ml TNF-α for 16 h. (B) Activation of HIV expression following superinfection by vectors expressing scrambled shRNA and CBF-1 shRNA. Black line: uninduced clone 2 cells; blue line: scrambled shRNA; purple line: CBF-1 shRNA; red line: induced by TNF-α. (C) Induction of luciferase expression in Jurkat pHR′P-Luc (HIV) and Jurkat pHR′P-SIN-18-Luc (CMV) by CBF-1 shRNA. The cells were infected with lentiviral vectors carrying shRNA to p50, p65, CBF-1, IKK-β or a scrambled shRNA control.
Knockdown of CBF-1 due to expression of the CBF-1 shRNA resulted in partial activation of the latent proviruses. At 48 h after infection, 33% of the cells showed increased HIV mCherry (RFP) expression (Figure 5A). In comparison, only 9% of control cells infected with the empty lentiviral vector (minus shRNA) showed enhanced mCherry expression. The partial activation of these cells is probably due to the scarcity of NF-κB in the unactivated cells. To demonstrate the requirement for NF-κB, and that none of these cells had lost the integrated latent HIV-1 proviruses, Jurkat clone 2 cells and its superinfected derivatives were induced with TNF-α. In all the three cell populations, more than 97% of the induced cells expressed mCherry at high levels. Comparable results are shown in Figure 5B, which compares the extent of activation of HIV gene expression in clone 2 cells following infection by vectors expressing a scrambled shRNA control and CBF-1 shRNA. Similarly, CBF-1 shRNA was able to increase luciferase expression in Jurkat cells latently infected with the pHR′P-Luc vector to 183% of the control values, whereas shRNA to p50 or p65 reduced proviral expression to 37 and 32%, respectively of the control values (Figure 5C).
We also evaluated the impact of CBF-1 knockdown on HIV gene expression by ChIP assays using primers for the promoter (Figure 6). Since the clone 2 cells are latently infected there was only a low level of RNAP II associated with the promoter either in the absence of activation signals or when clone 2 cells were infected with an empty shRNA lentiviral vector. Infection by the lentiviral vectors carrying CBF-1 shRNA resulted in a twofold increase in RNAP II at the promoter. Control ChIP experiments show that the knockdown of CBF-1 reduced it to undetectable levels at the HIV promoter. There was a parallel decline in CIR and HDAC-1 levels. In contrast, acetylated histone H3 levels increased when CBF-1 was knocked down. Thus, removal of CBF-1 leads to a loss of HDAC-containing corepressor complexes at HIV-1 LTR and consequently the acetylation of core histones and recruitment of RNAP II.
In the complementary experiments using either HeLa (Supplementary Figure S8) or Jurkat T cells (Supplementary Figures S7 and S9), we show that overexpression of CBF-1 led to a decline in RNAP II levels and a concomitant rise of CIR, mSIN3A and HDAC-1 levels at the promoter and the Nuc-1 region. In association with the HDAC-1 recruitment there was also a decline in acetylated histone H3 and H4 levels at the HIV LTR.
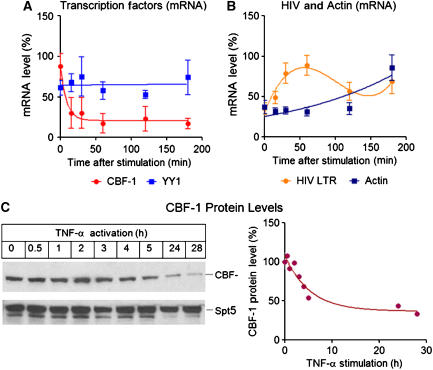
T-cell activation downregulates CBF-1 mRNA expression
An important property of any putative repressor used to establish HIV latency is that it must be removed following T-cell activation. To examine whether CBF-1 expression is altered during T-cell activation, we analyzed the cellular levels of CBF-1 mRNA following activation of Jurkat cells carrying a latent pHR′-luciferase provirus with antibodies to the CD3 T-cell receptor and CD28 co-receptors. As shown in Figure 7A, CBF-1 mRNA levels drop sharply (t1/2=15–30 min) and remain at low levels for at least up to 3 h of treatment. In contrast, HIV-1 transcript levels rise rapidly during the first 60 min of treatment, decline after NF-κB exit from the nucleus and then rise again after 3 h (Figure 7B). The rapid decline in CBF-1 mRNA levels in response to T-cell receptor stimulation places it in a small subset of transcription factors that are downregulated during T-cell activation (Argyropoulos et al, 2004). The most typical mRNA expression profile in activated T cells is illustrated by β-actin (Figure 7B), which shows relatively small changes in mRNA levels for the first 2 h and then begins to rise as the T cells become enlarged and more motile by 3 h. The YY1 gene shows a flat expression profile throughout the activation period (Figure 7A). Comparable results were obtained when cells were activated by TNF-α, or PMA/PHA treatment (Supplementary Figure S10). Consistent with the mRNA results, following T-cell activation, CBF-1 protein levels decline with t1/2=3–5 h (Figure 7C).
Figure 7.
CBF-1 mRNA and protein are rapidly downregulated in activated T cells. Jurkat cells were activated by treatment with anti-CD3 and anti-CD28 antibodies and the levels of mRNA were accessed by quantitative real-time PCR. (A) CBF-1 mRNA (red) and YY1 (light blue). (B) HIV mRNA (yellow) and β-actin mRNA (dark blue). (C) CBF-1 and Spt5 protein levels were measured by western blotting at various times following T-cell activation.
We conclude that in quiescent or unstimulated T cells, where CBF-1 levels are high and NF-κB levels are low, CBF-1 is able to bind to the HIV LTR and recruits HDACs containing corepressor complexes. This helps to establish repressive chromatin structures at LTR, which accelerate entry into latency. Following induction of NF-κB, nuclear levels of NF-κB rise, CBF-1 and its recruited corepressor complexes are replaced by NF-κB and its coactivators, and HIV transcription resumes.
Discussion
Silencing of HIV by CBF-1
In this paper, we have demonstrated that CBF-1 is a specific and highly potent inhibitor of HIV-1 transcription. CBF-1 is recruited to the HIV LTR by binding primarily to the two NF-κB elements present in the enhancer region of HIV-1 LTR. An additional CBF-1-binding site is located at position +148 to +153 downstream of the transcription start site, adjacent to Nuc-1. Promoters that lack CBF-1-binding sites, such as the CMV immediate-early promoter, are not repressed by CBF-1.
Direct evidence that endogenous levels of CBF-1 found in T-cell lines help to restrict HIV transcription in latently infected cells comes from experiments where intracellular CBF-1 levels were knocked down using lentiviral vectors expressing shRNA. For example, in Figure 6 we showed that when latent proviruses were induced by ablation of CBF-1 or activation by TNF-α, there is a decline in CBF-1, CIR and HDAC-1 levels and a parallel increase in RNAP II and acetylated histones at the HIV LTR. Thus, knockdown of CBF-1 partially relieves the chromatin blocks and results in significant activation of latent proviruses. However, maximal induction of the HIV LTR also requires the expression of NF-κB, indicating that additional restrictions on HIV transcription are present. In the complementary series of experiments shown in the Supplementary data, we have also shown that overexpression of CBF-1 decreases HIV gene expression in newly infected cells and enhances the recruitment of HDAC-1 to the promoter.
The recruitment of HDACs to the LTR seems to be the major mechanism by which CBF-1 blocks transcription. This is similar to the mechanisms that have been proposed previously for the blocking of HIV-1 transcription by YY1 and p50 (He and Margolis, 2002; Williams et al, 2006). In contrast to the report of Williams et al (2006), we have been unable to demonstrate that p50 acts as a repressor of HIV proviruses in the many different cell lines that we have examined. In our hands, p50 strongly activates transcription in HIV proviruses in HeLa cells and weakly activates HIV transcription in Jurkat T cells (Figure 1). We were also unable to demonstrate activation of latent HIV proviruses with shRNA to p50 under conditions where partial activation of the provirus could be observed using shRNA to CBF-1. It is possible that association of p50 with the HIV LTR is a specific property of the J-Lat 6.3 cell line that they employed in their studies. In contrast to p50, we have easily detected YY1 at the HIV LTR in latently infected T-cell lines. YY1 is known to inhibit HIV replication when overexpressed in trans (Coull et al, 2000). However, the presence of YY1 does not strictly correlate with the recruitment of HDACs in T cells. Following proviral induction by NF-κB, YY1 levels actually rise slightly. This is possibly due to the dual capacity of YY1 to act as a repressor and activator of transcription.
CBF-1 cofactor utilization
To mediate the repression of Notch-responsive genes, CBF-1 recruits different HDAC corepressor complexes through its repression domain. These include either SMRT/Sin3 (Hsieh and Hayward, 1995; Hsieh et al, 1996; Kao et al, 1998) or CIR/N-CoR/SMRT/SKIP (Hsieh et al, 1999). Both complexes contain HDACs that are able to deacetylate nearby histones. Alternatively, in certain cases, CBF-1 can restrict transcription by competitively inhibiting the interaction of TFIIA and TFIID at the promoter (Olave et al, 1998).
To determine the specific mechanisms used by CBF-1 to repress HIV transcription, and to define the composition of the corepressor complexes, we performed extensive ChIP analyses in several latently infected cell lines. Binding of CBF-1 to the HIV LTR led to the recruitment of CIR, Sin3a, HDAC-1 and a small amount of HDAC-2. The recruitment of the HDACs resulted in a dramatic loss of acetylated core histones at the HIV LTR when CBF-1 was present.
Indirect effects of CBF-1 could also have a negative impact on HIV transcription since many other NF-κB-regulated genes are also inhibited by CBF-1. For example, CBF-1 interacts with a dual NF-κB/CBF-1-binding site in both the IκBα promoter and the NF-κB2 promoter and inhibits their activity (Oswald et al, 1998; Oakley et al, 2003). Similarly, IL-6, a cytokine which is involved in T-cell activation, proliferation and survival is downregulated by CBF-1 (Liu et al, 2005).
Regulation of CBF-1 levels during T-cell activation
A unique feature of CBF-1 that makes it a particularly attractive candidate as a factor controlling HIV latency is that it is among the rare subset of genes that are downregulated during T-cell activation. Unactivated T cells (or primary T cells) show higher levels of CBF-1 mRNA and protein, but CBF-1 levels fall rapidly in response to proliferative stimulation. In contrast, under these conditions the levels of YY1 appear to be relatively constant (He and Margolis, 2002; Williams et al, 2006).
Role of CBF-1 in promoting HIV latency
The rarity of latently infected T cells in HIV-infected patients precludes direct analyses of the proteins present at the provirus. The biochemical analyses in this paper were therefore conducted using a series of clones carrying transcriptionally silent proviruses derived from lentiviral vectors carrying marker proteins. An important feature of the clones we have studied is that essentially the entire population can be reactivated following induction of NF-κB. Although Jurkat T cells are a transformed cell line, we believe that the underlying molecular mechanisms that lead to latency in Jurkat T cells are likely to be similar to those seen in HIV-1-infected patients since in both cases transcriptional reactivation of the latent proviruses requires induction of NF-κB (Brooks et al, 2003; Arlen et al, 2006) and inhibitors of HDACs are able to partially reactivate latent proviruses in cells isolated from patients as well as in transformed cell lines (Ylisastigui et al, 2004; Klichko et al, 2006).
Numerous studies have demonstrated that formation of a restrictive chromatin environment is a prerequisite for the entry of HIV into latency. Although it was originally believed that latency arises when HIV integrates into pre-existing regions of heterochromatin (Jordan et al, 2003), recent high-throughput sequencing studies of HIV integration sites of infected resting CD4+ T cells in HIV-infected patients with effective suppression of viremia have shown that HIV preferentially integrates into regions containing actively transcribed genes and is almost never found in heterochromatic regions (Han et al, 2004; Lewinski et al, 2005). Similarly, the latently infected Jurkat cells we have studied all carry integrants into actively transcribed genes (data not shown). It therefore seems likely that epigenetic shutdown mechanisms leading to HIV silencing are responsible for the induction of HIV latency.
Consistent with this idea, our ChIP experiments have shown that CBF-1 functions by recruiting corepressor complexes containing HDACs to induce transcriptional silencing. As shown in Figure 8, in quiescent or unstimulated T cells, where CBF-1 levels are high, the HIV LTR is repressed. After binding to the specific sequences at HIV-1 LTR, CBF-1 recruits HDAC-containing corepressor complexes. We have been able to confirm the results of du Chéné et al (2007) and Marban et al (2007) that latent HIV proviruses also carry proteins that are typically associated with heterochromatin, including methylated histones and HP1α and HP1γ (data not shown). It therefore seems likely that establishment of restrictive heterochromatic structures on the latent HIV provirus involves a series of sequential events starting with the recruitment of HDACs to the promoter via CBF-1, followed by methylation of histones by Suv39H1 and binding of the HP proteins to the methylated histones.
Figure 8.
Model for regulation of HIV latency by CBF-1. In latently infected cells, transcription is blocked by a repressive chromatin structure induced by CBF-1 binding to the HIV LTR. CBF-1 directly recruits histone deacetylases to the promoter, which repress transcription. Subsequently, histone methylases and HP1 proteins establish heterochromatic structures on the HIV LTR. Activation of HIV by stimulation of NF-κB leads to displacement of CBF-1 from the LTR and recruitment of histone acetyltransferases and chromatin-remodeling factors that lead to the formation of an active transcription unit.
An inevitable consequence of HIV silencing is the blocking of Tat production. As Tat levels become restricted, RNAP II elongation becomes increasingly inefficient and the virus is able to enter latency. Following induction of T cells carrying latent proviruses, NF-κB p65 is able to displace CBF-1 and its associated corepressor complexes from the HIV LTR. NF-κB p65 is then able to recruit transactivators such as the histone acetyl transferase (HAT) p300. Acetylation of core histones by HATs then leads to the recruitment of the SWI/SNF chromatin remodeling machinery and restores access of transcription machinery to the promoter, leading to the initial Tat- and Rev-independent rounds of HIV transcription. Once Tat and Rev are synthesized, a positive feedback mechanism is established, and the virus becomes committed to the syntheses of full length transcripts and productive growth.
Materials and methods
Lentiviral constructs
The lentiviral vectors pHR′ and pHR′SIN-18, a SIN vector (Dull et al, 1998) were modified by insertion of a polylinker (BamH1, SmaI, SalI, MscI, SpeI, SacII and XhoI) between the BamH1 and XhoI sites to create pHR′P and pHR′P-SIN-18. The PCR-amplified luciferase gene from pGL3 basic vector (Invitrogen) was inserted into pHR′P to generate pHR′P-Luc. In pHR′P-SIN-18-Luc, expression of the luciferase gene is under the control of the internal human CMV immediate-early promoter. pHR′P-SIN-Flag-CBF-1-IRES-Neo carries the Flag-CBF-1 insert cloned between the BamH1 and SalI sites of pHR′P-SIN-18 and an IRES and Neomycin gene cloned between the SalI and XhoI sites of the polylinker. pHR′-P-PNL-mCherry was produced by cloning the NotI to XhoI fragment from HIV-1 pNL4-3 into the pHR′P vector. The shRNA vector pHR′P-SIN-CMV-GFP was generated by cloning the CMV-GFP insert into the SacII to XhoI sites of the pHR′P-SIN-18 vector. shRNA inserts were initially cloned into the pSuper vector (Oligoengine). The shRNA plus the H1 promoter were then cloned into pHR′P-SIN-CMV-GFP between the BamH1 and SalI sites.
Generation of VSV-G-pseudotyped viral particles
293T and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. T-cell lines CEM and Jurkat were maintained in RPMI 1640 medium supplemented with 10% FCS.
Pseudotyped HIV-based lentiviral vector particles pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G) were produced using a three-plasmid co-transfection procedure (Naldini et al, 1996). The viruses were concentrated by ultracentrifugation, aliquoted and frozen at −80°C until use.
ChIP assays
ChIP was done as previously described (Kim et al, 2006). To activate cells, we used either 10 ng/ml TNF-α or 25 μl per 106 cells CD3/CD28 antibodies bound Dynal beads (Dynal Biotech). Most antibodies were purchased from Santa Cruz, including anti-RNAP II (N-20), CBF-1(H-50), CIR (C-19), mSIN3A (AK-11), HDAC-1 (H-51), HDAC-2 (H-54) and p65 (C-20). Anti-acetylated histone-3 and -4 antibodies were obtained from Upstate. Each sample (5%) was analyzed by quantitative real-time PCR to access the amount of sample immunoprecipitated by individual antibody. A non-antibody control value was subtracted from each sample value to remove the background counts.
Western blot
Western blotting was performed according to standard protocols. Anti-CBF-1 (Chemicon or Santa Cruz), anti-Flag (Sigma) and secondary HRPO-conjugated anti-rabbit or anti-mouse antibodies were (Dako) were used.
Electromobility shift assay
GST fusion CBF-1 protein produced in Escherichia coli was kindly provided by Katherine Jones. The gel shift probes were prepared by cleaving the HIV-1 LTR with either HindIII to BssHII (CBF-1 region: +76 to +255) or AvaI to PvuII (NF-κB region: −160 to −22). As a negative control, we employed the NarI to MscI fragment of the pcDNA3 vector (Invitrogen). The fragments were labeled with γ-32P ATP and T4 polynucleotide kinase. Binding reactions contained the 32P-labeled DNA fragment (10 000 c.p.m.) and either GST-CBF-1 or GST alone as a control. Reactions were performed in electromobility shift assay buffer (20 mM HEPES at pH 7.9, 0.2 mM EDTA, 20% glycerol, 100 mM KCl, 1 mM dithiothreitol), containing 2 μg poly (dI-dC), 1 μg bovine serum albumin (BSA), 10 μM ZnCl2, 0.5 mM phenylmethylsulfonyl fluoride and 100 ng/ml each of leupeptin and aprotinin for 15 min at room temperature. Reaction mixtures were fractionated on 5% non-denaturing polyacrylamide gels (acrylamide/bis-acrylamide, 37.5:1) containing 2.5% glycerol and TGE buffer (25 mM Tris, 192 mM glycine and 1 mM EDTA).
Quantitation of transcripts by real-time PCR
Total RNA was isolated from 2 × 106 cells using Trizol reagent (Gibco-BRL) according the manufacturer's instructions. Equal amounts of RNA (1–2 μg) were employed in cDNA synthesis using M-MLV reverse transcriptase (Invitrogen) for 1 h at 37°C in a total volume of 25 μl containing RT buffer, DTT, 80 U of RNasin (40 U/μl; Promega) and 1 μl of either oligo-dT or random hexamer primer. Purified cDNA (10%) was used as a template in real-time PCR reactions using a DNA engine Opticon-2 (MJ) Real-T-PCR machine. Data analysis utilized the Sequence Detector software (version 1.6.3 and 1.7) to determine threshold cycle, Ct, and reduced normalized fluorescence values, ΔRn, for each amplified product for each time point.
Luciferase assays
Cells transfected in six-well plates were harvested after 48 h, washed twice in phosphate-buffered saline, and then lysed in 100–200 μl of 1 × Passive Lysis Buffer (Promega) for 30 min at room temperature. From each lysate, 5 μl was assayed with 50 μl of enzyme substrate provided with Luciferase Assay System kit (Promega) using microplate luminometer with an injection unit (Bio-Rad).
Flow cytometry
Cells were subjected to flow cytometric analysis using FACScan flow cytometer (Becton Dickinson, Franklin Lakes, NJ) and data were analyzed with WinMDI software.
Supplementary Material
Supplementary Data
Acknowledgments
We thank Drs Katherine Jones, Didier Trono, Diana Hayward and Stanton Gerson for gifts of reagents. We thank our colleagues Young-Kyeung Kim, Richard Pearson, Julian Wong, Joseph Hokello, Julian Friedman, Zafiria Athanassiou and Kara Lassen for advice and assistance. The study was supported by NIH Grants AI067093 to JK and the Case CFAR (AI036219).
References
- Argyropoulos C, Nikiforidis GC, Theodoropoulou M, Adamopoulos P, Boubali S, Georgakopoulos TN, Paliogianni F, Papavassiliou AG, Mouzaki A (2004) Mining microarray data to identify transcription factors expressed in naive resting but not activated T lymphocytes. Genes Immun 5: 16–25 [DOI] [PubMed] [Google Scholar]
- Arlen PA, Brooks DG, Gao LY, Vatakis D, Brown HJ, Zack JA (2006) Rapid expression of human immunodeficiency virus following activation of latently infected cells. J Virol 80: 1599–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DG, Arlen PA, Gao L, Kitchen CM, Zack JA (2003) Identification of T cell-signaling pathways that stimulate latent HIV in primary cells. Proc Natl Acad Sci USA 100: 12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T-W, Davey RT Jr, Ostrowski M, Shawn Justement J, Engel D, Mullins JI, Fauci AS (2000) Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat Med 6: 757–761 [DOI] [PubMed] [Google Scholar]
- Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, Shi Y, Hansen U, Margolis DM (2000) The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol 74: 6790–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Chéné I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, Bertrand E, Marcello A, Emiliani S, Kiernan R, Benkirane M (2007) Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J 26: 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L (1998) A third-generation lentivirus vector with a conditional packaging system. J Virol 72: 8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S, Fischle W, Ott M, van Lint C, Amella CA, Verdin E (1998) Mutations in the Tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J Virol 72: 1666–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S, Van Lint C, Fischle W, Paras P Jr, Ott M, Brady J, Verdin E (1996) A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc Natl Acad Sci USA 93: 6377–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T (1997) CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA 94: 2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose R, Liou LY, Herrmann CH, Rice AP (2001) Induction of TAK (cyclin T1/P-TEFb) in purified resting CD4(+) T lymphocytes by combination of cytokines. J Virol 75: 11336–11343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu X, Pierson TC, Margolick JB, Siliciano RF, Siliciano JD (2004) Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol 78: 6122–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Margolis DM (2002) Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol Cell Biol 22: 2965–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A, Holloway A, Reeves R, Tremethick DJ (2004) Recruitment of SWI/SNF to the human immunodeficiency virus type 1 promoter. Mol Cell Biol 24: 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M (1995) Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373: 123–126 [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Hayward SD (1995) Masking of the CBF1/RBPJ kappa transcriptional repression domain by Epstein–Barr virus EBNA2. Science 268: 560–563 [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD (1996) Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein–Barr virus EBNA2. Mol Cell Biol 16: 952–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD (1999) CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA 96: 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22: 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev 12: 2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, Wu SY, Chiang CM, Karn J (2006) Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J 25: 3596–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita S, Su L, Amano M, Timmerman LA, Kaneshima H, Nolan GP (1997) The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 6: 235–244 [DOI] [PubMed] [Google Scholar]
- Klichko V, Archin N, Kaur R, Lehrman G, Margolis D (2006) Hexamethylbisacetamide remodels the human immunodeficiency virus type 1 (HIV-1) promoter and induces Tat-independent HIV-1 expression but blunts cell activation. J Virol 80: 4570–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22: 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsch O, Benveniste EN, Shaw GM, Levy DN (2002) Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J Virol 76: 8776–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsch O, Levy DN, Kosloff BR, Shaw GM, Benveniste EN (2003) CD154–CD40-induced reactivation of latent HIV-1 infection. Virology 314: 261–270 [DOI] [PubMed] [Google Scholar]
- Lai EC (2002) Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep 3: 840–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, Coffin JM, Bosch RJ, Margolis DM (2005) Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 366: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, Hannenhalli S, Verdin E, Berry CC, Ecker JR, Bushman FD (2005) Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J Virol 79: 6610–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Moore B, Cloyd MW (1996) Gradual shutdown of virus production resulting in latency is the norm during the chronic phase of human immunodeficiency virus replication and differential rates and mechanisms of shutdown are determined by viral sequences. Virology 225: 196–212 [DOI] [PubMed] [Google Scholar]
- Lin X, Irwin D, Kanazawa S, Huang L, Romeo J, Yen TSB, Peterlin BM (2003) Transcriptional profiles of latent human immunodeficiency virus in infected individuals: effects of Tat on the host and reservoir. J Virol 77: 8227–8236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HW, Cosa G, Landes CF, Zeng Y, Kovaleski BJ, Mullen DG, Barany G, Musier-Forsyth K, Barbara PF (2005) Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription. Biophys J 89: 3470–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusic M, Marcello A, Cereseto A, Giacca M (2003) Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J 22: 6550–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Parra M, Vries RG, Kauder SE, Verrijzer CP, Ott M, Verdin E (2006) The SWI/SNF chromatin-remodeling complex is a cofactor for tat transactivation of the HIV promoter. J Biol Chem 281: 19960–19968 [DOI] [PubMed] [Google Scholar]
- Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, Aunis D, Rohr O (2007) Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J 26: 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G, Baltimore DA (1987) An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326: 711–713 [DOI] [PubMed] [Google Scholar]
- Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D (1996) In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272: 263–267 [DOI] [PubMed] [Google Scholar]
- Oakley F, Mann J, Ruddell RG, Pickford J, Weinmaster G, Mann DA (2003) Basal expression of IkappaBalpha is controlled by the mammalian transcriptional repressor RBP-J (CBF1) and its activator Notch1. J Biol Chem 278: 24359–24370 [DOI] [PubMed] [Google Scholar]
- Olave I, Reinberg D, Vales LD (1998) The mammalian transcriptional repressor RBP (CBF1) targets TFIID and TFIIA to prevent activated transcription. Genes Dev 12: 1621–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Liptay S, Adler G, Schmid RM (1998) NF-kappaB2 is a putative target gene of activated Notch-1 via RBP-Jkappa. Mol Cell Biol 18: 2077–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza SM, Shen L-M, Mukhopadhyay R, Rosetti M, Pe'ery T, Mathews MB (2003) A naturally occurring substitution in human immunodeficiency virus Tat increases expression of the viral genome. J Virol 77: 8602–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scripture-Adams DD, Brooks DG, Korin YD, Zack JA (2002) Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J Virol 76: 13077–13082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treand C, du Chene I, Bres V, Kiernan R, Benarous R, Benkirane M, Emiliani S (2006) Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J 25: 1690–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Paras PJ, Van Lint C (1993) Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J 12: 3249–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, Saag MS, Shaw GM (1995) Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373: 117–122 [DOI] [PubMed] [Google Scholar]
- Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV (2005) Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122: 169–182 [DOI] [PubMed] [Google Scholar]
- Weinberger LS, Shenk T (2006) An HIV feedback resistor: auto-regulatory circuit deactivator and noise buffer. PLoS Biol 5: e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ, Lowe AD, Karn J (2001) Activation of HIV transcription in T-cells revisited: NF-κB p65 stimulates transcriptional elongation. J Virol 75: 8524–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC (2006) NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 25: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Kwon H, Chen LF, Greene WC (2007) Sustained induction of NF-kappaB is required for efficient expression of latent human immunodeficiency virus type 1. J Virol 81: 6043–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM (2004) Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 18: 1101–1108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data