Abstract
The ezrin/radixin/moesin (ERM) proteins are involved in actin filament/plasma membrane interaction that is regulated by Rho. We examined whether ERM proteins are directly phosphorylated by Rho- associated kinase (Rho-kinase), a direct target of Rho. Recombinant full-length and COOH-terminal half radixin were incubated with constitutively active catalytic domain of Rho-kinase, and ∼30 and ∼100% of these molecules, respectively, were phosphorylated mainly at the COOH-terminal threonine (T564). Next, to detect Rho-kinase–dependent phosphorylation of ERM proteins in vivo, we raised a mAb that recognized the T564-phosphorylated radixin as well as ezrin and moesin phosphorylated at the corresponding threonine residue (T567 and T558, respectively). Immunoblotting of serum-starved Swiss 3T3 cells with this mAb revealed that after LPA stimulation ERM proteins were rapidly phosphorylated at T567 (ezrin), T564 (radixin), and T558 (moesin) in a Rho-dependent manner and then dephosphorylated within 2 min. Furthermore, the T564 phosphorylation of recombinant COOH-terminal half radixin did not affect its ability to bind to actin filaments in vitro but significantly suppressed its direct interaction with the NH2-terminal half of radixin. These observations indicate that the Rho-kinase–dependent phosphorylation interferes with the intramolecular and/ or intermolecular head-to-tail association of ERM proteins, which is an important mechanism of regulation of their activity as actin filament/plasma membrane cross-linkers.
The ezrin/radixin/moesin (ERM)1 family consists of three closely related proteins, ezrin, radixin, and moesin (Bretscher et al., 1983; Pakkanen et al., 1987; Lankes et al., 1988; Tsukita et al., 1989; Sato et al., 1992). Sequencing analyses have revealed that the three ERM proteins are highly homologous; in the mouse, the levels of identity are 75, 72, and 80% for ezrin/radixin, ezrin/moesin, and radixin/moesin, respectively (Gould et al., 1989; Turunen et al., 1989; Funayama et al., 1991; Lankes and Furthmayr, 1991; Sato et al., 1992). The sequences of their NH2-terminal halves are highly conserved (∼85% identity for all pairs). A tumor suppressor molecule responsible for neurofibromatosis type 2 named merlin or schwannomin was identified and shown to have significant sequence similarity to ERM proteins; ∼49% identity overall and ∼65% identity in the NH2-terminal halves (Rouleau et al., 1993; Trofatter et al., 1993). This highly conserved NH2-terminal sequence is also found in the NH2-terminal ends of some membrane-associated proteins such as band 4.1 protein, talin, PTPH1, and PTPMEG, indicating the existence of a band 4.1 superfamily (Conboy et al., 1986; Rees et al., 1990; Gu et al., 1991; Yang and Tonks, 1991; Arpin et al., 1994; Takeuchi et al., 1994a ).
ERM family proteins are thought to function as general cross-linkers between the plasma membrane and actin filaments (Bretscher, 1983; Pakkanen et al., 1987; Tsukita et al., 1989; Algrain et al., 1993). The suppression of ERM protein expression with antisense oligonucleotides destroys microvilli, cell–cell, and cell–matrix adhesion sites (Takeuchi et al., 1994b ), and the introduction of dominant-negative constructs of radixin impairs cytokinesis (Henry et al., 1995). The highly conserved NH2-terminal halves of ERM proteins are responsible for their association with the plasma membrane. CD44 was identified as a binding partner of ERM proteins on the plasma membrane (Tsukita et al., 1994). Integral membrane proteins such as CD43, intercellular adhesion molecule (ICAM)-2, ICAM-3, and the H+/K+ ATPase pump have also been reported to be colocalized with ERM proteins (Hanzel et al., 1991; Yonemura et al., 1993; Helander et al., 1996; Serrador et al., 1997). On the other hand, the COOH-terminal halves of ERM proteins, especially the COOH-terminal 34 amino acids, interacts with actin filaments (Turunen et al., 1994; Pestonjamasp et al., 1995). The coexistence of plasma membrane–binding and actin filament–binding domains in individual molecules allows ERM proteins to function as plasma membrane/actin filament cross-linkers.
However, both the actin- and membrane-binding domains are thought to be masked in native full-length ERM proteins. As mentioned above, the COOH-terminal half of ezrin binds to actin filaments. However, the binding of native full-length ERM proteins to actin filaments has not been directly established under physiological conditions, although one recent paper describes that purified ezrin binds to nonmuscle β-actin filaments with high affinity (Yao et al., 1996). Similarly, at physiological ionic strength, full-length ERM proteins show very low affinity to the cytoplasmic domain of CD44 in vitro, whereas the NH2-terminal halves of ERM proteins lacking the COOH-terminal halves bind to CD44 with high affinity (Hirao et al., 1996). Furthermore, the NH2-terminal halves of ERM proteins can be directly associated with their COOH-terminal halves in vitro (Gary and Bretscher, 1993, 1995; Andréoli et al., 1994; Magendantz et al., 1995). These findings suggested an intramolecular and/or intermolecular head-to-tail association mechanism for ERM protein activation and inactivation (Berryman et al., 1995; Bretscher et al., 1995; Martin et al., 1995). Through head-to-tail association, the NH2- and COOH-terminal halves of native ERM proteins are thought to mutually suppress their functions, i.e., membrane and actin binding, respectively.
Some signal must release the mutual suppression in ERM proteins within cells, so that they can function as cross-linkers just beneath the plasma membrane. Previously we reported that Rho, a small GTP-binding protein, regulates the formation of CD44/ERM protein complex (Hirao et al., 1996). Takaishi et al. (1995) and Kotani et al. (1997) also suggested that in MDCK cells Rho regulates the association of ERM proteins with plasma membranes. Ridley and coworkers (Ridley and Hall, 1992; Ridley et al., 1992) found using serum-starved Swiss 3T3 cells that Rho plays a central role in the coordinated assembly of focal adhesions and stress fibers induced by growth factors such as lysophosphatidic acid (LPA). More recently, this group identified moesin as an essential factor for the Rho-dependent formation of stress fibers in serum-starved Swiss 3T3 cells (Mackay et al., 1997). However, how the Rho signaling pathway is involved in the regulation of the cross-linking activity of ERM proteins at the molecular level is still unclear.
Rho was reported to regulate the activities of some serine/threonine kinases including Rho-associated kinase (Rho-kinase)/ROKα (Leung et al., 1995; Matsui et al., 1996), p160 ROCK (Ishizaki et al., 1996), protein kinase N (Amano et al., 1996a ; Watanabe et al., 1996), and protein kinase C1 (Nonaka et al., 1995). On the other hand, ERM proteins are highly serine/threonine phosphorylated (Gould et al., 1986; Urushidani et al., 1989; Nakamura et al., 1995, 1996), and their phosphorylation has been suggested to be involved in the regulation of the ERM protein/membrane association (Chen et al., 1995). In this study, we examined whether ERM proteins are directly phosphorylated by Rho-kinase and whether their functions are regulated by Rho-kinase–dependent phosphorylation. First we incubated recombinant full-length radixin (F-rad) or COOH-terminal half radixin (C-rad) with the constitutively active catalytic domain of Rho-kinase and found that ∼100% of C-rad molecules were phosphorylated mainly at the COOH-terminal threonine residue (T564), which was identical to the phosphorylation site of moesin in thrombin-stimulated platelets (Nakamura et al., 1995, 1996). Furthermore, using mAb we found that this type of threonine phosphorylation was induced not only in radixin but also in ezrin and moesin in vivo in a Rho-dependent manner. Finally, this type of phosphorylation affected the intramolecular and/or intermolecular head-to-tail association of ERM proteins. These findings will lead to a better understanding of the Rho-dependent mechanism of regulation, not only ERM protein functions but also general actin/plasma membrane interactions.
Materials and Methods
Antibodies and Recombinant Proteins
The polyclonal antibody (pAb) TK89 was raised in rabbits against a synthesized peptide corresponding to mouse radixin sequence from amino acids 551–570. TK89 detected the COOH-terminal halves of all ERM proteins.
Recombinant F-rad, C-rad (311–583 amino acids), and glutathione- S-transferase (GST) fusion proteins with the catalytic domain of Rho-kinase (Rho-Kc; amino acids 6–553) were produced in Sf9 cells by recombinant baculovirus infection and purified as described previously (Amano et al., 1996b ; Hirao et al., 1996). The GST fusion protein with the NH2-terminal half of radixin (1–310 amino acids; GST–N-rad) was expressed in Escherichia coli. Purified GST–N-rad was then cleaved with thrombin to remove the GST according to the manufacturer's instructions. Purified recombinant proteins were concentrated with a Centricon-10 concentrator (Amicon, Beverly, MA).
In Vitro Kinase Reaction
The kinase reaction for Rho-Kc was carried out in 50 μl of reaction mixture containing 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 5 mM MgCl2, 250 μM γ-[32P]ATP (1–20 GBq/mmol), 2 pmol Rho-Kc, and various amounts of F-rad, C-rad, or their mixture. 0.2 pmol Rho-Kc was also used in some experiments, but to complete the kinase reaction as quickly as possible, 2 pmol Rho-Kc was used in most experiments. Initially, we produced F-rad and C-rad in E. coli. However, these recombinant proteins were not phosphorylated efficiently by Rho-Kc, and so we produced them in Sf9 cells by recombinant baculovirus infection. After 10 min of incubation at 30°C, the reaction mixture was boiled in SDS-PAGE sample buffer and resolved by SDS-PAGE. The 32P signals were analyzed by autoradiography (Fujix Bioimage Analyzer Bas 2000 System; Fuji Film Co. Ltd., Tokyo, Japan), and F-rad or C-rad was detected by silver staining.
Phosphopeptide Mapping
Two-dimensional mapping of N-1-tosylamide-2-phenylethylchloromethyl ketone (TPCK)-treated trypsin peptides from F-rad and C-rad was performed as described previously with slight modification (Tanabe et al., 1981). F-rad or C-rad (5 μg) was maximally phosphorylated as described above and resolved by SDS-PAGE. Each band was cut out from the gel, and the gel slices containing radioactive F-rad or C-rad were homogenized in 500 μl of 50 mM NH4HCO3 containing 25 μg trypsin, pH 8.4. After 20 h incubation at 37°C, the solution containing tryptic peptides was lyophilized and dissolved in 20 μl of TLE buffer (mixture of acetic acid, formic acid, and H2O at a 15:5:80 ratio). 10-μl aliquots of these solutions were spotted on silica gel–coated thin layer chromatography plates, and tryptic peptides were resolved by electrophoresis in the first dimension at 1,000 V for 1 h in TLE buffer using an AB Multiphor II (Pharmacia Biotech Sverige, Uppsala, Sweden) at 4°C, and by chromatography in the second dimension in thin layer chromatography buffer (mixture of butanol, pyridine, acetic acid, and H2O at a 32.5:25:5:20 ratio) for 4 h. The plates were then dried and the 32P signals were analyzed by autoradiography.
Determination of Phosphorylated Amino Acid Residues
C-rad (30 μg) was phosphorylated by Rho-Kc as described above, except that the reaction was carried out for 60 min in the presence of cold ATP instead of γ-[32P]ATP. As a control, nonphosphorylated C-rad was prepared in the same reaction buffer containing no ATP. Both phosphorylated and nonphosphorylated samples were separated by HPLC on a RESOURCE™ RPC column (1 ml; Pharmacia Biotech Sverige) preequilibrated with 0.1% trifluoroacetic acid (TFA). Elution was performed for 60 min with a linear gradient of acetonitrile (0–90%) containing 0.1% TFA at a flow rate of 1 ml/min. Purified proteins were evaporated and digested with 1 μg of lysyl endopeptidase in 260 μl of 100 mM Tris-HCl, pH 8.5, for 12 h at 37°C. The digested peptides were separated by HPLC on a TSKgel ODS-80Ts column (0.46 × 15 cm; Tosoh Co., Tokyo, Japan) preequilibrated with 0.1% TFA. Elution was performed for 75 min with a linear gradient of acetonitrile (0–50%) containing 0.1% TFA. Elution profiles at 206 nm were compared between phosphorylated and nonphosphorylated C-rad, and the phosphorylated amino acid residues were determined by amino acid sequence analysis according to the method developed previously (see Results for details; Kato et al., 1994; Fujita et al., 1996).
mAb Production
mAbs were raised using BDF1 mice according to the method previously described (Tsukita et al., 1994). A phosphopeptide (CRDKYKpTLRQIR) corresponding to amino acids 559–569 of radixin was synthesized and used as an antigen. One hybridoma clone producing an antibody (297S) that could distinguish between phosphorylated and nonphosphorylated C-rad was selected, expanded, and recloned.
Lysophosphatidic Acid Treatment of Serum-starved Swiss 3T3 Cells
Confluent serum-starved Swiss 3T3 cells were prepared according to the method developed by Ridley and Hall (1992) with slight modifications (Kumagai et al., 1993). Cells were seeded and cultured at a density of 3 × 105 in 6-cm culture dishes in DME supplemented with 10% FCS for 7–10 d. They were then transferred to FCS-free DME and culture was continued for 12 h. In some experiments, before serum starvation, cells were treated with 30 μg/ml C3 exoenzyme in the presence of 3 μl/ml lipofectamine (GIBCO BRL, Gaithersburg, MD) for 24 h in DME containing FCS. The serum-starved cells were stimulated with 1 μg/ml lysophosphatidic acid (LPA), incubated for various periods, and then treated with 75 μl of SDS-PAGE sample buffer. After sonication, samples were then resolved by SDS-PAGE and transfered to polyvinylidene difluoride (PVDF) membrane (Immobilon; Millipore Corp., Bedford, MA), followed by immunoblotting with TK89 pAb or 297S mAb.
Cosedimentation Experiments with Actin Filaments
Actin was purified from rabbit skeletal muscle as described previously (Tsukita et al., 1988). Gel-filtered G-actin was stored in G-buffer (2 mM Tris-HCl, pH 7.5, 0.2 mM ATP, 0.5 mM DTT, and 0.2% NaN3), and diluted at 20°C to 8 μM with F-buffer (20 mM Tris-HCl, pH 7.5, 75 mM KCl, 10 mM NaCl, 2 mM DTT, and 2.5 mM MgCl2) to initiate polymerization. Phosphorylated and nonphosphorylated C-rad were prepared as described above, except that the reaction was carried out for 60 min in the presence of cold ATP instead of γ-[32P]ATP, and then dialyzed against F-buffer, followed by centrifugation at 100,000 g for 30 min at 20°C. After actin filaments were polymerized for 30 min, various amounts of phosphorylated or nonphosphorylated C-rad in F-buffer (20 μl) were added to 20 μl of actin filament solution (or the solution containing 0.72 mg/ml BSA) and incubated for 30 min at 20°C. After centrifugation at 100,000 g for 30 min at 20°C in a Beckman TLA100 rotor (Beckman Instrs., Inc., Fullerton, CA) the supernatant and pellet were resolved by SDS-PAGE. Coomassie brilliant blue–stained gels were analyzed densitometrically using Adobe Photoshop™ 3.0J.
In some experiments (see Fig. 5 C), 3 pmol of partially [32P]phosphorylated F-rad and fully [32P]phosphorylated C-rad, which had been dialyzed against F-buffer, were centrifuged in the presence of 0.36 mg/ml BSA at 100,000 g or 10,000 g for 30 min at 20°C using siliconized tubes. Resultant supernatant and pellet were resolved in SDS-PAGE followed by Coomassie brilliant blue staining, immunoblotting with TK89, or autoradiography.
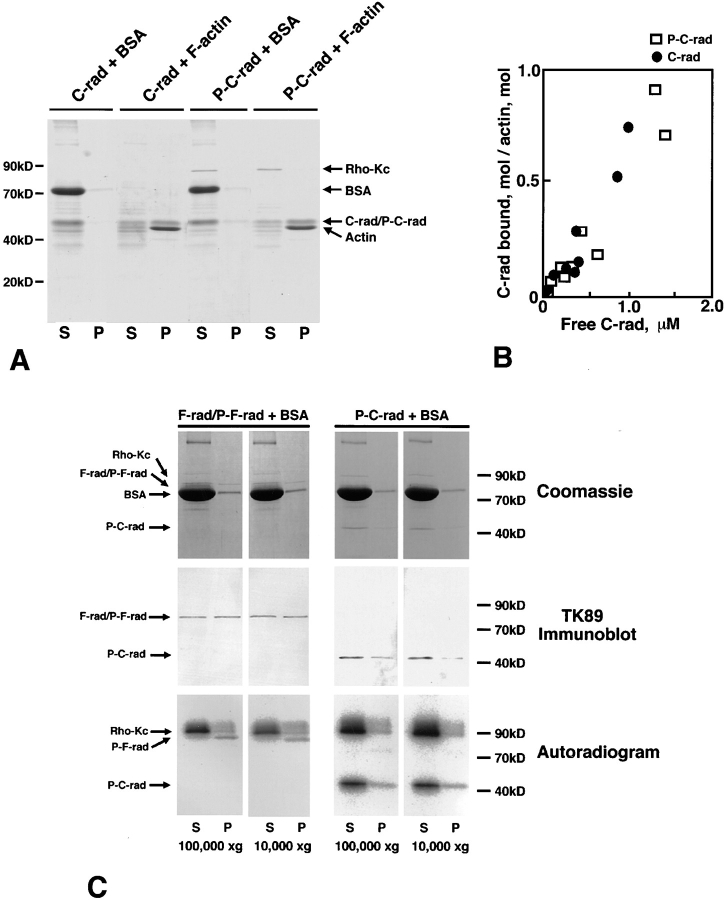
Figure 5.
Effects of T564 phosphorylation in radixin on the actin filament binding ability of C-rad. (A) Cosedimentation of non- or T564-phosphorylated C-rad with actin filaments. Non- (C-rad) or T564-phosphorylated C-rad (P-C-rad) was incubated with BSA (BSA) or with actin filaments (F-actin), then centrifuged at 100,000 g. Supernatant (S) and pellet (P) were resolved by SDS-PAGE followed by Coomassie brilliant blue staining. Both non- and T564-phosphorylated C-rad (C-rad/ P-C-rad) were cosedimented with actin filaments (Actin) to the same extent, but not with BSA (BSA). (B) Quantitative analysis. 2 μM F-actin was incubated with various amounts of non- or T564-phosphorylated C-rad and centrifuged, then the amounts of cosedimented non- (C-rad; filled circles) and T564-phosphorylated C-rad (P-C-rad; open squares) were quantified by densitometric scanning of Coomassie brilliant blue–stained gels. (C) Sedimentation of F-rad. Partially [32P]-phosphorylated F-rad (P-F-rad) and fully [32P]- phosphorylated C-rad (P-C-rad) were centrifuged in the presence of BSA at 100,000 or 10,000 g for 30 min. Supernatant (S) and pellet (P) were resolved by SDS-PAGE followed by Coomassie brilliant blue staining (Coomassie), immunoblot with TK89 (TK89 Immunoblot), or autoradiography (Autoradiogram). In the absence of actin filaments, F-rad, especially P-F-rad, were mostly recovered in pellet even at 10,000 g, whereas P-C-rad was mainly recovered in supernatant at 100,000 g as well as 10,000 g.
Alkaline Phosphatase Treatment
Fully phosphorylated C-rad (10 μg) was dialyzed against AP buffer (50 mM Tris-HCl, pH 8.2, 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, and 1 mM p-amidino PMSF), then incubated with 50 μl of glutathione–Sepharose 4B beads (Pharmacia Biotech Sverige) for 1 h at 4°C to absorb Rho-Kc (GST fusion protein). After the Rho-Kc–bound beads were removed by centrifugation, phosphorylated C-rad was concentrated with a Centricon-10 concentrator (Amicon Corp., Danvers, MA) up to 500 μg/ml. Calf intestine alkaline phosphatase (20 U/500 ng phosphorylated C-rad; Takara Shuzo Co., Ltd., Ohtsu, Japan) was then added and incubated for 1 h at 30°C.
Protein Iodination
Purified N-rad was iodinated with 125I using IODO-BEADS (Pierce Chemical Co., Rockford, IL). In brief, 100 μg of N-rad was incubated with IODO-BEADS for 4 min at 20°C in the presence of 0.5 mCi of [125I]NaI in 0.5 ml PBS, pH 7.5. The reaction was terminated by removal of solution from the IODO-BEADS, and labeled protein was separated from free 125I on a NAP-10 column (Pharmacia Biotech Sverige).
Gel Overlay Assay for Interdomain Interaction
Phosphorylated, nonphosphorylated C-rad, and alkaline phosphatase-treated phosphorylated C-rad were prepared as described above, except that the reaction was carried out for 60 min in the presence of cold ATP. The same amounts of each C-rad preparation (92 ng) was resolved by SDS-PAGE and transferred onto nitrocellulose membranes. Nitrocellulose membranes were incubated for 60 min with 1% nonfat dried milk, 0.1% Tween 20, 25 mM Tris-HCl, pH 7.5, and 150 mM NaCl, and then with 5 μg/ml purified 125I-labeled N-rad (107 cpm/pmol) in 1% nonfat dried milk, 0.1% Tween 20, 25 mM Tris-HCl, pH 7.5, and 150 mM NaCl, for 1 h at 20°C. After washing with 10 mM Tris-HCl and 250 mM NaCl, pH 7.5, C-rad and bound 125I-labeled N-rad were detected by immunoblotting with pAb TK89 (or mAb 297S) and autoradiography, respectively.
SDS–Polyacrylamide Gel Electrophoresis and Immunoblotting
One-dimensional SDS-PAGE (12.5%) was performed according to the method of Laemmli (1970). After electrophoresis, proteins were electrophoretically transferred from gels onto nitrocellulose membranes that were then incubated with the first antibody. Bound antibodies were visualized with alkaline phosphatase–conjugated goat anti–rabbit IgG and the appropriate substrates as described by the manufacturer (Amersham International, Buckinghamshire, UK).
Results
Phosphorylation of Radixin by Rho-Kinase In Vitro
To examine whether ERM proteins are phosphorylated by Rho-kinase in a cell-free system, F-rad and C-rad (amino acids 311–583) were produced in Sf9 cells by recombinant baculovirus infection. Purified F-rad, C-rad, and the mixture of F- and C-rad were then incubated with the Rho-kinase catalytic domain (Rho-Kc) in the presence of γ-[32P]ATP. Rho-Kc, which was also produced by recombinant baculovirus infection, was previously shown to be constitutively active (Amano et al., 1996b , 1997). Autoradiography revealed that both F-rad and C-rad were phosphorylated, and that the latter was phosphorylated more efficiently than the former (Fig. 1 A). As shown in Fig. 1 B, ∼1.3 mol of phosphate was maximally incorporated into 1 mol of C-rad in a time-dependent manner, whereas at most ∼0.3 mol of phosphate was detected per mole of F-rad.
Figure 1.
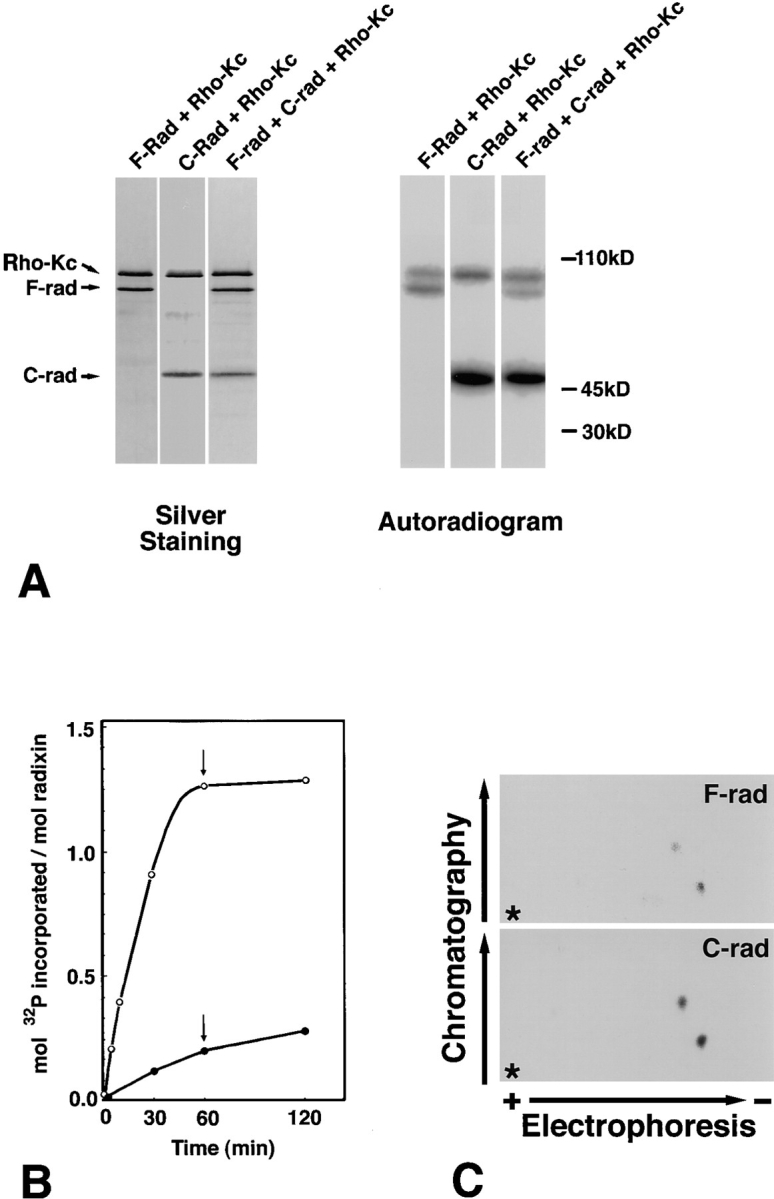
In vitro phosphorylation of the full-length radixin (F-rad) and the COOH-terminal half radixin (C-rad) by the constitutively active catalytic domain of Rho-kinase (Rho-Kc). (A) SDS-PAGE banding pattern of the reaction mixture (Silver Staining) and its accompanying autoradiogram (Autoradiogram). Purified 3 pmol F-rad (F-rad + Rho-Kc), C-rad (C-rad + Rho-Kc), or their mixture (F-rad + C-rad + Rho-Kc) was incubated with 2 pmol Rho-Kc for 10 min at 30°C. Phosphorylated proteins were resolved by SDS-PAGE followed by autoradiography. C-rad (C-rad) was phosphorylated more heavily than F-rad (F-rad). Rho-Kc (Rho-Kc) was autophosphorylated. (B) Time course and stoichiometry of the phosphorylation reaction. After 30, 60, and 120 min of incubation, the phosphorylation levels of F-rad (filled circle) and C-rad (open circle) were quantified. 2 pmol of Rho-Kc and 500 pmol of γ-[32P]ATP were added to the reaction mixture after 60 min of incubation (arrows). (C) Phosphopeptide mapping. Phosphorylated F-rad and C-rad were digested completely with TPCK-trypsin, subjected to two-dimensional peptide mapping (first dimension, Electrophoresis; second dimension, Chromatography) as described in Materials and Methods, and then analyzed by autoradiography. Two radioactive spots in F-rad and C-rad may correspond to the T564- and T573-containing tryptic fragments (see Fig. 2). To visualize the weak spot from F-rad the autoradiogram of C-rad was overexposed, but the intensity ratio of two spots from C-rad was the same as that from F-rad. Asterisks represent the origins of spotted samples.
Rho-Kinase–dependent Phosphorylation Sites of Radixin
We next compared the phosphorylation sites of F-rad and C-rad by phosphopeptide mapping. F-rad and C-rad were phosphorylated in vitro, digested completely with TPCK-treated trypsin, and subjected to two-dimensional peptide mapping followed by autoradiography. As shown in Fig. 1 C, phosphorylated F-rad and C-rad gave the identical phosphopeptide mapping pattern, indicating that the Rho-kinase–dependent phosphorylation sites of F-rad are located only in its COOH-terminal half.
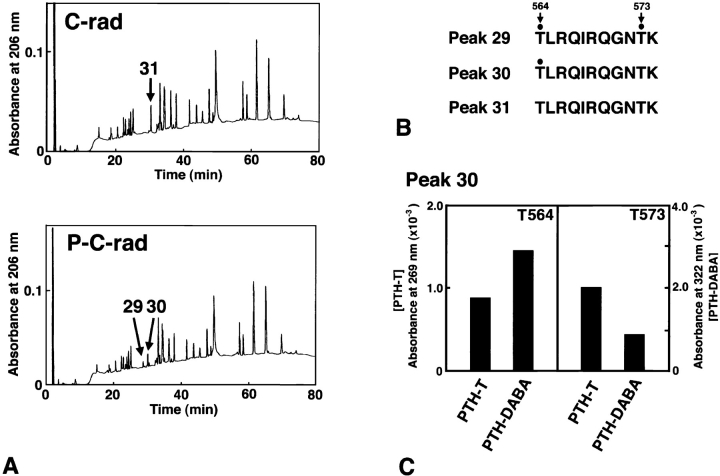
The phosphorylated amino acid residues in C-rad were then determined. Phosphorylated and nonphosphorylated C-rad were purified to ∼90% homogeneity by RESOURCE™RPC column chromatography. Purified proteins were completely digested with lysyl endopeptidase and applied to the TSKgel ODS-80Ts column. As shown in Fig. 2 A, peptides from phosphorylated as well as nonphosphorylated C-rad were separated into >20 peaks. In this chromatography procedure, phosphorylated peptides are eluted faster than nonphosphorylated peptides because of their hydrophilic modification. Peak 31 from nonphosphorylated C-rad was not detected in phosphorylated C-rad, and two peaks (30 and 29) that migrated slightly faster were detected only from phosphorylated C-rad. Sequencing showed that these three peaks had identical amino acid sequences and corresponded to the COOH-terminal 11 amino acid residues of radixin (amino acids 564–574). Of the two threonine residues in this sequence (T564 and T573), no threonine, only T564, and both T564 and T573 were phosphorylated in peaks 31, 30, and 29, respectively (Fig. 2 B and C). A peak of the phosphopeptide, in which only T573 was phosphorylated, was not detected. Quantitative analyses revealed that ∼100% of T564, but at most ∼40% of T573, was phosphorylated when C-rad was incubated with Rho-Kc for 1 h. Then we concluded that the major and primary phosphorylation site of radixin by Rho-kinase was T564 and referred to the Rho-Kc–phosphorylated C-rad as T564-phosphorylated C-rad.
Figure 2.
Determination of amino acid residues of radixin that are phosphorylated by Rho-kinase. (A) Comparison of elution profiles of lysyl endopeptidase-digested peptides of non- and fully phosphorylated C-rad from reverse-phase HPLC (C-rad and P-C-rad, respectively). Rho-kinase–dependent phosphorylation divided and shifted the peak 31 from nonphosphorylated C-rad into two peaks, peaks 30 and 29, from phosphorylated C-rad. For details see the text. (B) Amino acid sequences of peaks 29, 30, and 31. The PTH-threonine/PTH-dehydroaminobutyric acid ratio (see C) revealed that T564/T573 in peak 29 and T564 in peak 30 were phosphorylated (Kato et al., 1994; Fujita et al., 1996). (C) The PTH-threonine/PTH-dehydroaminobutyric acid ratio of T564 and T573 in peak 30. The amounts of PTH-threonine (PTH-T) and PTH-dehydroaminobutyric acid (PTH-DABA) were determined from the absorbance at 269 and 322 nm, respectively. When the threonine residue (T564 in this case) was phosphorylated, the PTH-T/PTH-DABA ratio fell to below 1.0.
Rho-dependent Phosphorylation of the COOH-terminal Threonine Residue of ERM Proteins In Vivo
We then attempted to determine whether T564 of radixin was phosphorylated in a Rho-dependent manner in vivo. Rho was reported to be activated when serum-starved confluent Swiss 3T3 are stimulated with LPA, resulting in the formation of focal contacts and stress fibers (Ridley and Hall 1992; Ridley et al., 1992). Using this system with 32P-labeled Swiss 3T3 cells, we first examined whether ERM proteins were phosphorylated in a Rho-dependent manner. However, even under the serum-starved condition, ERM proteins were fairly phosphorylated, and the Rho-dependent increase of ERM phosphorylation was failed to be detected (data not shown).
Thus, to selectively detect T564 phosphorylation of radixin, we raised an mAb that can distinguish T564-phosphorylated radixin from the nonphosphorylated molecule. As an antigen, we synthesized a phosphopeptide corresponding to the COOH-terminal amino acids 559–569 of radixin in which T564 is phosphorylated. Since this amino acid sequence was completely conserved among ERM proteins, it was expected that mAb specific for this antigen would recognize not only T564-phosphorylated radixin but also T567-phosphorylated ezrin and T558-phosphorylated moesin. After intensive screening, one mAb, 297S, was obtained. As shown in Fig. 3, this mAb specifically recognized T564-phosphorylated C-rad, but not nonphosphorylated C-rad. When the whole cell lysate of a semiconfluent culture of Swiss 3T3 cells was immunoblotted with mAb 297S, ezrin and moesin as well as radixin were clearly detected. Since these cells were cultured in the presence of serum, we next examined the phosphorylation level of respective T567, T564, and T558 of ERM proteins in the serum-starved cells.
Figure 3.
Production of mAb 297S that distinguishes T564-phosphorylated from nonphosphorylated radixin. Nonphosphorylated C-rad (100 ng; C-rad), T564-phosphorylated C-rad (100 ng; P-C-rad), and whole-cell lysate of Swiss 3T3 cells under conventional culture conditions (25 μg; Swiss 3T3 lysate) were immunoblotted with pAb TK89 (pAb TK89) or mAb 297S (mAb 297S). The former antibody recognized radixin as well as ezrin and moesin irrespective of their phosphorylation state, whereas the latter distinguished T564-phosphorylated C-rad from the nonphosphorylated molecule. The mAb 297S recognized ezrin and moesin that were phosphorylated at the corresponding threonine residues, T567 and T558, respectively.
In serum-starved confluent Swiss 3T3 cells, we quantitatively determined the amount of total and T564-phosphorylated radixin/T558-phosphorylated moesin by scanning densitometry of pAb TK89 and mAb 297S immunoblots, respectively, using purified phosphorylated C-rad to generate standard curves. The phosphorylation level of T567 in ezrin was difficult to be quantitatively analyzed due to its low expression level in Swiss 3T3 cells. We found that even under the serum-starved condition ∼25% of radixin/ moesin were phosphorylated at their COOH-terminal threonine residue. Immunoblotting with mAb 297S revealed that within 30 s after LPA stimulation, the COOH-terminal threonine residue of radixin/moesin was rapidly phosphorylated (two- to threefold over the basal level) followed by rapid dephosphorylation (Fig. 4, A and B). This LPA-induced rapid phosphorylation of radixin/moesin was significantly suppressed in the presence of C3 toxin, a potent inhibitor of Rho (Aktories et al., 1988; Kikuchi et al., 1988; Narumiya et al., 1988; Braun et al., 1989; Fig. 4 C). Taking it into consideration that the phosphorylation level of T567 in ezrin also appeared to rapidly increase then decrease within 2 min after LPA stimulation (Fig. 4 A), we concluded that T567 (ezrin), T564 (radixin), and T558 (moesin) were phosphorylated in vivo in a Rho-dependent manner. These findings suggest that the activation of Rho causes the phosphorylation of ERM proteins through activation of Rho-kinase.
Figure 4.
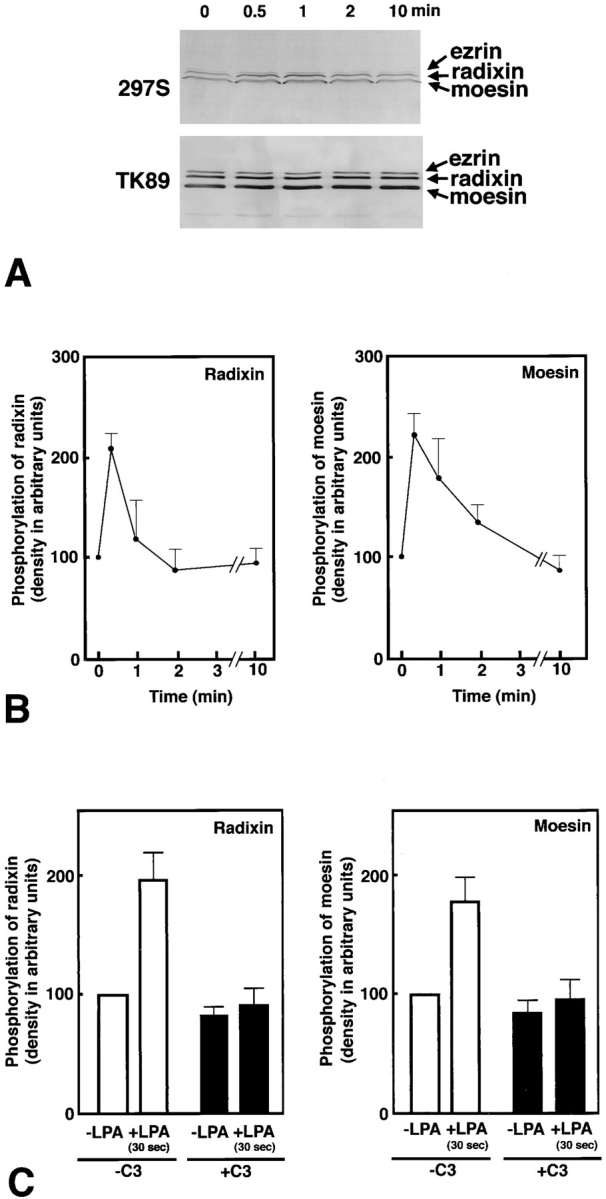
LPA-induced threonine phosphorylation of ERM proteins in vivo. (A) Serum-starved Swiss 3T3 cells were stimulated with LPA, and at 0, 0.5, 1, 2, or 10 min of incubation, whole-cell lysate was resolved by SDS-PAGE followed by immunoblotting with mAb 297S (297S) or with pAb TK89 (TK89). 297S were specific for T567-, T564-, and T558-phosphorylated ezrin, radixin, and moesin, respectively, whereas TK89 recognized ERM proteins irrespective of their phosphorylation state. (B) Quantitative analyses of changes in the phosphorylation levels of T564 in radixin and T558 in moesin. By immunoblotting with TK89 in combination with scanning densitometry, the precisely equal amount of ERM proteins were reelectrophoresed per each lane for the following quantification. The amount of T564-phosphorylated radixin and T558-phosphorylated moesin was quantitatively determined by scanning densitometry of 297S-immunoblots (see A) using purified phosphorylated C-rad to generate a standard curve. The curve was linear over the concentration range used here. The mean densities of 297S-immunoblot bands of radixin and moesin at 0 min were set at 100 arbitrary units. The phosphorylation levels of T564 in radixin and T558 in moesin rapidly increased then decreased within 2 min after LPA stimulation. Although the change in the phosphorylation level of T567 in ezrin was difficult to be quantitatively followed due to its low expression level in Swiss 3T3 cells, its phosphorylation level also appeared to rapidly increase then decrease within 2 min after LPA stimulation (see A). The data represent the means ± SEM of four determinations. The phosphorylation levels of radixin and moesin at 30 s were significantly different from those at 0 s (P < 0.005) and at 2 min (P < 0.05) as determined by the paired t test. (C) Suppression of LPA-induced T564 phosphorylation of radixin and T558 phosphorylation of moesin by C3 exoenzyme. Non- and C3 exoenzyme–pretreated serum-starved cells were stimulated with 1 μg/ml LPA for 30 s, and the phosphorylation levels of T564 of radixin and T558-moesin in these cells were quantitatively compared to those in LPA-nontreated serum-starved cells by immunoblotting with mAb 297S. The data represent the means ± SEM of four determinations. In the absence of C3 exoenzyme (-C3), the phosphorylation levels of radixin and moesin in 30 s LPA-treated cells were significantly different from those at 0 s (P < 0.05) as determined by the paired t test, while in the presence of C3 exoenzyme (+C3) this LPA-induced increase of the phosphorylation levels of radixin or moesin was not detected. In this experiment, probably due to lipofectamine, the Rho-dependent increase of ERM phosphorylation was varied and suppressed to some extent as compared to B.
Effects of T564 Phosphorylation on Actin-binding Ability of Radixin
T567 (ezrin), T564 (radixin), and T558 (moesin) are located in the putative actin-binding domain (KYKTL) of ERM proteins (Turunen et al., 1994; Pestonjamasp et al., 1995). Thus, we compared the actin-binding ability of T564-phosphorylated C-rad with that of the nonphosphorylated molecule in vitro by actin filament cosedimentation analysis. As shown in Fig. 5 A, when nonphosphorylated and fully phosphorylated C-rad were incubated with skeletal muscle actin filaments followed by centrifugation, both were sedimented in equal amounts. Quantitative analysis indicated no significant difference in actin-binding ability between non- and fully phosphorylated C-rad, indicating that the phosphorylation of T564 does not affect the direct binding of C-rad to actin filaments (Fig. 5 B).
We also attempted to examine the actin-binding ability of partially phosphorylated F-rad. However, as shown in Fig. 5 C, approximately half of the amount of F-rad, especially most of phosphorylated F-rad, was precipitated in the absence of actin filaments even at 10,000 g centrifugation, whereas phosphorylated C-rad was mostly recovered in the 100,000 g supernatant. This indicated that the actin filament cosedimentation analysis was not potent to assess the actin-binding ability of F-rad.
Effects of T564 Phosphorylation on Interdomain Interaction of Radixin
The NH2-terminal halves of ERM proteins directly bind to their COOH-terminal halves, and this interdomain interaction has been reported to be important in the regulation of cross-linking activity of ERM proteins (Berryman et al., 1995; Bretscher et al., 1995). Using the gel overlay assay, domains responsible for the interdomain interaction were narrowed down in ezrin to the NH2-terminal amino acids 1–296 and the COOH-terminal amino acids 479–585 (Gary et al., 1995). T567 is located in the latter domain. Thus, we examined whether the T564 phosphorylation affects the interdomain interaction in radixin. The same amounts of non- and fully phosphorylated C-rad were electrophoresed and transferred onto nitrocellulose membranes and then incubated with the iodinated NH2-terminal half of radixin (125I-N-rad) that was purified from recombinant GST fusion protein produced in E. coli. As shown in Fig. 6 A, 125I-N-rad bound specifically to nonphosphorylated C-rad but not to phosphorylated C-rad. When phosphorylated C-rad was pretreated with alkaline phosphatase, it was dephosphorylated, which restored its binding ability to 125I-N-rad. The specific interaction between nonphosphorylated C-rad and 125I-N-rad was further confirmed by a dose–response experiment (Fig. 6 B). We thus concluded that the phosphorylation of T564 affected the direct binding between the NH2- and COOH-terminal halves of ERM proteins.
Figure 6.
Effects of T564 phosphorylation in radixin on the direct interaction between its NH2- and COOH-terminal halves. (A) The same amounts (92 ng) of nonphosphorylated C-rad (C-rad), T564-phosphorylated C-rad (P-C-rad), and alkaline phosphatase-treated T564-phosphorylated C-rad (P-C-rad/AP) were electrophoresed and then transferred onto nitrocellulose membranes. They were immunoblotted with pAb TK89 (TK89) or mAb 297S (297S) to check the amounts and the phosphorylation level of each sample, respectively (Immunoblot). They were then incubated with the iodinated NH2-terminal half of radixin (1 25I-N-rad) that was purified from GST fusion protein (Overlay). Bound 125I-N-rad was detected by autoradiography. 125I-N-rad specifically bound to nonphosphorylated C-rad but not to T564-phosphorylated C-rad. (B) Various amounts (7.5–120 ng) of non- (C-rad) and T564-phosphorylated C-rad (P-C-rad) were electrophoresed, transferred onto nitrocellulose membranes, and incubated with 125I-N-rad. The amount of bound 125I-N-rad and electrophoresed C-rad/P-C-rad was detected by autoradiography (125I-N-rad Overlay) and Coomassie brilliant blue staining (Coomassie), respectively. 125I-N-rad specifically bound to nonphosphorylated C-rad in a dose-dependent manner.
Discussion
C-rad was fully phosphorylated in vitro mainly at its COOH-terminal threonine (T564) by the constitutively active of Rho-kinase. For in vivo analysis of this type of phosphorylation, we produced a mAb (297S) that distinguished T564-phosphorylated from nonphosphorylated radixin. Because the amino acid sequence around T564 of radixin is completely conserved among ERM proteins, this mAb also recognized T567-phosphorylated ezrin and T558-phosphorylated moesin. Immunoblots of LPA-stimulated serum-starved Swiss 3T3 cells with this mAb revealed that not only T564 of radixin, but also T567 of ezrin and T558 of moesin, were phosphorylated in vivo in a Rho-dependent manner. Of course, these findings do not directly prove that it is the Rho-kinase that phosphorylates ERM proteins in vivo. However, considering that Rho-kinase is one of the major targets for Rho in terms of the reorganization of actin-based cytoskeletons (Leung et al., 1996; Amano et al., 1997; Ishizaki et al., 1997), we were led to conclude that, when Rho is activated in vivo, radixin, as well as ezrin and moesin, are similarly phosphorylated through the activation of Rho-kinase. This conclusion favors the notion that ERM proteins are functionally redundant (Takeuchi et al., 1994b ; Hirao et al., 1996; Tsukita et al., 1997a ,b). These observations are also consistent with the previous report by Nakamura et al. (1995, 1996) that in platelets, thrombin stimulation induced the T558 phosphorylation of moesin. As moesin is predominant among ERM proteins in platelets (Nakamura et al., 1995) and Rho signaling is involved in the thrombin-induced activation of platelets (Morii et al., 1992), it is likely that T558 of moesin in platelets is phosphorylated by Rho-kinase.
Two possible functions have been proposed for the COOH-terminal highly conserved amino acid sequence around ezrin T567, radixin T564, or moesin T558. First, this domain is responsible for actin filament binding of ERM proteins. The actin-binding site was narrowed down in ezrin to the COOH-terminal 34 amino acids. The sequence around T567, KYKTL, in the COOH-terminal 34 amino acids was regarded as a consensus sequence for actin binding (KYKXL) that was also found in other actin-binding proteins such as the myosin heavy chain and β subunit of Cap-Z (Turunen et al., 1994), although there has been no evidence showing the direct interaction of actin filaments with KYKTL in ERM proteins. It was thus expected that phosphorylation of the KYKTL site would affect actin binding of the COOH halves of ERM proteins, although T is not necessarily required as an actin-binding consensus sequence. As shown in Fig. 5, nonphosphorylated C-rad was cosedimented with actin filaments, but T564-phosphorylated C-rad was also cosedimented to the same extent, indicating that KYK-pT-L shows the same affinity to actin filaments as KYKTL.
Second, the COOH-terminal end domain is responsible for the head-to-tail association of ERM proteins. As described in the introduction, the intra- or intermolecular head-to-tail association is thought to be very important for the regulation of ERM protein activity (Berryman et al., 1995; Bretscher et al., 1995). When the NH2- and COOH-terminal halves bind to each other to form closed forms (and/or oligomers), ERM proteins are inactive as cross-linkers between actin filaments and plasma membranes. When some signal interferes with this binding, ERM proteins are activated (opened). In this study, we found that the T564 phosphorylation of radixin markedly suppressed its head-to-tail association. This suggests that the T564-phosphorylation of radixin (and probably also the phosphorylation of ezrin T567 and moesin T558) keeps them open and active. If the life time of the opened form of ERM proteins is prolonged by this phosphorylation, the actin filament/plasma membrane association is upregulated, which is consistent with the previous observation that the CD44/ERM protein complex is stabilized by the activation of Rho (Hirao et al., 1996). We also found previously that Rho-GDI (GDP dissociation inhibitor; Araki et al., 1990; Takai et al., 1995) was coimmunoprecipitated with the CD44/ERM protein complex (Hirao et al., 1996). Furthermore, it was recently shown that Rho-GDI carrying the GDP-bound form Rho (GDP-Rho) directly binds to the NH2-terminal half of ERM proteins, and that this binding dissociates the Rho-GDI/GDP-Rho complex to release free GDP-Rho (Takahashi et al., 1997). Thus we speculate that GDP-Rho, which is recruited to ERM proteins, is converted to GTP-Rho and that GTP-Rho activates Rho-kinase to phosphorylate ERM proteins. The phosphorylated ERM proteins with opened conformation may function as actin filament/plasma membrane cross-linkers.
Rho-kinase phosphorylated T564 in ∼100% of C-rad molecules but in at most ∼30% of F-rad in vitro, indicating that Rho-kinase phosphorylates the opened form of radixin more efficiently than the closed form at least in vitro. Therefore, we speculate that the Rho-kinase–dependent phosphorylation of ERM proteins does not activate (open) ERM proteins but stabilizes the activated (opened) conformation. However, it is also possible that Rho-kinase can open ERM proteins in vivo in the presence of other Rho-independent kinases, other targets for Rho and ERM-binding proteins such as EBP50 (Zhang et al., 1995; Chong et al., 1994; Bowman et al., 1993; Malcolm et al., 1994; Madaule et al., 1995; Amano et al., 1996a ; Kimura et al., 1996; Reid et al., 1996; Watanabe et al., 1996, 1997; Reczek et al., 1997). Actually, ezrin is effectively tyrosine phosphorylated by EGF and HGF stimulation (Krieg et al., 1992; Crepaldi et al., 1997) and functions as a protein kinase A–anchoring protein (Dransfield et al., 1997). It is also serine/threonine phosphorylated in vivo by protein kinase A probably at residues other than those phosphorylated by Rho-kinase (Urushidani et al., 1989).
If the phosphorylation of respective T567, T564, and T558 of ERM proteins keeps them open without affecting the actin-binding ability of their COOH-terminal regions, it was expected that Rho-kinase–dependent phosphorylation would upregulate the actin-binding ability of F-rad. As shown in Fig. 5 C, however, even in the absence of actin filaments, F-rad, especially T564-phosphorylated F-rad, was mostly recovered in the low speed pellet, making it difficult to evaluate the actin-binding ability of F-rad in vitro by the cosedimentation analysis. Most recently, another F-actin–binding domain was identified in the NH2-terminal domain of ezrin (Martin et al., 1997; Roy et al., 1997). Furthermore, this domain also bound to G-actin, which was consistent with our initial observation of the barbed-end capping activity of purified native radixin (Tsukita et al., 1989). Therefore, to understand the regulatory mechanism of the physiological functions of ERM proteins, detailed comparison of the F-actin– and/or G-actin– binding ability between nonphosphorylated and phosphorylated full-length ERM proteins will be required in the next step.
The relationship between Rho-signaling and the actin-based cytoskeleton was first noted by Ridley and coworkers (Ridley and Hall, 1992; Ridley et al., 1992). They showed using serum-starved Swiss 3T3 cells that Rho plays a central role in the coordinated assembly of focal adhesions and stress fibers induced by growth factors. However, its molecular mechanism has still yet to be elucidated in detail. Constitutively active Rho-kinase induces the formation of focal adhesions and stress fibers (Leung et al., 1996; Amano et al., 1997; Ishizaki et al., 1997). Rho-kinase was reported to activate myosin ATPase, which is thought to be important for stress fiber formation (Amano et al., 1996a ; Kimura et al., 1996). On the other hand, moesin was identified as an essential factor for the Rho-dependent formation of stress fibers in serum-starved Swiss 3T3 cells (Mackay et al., 1997). This study showed that the Rho-dependent threonine phosphorylation of ERM proteins occurred very rapidly before the coordinated assembly of focal adhesions and stress fibers in serum-starved Swiss 3T3 cells. Further analyses of the physiological relationship between the Rho-kinase–dependent activation of myosin ATPase and ERM protein phosphorylation will lead to a better understanding of the Rho-dependent regulation of the actin-based cytoskeleton.
Acknowledgments
We would like to thank Professor Y. Takai (Department of Molecular Biology and Biochemistry, Osaka University, Japan) and all the members of our laboratory (Department of Cell Biology, Faculty of Medicine, Kyoto University, Japan) for their helpful discussions throughout this study. Our thanks are also due to Dr. A. Hall for his generous gift of pGEX-C3. We are grateful to Miss M. Sato and Mr. T. Nakagawa for their excellent technical assistance.
This work was supported in part by a Grant-in-Aid for Cancer Research and a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Science and Culture of Japan (to S. Tsukita).
Abbreviations used in this paper
- C-rad
COOH-terminal half recombinant radixin
- ERM
ezrin/radixin/moesin
- F-rad
full-length recombinant radixin
- GST
glutathione-S-transferase
- LPA
lysophosphatidic acid
- N-rad
NH2-terminal half recombinant radixin
- pAb
polyclonal antibody
- Rho-Kc
recombinant Rho-kinase catalytic domain
- Rho-kinase
Rho- associated kinase
- TFA
trifluoroacetic acid
References
- Aktories K, Rosener S, Blaschke U, Chhatwal GS. Botulinum ADP-ribosyltransferase C3: purification of the enzyme and characterization of the ADP-ribosylation reaction in platelet membranes. Eur J Biochem. 1988;172:445–450. doi: 10.1111/j.1432-1033.1988.tb13908.x. [DOI] [PubMed] [Google Scholar]
- Algrain M, Turunen O, Vaheri A, Louvard D, Arpin M. Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. J Cell Biol. 1993;120:129–139. doi: 10.1083/jcb.120.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M, Mukai H, Ono Y, Chihara K, Matsui T, Hamajima Y, Okawa K, Iwamatsu A, Kaibuchi K. Identification of a putative target for Rho as the serine-threonine kinase protein kinase N. Science. 1996a;271:648–650. doi: 10.1126/science.271.5249.648. [DOI] [PubMed] [Google Scholar]
- Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) . J Biol Chem. 1996b;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- Amano M, Chihara K, Kimura K, Fukata Y, Nakamura N, Matsuura Y, Kaibuchi K. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science. 1997;275:1308–1311. doi: 10.1126/science.275.5304.1308. [DOI] [PubMed] [Google Scholar]
- Andréoli C, Martin M, Le-Borgne R, Reggio H, Mangeat P. Ezrin has properties to self-associate at the plasma membrane. J Cell Sci. 1994;107:2509–2521. doi: 10.1242/jcs.107.9.2509. [DOI] [PubMed] [Google Scholar]
- Araki S, Kikuchi A, Hata Y, Isomura M, Takai Y. Regulation of reversible binding of smg p25A, a ras p21-like GTP-binding protein, to synaptic plasma membranes and vesicles by its specific regulatory protein, GDP-dissociation inhibitor. J Biol Chem. 1990;265:13007–13015. [PubMed] [Google Scholar]
- Arpin M, Algrain M, Louvard D. Membrane-actin microfilament connections: an increasing diversity of players related to band 4.1. Curr Opin Cell Biol. 1994;6:136–141. doi: 10.1016/0955-0674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- Berryman M, Gary R, Bretscher A. Ezrin oligomers are major cytoskeletal components of placental microvilli: a proposal for their involvement in cortical morphogenesis. J Cell Biol. 1995;131:1231–1242. doi: 10.1083/jcb.131.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman EP, Uhlinger DJ, Lambeth JD. Neutrophil phospholipase D is activated by a membrane-associated Rho family small molecular weight GTP-binding protein. J Biol Chem. 1993;268:21509–21512. [PubMed] [Google Scholar]
- Braun U, Habermann B, Just I, Aktories K, Vandekerckhove J. Purification of the 22kDa protein substrate of botulinum ADP-ribosyltransferase C3 from porcine brain cytosol and its characterization as a GTP-binding protein highly homologous to the Rho gene product. FEBS Lett. 1989;243:70–76. doi: 10.1016/0014-5793(89)81220-7. [DOI] [PubMed] [Google Scholar]
- Bretscher A. Purification of an 80,000-dalton protein that is a component of isolated microvillus cytoskeleton, and its localization in nonmuscle cells. J Cell Biol. 1983;97:425–432. doi: 10.1083/jcb.97.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Gary R, Berryman M. Soluble ezrin purified from placenta exists as stable monomers and elongated dimers with masked C-terminal ezrin-radixin-moesin association domains. Biochemistry. 1995;34:16830–16837. doi: 10.1021/bi00051a034. [DOI] [PubMed] [Google Scholar]
- Chen J, Cohn JA, Mandel LJ. Dephosphorylation of ezrin as an early event in renal microvillar breakdown and anoxic injury. Proc Natl Acad Sci USA. 1995;92:7495–7499. doi: 10.1073/pnas.92.16.7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong LD, Traynor-Kaplan A, Bokoch GM, Schwartz MA. The small GTP-binding protein Rho regulates a phosphatidylinositol 4-phosphate 5-kinase in mammalian cells. Cell. 1994;79:507–513. doi: 10.1016/0092-8674(94)90259-3. [DOI] [PubMed] [Google Scholar]
- Conboy J, Kan YW, Shohet SB, Mohandas N. Molecular cloning of protein 4.1, a major structural element of the human erythrocyte membrane skeleton. Proc Natl Acad Sci USA. 1986;83:9512–9516. doi: 10.1073/pnas.83.24.9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepaldi T, Gautreau A, Comoglio PM, Louvard D, Arpin M. Ezrin is an effector of hepatocyte growth factor-mediated migration and morphogenesis in epithelial cells. J Cell Biol. 1997;138:423–434. doi: 10.1083/jcb.138.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dransfield DT, Bradford AJ, Smith J, Martin M, Roy C, Mangeat PH, Goldenring JR. Ezrin is a cyclic AMP-dependent protein kinase anchoring protein. EMBO (Eur Mol Biol Organ) J. 1997;16:35–43. doi: 10.1093/emboj/16.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Sasaki T, Fukui K, Kotani H, Kimura T, Hata Y, Südhof TC, Scheler RH, Takai Y. Phosphorylation of Munc-18/n-Sec1/ rbSec1 by protein kinase C. J Biol Chem. 1996;271:7265–7268. doi: 10.1074/jbc.271.13.7265. [DOI] [PubMed] [Google Scholar]
- Funayama N, Nagafuchi A, Sato N, Sa, Tsukita, Sh, Tsukita Radixin is a novel member of the band 4.1 family. J Cell Biol. 1991;115:1039–1048. doi: 10.1083/jcb.115.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary R, Bretscher A. Heterotypic and homotypic associations between ezrin and moesin, two putative membrane-cytoskeletal linking proteins. Proc Natl Acad Sci USA. 1993;90:10846–10850. doi: 10.1073/pnas.90.22.10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary R, Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol Biol Cell. 1995;6:1061–1075. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Cooper JA, Bretscher A, Hunter T. The protein-tyrosine kinase substrate, p81, is homologous to a chicken microvillar core protein. J Cell Biol. 1986;102:660–669. doi: 10.1083/jcb.102.2.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Bretscher A, Esch FS, Hunter T. cDNA cloning and sequencing of the protein-tyrosine kinase substrate, ezrin, reveals homology to band 4.1. EMBO (Eur Mol Biol Organ) J. 1989;8:4133–4142. doi: 10.1002/j.1460-2075.1989.tb08598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, York JD, Warshawsky I, Majerus PW. Identification, cloning, and expression of a cytosolic megakaryocyte protein-tyrosine-phosphatase with sequence homology to cytoskeletal protein 4.1. Proc Natl Acad Sci USA. 1991;88:5867–5871. doi: 10.1073/pnas.88.13.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzel D, Reggio H, Bretcher A, Forte JG, Mangeat P. The secretion-stimulated 80K phosphoprotein of parietal cells is ezrin, and has properties of a membrane cytoskeletal linker in the induced apical microvilli. EMBO (Eur Mol Biol Organ) J. 1991;10:2363–2373. doi: 10.1002/j.1460-2075.1991.tb07775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander ST, Carpén O, Turunen O, Kovanen PE, Vaheri A, Timonen T. ICAM-2 redistributed by ezrin as a target for killer cells. Nature. 1996;382:265–268. doi: 10.1038/382265a0. [DOI] [PubMed] [Google Scholar]
- Henry MD, Gonzalez C, Agosti, Solomon F. Molecular dissection of radixin: distinct and interdependent functions of the amino- and carboxy-terminal domains. J Cell Biol. 1995;129:1007–1022. doi: 10.1083/jcb.129.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Sh, Tsukita, Sa, Tsukita Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki T, Maekawa M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N, Narumiya S. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO (Eur Mol Biol Organ) J. 1996;15:1885–1893. [PMC free article] [PubMed] [Google Scholar]
- Ishizaki T, Naito M, Fujisawa K, Maekawa M, Watanabe N, Saito Y, Narumiya S. p160ROCK, a Rho-associated coiled-coil forming protein kinase, works downstream of Rho and induces focal adhesions. FEBS Lett. 1997;404:118–124. doi: 10.1016/s0014-5793(97)00107-5. [DOI] [PubMed] [Google Scholar]
- Kato M, Sasaki T, Imazumi K, Takahashi K, Araki K, Shirataki H, Matsuura Y, Ishida A, Fujisawa H, Takai Y. Phosphorylation of rabphilin-3A by calmodulin-dependent protein kinase II. Biochem Biophys Res Commun. 1994;205:1776–1784. doi: 10.1006/bbrc.1994.2875. [DOI] [PubMed] [Google Scholar]
- Kikuchi A, Yamamoto K, Fujita T, Takai Y. ADP-ribosylation of the bovine brain Rho protein by botulinum toxin type C1. J Biol Chem. 1988;263:16303–16308. [PubMed] [Google Scholar]
- Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- Kotani H, Takaishi K, Sasaki T, Takai Y. Rho regulates association of both the ERM family and vinculin with the plasma membrane in MDCK cells. Oncogene. 1997;14:1705–1713. doi: 10.1038/sj.onc.1200998. [DOI] [PubMed] [Google Scholar]
- Krieg J, Hunter T. Identification of the two major epidermal growth factor-induced tyrosine phosphorylation sites in the microvillar core protein ezrin. J Biol Chem. 1992;267:19258–19265. [PubMed] [Google Scholar]
- Kumagai N, Morii N, Fujisawa K, Nemoto Y, Narumiya S. ADP-ribosylation of rho p21 inhibits lysophosphatidic acid-induced protein tyrosine phosphorylation and phosphatidylinositol 3-kinase activation in cultured Swiss 3T3 cells. J Biol Chem. 1993;268:24535–24538. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lankes W, Griesmacher A, Grünwald J, Schwarts-Albiez R, Keller R. A heparin-binding protein involved in inhibition of smooth-muscle cell proliferation. Biochem J. 1988;251:831–842. doi: 10.1042/bj2510831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankes W, Furthmayr H. Moesin: a member of the protein 4.1-talin-ezrin family of proteins. Proc Natl Acad Sci USA. 1991;88:8297–8301. doi: 10.1073/pnas.88.19.8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T, Manser E, Tan L, Lim L. A novel serine/threonine kinase binding the ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J Biol Chem. 1995;270:29051–29054. doi: 10.1074/jbc.270.49.29051. [DOI] [PubMed] [Google Scholar]
- Leung T, Chen X, Manser E, Lim L. The p160 RhoA-binding kinase ROKα is a member of a kinase family and is involved in the reorganization of the cytoskeleton. Mol Cell Biol. 1996;16:5313–5327. doi: 10.1128/mcb.16.10.5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJG, Esch F, Furthmayr H, Hall A. Rho- and Rac-dependent assembly of focal adhesion complexes and actin filaments in permeabilized fibroblasts: an essential role for ezrin/radixin/moesin proteins. J Cell Biol. 1997;138:927–938. doi: 10.1083/jcb.138.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule P, Furuyashiki T, Reid T, Ishizaki T, Watanabe G, Morii N, Narumiya S. A novel partner for the GTP-bound forms of rho and rac. . FEBS Lett. 1995;377:243–248. doi: 10.1016/0014-5793(95)01351-2. [DOI] [PubMed] [Google Scholar]
- Malcolm KC, Ross AH, Qiu RG, Symons M, Exton JH. Activation of rat liver phospholipase D by the small GTP-binding protein RhoA. J Biol Chem. 1994;269:25951–25954. [PubMed] [Google Scholar]
- Magendantz M, Henry MD, Lander A, Solomon F. Interdomain interaction of radixin in vitro. . J Biol Chem. 1995;270:25324–25327. doi: 10.1074/jbc.270.43.25324. [DOI] [PubMed] [Google Scholar]
- Martin M, Andréoli C, Sahuquet A, Montcourrier P, Algrain M, Mangeat P. Ezrin NH2-terminal domain inhibits the cell extension activity of the COOH-terminal domain. J Cell Biol. 1995;128:1081–1093. doi: 10.1083/jcb.128.6.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Roy C, Montcourrier P, Sahuquet A, Mangeat P. Three determinants in ezrin are responsible for cell extension activity. Mol Biol Cell. 1997;8:1543–1557. doi: 10.1091/mbc.8.8.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, Ito M, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Rho-associated kinase, a novel serine/threonine kinase, as a putative target for the small GTP binding protein Rho. EMBO (Eur Mol Biol Organ) J. 1996;15:2208–2216. [PMC free article] [PubMed] [Google Scholar]
- Morii N, Teru-uchi T, Tominaga T, Kumagai N, Kozaki S, Ushikubi F, Narumiya S. A rhogene product in human blood platelets. J Biol Chem. 1992;267:20921–20926. [PubMed] [Google Scholar]
- Nakamura F, Amieva MR, Furthmayr H. Phosphorylation of threonine 558 in the carboxyl-terminal actin-binding domain of moesin by thrombin activation of human platelets. J Biol Chem. 1995;270:31377–31385. doi: 10.1074/jbc.270.52.31377. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Amieva MR, Hirota C, Mizuno Y, Furthmayr H. Phosphorylation of 558T of moesin detected by site-specific antibodies in RAW264.7 macrophages. Biochem Biophys Res Commun. 1996;226:650–656. doi: 10.1006/bbrc.1996.1410. [DOI] [PubMed] [Google Scholar]
- Narumiya S, Sekine A, Fujiwara M. Substrate for botulinum ADP-ribosyltransferase, Gb, has an amino acid sequence homologous to a putative Rho gene product. J Biol Chem. 1988;263:17255–17257. [PubMed] [Google Scholar]
- Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO (Eur Mol Biol Organ) J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakkanen R, Hedman K, Turunen O, Wahlström T, Vaheri A. Microvillus-specific Mr 75,000 plasma membrane protein of human choriocarcinoma cells. J Histochem Cytochem. 1987;35:809–816. doi: 10.1177/35.8.3298422. [DOI] [PubMed] [Google Scholar]
- Pestonjamasp K, Amieva MR, Strassel CP, Nauseef WM, Furthmayr H, Luna EJ. Moesin, ezrin, and p205 are actin-binding proteins associated with neutrophil plasma membranes. Mol Biol Cell. 1995;6:247–259. doi: 10.1091/mbc.6.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D, Berryman M, Bretscher A. Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees DJG, Ades SE, Singer SJ, Hynes RO. Sequence and domain structure of talin. Nature. 1990;347:685–689. doi: 10.1038/347685a0. [DOI] [PubMed] [Google Scholar]
- Reid T, Furuyashiki T, Ishizaki T, Watanabe G, Watanabe N, Fujisawa K, Morii N, Madaule P, Narumiya S. Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the rho-binding domain. J Biol Chem. 1996;271:13556–13560. doi: 10.1074/jbc.271.23.13556. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Rouleau GA, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang-Xuan K, Demczuk S, Desmaze C, Plougastel B, et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- Roy C, Martin M, Mangeat P. A dual involvement of the amino-terminal domain of ezrin in F- and G-actin binding. J Biol Chem. 1997;272:20088–20095. doi: 10.1074/jbc.272.32.20088. [DOI] [PubMed] [Google Scholar]
- Sato N, Funayama N, Nagafuchi A, Yonemura S, Sa, Tsukita, Sh, Tsukita A gene family consisting of ezrin, radixin, and moesin. Its specific localization at actin filament/plasma membrane association sites. J Cell Sci. 1992;103:131–143. doi: 10.1242/jcs.103.1.131. [DOI] [PubMed] [Google Scholar]
- Serrador JM, Alonso-Lebrero JL, del Pozo MA, Furthmayr H, Schwartz-Albiez R, Calvo J, Lozano F, Sánchez-Madrid F. Moesin interacts with the cytoplasmic region of intercellular adhesion molecule-3 and is redistributed to the uropod of T lymphocytes during cell polarization. J Cell Biol. 1997;138:1409–1423. doi: 10.1083/jcb.138.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Tanaka K, Nakanishi H. Rho as a regulator of the cytoskeleton. Trends Biochem Sci. 1995;20:227–231. doi: 10.1016/s0968-0004(00)89022-2. [DOI] [PubMed] [Google Scholar]
- Takaishi K, Sasaki T, Kameyama T, Sa, Tsukita, Sh, Tsukita, Takai Y. Translocation of activated Rho from the cytoplasm to membrane ruffling area, cell-cell adhesion sites and cleavage furrows. Oncogene. 1995;11:39–48. [PubMed] [Google Scholar]
- Takahashi K, Sasaki T, Mammoto A, Takaishi K, Kameyama T, Sa, Tsukita, Sh, Tsukita, Takai Y. Direct interaction of Rho GDP dissociation inhibitor with ezrin/radixin/moesin initiates the activation of the Rho small G protein. J Biol Chem. 1997;272:23371–23375. doi: 10.1074/jbc.272.37.23371. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Kawashima A, Nagafuchi A, Sh, Tsukita Structural diversity of band 4.1 superfamily members. J Cell Sci. 1994a;107:1921–1928. doi: 10.1242/jcs.107.7.1921. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Sato N, Kasahara H, Funayama N, Nagafuchi A, Yonemura S, Sa, Tsukita, Sh, Tsukita Perturbation of cell adhesion and microvilli formation by antisense oligonucleotides to ERM family members. J Cell Biol. 1994b;125:1371–1384. doi: 10.1083/jcb.125.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Yamaguchi M, Matsukage A, Takahashi T. Structural homology of DNA polymerase β from various mammalian cells. J Biol Chem. 1981;256:3098–3102. [PubMed] [Google Scholar]
- Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;72:791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- Tsukita Sa, Mimura N, Tsukita Sh, Khono K, Ohtaki T, Oshima T, Ishikawa H, Asano A. Characteristic structures of actin gels induced with hepatic actinogelin or with chicken gizzard alpha-actinin: implication for their function. Cell Motil Cytoskel. 1988;10:451–463. doi: 10.1002/cm.970100402. [DOI] [PubMed] [Google Scholar]
- Tsukita Sa, Hieda Y, Tsukita Sh. A new 82 kD-barbed end capping protein (radixin) localized in the cell-to-cell adherens junction: purification and characterization. J Cell Biol. 1989;108:2369–2382. doi: 10.1083/jcb.108.6.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita Sa, Oishi K, Sato N, Sagara J, Kawai A, Tsukita Sh. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol. 1994;126:391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita Sa, Yonemura S, Tsukita Sh. ERM proteins: head-to-tail regulation of actin-plasma membrane interaction. Trends Biochem Sci. 1997a;22:53–58. doi: 10.1016/s0968-0004(96)10071-2. [DOI] [PubMed] [Google Scholar]
- Tsukita Sa, Yonemura S, Tsukita Sh. ERM (ezrin/radixin/moesin) family: from cytoskeleton to signal transduction. Curr Opin Cell Biol. 1997b;9:70–75. doi: 10.1016/s0955-0674(97)80154-8. [DOI] [PubMed] [Google Scholar]
- Turunen O, Winqvist R, Pakkanen R, Grzeschik KH, Wahlström T, Vaheri A. Cytovillin, a microvillar Mr 75,000 protein. cDNA sequence, pro-karyotic expression, and chromosomal localization. J Biol Chem. 1989;264:16727–16732. [PubMed] [Google Scholar]
- Turunen O, Wahlström T, Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J Cell Biol. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urushidani T, Hanzel DK, Forte JG. Characterization of an 80-kDa phosphoprotein involved in parietal cell stimulation. Am J Physiol. 1989;256:G1070–G1081. doi: 10.1152/ajpgi.1989.256.6.G1070. [DOI] [PubMed] [Google Scholar]
- Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Morii N, Mukai H, Ono Y, Kakizuka A, Narumiya S. Protein kinase N (PKN) and PKN-related protein rhophilin as targets of small GTPase Rho. Science. 1996;271:645–648. doi: 10.1126/science.271.5249.645. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO (Eur Mol Biol Organ) J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Tonks NK. Isolation of a cDNA clone encoding a human protein-tyrosine phosphatase with homology to the cytoskeletal-associated proteins band 4.1, ezrin, and talin. Proc Natl Acad Sci USA. 1991;88:5949–5953. doi: 10.1073/pnas.88.14.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Cheng L, Forte JG. Biochemical characterization of ezrin-actin interaction. J Biol Chem. 1996;271:7224–7229. doi: 10.1074/jbc.271.12.7224. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Nagafuchi A, Sato N, Sh, Tsukita Concentration of an integral membrane protein, CD43 (leukosialin, sialophorin), in the cleavage furrow through the interaction of its cytoplasmic domain with actin-based cytoskeletons. J Cell Biol. 1993;120:437–449. doi: 10.1083/jcb.120.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, King WG, Dillon S, Hall A, Feig L, Rittenhouse SE. Activation of platelet phosphatidylinositide 3-kinase requires the small GTP-binding protein Rho. J Biol Chem. 1995;268:22251–22254. [PubMed] [Google Scholar]