Abstract
Minichromosome maintenance (MCM) proteins are essential eukaryotic DNA replication factors. The binding of MCMs to chromatin oscillates in conjunction with progress through the mitotic cell cycle. This oscillation is thought to play an important role in coupling DNA replication to mitosis and limiting chromosome duplication to once per cell cycle. The coupling of DNA replication to mitosis is absent in Drosophila endoreplication cycles (endocycles), during which discrete rounds of chromosome duplication occur without intervening mitoses. We examined the behavior of MCM proteins in endoreplicating larval salivary glands, to determine whether oscillation of MCM–chromosome localization occurs in conjunction with passage through an endocycle S phase. We found that MCMs in polytene nuclei exist in two states: associated with or dissociated from chromosomes. We demonstrate that cyclin E can drive chromosome association of DmMCM2 and that DNA synthesis erases this association. We conclude that mitosis is not required for oscillations in chromosome binding of MCMs and propose that cycles of MCM–chromosome association normally occur in endocycles. These results are discussed in a model in which the cycle of MCM–chromosome associations is uncoupled from mitosis because of the distinctive program of cyclin expression in endocycles.
Polyploidy is observed during the normal development of many organisms (Wilson, 1987). A well-documented means for attaining polyploidy in both plants and animals is endoreplication, in which successive rounds of chromosome duplication occur in the absence of cell division. For example, endosperm development in maize, trichome cell formation in Arabidopsis, and trophoblast differentiation in mammals involve cycles of endoreplication (Hulskamp et al., 1994; Grafi and Larkins, 1995; Varmuza et al., 1996). During Drosophila development, endoreplication is first observed in specific tissues of the embryo and continue in most larval cells (Rudkin, 1972; Ashburner, 1989; Smith and Orr-Weaver, 1991). In the salivary glands of Drosophila melanogaster, 8–10 discrete rounds of S phases occur during embryonic and larval periods to produce polytene chromosomes (Rudkin, 1972). Most of the genome is duplicated once during each of these S phases, and each S phase is followed by a gap phase. Duplication of the genome, however, is not complete in each S phase and certain specific regions are underrepresented in the final polytene state (Rudkin, 1969; Hammond and Laird, 1985).
In mitotic cycles, chromosome duplication (S phase) must be coupled to chromosome segregation (mitosis) to preserve the integrity of the genome through many cell divisions. Mammalian cell fusion experiments indicate that one basis for this coupling is an oscillation in the ability of a nucleus to replicate its DNA (Rao and Johnson, 1970; for review see Heichman and Roberts, 1994). A postmitotic nucleus (in G1) is competent to replicate DNA, but this competence is lost with progress through S phase, and a postreplication nucleus (in G2) is unable to replicate DNA. Subsequent passage through mitosis restores the ability to replicate DNA, thereby coupling mitosis and DNA replication. Thus, loss of competence to replicate DNA during S phase and subsequent restoration in M ensures that the genome is replicated only once in a given cell cycle.
In endocycles of Drosophila salivary gland cells, successive S phases take place in the absence of mitosis. Thus, passage through mitosis is clearly not a requirement for DNA replication in salivary gland cells. Endocycles, therefore, offer a unique opportunity to examine the regulation of DNA replication in the absence of the mitotic program. In addition, polytene chromosomes, because of their immense size, allow a visual examination of protein–chromatin interactions in interphase, in the absence of chromatin condensation.
In a variety of experimental systems, the competence to replicate DNA is characterized by the presence of chromatin-bound minichromosome maintenance (MCM)1 proteins (for reviews see Tye, 1994; Kearsey et al., 1996; Romanowski and Madine, 1996; Chong et al., 1996). MCMs are essential DNA replication factors that are conserved from archaebacteria to yeast and human. In mitotic cycles, MCMs associate with chromatin before DNA replication, dissociate from chromatin with progress through S phase, and are absent from postreplication chromatin (Kimura et al., 1994; Madine et al., 1995; Krude et al., 1996; Coue et al., 1996). Passage through mitosis restores chromatin association of MCMs and resets the cycle. Thus, a cycle of MCM–chromatin association closely resembles the cycle of competence to replicate DNA and perhaps acts to couple DNA replication to mitosis.
To study the regulation of DNA replication in the absence of mitosis, we have examined three Drosophila MCMs during endoreplication cycles. In polytene cells of larval salivary glands, Drosophila MCM proteins were detected in two distinct states: associated with chromosomes or dissociated from chromosomes. We demonstrate that Drosophila cyclin E, which is essential for embryonic S phases, can promote chromosome association of DmMCM2. DNA synthesis erases this association, thereby resetting the cycle.
Materials and Methods
Antibody Staining
For immunostaining of salivary glands, w67 or Sevelin embryos were collected for 4 h on grape juice agar plates and aged for 4 d at 25°C to reach feeding third instar. Salivary glands were dissected in Schneider's insect culture medium and fixed for 10 min in PBS + 0.2% Tween (PBT) containing 10% formaldehyde. Fixed tissues were blocked in PBT containing 3% normal goat or donkey serum (Vector Labs, Inc., Burlingame, CA) and probed with purified primary antibodies diluted 1:50–1:500 in blocking solution. In our attempts to first spread the polytene chromosomes before antibody staining, we found that protocols for spreading of chromosomes, especially the treatment with acetic acid, are incompatible with immunostaining of MCMs. Incompatibility of acetic acid–containing protocols and antibody staining for MCMs also extends to the embryo.
Specificity of each MCM antibody has been described previously (Su et al., 1996). Primary antibodies were detected with a rhodamine-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). DNA was stained with 10 μg/ml bisbenzamid (Hoechst 33258; Aldrich, Milwaukee, WI). Images were obtained using a Leica fluorescence microscope (Deerfield, IL) attached to a CCD camera. Salivary gland images were processed using Delta Vision software (Applied Precision, Issaquah, WA).
For immunoblotting, salivary glands were dissected and cleaned of most associated tissues before homogenization in PBT. Homogenates were boiled in SDS-containing sample buffer, loaded onto denaturing gels, and immunoblotted using standard methods. Rabbit polyclonal antisera to cyclin E have been described before (Sauer et al., 1995) and were used at 1:1,500 dilution.
Heat Induction and Bromo-deoxyuridine Incorporation
Heat-inducible transgenes for cyclin E have been described previously (Knoblich et al., 1994; Duronio et al., 1995) and were carried as a homozygous line (E3). w67, the isogenic stock used for carrying the transgenes, was used as a control in all heat shock experiments. For heat shock experiments, feeding third-instar larvae were incubated inside Eppendorf tubes (Madison, WI) in a 37°C water bath for 30 min. After recovery at room temperature (rt) for various times, salivary glands were dissected in Schneider's insect culture medium and fixed as above for DmMCM2 staining. For detection of DNA synthesis, dissected glands were labeled for 15–20 min in Schneider's medium containing 1 mg/ml bromo-deoxyuridine (BrdU) (Sigma Chemical Co., St. Louis, MO) before fixing. Fixed glands were either probed with anti-DmMCM2 antibodies or processed for BrdU detection by acid denaturation according to published protocols. In our attempts to double-stain for DmMCM2 and BrdU simultaneously, it was difficult to score MCM localization in such double-stained glands, perhaps because denaturation of DNA that was required for immunological detection of incorporated BrdU compromised the integrity of chromosomes and, hence, the localization of MCMs.
For double heat shock experiments, the first heat shock was performed as above. At 2 or 3 h after recovery at rt, larvae were subjected to a second identical heat shock. After a recovery of 20–30 min at rt after the second heat shock, salivary glands were dissected, fixed, and stained for DmMCM2, as above.
Inhibition of DNA Synthesis
To block DNA synthesis after heat induction, dissected glands were incubated in Schneider medium containing 50 μg/ml aphidicolin (Sigma Chemical Co.). For delivery by feeding, aphidicolin was added to 500 μg/ml in baker's yeast paste and fed to Sevelin larvae for 15 h (late second instar to early third instar) before dissection.
Results
MCMs in Polytene Nuclei Exist in Two States: Associated with or Dissociated from Chromosomes
Of the five known members of the Drosophila MCM family (Su et al., 1997), three that have been examined are detected in endocycling tissues by Western blotting (Feger et al., 1995; Su et al., 1996; note that the names used here follow Chong et al., 1996). To analyze their subcellular localization, we immunolocalized DmMCM2, DmMCM4 (previously called Dpa), and DmMCM5 (previously called DmCDC46) in larval salivary glands. All three MCMs were detected as nuclear proteins in polytene nuclei of salivary glands during the three larval instars (shown for DmMCM2 in Fig. 1). In a majority of polytene nuclei from second- and third- instar larvae (>80%), most of the nuclear MCM stain was excluded from regions occupied by DNA and by the nucleolus (Fig. 1, 3 and 5). We infer that most of the nuclear MCMs are not associated with chromosomes in these nuclei, and refer to this pattern of staining as “nucleoplasmic.” In contrast, in a small fraction of nuclei (∼10% in mid-third instar w67 or Sevelin larvae), nuclear MCM staining was coincident with the DNA (Fig. 1, 2 and 4). We interpret this state as association of MCMs with polytene chromosomes as “chromosomal.” Note that nuclei with chromosomal and nucleoplasmic MCMs can be distinguished when viewed in cross-section, because, in the former, MCM stain fills the nonnucleolar region of the nucleus, whereas in the latter, MCM stain appears netlike (compare Fig. 1, 4 and 5; schematized in Fig. 3).
Figure 1.
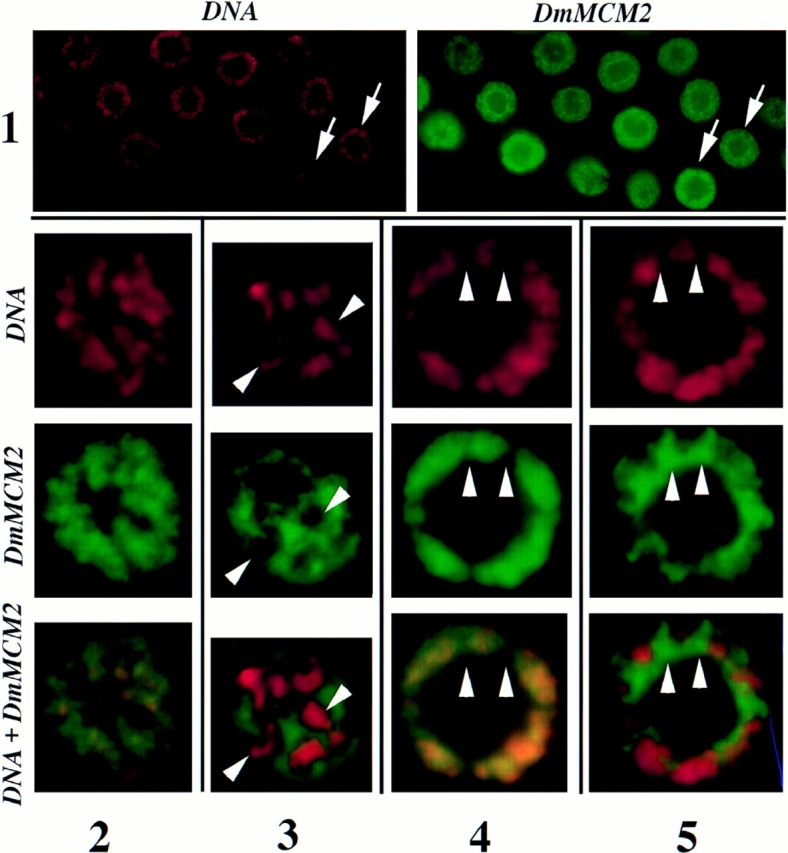
Chromosomal and nucleoplasmic localization of DmMCM2 polytene nuclei. Salivary glands from third-instar larvae were fixed and stained for DmMCM2 and DNA. Note that all nuclei contain DmMCM2 (1) despite differences in the pattern of staining (for example, nuclei indicated by arrows). 2–5 show, at higher magnification, representative nuclei showing two distinct types of nuclear DmMCM2 staining. 2 and 3 are grazing optical sections and 4 and 5 are optical sections across the nucleus. The “space” in the center of each nucleus is the nucleolus. Note the colocalization of DmMCM2 and DNA signals in 2 and 4, such that distinct green or red regions are not seen. For example, regions indicated by arrowheads in 4 lack the DNA stain as well as the DmMCM2 signal. In contrast, DmMCM2 stain is excluded from the chromosomes in 3 and 5, such that distinct green or red regions are visible. For example, regions indicated by arrowheads in 3 have the DNA stain but lack the DmMCM2 signal, and regions indicated by arrowheads in 5 lack the DNA stain but have the DmMCM2 signal.
Figure 3.
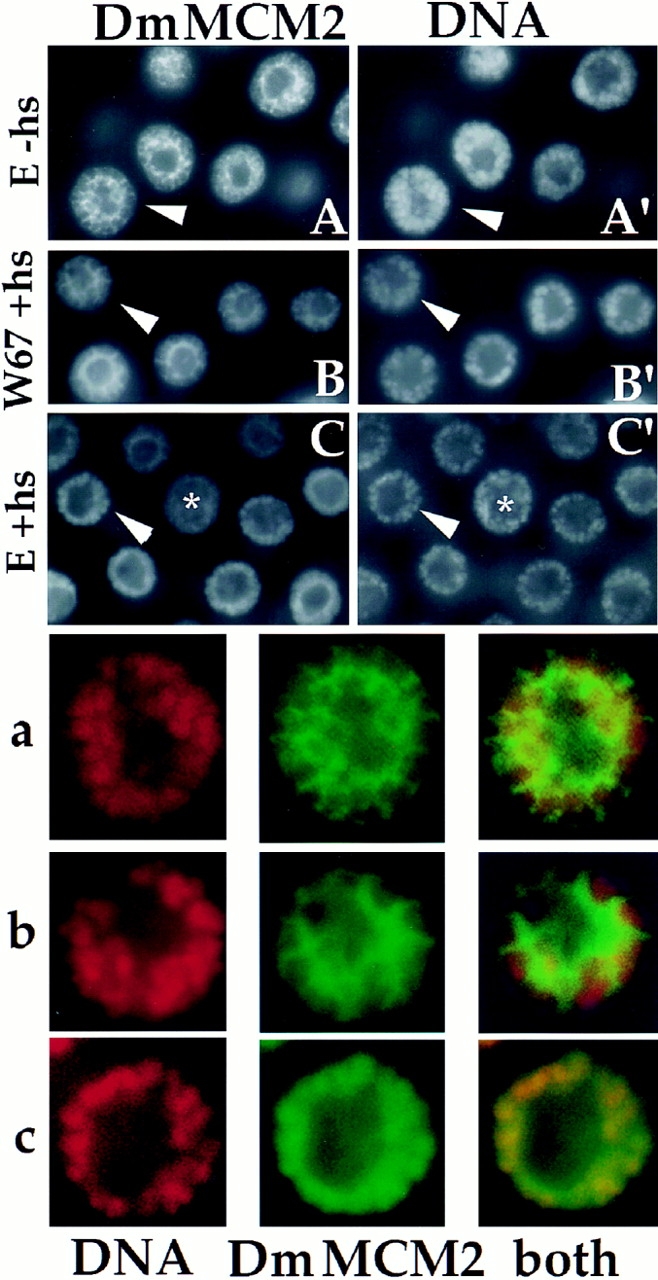
Induction of DmMCM2–chromosome association by cyclin E. Salivary glands were dissected from transgenic larvae before heat shock (E-hs) and at 25 min after heat shock to induce cyclin E (E +hs), or from heat-shocked controls lacking the transgene (W67 +hs), fixed, and stained for DmMCM2 and DNA. Nuclei indicated by arrowheads are magnified and shown in a (from A and A′), b (from B and B′) and c (from C and C′). In A and B, DmMCM2 did not colocalize with DNA in most nuclei and appears netlike when viewed in cross-section (similar to Fig. 1, lane 5 and schematized in Fig. 4 as nucleoplasmic). In contrast, in C, colocalization of DmMCM2 and DNA results in uniform DmMCM2 stain in the nonnucleolar region of many nuclei (similar to Fig. 1, 4 and schematized in Fig. 4 as chromosomal).
Polytene chromosomes are not condensed but are interphase chromosomes that are nevertheless visible because of their immense size. Thus, polytene cells allow us to detect two interphase states of MCM–chromosome interaction in vivo. These results parallel findings in mitotically proliferating mammalian cells, which show that MCMs are nuclear throughout interphase, but can be either associated with or dissociated from chromatin (Kimura et al., 1994; Todorov et al., 1995). A transition from a dissociated to an associated state, however, is thought to depend on passage through mitosis (for reviews see Tye, 1994; Kearsey et al., 1996; Romanowski and Madine, 1996; Chong et al., 1996). Our detection of these two states in polytene cells suggests that the change from one state to the other occurs in the absence of mitosis in endocycling cells. Data presented below support this idea.
MCM–Chromosome Association during S Phase in Polytene Cells
We next asked if transitions between the two states of MCM–chromosome association occur in conjunction with progress through an endocycle. Endocycles of larval salivary glands are asynchronous and do not follow an obvious stereotypical pattern. Consequently, we cannot define the stage of a given cell with respect to its position in S or gap phase based on developmental stage. Therefore, we sought to induce a synchronous S phase in the salivary glands to examine changes in MCM staining that may accompany DNA synthesis.
Analysis of Drosophila embryos indicate that cyclin E is essential for embryonic endocycles. It is expressed in transient pulses each of which overlaps the beginning of each endocycle S phase, and cyclin E mutant embryos fail to undergo endoreplication (Knoblich et al., 1994; Duronio and O'Farrell, 1995). Moreover, production of Drosophila cyclin E from a heat-inducible transgene can drive endocycling cells of the embryo into a synchronous S phase (Knoblich et al., 1994; Duronio and O'Farrell, 1995). We asked if a similar production of cyclin E can induce synchronous DNA synthesis in endocycling cells of early third-instar larvae. To induce cyclin E, larvae carrying the appropriate transgene were subjected to a 37°C heat pulse for 30 min. Salivary glands were dissected at various times after heat shock to monitor cyclin E induction by immunoblotting (Fig. 2 A), or to detect DNA synthesis by labeling with a nucleotide analogue, BrdU, for 15 min. The labeled glands were then fixed, and incorporated BrdU was detected by immunostaining (Fig. 2 B).
Figure 2.
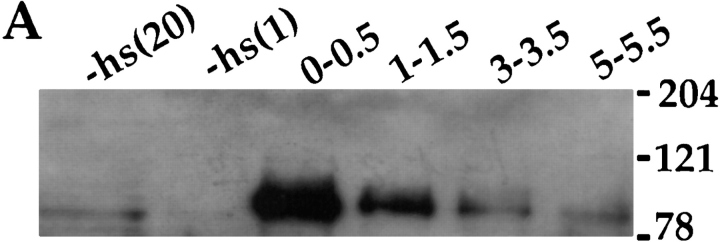
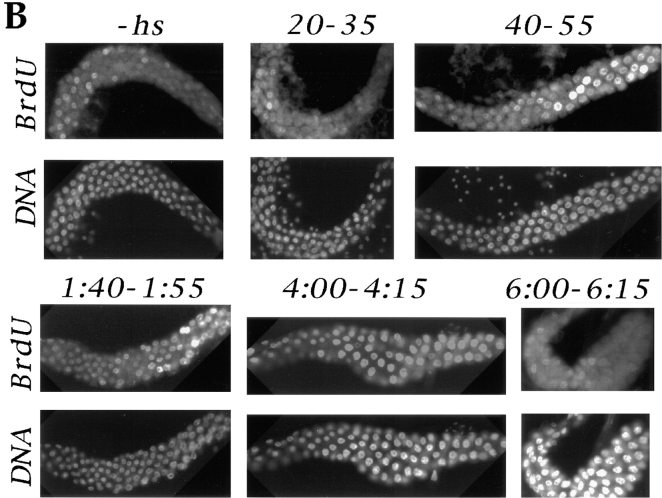
Induction of DNA replication by cyclin E. (A) The detection of cyclin E in salivary gland extracts by immunoblotting. cyclin E was produced by heat-shocking feeding-stage third-instar larvae carrying the appropriate transgene. Salivary glands were dissected from control larvae (−hs lanes) or from heat-shocked larvae at various times after heat shock (in h, indicated above each lane). Extracts were separated on denaturing gels and immunoblotted using a previously characterized antiserum against cyclin E (Sauer et al., 1995). “−hs” lanes contain extract from either 20 pairs or one pair of salivary glands as indicated. Each of the other lanes contain extract from one pair of salivary glands. The positions of molecular mass markers, in kD, are indicated on the side. (B) Induction of DNA synthesis by cyclin E. cyclin E was produced by heat-shocking feeding-stage third-instar larvae carrying the appropriate transgene. Salivary glands were dissected and labeled with a nucleotide analog, BrdU, at various times after heat shock as indicated above the lanes (min or h:min). Incorporated BrdU was detected immunologically and DNA was stained with Hoechst 33258. −hs: no heat shock control.
In control glands (from w67 larvae with or without heat shock, or transgenic larvae that have not been heat shocked), ∼20% of nuclei in each salivary gland showed BrdU signal that exceeded the cytoplasmic background staining (Fig. 2 B, −hs). The intensity of BrdU signal varied within each gland, perhaps due to S phase asynchrony. About 80% of the cells in these controls did not show significant BrdU signal and are presumably in gap phase. Beginning about 1 h after heat shock, more nuclei with detectable BrdU signal were seen per gland from transgenic larvae (Fig. 2 B, 40-55). This was followed by a period, lasting up to 4 h after heat shock, during which most polytene cells of the salivary glands incorporated BrdU. The intensity of BrdU signal varied within each gland at these time points. This could be caused by asynchrony already present in the gland before heat shock, or by variations in the effectiveness of heat shock with respect to cyclin E production and consequent DNA synthesis. By 6 h after heat shock, most of the nuclei were no longer incorporating BrdU. These data indicate that expression of cyclin E induces DNA synthesis in most nuclei of salivary glands and that the induced S phase lasts between 3 and 6 h. Despite apparent variations in the kinetics of S phase progression, this protocol allowed us to examine populations of polytene nuclei that are about to enter S phase, those that have entered S phase, those that are finishing S phase, and those that have finished S phase. Heat shock did not induce DNA synthesis in w67 controls (not shown).
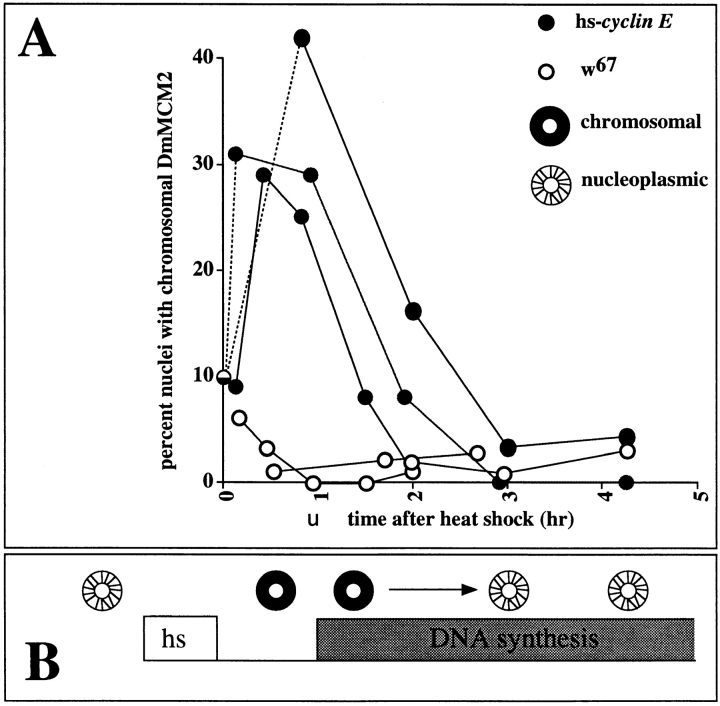
To address the behavior of MCMs with respect to S phase, we examined the subcellular localization of DmMCM2 in salivary glands after cyclin E induction. Parallel samples from experiments described above were fixed and immunostained for DmMCM2 (see Materials and Methods; Fig. 3). In non–heat-shocked glands, as well as in heat-shocked controls lacking the hs–cyclin E transgene, ∼10% of nuclei in each salivary gland showed chromosomal DmMCM2 stain (Figs. 3 and 4; see above also). In response to heat induction, the fraction of nuclei with chromosomal MCM staining increased in transgenic larvae but not in controls (Figs. 3 and 4). This increase was rapid. A major increase was often detected at the first time point, 8 min after heat shock, and maximal chromosomal staining occurred within 1 h after the heat shock period. Note that peak levels of chromosomal staining occurred before detectable induction of BrdU incorporation (Fig. 4 A, arrowhead). As DNA synthesis commenced, the fraction of nuclei with chromosomal DmMCM2 decreased and eventually reached undetectable levels as cells progressed to late S phase. We did not observe nuclei with chromosomal DmMCM2 staining after BrdU incorporation ceased (∼6 h after heat shock). From these data we infer that cyclin E induced the movement of DmMCM2 from the nucleoplasm to the chromosomes, and that progress through S phase is accompanied by dissociation of DmMCM2 from chromosomes (Fig. 4 B).
Figure 4.
Quantification of DmMCM2–chromosome association. A shows the time course of changes in DmMCM2 localization after heat shock of larvae with the inducible cyclin E transgene (filled circles) or of w67 controls (open circles). DmMCM2 was detected immunologically in parallel samples from experiments such as those in Fig. 2. Salivary glands were not analyzed for BrdU and DmMCM2 simultaneously for technical reasons outlined in Materials and Methods. The percentage of nuclei showing chromosomal-associated DmMCM2 staining in each salivary gland was plotted against time after heat shock. Data from three of four experiments performed are shown. The value for the no– heat shock control for cyclin E transgenic larvae is shown at t = 0 (half-filled circle). Dashed lines represent extrapolation to the no–heat shock value. At least three and up to eight glands of ∼120 nuclei each are counted for each time point. Induced DNA synthesis was detectable at about 1 h after heat shock (Fig. 2; arrowhead). Slight variations in the time of onset of MCM–chromosome association is seen, as in the examples shown here. But in all cases, maximal chromosome association occurred before induction of BrdU incorporation. B shows the inferred pattern of DmMCM2 localization in a polytene nucleus during a cyclin E–induced S phase.
We scored chromosomal DmMCM2 staining in about a third of the nuclei in each gland after induction of cyclin E (Fig. 4 A). We believe this number to be an underestimate because we score only nuclei in which most, if not all, nuclear DmMCM2 colocalized with DNA as “chromosomal” (such as in Fig. 1, 2 and Fig. 3, C and C′, arrowheads). Nuclei in which only a fraction of total nuclear DmMCM2 localized onto chromosomes after cyclin E expression would not have been scored as “chromosomal” (such an ambiguous nucleus is indicated by an asterisk in Fig. 3, C and C′). In addition to the stringency in scoring, two additional factors, addressed below, may help explain why only a third of the nuclei were scored as having chromosomal DmMCM2 staining after cyclin E induction.
At the beginning of the heat pulse used to induce cyclin E, most of the cells are in gap phase (Fig. 2 B) and showed nucleoplasmic MCM staining (Figs. 3 and 4). Induction of cyclin E led to an increase in MCM–chromosome association. Thus, cells in gap phase can load DmMCM2 onto chromosomes in response to cyclin E. To ask if cells in S phase have a similar capacity, we expressed cyclin E in synchronously replicating salivary gland cells. To do this, we first synchronized endocycles by induction of cyclin E. This was followed by a second heat pulse 4 h later, when DmMCM2 was nucleoplasmic in most nuclei, and nuclei were still replicating as a consequence of the first heat pulse (see Materials and Methods). We then examined the subcellular localization of DmMCM2 after the second heat pulse. We found that DmMCM2 remained nucleoplasmic. No nuclei with chromosome-associated DmMCM2 were seen in seven glands (∼800 nuclei in total). Thus, induction of S phase is followed by a period during which heat induction of cyclin E is not able to induce MCM relocalization onto chromosomes under our experimental conditions. Because about a fifth of the polytene cells in the undisturbed glands are in S phase (for example, Fig. 2), this refractory period could account for some of the nuclei that failed to induce chromosomal localization of DmMCM2. In addition, some nuclei, particularly those that already had chromosomal localized MCMs, may have advanced rapidly into S phase and lost chromosomal MCM staining before analysis. Hence, although the difference between the number of nuclei with chromosomal DmMCM2 staining after cyclin E induction (about a third) and the number that synthesized DNA (almost all) raises the formal possibility that some nuclei synthesized DNA without first loading DmMCM2 onto chromatin, we believe this to be unlikely.
From these data we conclude that cyclin E can drive gap to S transition in polytene cells and the transfer of DmMCM2 from the nucleoplasm to the chromosomes.
DNA Synthesis Is Required for Dissociation of DmMCM2 from Chromosomes
We observed that during a cyclin E–induced S phase, DmMCM2 dissociated from polytene chromosomes in conjunction with progression of DNA synthesis. To test whether DNA synthesis is required for the dissociation of DmMCM2 from chromosomes, we blocked DNA synthesis with an inhibitor, aphidicolin (Schubiger and Edgar, 1994), and examined the consequences on DmMCM2 staining. Two protocols were used to administer aphidicolin. First, immediately after heat induction of cyclin E, salivary glands were dissected and placed in culture medium containing the inhibitor. At 2 or 3 h after heat shock, salivary glands were fixed and stained for DmMCM2. We found that, in the presence of aphidicolin, chromosome-associated DmMCM2 was retained for up to 3 h after induction of S phase by cyclin E; 23% of nuclei were observed with chromosomal DmMCM2 staining in eight glands. This number should be compared with 0%, at a similar time after heat shock, in the absence of aphidicolin (Fig. 4 A, filled circles at 2- and 3-h time points).
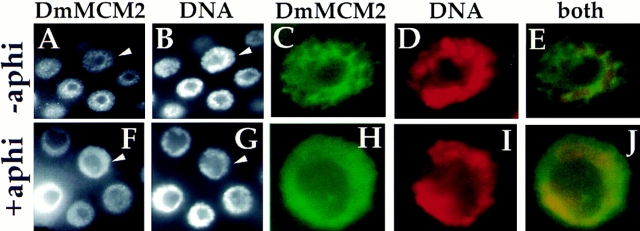
Although the aphidicolin treatment clearly stabilized chromosomal DmMCM2, the proportion of nuclei showing chromosomal staining varied from gland to gland (not shown), suggesting that inhibition of DNA synthesis may not have been efficient. This may be due to a substantial time, required for dissection, between the beginning of heat induction of cyclin E and the delivery of aphidicolin to the samples. As an alternative way to inhibit DNA synthesis, we added aphidicolin to the food of feeding third-instar larvae for 15 h. During this time, we expect that a large fraction of polytene nuclei would attempt endoreplication (Rudkin, 1972). We reasoned that the MCMs would bind to chromosomes as cells prepare to enter S phase (e.g., Fig. 4 B), but that the presence of aphidicolin would then block DNA synthesis. If DNA synthesis was required for dissociation of MCMs from chromosomes, we would essentially “trap” MCMs on polytene chromosomes of larvae that have been fed aphidicolin, but not in control larvae. After aphidicolin feeding, salivary glands were dissected, fixed, and stained for DmMCM2. Efficient retention of DmMCM2 on chromosomes was seen using this procedure (Fig. 5): 65% of nuclei in salivary glands of aphidicolin-fed larvae showed chromosomal DmMCM2 (data from four glands), compared to <5% in controls. Note that we do not expect to see retention of chromosomal MCMs in all the nuclei. This is because at the beginning of the experiment some of the nuclei would have been in S phase with MCMs dissociated from chromosomes and may have remained arrested in this state in the presence of aphidicolin. In addition, because of asynchrony in endocycles, some of the nuclei may not have gone through the period of loading MCMs onto DNA during the course of aphidicolin feeding. On the basis of these data, we conclude that DNA replication is required for dissociation of DmMCM2 from the chromosomes.
Figure 5.
DmMCM2 is retained on chromosomes when DNA synthesis is inhibited. Larval salivary glands from control larvae (−aphi) and larvae on aphidicolin-containing diet (+aphi) were dissected, fixed, and stained for DmMCM2 and DNA as indicated. The nucleus indicated with an arrowhead in A and B is magnified and shown in C–E. The nucleus indicated with an arrowhead in F and G is magnified and shown in H–J. In F and H, colocalization of DmMCM2 and DNA results in uniform DmMCM2 stain in the nonnucleolar region of nuclei when viewed in cross-section (similar to Fig. 1, 4 and schematized in Fig. 4 as chromosomal). In contrast, in most nuclei in A and in the nucleus in C, DmMCM2 did not colocalize with DNA and appears netlike (as in Fig. 1, 5 and schematized in Fig. 4 as nucleoplasmic).
Discussion
For a nucleus to replicate its DNA, two components are thought to be needed: signals that trigger DNA synthesis and the competence to respond to these signals, referred to as “replication competence” (for reviews see Heichman and Roberts, 1994; Su et al., 1995; Wuarin and Nurse, 1996). The replication competence of a nucleus oscillates during progression through a mitotic cycle. Importantly, coupling between mitosis and the acquisition of replication competence limits DNA replication to once per mitotic cell cycle. Coordinating DNA replication with mitotic cell cycle progress is thought to involve MCM proteins that cycle between chromosome-bound and free states in conjunction with cell cycle progression (for reviews see Tye, 1994; Chong et al., 1996; Kearsey et al., 1996; Romanowski and Madine, 1996). However, exceptions to the coupling of S phase to mitosis occur. In Drosophila endoreplication cycles, DNA is replicated in discrete S phases that are separated by gap phases. We have examined the behavior of MCMs in Drosophila endocycles to ask if these proteins cycle between chromosome-bound and free states in conjunction with progress of an endocycle. Analysis of larval salivary glands showed that (a) cyclin E can induce polytene cells in gap phase to synthesize DNA, (b) MCM proteins can be either chromosome associated or dissociated in polytene cells and cyclin E can promote chromosome association of DmMCM2 in permissive stages of the cycle, and (c) DNA replication results in dissociation of DmMCM2 from chromosomes. These findings demonstrate that localization of MCMs to chromatin in endocycles is coupled to S phase, as in mitotic cycles, despite the absence of mitosis in endocycles. The observed induction of MCM–chromosome association in response to cyclin E is novel and is compatible with other observations in mitotic cycles and endocycles as described below.
Endocycling Cells Can Enter S Phase in Response to cyclin E
In mammalian cells undergoing mitotic proliferation, G2 nuclei are not competent to replicate their DNA in response to signals that drive G1 nuclei into S phase. Data from diverse experimental systems indicate that cyclin E:cdk activity contributes to signals that drive the G1 to S transition (Knoblich et al., 1994; Duronio and O'Farrell, 1995; Jackson et al., 1995; Ohtsubo et al., 1995; Richardson et al., 1995; Sauer et al., 1995). Although discrete gap phases occur in endocycles, it is not clear whether the endocycle gap phase is analogous to either G1 or G2 of mitotic cells. We find that endocycle nuclei in gap phase replicate their DNA in response to cyclin E. Thus, these nuclei are competent to replicate, and in this way, they resemble G1 nuclei. There does not appear to be a significant population of nuclei that lack replication competence; virtually all the nuclei of the salivary glands incorporated BrdU after induction of cyclin E.
The ability of endocycle cells in gap phase to enter S phase in response to cyclin E may be due to the absence of mitotic cyclins, cyclin A and cyclin B, in these cells (Lehner and O'Farrell, 1989, 1990). Work in yeast and Drosophila supports a model in which replication in a G2 nucleus is inhibited by the presence of mitotic cyclin:cdks (for reviews see Diffley, 1996; Nasmyth, 1996); for example, Drosophila cyclin A and cdc2 mutants rereplicate DNA in G2 (Sauer et al., 1995; Hayashi, 1996). Moreover, ectopic expression of cyclin A can block embryonic endocycles (Sprenger, F., N. Yakubovich, and P.H. O'Farrell, manuscript in preparation), consistent with the idea that absence of mitotic cyclin:cdks normally contributes to the ability of endocycle cells in gap phase to enter S.
cyclin E Can Drive MCM-Chromosome Association
Numerous observations suggest that MCM association with chromatin is essential for DNA replication. The phenotype of mutations and the results of antibody injection and immunodepletion in yeasts, human cells, Xenopus, and Drosophila suggest a requirement for MCMs in DNA replication (for reviews see Tye, 1994; Chong et al., 1996). Importantly, work in Xenopus suggests that MCMs carry out their essential function when bound to chromatin; an MCM-containing complex can be experimentally induced to bind chromosomes and allow DNA replication in G2 nuclei that normally lack bound MCMs (Chong et al., 1995; Madine et al., 1995). Consistent with the idea that chromatin binding of MCMs is required for S phase, binding of MCMs to chromatin occurs before initiation of replication in mammalian cultured cells (Kimura et al., 1994) and in the Drosophila embryo (Su and O'Farrell, 1997). We also found that MCM–chromosome association increased in response to cyclin E and that this increase preceded BrdU incorporation in salivary gland endoreplication.
Several proposals could explain the ability of cyclin E to promote binding of DmMCM2 to chromosomes. Cyclin E might control the binding of MCMs to chromosomes via direct modification of MCMs. Alternatively, assembly of MCMs onto chromosomes may depend upon prior assembly of other replication proteins such as CDC6 and ORC2, as previously shown in Xenopus extracts (Coleman et al., 1996; Romanowski et al., 1996). Cyclin E might act to control the assembly of these, either by modification or by directing their expression; in Saccharomyces cerevisiae, Drosophila, and vertebrates, G1 cyclins activate an S phase transcription program that may contribute functions to the binding of MCMs to chromosomes (Duronio and O'Farrell, 1995; Piatti et al., 1995; Coleman et al., 1996). Examining molecular changes in MCMs and analyzing the requirement for protein synthesis and transcription in MCM–chromosome association should help address some of these proposals.
Although we used heat induction to demonstrate the ability of cyclin E to drive MCM–chromosome association, the natural expression pattern of cyclin E suggests that it performs a similar task in unperturbed endocycles. First, cyclin E is required for endocycle S phases. Second, cyclin E is expressed in pulses that match the spatial and temporal pattern of endocycle S phases in the embryo (Duronio and O'Farrell, 1994). Moreover, the rise in cyclin E transcript overlaps the beginning of each S phase (Duronio and O'Farrell, 1995). We suggest that in the natural cycle, as in the cycle we provoked, cyclin E first induces an assembly of MCMs, and possibly other replication factors, onto chromatin and, subsequently, synthesis of DNA. DNA synthesis then results in dissociation of MCMs, and possibly other factors, from chromatin and resets the cycle until the next pulse of cyclin E.
In mitotic cycles, chromosome association of MCMs accompanies passage through mitosis. In endocycles, cyclin E is able to promote association of MCMs with chromosomes in the absence of mitosis. Thus, we conclude that passage through mitosis is not a requirement for MCM– chromosome association. One aspect of mitosis, nuclear envelope breakdown (NEB), has been implicated in chromatin binding of a MCM-containing protein complex in Xenopus extracts (Blow and Laskey, 1986; Chong et al., 1995; Madine et al., 1995). Evidence for NEB in Drosophila endocycles has not been reported. In our monitoring of live larvae carrying the nuclear-localized green fluorescent protein (Davis et al., 1996), we did not observe any evidence of NEB in polytene nuclei of salivary glands (Su, T.T., and P.H. O'Farrell, unpublished observations). NEB, we propose, plays no role in MCM–chromosome interactions in endocycles and is not required for chromosome binding of MCMs or for DNA replication.
DNA Synthesis Is Required for the Dissociation of DmMCM2 from Chromosomes
In mitotic cycles, DNA synthesis causes dissociation of MCMs from chromatin during S phase (Chong et al., 1995; Kubota et al., 1995; Madine et al., 1995; Coue et al., 1996). It is thought that cyclin accumulation inhibits reassociation of dissociated MCMs (for reviews see Diffley, 1996; Nasmyth, 1996). This arrangement could prevent MCM reassociation and rereplication from the time of initial accumulation of cyclins at the entry into S phase until their destruction upon exit from mitosis.
In endocycles, MCMs do not reassociate with chromatin during S phase. Because of their absence in endocycles, mitotic cyclins cannot be responsible for inhibiting the reassociation of MCMs with chromosomes during endocycle S phases. We speculate below that cyclin E may contribute to this inhibition.
In many organisms, cdks can have both a positive and a negative role with regard to DNA replication (for reviews see Su et al., 1995; Diffley, 1996; Nasmyth, 1996). Although their absence appears to be required for the acquisition of competence to replicate DNA, their presence is required for DNA synthesis from assembled replication complexes. The ability of cyclin E:cdk2 to promote DNA replication in vertebrates and Drosophila is well documented (Knoblich et al., 1994; Duronio and O'Farrell, 1995; Jackson et al., 1995; Ohtsubo et al., 1995; Richardson et al., 1995; Sauer et al., 1995). Two observations suggest that cyclin E:Cdk2 can also prevent rereplication. First, cyclin E:cdk2, at sufficiently high levels, has been shown to block an early step required for DNA replication in Xenopus extracts (Hua et al., 1997). Second, whereas cyclin E is required for endoreplication and is expressed in pulses, continuous expression of cyclin E in Drosophila salivary gland blocks endoreplication (Follette et al., 1998). Thus, not just the presence of cyclin E but “oscillations” in its levels appear essential for endocycles. We suggest that pulses of cyclin E may be coupled to MCM behavior and endoreplication in the following manner. Expression of cyclin E first drives chromosome binding of MCMs, followed by entry to S phase. Subsequently, DNA synthesis causes dissociation of MCMs from chromosomes; reassociation is prevented by the presence of cyclin E. Decay of cyclin E would then produce a “permissive” state for MCM binding to chromatin, but the actual binding would await the appearance of a new round of cyclin E expression. In such a scenario, cyclin E has opposing effects on MCM–chromosome association depending on the state of the nuclei; postreplication nuclei that have experienced a drop in cyclin E are in a permissive state for MCM-loading in response to the next pulse of cyclin E. In contrast, nuclei in S phase have yet to experience a drop in cyclin E and thus are unable to load MCMs onto chromosomes. Defining the permissive state for MCM–chromosome binding in molecular terms is an important future goal. In this regard, analysis of factors known to be required for MCM–chromatin binding, e.g., ORC and CDC6, in the endocycle should be informative.
Application of Our Observations to Mitotic Cycles
In addition to providing an explanation to MCM behavior in endocycles, the proposed coupling of MCM–chromatin association with cyclin:cdk oscillations, rather than NEB, can explain observations made in mitotic cells. Here, cdks that are present during S and G2 phases would prevent association of MCMs to chromatin; loss of cyclins at mitosis would permit reassociation. This is consistent with observations in human cells that MCMs are not associated with chromatin in G2, and generally are tightly associated in G1 (Kimura et al., 1994). The notion that NEB is not necessary for MCM–chromatin association is consistent with the findings that Xenopus MCMs can cross the nuclear envelope (Madine et al., 1995), and that treatment of human G2 cells with kinase inhibitors allows rereplication in the absence of nuclear permeabilization (Coverley et al., 1996).
In addition, we propose that loss of mitotic cyclins permits the association, but that final association will depend on other conditions. This is based on the observations presented here; in endocycles, MCMs do not bind chromosomes constitutively despite the absence of mitotic cyclins. Thus, absence of mitotic cyclins is not sufficient for chromosome binding of MCMs. We find that cyclin E can provide activities that are required for chromosome binding of MCMs, in the absence of mitotic cyclins. This latter aspect of our model can explain recent observations in Drosophila embryonic mitotic cycles (Su and O'Farrell, 1997). MCMs associate with anaphase chromosomes upon the destruction of mitotic cyclins in some cell cycles but not in others. Specifically, association to anaphase chromosomes occurs in the early embryonic cycles in which Cyclin E is present, but not in anaphase of cycle 16 when Cyclin E is inactivated. Mutation of dacapo, which encodes a cdk inhibitor that is essential for the inactivation of Cyclin E (Lane et al., 1997; de Nooij et al., 1997), restores MCM binding to chromosomes in mitosis 16. We suggest that in some settings, the reassociation of MCMs after mitosis depends on G1 cyclins.
The Role of MCMs in Endocycles
The observations reported here demonstrate that a cycle of MCM–chromatin association is coupled to endocycle S phases and suggest that MCMs might play a role in endoreplication. Although MCMs are essential for viability in Drosophila, analysis of MCM mutants have not clearly defined the role of MCMs in DNA synthesis. In homozygous MCM mutants, defects in mitotic proliferation are not seen until late embryogenesis, and endoreplication continues to the end of larval growth (Feger et al., 1995; Treisman et al., 1995). The persistence of maternally supplied proteins, however, has been implicated in the lack of an earlier defect, as has been seen for mutations in other replication functions (Henderson et al., 1994; Knoblich et al., 1994; Feger et al., 1995; Treisman et al., 1995). Despite the continuation of endocycles in MCM mutants, Drosophila MCMs may play essential roles in endocycling cells. Polytene chromosomes of DmMCM2 mutants are more fragile than wild-type (Treisman et al., 1995), suggesting the presence of underreplicated regions. Our observation that DmMCM2–chromosome interactions change in conjunction with DNA synthesis is consistent with a role for MCMs in endocycles.
In summary, our data indicate that changes in the interaction of a group of replication factors, MCMs, with chromosomes can occur independently of mitosis. Moreover, we found that the absence of the mitotic program is not sufficient for chromosome binding of MCMs, but that this association requires additional activities that are inducible by a G1 cyclin. In many respects, MCM behavior in endocycles resembles what has been observed in mitotic cycles: MCM–chromosome association precedes DNA synthesis, DNA synthesis is required for dissociation of MCM from chromosomes, and MCMs remain dissociated from chromosomes later in S phase and in the following gap phase. Endocycles, therefore, represent an alternate system for studies of replication regulation, without the restrictions imposed by the mitotic program.
Acknowledgments
We thank P.J. Follette, S.D. Campbell, C. Detweiler, and S.J. Vidwans (all from University of California San Francisco, San Francisco, CA) for critical reading of the manuscript and C. Lehner (University of Bayreuth, Bayreuth, Germany) for the anti-cyclin E sera.
Abbreviations used in this paper
- BrdU
bromo-deoxyuridine
- MCM
minichromosome maintenance
- NEB
nuclear envelope breakdown
- PBT
PBS + 0.2% Tween
- rt
room temperature
Footnotes
This project was funded by National Institute of Health (NIH) grant (R01-GM57193) to P.H. O'Farrell and an NIH postdoctoral fellowship (GM15032) to T.T. Su.
Address all correspondence to Patrick O'Farrell, Department of Biochemistry and Biophysics, University of California San Francisco, San Francisco, California 94143-0448. Tel.: (415) 476-4707/0525. Fax: (415) 502-5143/5145. E-mail: ofarrell@cgl.ucsf.edu; tintin@cgl.ucsf.edu
References
- Ashburner, M. 1989. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1331 pp.
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Chong JPJ, Thommes P, Blow JJ. The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- Chong JP, Mahbubani HM, Khoo CY, Blow JJ. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Coue M, Kearsey SE, Mechali M. Chromotin binding, nuclear localization and phosphorylation of Xenopus cdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO (Eur Mol Biol Organ) J. 1996;15:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Coverley D, Wilkinson HR, Downes CS. A protein kinase-dependent block to reinitiation of DNA replication in G2 phase in mammalian cells. Exp Cell Res. 1996;225:294–300. doi: 10.1006/excr.1996.0179. [DOI] [PubMed] [Google Scholar]
- Davis I, Girdham CH, O'Farrell PH. A nuclear GFP that marks nuclei in living Drosophila embryos; maternal supply overcomes a delay in the appearance of zygotic fluorescence. Dev Biol. 1995;170:726–729. doi: 10.1006/dbio.1995.1251. [DOI] [PubMed] [Google Scholar]
- de Nooij JC, Letendre MA, Hariharan IK. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell. 1996;87:1237–1247. doi: 10.1016/s0092-8674(00)81819-x. [DOI] [PubMed] [Google Scholar]
- Diffley JF. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH. Developmental control of a G1-S transcriptional program in Drosophila. Development (Camb) 1994;120:1503–1515. doi: 10.1242/dev.120.6.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH. Developmental control of the G1 to S transition in Drosophila: cyclin E is a limiting downstream target of E2F. Genes Dev. 1995;9:1456–1468. doi: 10.1101/gad.9.12.1456. [DOI] [PubMed] [Google Scholar]
- Feger G, Vaessin H, Su TT, Wolff E, Jan LY, Jan YN. dpa, a member of the MCM family, is required for mitotic DNA replication but not endoreplication in Drosophila. EMBO (Eur Mol Biol Organ) J. 1995;14:5387–5398. doi: 10.1002/j.1460-2075.1995.tb00223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follette, P.J., R.J. Duronio, and P.H. O'Farrell. 1998. Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Curr. Biol. In Press. [DOI] [PMC free article] [PubMed]
- Grafi G, Larkins BA. Endoreduplication in maize endosperm: involvement of M phase-promoting factor inhibition and induction of S phase-related kinase. Science. 1995;269:1262–1264. doi: 10.1126/science.269.5228.1262. [DOI] [PubMed] [Google Scholar]
- Hammond MP, Laird CD. Control of DNA replication and spatial distribution of defined DNA sequences in salivary gland cells of Drosophila melanogaster. . Chromosoma (Berl) 1985;91:279–286. doi: 10.1007/BF00328223. [DOI] [PubMed] [Google Scholar]
- Heichman KA, Roberts JM. Rules to replicate by. Cell. 1994;79:557–562. doi: 10.1016/0092-8674(94)90541-x. [DOI] [PubMed] [Google Scholar]
- Hayashi S. A Cdc2 dependent checkpoint maintains diploidy in Drosophila. Development (Camb) 1996;122:1051–1058. doi: 10.1242/dev.122.4.1051. [DOI] [PubMed] [Google Scholar]
- Henderson DS, Banga SS, Grigliatti TA, Boyd JB. Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophila gene encoding PCNA. EMBO (Eur Mol Biol Organ) J. 1994;13:1450–1459. doi: 10.1002/j.1460-2075.1994.tb06399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XH, Yan H, Newport J. A role for Cdk2 kinase in negatively regulating DNA replication during S phase of the cell cycle. J Cell Biol. 1997;137:183–192. doi: 10.1083/jcb.137.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulskamp M, Misra S, Jurgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-x. [DOI] [PubMed] [Google Scholar]
- Jackson PK, Chevalier S, Philippe M, Kirschner MW. Early events in DNA replication require cyclin E and are blocked by p21CIP1. J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey SE, Labib K, Maiorano D. Cell cycle control of eukaryotic DNA replication. Curr Opin Genet Dev. 1996;6:208–214. doi: 10.1016/s0959-437x(96)80052-9. [DOI] [PubMed] [Google Scholar]
- Kimura H, Nozaki N, Sugimoto K. DNA polymerase alpha associated protein P1, a murine homolog of yeast MCM3, changes its intranuclear distribution during the DNA synthetic period. EMBO (Eur Mol Biol Organ) J. 1994;13:4311–4320. doi: 10.1002/j.1460-2075.1994.tb06751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA, Sauer K, Jones L, Richardson H, Saint R, Lehner CF. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Krude T, Musahl C, Laskey RA, Knippers R. Human replication proteins hCdc21, hCdc46 and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. J Cell Sci. 1996;109:309–318. doi: 10.1242/jcs.109.2.309. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Lane ME, Sauer K, Wallace K, Jan YN, Lehner CF, Vaessin H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- Lehner CF, O'Farrell PH. Expression and function of Drosophila Cyclin A during embryonic cell cycle progression. Cell. 1989;56:957–968. doi: 10.1016/0092-8674(89)90629-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF, O'Farrell PH. The roles of Drosophila cyclins A and B in mitotic control. Cell. 1990;61:535–547. doi: 10.1016/0092-8674(90)90535-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine MA, Khoo C-Y, Mills AD, Laskey RA. An MCM3 protein complex is required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- Madine MA, Khoo C-Y, Mills AD, Mushal C, Laskey RA. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr Biol. 1995;5:1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO (Eur Mol Biol Organ) J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PN, Johnson RT. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- Richardson H, O'Keefe LV, Marty T, Saint R. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development (Camb) 1995;121:3371–3379. doi: 10.1242/dev.121.10.3371. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA. Mechanisms restricting DNA replication to once per cell cycle: MCMs, pre-replicative complexes and kinases. Trends Cell Biol. 1996;6:184–188. doi: 10.1016/0962-8924(96)10015-5. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rudkin, G.T. 1969. Non replicating DNA in Drosophila. Genetics. 61(Suppl.): 227–238. [PubMed]
- Rudkin, G.T. 1972. Replication in polytene chromosomes. In Developmental Studies in Giant Chromosomes. W. Beerman, editor. Springer-Verlag, Berlin, New York, Heidelberg. 59–85.
- Sauer K, Knoblich JA, Richardson H, Lehner CF. Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev. 1995;9:1327–1339. doi: 10.1101/gad.9.11.1327. [DOI] [PubMed] [Google Scholar]
- Schubiger G, Edgar B. Using inhibitors to study embryogenesis. Methods Cell Biol. 1994;44:697–713. doi: 10.1016/s0091-679x(08)60939-5. [DOI] [PubMed] [Google Scholar]
- Smith AV, Orr-Weaver TL. The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development (Berl) 1991;112:997–1008. doi: 10.1242/dev.112.4.997. [DOI] [PubMed] [Google Scholar]
- Su TT, O'Farrell PH. Chromosome association of minichromosome maintenance proteins in Drosophilamitotic cycles. J Cell Biol. 1997;139:13–22. doi: 10.1083/jcb.139.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT, Follette PJ, O'Farrell PH. Qualifying for the license to replicate. Cell. 1995;81:825–828. doi: 10.1016/0092-8674(95)90000-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT, Feger G, O'Farrell PH. Drosophila MCM protein complexes. Mol Biol Cell. 1996;7:319–329. doi: 10.1091/mbc.7.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT, Yakubovich N, O'Farrell PH. Cloning of DrosophilaMCM homologs and analysis of their requirement during embryogenesis. Gene. 1997;192:283–289. doi: 10.1016/s0378-1119(97)00107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov IT, Attaran A, Kearsey SE. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman JE, Follette PJ, O'Farrell PH, Rubin GM. Cell proliferation and DNA replication defects in a Drosophila MCM2 mutant. Genes and Dev. 1995;9:1709–1715. doi: 10.1101/gad.9.14.1709. [DOI] [PubMed] [Google Scholar]
- Tye B-K. The MCM2-3-5 proteins: are they replication licensing factors? . TIBS (Trends Cell Biol) 1994;4:160–166. doi: 10.1016/0962-8924(94)90200-3. [DOI] [PubMed] [Google Scholar]
- Varmuza S, Prideaux V, Kothary R, Rossant J. Polytene chromosomes in mouse trophoblast giant cells. Development. 1996;102:127–134. doi: 10.1242/dev.102.1.127. [DOI] [PubMed] [Google Scholar]
- Wilson, E.B. 1987. The Cell in Development and Heredity. Garland Publishing Inc., New York. 1232 pp.
- Wuarin J, Nurse P. Regulating S phase: CDKs, licensing and proteolysis. Cell. 1996;85:785–787. doi: 10.1016/s0092-8674(00)81261-1. [DOI] [PubMed] [Google Scholar]